Abstract
Nuclear factor Y (NF-Y) is a ubiquitous transcription factor composed of three distinct subunits (NF-YA, NF-YB, and NF-YC). We found that the Arabidopsis thaliana NFYA5 transcript is strongly induced by drought stress in an abscisic acid (ABA)-dependent manner. Promoter:β-glucuronidase analyses showed that NFYA5 was highly expressed in vascular tissues and guard cells and that part of the induction by drought was transcriptional. NFYA5 contains a target site for miR169, which targets mRNAs for cleavage or translational repression. We found that miR169 was downregulated by drought stress through an ABA-dependent pathway. Analysis of the expression of miR169 precursors showed that miR169a and miR169c were substantially downregulated by drought stress. Coexpression of miR169 and NFYA5 suggested that miR169a was more efficient than miR169c at repressing the NFYA5 mRNA level. nfya5 knockout plants and plants overexpressing miR169a showed enhanced leaf water loss and were more sensitive to drought stress than wild-type plants. By contrast, transgenic Arabidopsis plants overexpressing NFYA5 displayed reduced leaf water loss and were more resistant to drought stress than the wild type. Microarray analysis indicated that NFYA5 is crucial for the expression of a number of drought stress–responsive genes. Thus, NFYA5 is important for drought resistance, and its induction by drought stress occurs at both the transcriptional and posttranscriptional levels.
INTRODUCTION
Drought stress is a major environmental factor limiting crop productivity worldwide. To reduce the adverse effects of drought stress, plants have evolved multifaceted strategies, including morphological, physiological, and biochemical adaptations (Ingram and Bartels, 1996; Xiong et al., 2002; Zhu, 2002; Shinozaki et al., 2003; Bohnert et al., 2006). Some of these strategies aim to avoid dehydration stress by increasing water uptake or reducing water loss, while other strategies seek to protect plant cells from damage when water is depleted and tissue dehydration becomes inevitable (Verslues et al., 2006). Changes in gene expression play an important role in plant drought stress response, and many stress-induced genes are known or presumed to play roles in drought resistance. For many of these genes, the hormone abscisic acid (ABA) is a key signaling intermediate controlling their expression. This has been shown in large part by analysis of ABA-deficient and ABA-insensitive mutants in Arabidopsis thaliana (Koornneef et al., 1998).
A number of stress-regulated genes encode regulatory proteins, such as transcription factors that are important in regulating the expression of downstream genes (Singh et al., 2002). In Arabidopsis, members of the AP2/ERF, bZIP, NAC, HD-ZIP, and MYB/MYC families, as well as several classes of zinc finger domain proteins, are induced by drought stress (Shinozaki et al., 2003; Zhang et al., 2004). In several cases, it has been shown that altering the expression of a transcription factor can alter stress resistance by activating downstream target genes. Examples of this are the CBFs/DREBs, NACs, and RING-H2 zinc finger proteins (Jaglo-Ottosen et al., 1998; Kasuga et al., 1999; Hu et al., 2006; Ko et al., 2006).
It is well known that regulation of gene expression at the transcriptional level plays a crucial role in the development and physiological status of plants. With the discovery of small RNAs, increased attention has been focused on the importance of posttranscriptional gene regulation by small RNAs (Carrington and Ambros, 2003; Bartel, 2004; Tang, 2005). These small RNAs include 20- to 24-nucleotide microRNAs, 21-nucleotide transacting small interfering RNAs (siRNAs), ∼24-nucleotide repeat-associated siRNAs and 21- or 24-nucleotide natural antisense transcript-generated small interfering RNAs (nat-siRNAs). MicroRNAs (miRNAs) are processed from hairpin precursors by the ribonuclease III–like enzyme Dicer and differ from siRNAs, which are not generated from long double-stranded RNAs.
Plant miRNAs are involved in various developmental processes, including flowering, leaf and root development, embryo development, and auxin signaling (Carrington and Ambros, 2003; Bartel, 2004; Allen et al., 2005; Jones-Rhoades et al., 2006). Recently, studies in Medicago truncatula found that symbiotic nodule development is regulated by miR169 (Combier et al., 2006). miRNAs also play important roles in plant responses to abiotic stresses, such as sulfate and phosphate nutrient deprivation, and oxidative stress (Jones-Rhoades and Bartel, 2004; Fujii et al., 2005; Sunkar et al., 2006; Sunkar and Zhu, 2007). nat-siRNAs were demonstrated to regulate salt tolerance and disease resistance in Arabidopsis (Borsani et al., 2005; Katiyar-Agarwal et al., 2006). However, despite the importance of drought resistance, thus far no small RNAs have been reported to regulate drought stress responses.
Nuclear factor Y (NF-Y) is a ubiquitous transcription factor with high affinity and sequence specificity for the CCAAT box, a cis-element present in ∼25% of eukaryotic gene promoters. NF-Y is a heterotrimeric complex composed of NF-YA (also known as CBF-B or HAP2), NF-YB (CBF-A or HAP3), and NF-YC (CBF-C or HAP5). In mammals, NF-YB and NF-YC tightly dimerize through a histone fold motif, then NF-YA associates prior to DNA binding, with the sequence-specific interaction of the trimer mediated by NF-YA (Mantovani, 1999). NF-YA and NF-YC subunits contain large domains rich in glutamines and hydrophobic residues that are important for activating transcription (Mantovani, 1999). In animals and yeast, each subunit of NF-Y is encoded by a single gene, whereas the Arabidopsis genome encodes 10 NF-YAs, 13 NF-YBs, and 13 NF-YCs (Gusmaroli et al., 2002). It has been demonstrated that NFYB9 (LEC1) plays a pivotal role in embryo development (Lee et al., 2003). Recently, overexpression of NFYB1 in Arabidopsis and maize (Zea mays) was shown to significantly improve drought resistance and yield under drought stress conditions (Nelson et al., 2007). However, the biological roles of most of the NF-Y family members in plants are not understood.
Here, we show that expression of NFYA5, a member of the Arabidopsis NF-YA family, is strongly induced by drought stress and ABA treatments. Promoter:β-glucuronidase (GUS) analysis suggested that part of this induction occurred at the transcriptional level; however, transcriptional regulation alone could not explain the high level of NFYA5 transcript accumulation seen after stress or ABA treatment. We found that NFYA5 contained a target site for miR169, and miR169 expression was downregulated by drought. When we analyzed the expression of miR169 precursors, we found that two of them, miR169a and miR169c, were downregulated by drought stress. Coexpression of miR169 and NFYA5 mRNAs suggested that miR169a was more efficient than miR169c at downregulating the NFYA5 mRNA.
Thus, the results suggest that downregulation of miR169a by drought stress contributes to the high level induction of NFYA5 by drought and ABA. NFYA5 was highly expressed in vascular tissues and guard cells, and analysis of nfya5 knockout plants and miR169a or NFYA5 overexpression lines showed that NFYA5 was important in controlling stomatal aperture and drought resistance. Taken together, our results show that NFYA5 is important for drought resistance, and it is regulated by drought stress at both transcriptional and posttranscriptional levels.
RESULTS
NFYA5 Expression Is Induced by Drought Stress and ABA Treatments
We investigated NFYA5 initially because it is annotated to overlap with another gene (At1g54150) on the antisense strand in their 3′ untranslated region (UTR) regions to form a natural cis-antisense transcript (NAT) gene pair. Previously, we reported a new type of endogenous siRNA derived from a NAT pair formed by D1-pyrroline-5-carboxylate dehydrogenase and SRO5 and described its role in salt tolerance in Arabidopsis (Borsani et al., 2005). The NFYA5 transcript was strongly induced by drought stress (Figure 1A). In the publicly available Arabidopsis eFP Browser microarray database (http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi), NFYA5 expression was also strongly induced by osmotic (300 mM mannitol) or salt (150 mM NaCl) stress. By contrast, NFYA5 expression was not induced by drought treatment (15-min exposure to an air stream of 18-d-old seedlings grown on rafts floating on liquid Murashige and Skoog [MS] medium), possibly because the brief air exposure was not severe enough to activate drought responsive genes.
Figure 1.
Regulation of NFYA5 Expression by Drought Stress and ABA.
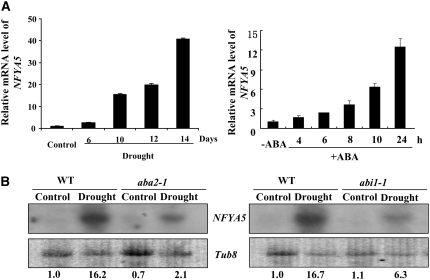
(A) Real-time PCR assay of the accumulation of NFYA5 gene transcript in Arabidopsis plants in response to drought stress (withholding water from 3-week-old soil-grown plants for the indicated durations) and to ABA (2-week-old seedlings on agar medium). The expression levels were normalized to that of Tub8, and the level of NFYA5 transcript in the controls was set at 1.0. Error bars represent se for three independent experiments.
(B) Detection of NFYA5 mRNA in ABA-deficient (aba2-1) or signaling (abi1-1) mutants. The wild type and mutants were grown with sufficient water for 3 weeks, and then water was withheld for 10 d. Twenty micrograms of total RNA from each sample was loaded and hybridized with a 32P-labeled full-length NFYA5 cDNA probe. Tub8 was used as a loading control, and numbers below each lane indicate the expression level of NFYA5 relative to Tub8.
ABA accumulation is required for some drought stress–induced upregulation of gene expression (Shinozaki and Yamaguchi-Shinozaki, 1996; Zhu, 2002). Thus, we tested the response of NFYA5 to ABA treatment. NFYA5 expression increased ∼13-fold by 24 h after the application of 100 μM ABA. To verify that ABA is required for the drought-induced increase of NFYA5 expression, we examined an ABA-deficient (aba2-1) mutant and an ABA-insensitive (abi1-1) mutant (Koornneef et al., 1998) In both the Columbia (Col-0) and Landsberg erecta (Ler) wild types, the expression of NFYA5 was strongly induced by withholding watering for 10 d; however, drought-induced accumulation of NFYA5 mRNA was substantially reduced in aba2-1 and abi1-1 (Figure 1B). This result suggests that NFYA5 expression is at least partly dependent on ABA signaling.
Transcriptional Level Induction by Drought Stress and ABA and Tissue Expression Pattern of NFYA5
Although the real-time PCR and RNA gel blot assays both showed a clear induction of NFYA5 RNA accumulation in response to drought stress or ABA, such assays could not address whether this increase was caused by increased promoter activity or altered RNA stability. To address this question, we constructed a promoter:GUS fusion using a 1.7-kb fragment upstream from the initiation codon of the NFYA5 gene. Analysis of GUS staining patterns in several transgenic lines showed that GUS staining increased in response to ABA treatment (Figure 2A). Drought treatment (water withholding for 10 d) increased the GUS activity in leaves from 280 ± 9 to 450 ± 25 (SE, n = 4). This was much less than the ∼15-fold upregulation of NFYA5 mRNA level found by quantitative PCR analysis (Figure 1A). Thus, in addition to transcriptional induction, posttranscriptional regulation of NFYA5 may also play a role.
Figure 2.
NFYA5 Expression Pattern and Transcriptional Regulation.
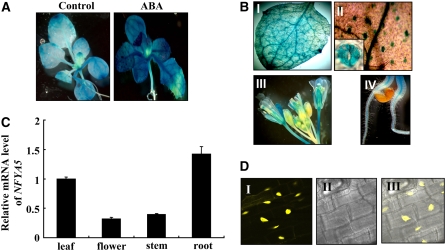
(A) GUS activity in 2-week-old transgenic seedlings on MS-agar medium that were exposed to ABA treatment for 8 h or immersed in water for 8 h (control).
(B) NFYA5p:GUS expression pattern in various tissues. The staining was prominent in the vascular tissues (I) and guard cells (II) of leaves. The staining was also visible in floral tissues of the inflorescence (III) and root vascular system (IV).
(C) Tissue pattern of NFYA5 transcript accumulation. Total RNA was isolated from various tissues of 4-week-old wild-type plants grown under long-day growth conditions. Real-time RT-PCR quantifications were normalized to the expression of 18S rRNA. Error bars represent se for three independent experiments.
(D) Subcellular localization of NFYA5. The NFYA5-YFP fusion construct was expressed in transgenic Arabidopsis under the control of the cauliflower mosaic virus 35S promoter, and the plant roots were observed under a confocal microscope. The photographs were taken in the dark field for yellow fluorescence (I), in the bright field for the morphology of the cells (II), and in combination (III).
We also used the promoter:GUS transgenic lines to examine the expression pattern of NFYA5. NFYA5 expression was high in leaf tissues, with prominent expression in both the leaf vascular system and, importantly, a high level of expression in guard cells (Figure 2B, panels I and II). GUS staining was also observed in floral tissues and the root vascular system (Figure 2B, panels III and IV). Quantitative PCR analysis of NFYA5 mRNA levels in different tissues was consistent with the tissue pattern of GUS staining and showed that expression was highest in leaf and root tissues with significant expression also occurring in floral and stem tissues (Figure 2C).
In Arabidopsis, the A subunit of the NF-Y complex is encoded by a 10-member gene family. Though other regions of the proteins vary, the NF-YAs contain a highly conserved core region that consists of two functional subdomains: an NF-YB/NF-YC binding subdomain and a DNA binding subdomain, which are connected by a small linker (Romier et al., 2003; Wenkel et al., 2006; see Supplemental Figure 1 online). The Psort II program predicted a nuclear localization of NFYA5 protein with 70% certainty. To confirm the subcellular localization of NFYA5 protein, a translational fusion between yellow fluorescent protein (YFP) and the C terminus of NFYA5 was transformed into Arabidopsis. Cells expressing the NFYA5-YFP fusion protein showed that the YFP signal appeared only in the nucleus (Figure 2D).
Drought Stress Downregulates a miRNA That Targets the 3′ UTR of NFYA5
Because the high level of NFYA5 mRNA accumulation in Arabidopsis under drought stress could not be explained solely by the promoter activity of NFYA5, this strongly suggested the presence of another regulatory mechanism operating at the posttranscriptional level. One possibility is regulation by small RNAs. To investigate this possibility, we searched the Arabidopsis MPSS Plus Database (http://mpss.udel.edu/at/) and found two small RNA signatures (17 nucleotides) that matched the 3′ end of At1g54150 and are complementary to the 3′ UTR of NFYA5. The small RNA signatures in the Arabidopsis MPSS Plus Database were identical to part of ASRP1815 (19 nucleotides) in the ASRP small RNA database (http://asrp.cgrb.oregonstate.edu). Thus, we designed an oligonucleotide probe complementary to ASRP1815 (Figure 3A).
Figure 3.
Drought Stress Downregulates a 21-Nucleotide Small RNA That Is Complementary to NFYA5 mRNA.
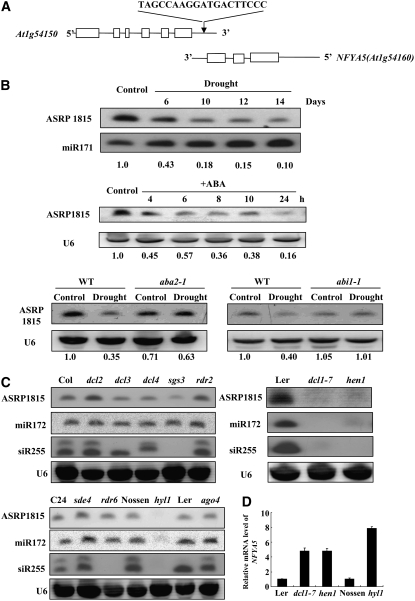
(A) Diagram of the cis-antisense gene pair of NFYA5 and At1g54150. Exons are boxed, and lines between boxes represent introns. Arrow indicates target position of the small RNA.
(B) Regulation of the small RNA by drought stress and ABA treatment. miR171 or U6 RNA was probed as a loading control. Numbers below each lane indicate relative expression.
(C) Accumulation of the small RNA in various RNA silencing mutants. Forty micrograms of small RNA from each sample was loaded per lane and hybridized with a 32P-labeled oligonucleotide probe corresponding to the sequence of ASRP1815. miR172, siR255, and U6 were probed as loading controls.
(D) NFYA5 mRNA levels in dcl1-7, hen1, and hyl1 and their corresponding wild types. The expression levels were normalized to that of Tub8. Error bars represent se for three independent experiments.
Using this oligonucleotide probe, we detected a 21-nucleotide small RNA in plants grown under normal growth conditions. In plants subjected to drought stress, the level of the ASRP1815 small RNA decreased substantially, to ∼10% of the level in unstressed plants after 14 d (Figure 3B). In this same treatment, the expression of NFYA5 increased substantially in response to the drought stress. This is consistent with the possibility that lower expression of ASRP1815 under drought may reduce the small RNA–directed degradation of NFYA5 mRNA. Also consistent with ABA-induced NFYA5 mRNA accumulation, we found that ABA treatment suppressed the level of ASRP1815 to ∼16% of the control level (Figure 3B). In both the Col-0 and Ler wild types, the expression of ASRP 1815 was strongly suppressed by drought stress; however, ASRP 1815 expression in the aba2-1 and abi1-1 mutants was not substantially affected by drought (Figure 3B). Therefore, drought stress–induced suppression of ASRP 1815 was dependent on ABA signaling.
The fact that NFYA5 and At1g54150 form a NAT gene pair raised the possibility that they may generate a nat-siRNA that regulates NFYA5. The biogenesis of nat-siRNAs requires DCL2 or DCL1, RDR6, SGS3, and NRPD1a (Borsani et al., 2005; Katiyar-Agarwal et al., 2006). To test whether ASRP1815 was a nat-siRNA, we examined its biogenesis in mutants defective in various proteins known to be required for biogenesis of specific types of small RNAs. ASRP1815 was still produced in dcl3, rdr2, dcl4, dcl2, rdr6, sgs3, or sde4/nrpd1a (Figure 3C). This suggested that it was not a heterochromatic siRNA, transacting siRNA, or nat-siRNA of the types described previously. Instead, ASRP1815 was absent in hen1, dcl1-7, and hyl1 (Figure 3C). The requirement of these components suggests that ASRP1815 is probably a miRNA. Disruption of NFYA5 expression had little effect on ASRP1815 accumulation (see Supplemental Figure 2 online), consistent with the notion that ASRP1815 is not a nat-siRNA.
We tested the levels of NFYA5 mRNA in the mutants where the ASRP1815 small RNA was absent. The level of NFYA5 mRNA was higher in hen1, dcl1-7, and hyl1 than that in the wild type (Figure 3D). This result suggested that ASRP1815 indeed downregulates NFYA5 expression.
NFYA5 Is Mainly Regulated by miR169a
The requirement of HEN1, DCL1, and HYL1 suggested that we were detecting a miRNA with the ASRP1815 probe. Thus, we searched for sequences homologous to ASRP1815 in small RNA databases and found that ASRP1815 is homologous to sequences of the miR169 (21 nucleotides) family. Indeed, NFYA5 is one of the predicted targets of the miR169 family (Jones-Rhoades and Bartel, 2004). The family of miR169 in Arabidopsis contains 14 members, and this miRNA family is conserved in Oryza sativa and Populus trichocarpa (Bonnet et al., 2004; Jones-Rhoades and Bartel, 2004; Sunkar and Zhu, 2004; Sunkar et al., 2005). Based on sequence of the miRNA produced, the MIR169a/b/c/h/i/j/k/l/m/n family members are predicted to target NFYA5 and can be divided into three subgroups. MIR169a represents the first subgroup, MIR169b and MIR169c form the second group, and the third group is made up of MIR169h/i/j/k/l/m/n. The main difference among the subgroups is the sequence at the 3′ end: the last two nucleotides of MIR169a are G and A, while those of MIR169b/c and MIR169 h/i/j/k/l/m/n are GG and UG, respectively. Also, the 5′ end of MIR169 h/i/j/k/l/m/n is UA, which is different from CA of other two subgroups.
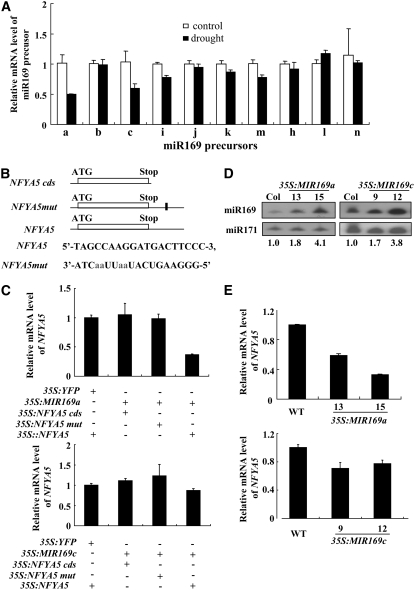
Because of their sequence similarity, the MIR169 family members cannot be differentiated in small RNA gel blots because of cross-hybridization. To determine which of the MIR169 loci may regulate NFYA5, we performed real-time RT-PCR using miR169 locus-specific primers to determine if expression of any of the miR169 loci is regulated by drought stress. RNA was extracted from soil-grown Arabidopsis plants that had been subjected to water withholding for 10 d. Only MIR169a and MIR169c exhibited substantial changes in transcript abundance in response to drought stress (Figure 4A). Both MIR169a and MIR169c were downregulated by drought stress, consistent with the downregulation of the mature miRNA by drought. Thus, we undertook further experiments to determine if MIR169a and MIR169c could downregulate NFYA5 mRNA.
Figure 4.
NFYA5 Is Mainly Regulated by miR169a.
(A) Detection of precursor transcripts of the MIR169 family in response to drought stress by real-time RT-PCR. Quantifications were normalized to the expression of Tub8. Error bars represent se for three independent experiments.
(B) Diagram of NFYA5 expression constructs. The introduced mutations in the target site of NFYA5 are shown in lowercase letters. Black box in the 3′ UTR indicates the miRNA target site.
(C) Coexpression of various combinations of miR169 and NFYA5 expression constructs in N. benthamiana. As a control, NFYA5 was also coexpressed with an unrelated YFP construct. Real-time RT-PCR quantifications were normalized to the expression of 18S rRNA of tobacco. Error bars represent se for three independent experiments.
(D) Overexpression of miR169a and miR169c in transgenic Arabidopsis. RNA gel blot analysis of miR169a and miR169c levels in the wild type and two representative transgenic lines. miR171 is shown as a loading control. Numbers below each lane indicate relative expression.
(E) Detection of corresponding NFYA5 gene transcripts in 35S:MIR169 transgenic plant lines by real-time RT-PCR. Quantifications were normalized to the expression of Tub8. Error bars represent se for three independent experiments.
In many cases, it has been assumed that members of a miRNA family have mostly redundant functions; however, Sieber et al. (2007) recently provided evidence that closely related miRNAs that were predicted to target the same genes had in fact different functions during development. To test whether MIR169a or MIR169c may play a specific role in regulating NFYA5 expression, we performed transient coexpression assays in Nicotiana benthamiana. Because the target site of miR169 is located in the 3′ UTR of NFYA5, we tested three NFYA5 constructs: a full-length NFYA5 including the 3′ UTR, a construct without 3′ UTR, and another construct that was mutated in the 3′ UTR to introduce four mismatches between it and miR169 (Figure 4B). Both the NFYA5 and miR169a or miR169c constructs were expressed under control of the 35S promoter. After 2 d of coexpression in N. benthamiana, RNA was extracted and NFYA5 expression analyzed by quantitative RT-PCR. mRNA levels from the constructs that lacked a functional miR169 target site, NFYA5cds and NFYA5mut, were not affected by coexpression with miR169 (Figure 4C). However, the level of NFYA5 mRNA containing a miR169 target site was decreased significantly (37% of the control level) when coexpressed with MIR169a (Figure 4C). Interestingly, coexpression with miR169c caused only a 13% decrease in the level of NFYA5 transcript. The results suggested that the degradation of NFYA5 mRNA was mainly directed by miR169a. Small RNA gel blots prepared from the same samples and probed with oligonucleotides complementary to miR169 clearly showed that a 21-nucleotide small RNA was highly expressed in all of the coexpression samples (see Supplemental Figures 3A and 3B online). Thus, the failure of strong miR169c-directed cleavage of AtNFYA5 was not due to a lack of miR169c expression.
To confirm these results, we also overexpressed the precursors of MIR169a and MIR169c in Arabidopsis and chose lines with similar miR169 expression levels to quantify the effect of miR169a and miR169c overexpression on NFYA5 (Figure 4D). In agreement with the transient coexpression assay results, overexpression of miR169a caused a larger decrease in the level of NFYA5 mRNA than overexpression of miR169c (Figure 4E). We also chose two other members of the miR169 family, miR169b and miR169 h, and overexpressed them in Arabidopsis (see Supplemental Figures 2B and 2C online). The relative mRNA levels of NFYA5 did not change substantially despite the overexpression of the miRNA (see Supplemental Figures 2D and 2E online). These results again strongly suggest MIR169a as the major miRNA locus important for the regulation of AtNFYA5 expression.
35S:MIR169a and nfya5 Loss-of-Function Mutant Plants Are Hypersensitive to Drought Stress
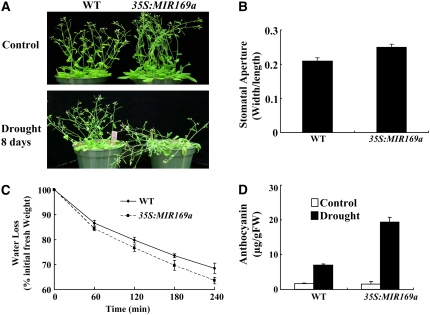
Drought-responsive gene regulation and ABA signaling are crucial for drought resistance (Pei et al., 1998; Zhu, 2002). The drought- and ABA-inducible expression of NFYA5 and its strong expression in guard cells prompted us to analyze its potential role in drought resistance. First, we tested plants overexpressing miR169a (35S:MIR169a, line #15), in which the level of NFYA5 mRNA was ∼33% of the wild type (Figure 4E). Wild-type and 35S:MIR169a-15 plants were grown for 3 weeks in soil and were then subjected to water withholding for 8 d. 35S:MIR169a-15 plants showed leaf rolling and the leaves became purple, whereas the wild-type plants were still turgid and their leaves remained green (Figure 5A). This result suggested that 35S:MIR169a-15 plants may have depleted the soil water more rapidly than the wild type and thus wilted more quickly. To investigate this possibility, leaves from 35S:MIR169a-15 and wild-type plants grown in soil were used for stomatal aperture measurements. The stomatal aperture index of 35S:MIR169a-15 leaves was 0.25, which was ∼20% greater than that of the wild type (Figure 5B). Consistent with these results, detached leaves of 35S:MIR169a-15 plants consistently lost water more quickly than those of the wild type (Figure 5C), suggesting that the more rapid appearance of wilting after water withholding in 35S:MIR169a-15 could be attributed at least in part to an inability of these plants to efficiently close their stomata and reduce transpiration. Because ABA is a regulator of stomatal aperture and transpiration, this phenotype is consistent with the ABA-induced expression of NFYA5 and suggests that NFYA5 may be important for ABA response in guard cells. Another indicator of stress sensitivity is the accumulation of the purple flavonoid pigment anthocyanin in leaves. The anthocyanin levels in 35S:MIR169a-15 plants after withholding water for 8 d was 19.4 μg g−1 fresh weight, which was ∼3 times as much as that of the wild type, again supporting that 35S:MIR169a-15 was more sensitive to drought stress. The results suggest that adequate expression of NFYA5 is required for drought resistance.
Figure 5.
35S:MIR169a Plants Are More Sensitive to Drought Stress.
(A) 35S:MIR169a-overexpressing Arabidopsis plants are more sensitive to drought stress. Wild type (Col) and 35S:MIR169a plants were grown in soil with sufficient water for 3 weeks, and then the water was withheld for 8 d. A representative picture is shown. Control, without water withholding.
(B) Measurement of stomatal aperture in wild-type and 35S:MIR169a plants. Data are mean ratios of width to length ± se of three independent experiments (n = 40 to 50).
(C) Water loss from detached leaves of wild-type and 35S:MIR169a plants. Water loss is expressed as the percentage of initial fresh weight. Values are means from 10 leaves for each of four independent experiments.
(D) Anthocyanin content in leaves of Arabidopsis with or without drought treatment for 8 d. Error bars represent se for four independent experiments. FW, fresh weight.
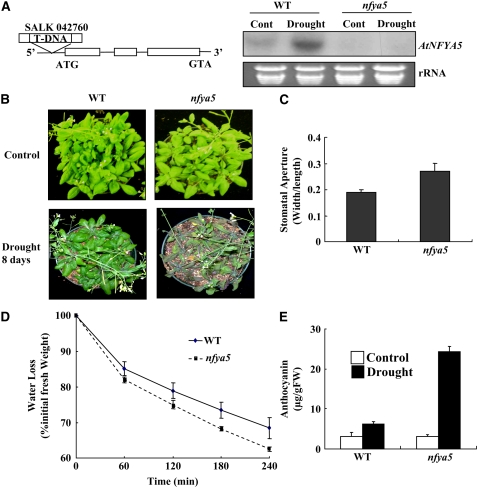
We searched the publicly available T-DNA collections and obtained a T-DNA insertion mutant (SALK_042760 in the Col background) from the ABRC to further investigate the function of NFYA5. Plants homozygous for the T-DNA insertion were identified by PCR, and sequencing of the T-DNA flanking region confirmed the insertion site in the promoter region of NFYA5 (Figure 6A). RNA gel blot analysis showed that the NFYA5 transcript was absent in the T-DNA line designated as nfya5 (Figure 6A). In agreement with the phenotypes of 35S:MIR169a-15 plants, nfya5 knockout mutant plants were also hypersensitive to drought stress (Figure 6B). The stomatal aperture index of nfya5 leaves was 0.27, which was 42% greater than that of wild-type leaves (Figure 6C). Consistent with these results, detached leaves of nfya5 lost water more quickly than those of wild-type leaves (Figure 6D). The anthocyanin levels in nfya5 leaves after withholding water for 8 d was 4 times as much as that of the wild type, again supporting that nfya5 plants were more sensitive to drought stress. These results show that NFYA5 is necessary for drought resistance.
Figure 6.
nfya5 Mutant Plants Are More Sensitive to Drought Stress.
(A) Schematic diagram of the T-DNA insertion site in the NFYA5 locus and detection of NFYA5 mRNA by RNA gel blot analysis. Exons are boxed, and lines between boxes represent introns. Twenty micrograms of total RNA from each sample was loaded and hybridized with 32P-labeled full-length NFYA5 probe. The corresponding ethidium bromide–stained rRNA is shown as a loading control.
(B) nfya5 mutant plants are more sensitive to drought stress. Wild-type (Col) and nfya5 plants were grown in soil with sufficient water for 3 weeks, and then the water was withheld for 8 d. A representative picture is shown.
(C) Measurement of stomatal aperture in wild-type and nfya5 mutant plants. Data are mean ratios of width to length ± se of three independent experiments (n = 40 to 50).
(D) Water loss from detached leaves of wild-type and nfya5 mutant plants. Water loss was expressed as the percentage of initial fresh weight. Values are means from 10 leaves for each of four independent experiments.
(E) Anthocyanin content in leaves of Arabidopsis with or without drought treatment for 8 d. Error bars represent se for four independent experiments.
Overexpression of NFYA5 Improves Drought Resistance
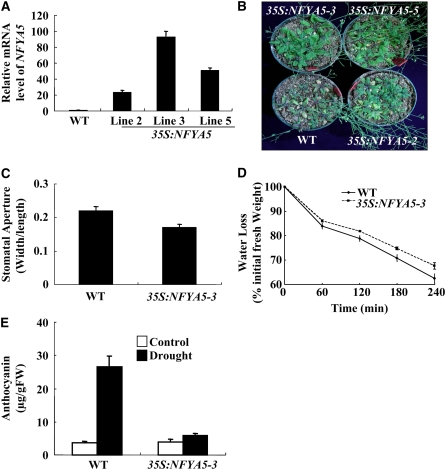
To further characterize the function of NFYA-5 in drought resistance, we generated transgenic Arabidopsis plants overexpressing the gene (without the 3′ UTR) under control of the constitutive cauliflower mosaic virus 35S promoter. Three transgenic lines (#2, 3, and 5) were chosen for further analysis based on their high level of NFYA5 expression (Figure 7A). To evaluate the effects of NFYA5 overexpression, 3-week-old soil-grown wild-type and 35S:NFYA5 plants were subjected to water withholding for 14 d. At the 14th day of water withholding, most of the wild-type plants appeared dehydrated, but the 35S:NFYA5 plants appeared less dehydrated than the wild type (Figure 7B). In contrast with nfya5, the stomatal aperture of 35S:NFYA5-3 was smaller than that of the wild type (Figure 7C). Detached leaves of 35S:NFYA5-3 lost water more slowly than those of the wild type (Figure 7D), and the anthocyanin levels in 35S:NFYA5-3 after withholding water for 14 d were much lower (Figure 6E). These results show that overexpression of NFYA5 improves plant drought resistance.
Figure 7.
Improved Drought Resistance in 35S:NFYA5 Plants.
(A) Detection of NFYA5 mRNA in 35S:NFYA5 transgenic Arabidopsis. Real-time RT-PCR quantifications were normalized to the expression of Tub8. Error bars represent se (n = 3).
(B) Drought resistance of 35S:NFYA5 plants (lines 2, 3, and 5). Wild-type and 35S:NFYA5 Arabidopsis plants were grown in soil with sufficient water for 3 weeks, and then the water was withheld for 14 d. A representative picture is shown.
(C) Measurement of stomatal aperture in wild-type and 35S:NFYA5-3 transgenic plants. Data are mean ratios of width to length ± se of three independent experiments (n = 40 to 50).
(D) Water loss from detached leaves of wild-type and 35S:NFYA5-3 plants. Water loss was expressed as the percentage of initial fresh weight. Values are means from 10 leaves for each of four independent experiments.
(F) Anthocyanin content in leaves of Arabidopsis with or without drought treatment for 14 d. Error bars represent se for four independent experiments.
NFYA5 Regulates the Expression of Stress-Responsive Genes
The nuclear localization and DNA binding domain of NFYA5 suggest that the protein may act in regulating the expression of other genes important for drought resistance. To test this possibility, we performed microarray experiment using Affymetrix Arabidopsis ATH1 Genechips. Approximately 130 genes showed statistically significant changes in expression in 35S:NFYA5 compared with wild-type seedlings under nonstress conditions (see Supplemental Table 1 online). Out of the affected genes, 28 showed a twofold or more change in expression (17 increased and 11 decreased) in the transgenic plants (Table 1). Most of these genes have known or presumed function associated with abiotic stress responses, and a number of them appear to be involved in oxidative stress (e.g., a subunit of cytochrome b6-f complex, glutathione S-transferase [GST], peroxidases, and an oxidoreductase family protein). Out of the genes affected by NFYA5 ectopic expression, most of them contain the CCAAT motif in their promoter regions (Table 1), as expected since this short sequence motif can be found in a substantial fraction of gene promoters in general. Using the AlignACE program (Hughes et al., 2000), we found a consensus cis-regulatory element, TX(C/A)TTXGX(C/A)CAXT, that contains the CCAAT motif in the promoters of a subset of the genes showing increased expression in the NFYA5 overexpression plants (Table 1). It is possible that these genes are the direct targets of NFYA5.
Table 1.
Genes with Expression Changes (pfp < 0.05) of at Least Twofold in the 35S:NFYA5 Transgenic Plants from Microarray Analysis
| Affy ID | Gene ID | Description | Number of CCAAT Motifs | Number of Novel Elements | Fold Change (OX/WT)a |
|---|---|---|---|---|---|
| 263158_at | AT1G54160 | NFYA5 | 0 | 0 | 12.72 |
| 245275_at | AT4G15210 | Cytosolic β-amylase expressed in rosette leaves and inducible by sugar | 1 | 0 | 3.34 |
| 244966_at | ATCG00600 | Cytochrome b6f complex, subunit V | 2 | 1 | 3.33 |
| 262517_at | AT1G17180 | Glutathione transferase belonging to the tau class of GSTs | 4 | 0 | 3.24 |
| 267565_at | AT2G30750 | Putative cytochrome P450 | 0 | 0 | 3.08 |
| 261021_at | AT1G26380 | FAD binding domain-containing protein | 1 | 0 | 3.02 |
| 266098_at | AT2G37870 | Protease inhibitor/seed storage/lipid transfer protein family protein | 2 | 1 | 2.81 |
| 260568_at | AT2G43570 | Chitinase, putative | 5 | 1 | 2.77 |
| 262518_at | AT1G17170 | Glutathione transferase belonging to the tau class of GSTs | 1 | 2 | 2.46 |
| 247224_at | AT5G65080 | MADS domain protein | 5 | 1 | 2.38 |
| 267101_at | AT2G41480 | Peroxidase | 0 | 0 | 2.29 |
| 244993_s_at | ATCG01000 ATCG01130 | [ATCG01000]hypothetical protein [ATCG01130]hypothetical protein | 44 | 0 | 2.28 |
| 249481_at | AT5G38900 | DSBA oxidoreductase family protein | 3 | 1 | 2.23 |
| 263497_at | AT2G42540 | COR15A | 1 | 0 | 2.20 |
| 263495_at | AT2G42530 | Cold-responsive protein | 7 | 0 | 2.14 |
| 247718_at | AT5G59310 | Member of the lipid transfer protein family | 3 | 0 | 2.01 |
| 254232_at | AT4G23600 | CORI3 | 4 | 0 | 2.00 |
| 246375_at | AT1G51830 | ATP binding/kinase/protein Ser/Thr kinase | 2 | 0 | 0.50 |
| 251226_at | AT3G62680 | PRP3; structural constituent of cell wall | 2 | 0 | 0.48 |
| 254828_at | AT4G12550 | AIR1; lipid binding | 1 | 0 | 0.48 |
| 264577_at | AT1G05260 | RCI3; peroxidase | 1 | 0 | 0.47 |
| 258338_at | AT3G16150 | l-Asparaginase, putative/l-Asn amidohydrolase, putative | 3 | 0 | 0.47 |
| 254044_at | AT4G25820 | XTR9 | 2 | 0 | 0.46 |
| 266353_at | AT2G01520 | Major latex protein-related | 3 | 0 | 0.46 |
| 258473_s_at | AT3G02620, AT3G02610 | [AT3G02620] acyl-desaturase, putative/stearoyl-ACP desaturase, putative; [AT3G02610]acyl-desaturase | 31 | 0 | 0.45 |
| 254644_at | AT4G18510 | CLE2; receptor binding | 4 | 0 | 0.42 |
| 253767_at | AT4G28520 | CRU3; nutrient reservoir | 3 | 0 | 0.41 |
| 249082_at | AT5G44120 | CRA1; nutrient reservoir | 0 | 0 | 0.11 |
OX/WT, overexpression line/wild type.
We confirmed the microarray results by real-time RT-PCR. In agreement with our microarray data, the real time RT-PCR assay showed that At4g15210 (cytosolic β-amylase), At2g37870 (protease inhibitor), At1g17170 (glutathione transferase), At2g42530 (cold-responsive protein), and At2g42540 (COR15A) were expressed at higher levels in 35S:NFYA5 plants under normal conditions (see Supplemental Figure 4 online), suggesting constitutive expression of stress-responsive genes in 35S:NFYA5. Under dehydration conditions, these genes were strongly induced in the wild type and 35S:NFYA5; however, dehydration-induced accumulation of the majority of these genes was substantially reduced in nfya5, suggesting that, for many of these genes, NFYA5 is required for optimal induction by dehydration stress.
DISCUSSION
Gene regulation under drought stress is mediated by multiple transcriptional cascades (Zhu, 2002; Yamaguchi-Shinozaki and Shinozaki, 2006). In each of these cascades, a transcription factor gene is induced, which in turn activates or represses downstream target genes important for drought resistance. NFYA5 may define one of these drought stress–responsive transcriptional cascades. This transcriptional cascade is critical for drought resistance because 35S:MIR169a and nfya5 mutant plants are hypersensitive to drought stress, whereas overexpression of NFYA5 improves drought resistance. A related transcription factor, NFYB1, was recently reported to confer drought tolerance not only in Arabidopsis but also in maize, when overexpressed. Field test results showed the utility of NFYB1 overexpression in stabilizing crop yield under drought conditions (Nelson et al., 2007). The physiological pathway by which NFYB1 improves drought tolerance is not known; however, its target genes appear largely different from that of CBF4, another drought-induced transcription factor. Another At NFYB family member, LEC1, is known to be essential for embryo development and dessication tolerance (Lotan et al., 1998). Our results suggest that part of the role of NFYA5 in drought resistance involves its expression in guard cells and control of stomatal aperture. In addition, NFYA5 is broadly expressed in various tissues. In non-guard cells, NFYA5 is likely important for dehydration tolerance via its role in activating target stress-responsive genes, such as genes involved in oxidative stress responses. The candidate target genes of At NFYB1 do not have obvious associations with stress tolerance, and some of them appear to be related to polysaccharide metabolism (Nelson et al., 2007). The lack of substantial overlap between the target genes of NFYA5 and At NFYB1 indicates that the two transcription factors may be involved in separate gene regulons.
Our microarray analysis showed that NFYA5 overexpression caused only moderate changes in the expression of a relatively small number of genes under the conditions tested. It is possible that the NFYA5 protein may need to be modified under drought stress, or additional factors such as other NF-Y subunit genes may have to be overexpressed together, to have a more pronounced effect on target genes. Alternatively, substantial changes in target gene expression in certain tissues or cells may have been masked by our analysis of expression changes using whole seedlings.
Transcriptional induction may explain part of NFYA5 transcript accumulation under drought stress. ABA is involved in the transcriptional regulation since it is required for NFYA5 transcript accumulation and it activates NFYA5 promoter activity. Using the PlantCARE program (http://bioinformatics.psb.ugent.be/webtools/plantcare), two ABA-responsive element sequences could be found in the 1.7-kb promoter region of At NFYA5. Although the upstream transcription factor controlling NFYA5 transcription under drought stress is not known, it might be one of the ABA-responsive element binding proteins.
An interesting feature of NFYA5 regulation under drought stress is the involvement of a miRNA. Our results suggest that the accumulation of NFYA5 transcript is suppressed by miR169. Drought stress downregulates miR169 expression, thus relieving miR169 repression of NFYA5. miR169 is encoded by many loci. Only two of the loci, MIR169a and MIR169c, are substantially downregulated by drought stress. Based on the sequence read frequency data at the Arabidopsis ASRP small RNA database (http://asrp.cgrb.oregonstate.edu/db/microRNA.html?fid=12/), miR169a constitutes ∼90% of the total miR169 population. Therefore, a substantial downregulation of miR169a would result in a reduction in overall miR169 level. Our data indicate that miR169a rather than miR169c plays a major role in repressing NFYA5 transcript accumulation, despite the fact that both miRNAs have three mismatches with NFYA mRNA. ABA is required for the downregulation of MIR169a and MIR169c by drought stress. Therefore, ABA is involved in both the transcriptional and posttranscriptional regulation of NFYA5. The downregulation of MIR169a and MIR169c by ABA and drought stress likely involves transcriptional repression at the two loci. It has been proposed that the induction of miRNAs and siRNAs leads to the downregulation of negative regulators of stress tolerance, whereas the suppression of miRNAs and siRNAs allows certain positive regulators to accumulate and function under stress (Sunkar et al., 2007). Consistent with this hypothesis, our results here support that the downregulation of miR169 contributes to the transcript accumulation of NFYA5, a critical positive regulator of drought stress resistance.
The importance of transcriptional regulation for plant stress resistance has been abundantly documented (Zhu, 2002; Yamaguchi-Shinozaki and Shinozaki, 2006). Much less is known about posttranscriptional regulation of stress-responsive genes. NFYA5 is regulated by drought stress not only transcriptionally but also posttranscriptionally via a miRNA. This dual regulation is consistent with the critical importance of NFYA5 for drought resistance. Both NFYA5 and miR169 are highly conserved in rice (Jones-Rhoades and Bartel, 2004), so the dual modes of regulation of NFYA5 may also apply to other plants.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana mutants, rdr2-1, dcl2-1, dcl3-1, and dcl4-1, were kindly provided by James Carrington (Center for Gene Research and Biotechnology, Oregon State University). dcl1-7 and hen1-1 were kindly provided by Xuemei Chen (University of California, Riverside). rdr6 and sde4/nrpd1a were kindly provided by David Baulcombe (John Innes Center for Plant Science Research, Sainsbury Laboratory, UK). sgs3 was kindly provided by Herve Vaucheret (Laboratoire de Biologie Cellulaire, Institut National de la Recherche Agronomique, Versailles, France). hyl1 was a gift from Nina Federoff (The Huck Institute of Life Science, Pennsylvania State University). These mutants were in the Col-0, Ler, Nosssen-0, or C24 genetic backgrounds as indicated in the text and figures. aba2-1[CS156], abi1-1[CS22], and the T-DNA insertion mutant of NFYA5 (SALK_042760) were obtained from the ABRC. Arabidopsis seedlings in MS nutrient agar medium were grown under continuous light (70 μmol m−2 s−1) at 23 ± 1°C. Soil-grown (Metromix 350) Arabidopsis and Nicotiana benthamiana plants were grown under a 16-h-light/8-h-dark photoperiod at 23 ± 1°C. For dehydration treatment, 2-week-old seedlings were pulled out of agar medium and left to dry on Whatman 3MM paper on a laboratory bench for durations as indicated. For drought treatment, plants were grown in soil with sufficient water for 3 weeks, and then the water was withheld for durations as indicated.
Constructs and Generation of Transgenic Plants
Site-directed mutagenesis was performed to generate NFYA5 mutated in the region complementary to the small RNA by the QuickChange II site-directed mutagenesis kit (Stratagene). This fragment was sequenced to ensure that only the desired mutations were introduced. NFYA5 with and without its 3′ UTR was amplified with the primers indicated (with/without 3′ UTR forward 5′-CACCATGCAAGTCTTTCAAAGGAAAG-3′, with 3′ UTR reverse 5′-GTAATGCAATTGTACTCTCGAG-3′, and without 3′ UTR reverse 5′-TCAAGTCCCTGACATGAGAGCTGAGG-3′). These constructs were cloned into the plant expression GATEWAY destination vector pMDC32.
To generate pMDC32:miR169 constructs, a 200-bp fragment surrounding the miRNA sequence including the fold-back structure was amplified from genomic DNA with the following primers: miR169a forward 5′-CACCTGGGTATAGCTAGTGAAACGCG-3′ and reverse 5′-CCTTAGCTTGAGTTCTTGCGA-3′, miR169b forward 5′-CACCCCCAACGGAGTAGAATTG-3′ and reverse 5′-CTCATACGGTCGATGTAATCCGT-3′, miR169c forward 5′-CACCTCGTCCATTATGAGTATT-3′ and reverse 5′-CTAATATGATATGAATATGGATGA-3′, miR169h forward 5′-CACCTCATATAA GAGAAAATGGTG-3′ and reverse 5′-CCAAAAAAGAGAAATGTGAATGAG-3′. The amplified fragments were introduced into the pENTR/D-TOPO vector (Invitrogen) and cloned into pMDC32 by LR reactions (Invitrogen).
For the NFYA5 promoter:GUS construct, a 1.7-kb fragment upstream from the initiation codon was amplified with the forward primer 5′-CACCTGTATGACATATTCTGTGTGGAG-3′ and reverse primer 5′-TGCAAATTGGGTATTGGCTATG-3′ and cloned into the pMDC164 vector following Gateway recombination.
A fusion of YFP to the C-terminal end of NFYA5 was generated and introduced to pEarleyGate 101 vector by Gateway recombination. YFP images were collected on a Leica SP2 confocal microscope.
RNA Analysis
Total RNA was extracted from the wild type, mutants, and transgenic plants with Trizol reagent (Invitrogen). For enrichment of small RNAs, high molecular weight RNA was selectively precipitated by the addition of 1 volume of 20% PEG-1 M NaCl (Llave et al., 2002). High molecular weight RNA was separated on 1.2% formaldehyde-MOPS agarose, and low molecular weight RNA was fractioned on 17% denaturing polyacrylamide gels. The blots were probed and washed as described (Borsani et al., 2005).
For real-time RT-PCR, 5 μg of total RNA isolated with the RNeasy plant mini kit was used for the first-strand cDNA synthesized using SuperScript III first-strand synthesis supermix (Invitrogen). The cDNA reaction mixture was diluted three times, and 5 μL was used as template in a 25-μL PCR reaction. PCR was performed after a preincubation at 95°C for 3 min and was followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 55°C for 40 s, and extension at 72°C for 40 s. All the reactions were performed in the iQ5 real-time PCR detection system using iQ SYBR green supermix (Bio-Rad). Primers specific for the precursor of miR169 were used to detect expression levels of miR169 (see Supplemental Table 1 online). Primers were also designed to detect the transcription level of NFYA5. Each experiment was replicated three times. The comparative Ct method was applied. The primers used in this experiment were listed in Supplemental Table 2 online.
Transient Expression in N. benthamiana
Site-directed mutagenesis constructs, with and without 3′ UTR constructs, were transformed into Agrobacterium tumefaciens strain 3301. Overnight cultures were harvested and mixed at a 1:1 ratio with various combinations. After 1 h of incubation at room temperature in 10 mM MgCl2, 10 mM MES, pH 5.6, and 150 μM acetosyringone, Agrobacterium suspension was coinfiltrated into 3-week-old N. benthamiana leaves. Leaves were harvested 2 d after the infiltration and small RNA extraction and blotting performed as described above.
Stomatal Aperture Analysis
Rosette leaves from 3-week-old soil-grown plants at similar developmental stages were harvested. Leaves were frozen immediately in liquid nitrogen and observed for guard cells by environmental scanning electron microscopy (Hitachi; TM 1000). Width and length of stomotal pores were measured for statistical analysis (Lemichez et al., 2001).
Water Loss Measurement
For water loss measurement, six leaves per individual of mutant and wild-type plants growing under normal conditions for 3 weeks were excised, and fresh weight was determined at designated time intervals. Four replicates were done for each line. Water loss was represented as the percentage of initial fresh weight at each time point.
Anthocyanin Content Measurement
Anthocyanin contents were measured as described by Rabino and Mancinelli (1986) and Sunkar et al. (2006). The pigments were extracted with 99:1 methanol:HCl (v/v) at 4°C, and the OD530 and OD657 for each sample were measured and OD530 – 0.25 × OD657 was used to compensate for the contribution of chlorophyll and its products to the absorption at 530.
GUS Activity Assay
Histochemical localization of GUS staining was performed by incubating the transgenic plants in 1 mg mL−1 5-bromo-4-chloro-3 indolyl β-d-glucuronic acid, 0.1 M Na2HPO4 buffer, pH 7.0, 0.5 mM K3(Fe[CN]6), and 10 mM EDTA overnight at 37°C, followed by clearing with 70% ethanol. GUS activity was assayed according to the procedure of Jefferson (1987). One hundred milligrams of frozen tissues were homogenized in 100 μL of extraction buffer (50 mM NaPO4, pH 7.0, 1 mM Na2EDTA, 0.1% [v/v] Triton X-100, 0.1% [w/v] sodium lauryl sarcosine, and 10 mM DTT) and centrifuged for 10 min at 4°C at 13,000 rpm. The fluorogenic assay was incubated in a 0.5-mL volume extraction buffer supplied with 1 mM 4-methylumbelliferyl-β-d-glucuronide (Sigma-Aldrich) for 2 h and then stopped by 0.2 M Na2CO3. Protein concentration was determined according to the Bio-Rad protocol provided with the protein assay kit. GUS activity was calculated as picomoles MU per minute per milligram of protein.
Microarray Analysis
For Affymetrix GeneChip array analysis, wild-type and 35S:NFYA5 seedlings were grown on MS plates for 15 d at 22°C with a cycle of 16 h light and 8 h darkness. Total RNA was extracted using an RNeasy plant mini kit (Qiagen) and was then used for preparation of biotin-labeled complementary RNA targets. Microarray analysis was performed as described by Breitling et al. (2004). Two biological replicates were used for each genotype. We normalized expression profiles with the RMA method (Irizarry et al., 2003). A list of genes with statistically significant changes in expression between the genotypes was generated by the RankProd method in which multiple testing was taken into account by the use of pfp (percentage of false prediction) (pfp < 0.05) (Gentleman et al., 2004; Hong et al., 2006). For predicting the consensus novel cis-regulatory element, we used the AlignACE program (Hughes et al., 2000). We applied the program to 1-kb upstream promoter sequences of up- or downregulated genes. Out of several candidate consensus elements, we chose one that contains a weak CCAAT consensus motif that was found within promoters of upregulated genes.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: NFYA5 (At1g54160), MIR169a (At3g13405), DCL3 (At3g43920), RDR2 (At4g11130), DCL4 (At5g20320), DCL2 (At3g03300), RDR6 (At3g49500), SGS3 (At5g23570), HEN1 (At4g20910), DCL1 (At1g01040), HYL1 (At1g09700), SDE4 (At1g63020), AGO4 (At2g27040), nfya5 (SALK_042760), aba2-1 [CS156], and abi1-1 [CS22]. The microarray data discussed in this publication have been deposited in the National Center for Biotechnology Information's Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE12029 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE12029).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequence Alignment of the Conserved Domains in Arabidopsis NFYA Family Members.
Supplemental Figure 2. Analysis of miR169 and NFYA5 mRNA Levels in nfya5, 35S:MIR169b, and 35S:MIR169h Transgenic Plant Lines.
Supplemental Figure 3. Coexpression of Various Combinations of miR169 and NFYA5 Constructs in N. benthamiana.
Supplemental Figure 4. Analysis of Transcript Levels in the Wild-Type and 35S:NFYA5 Transgenic Plants.
Supplemental Table 1. List of Genes with Significant Expression Changes in the 35S:NFYA5 Transgenic Plants form Microarray Analysis.
Supplemental Table 2. List of Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Paul E. Verslues for critical reading and editing of the manuscript and Rebecca Stevenson for technical assistance. This work was supported by National Institutes of Health Grants R01GM070795 and R01GM059138 (J.-K.Z.) and the National Science Foundation of China Grant 30500308 (W.-X.L.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Jian-Kang Zhu (jian-kang.zhu@ucr.edu).
Online version contains Web-only data.
References
- Allen, E., Xie, Z.X., Gustafson, A.M., and Carrington, J.C. (2005). MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121 207–221. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116 281–297. [DOI] [PubMed] [Google Scholar]
- Bohnert, H.J., Gong, Q., Li, P., and Ma, S. (2006). Unraveling abiotic stress tolerance mechanisms – Getting genomics going. Curr. Opin. Plant Biol. 9 180–188. [DOI] [PubMed] [Google Scholar]
- Bonnet, E., Wuyts, J., Rouze, P., and de Peer, Y.V. (2004). Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc. Natl. Acad. Sci. USA 101 11511–11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani, O., Zhu, J., Verslues, P.E., Sunkar, R., and Zhu, J.K. (2005). Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123 1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling, R., Armengaud, P., Amtmann, A., and Herzyk, P. (2004). Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573 83–92. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C., and Ambros, V. (2003). Role of microRNAs in plant and animal development. Science 301 336–338. [DOI] [PubMed] [Google Scholar]
- Combier, J.P., Frugier, F., Billy, F., Boualem, A., El-Yahyaoui, F., Moreau, S., Vernié, T., Ott, T., Gamas, P., Crespi, M., and Niebel, A. (2006). MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 20 3084–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R., Domrachev, M., and Lash, A.E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, H., Chiou, T.J., Lin, S.I., Aung, K., and Zhu, J.K. (2005). A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 15 2038–2043. [DOI] [PubMed] [Google Scholar]
- Gentleman, R.C., et al. (2004). Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 5 R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusmaroli, G., Tonelli, C., and Mantovani, R. (2002). Regulation of novel members of the Arabidopsis thaliana CCAAT-binding nuclear factor Y subunits. Gene 283 41–48. [DOI] [PubMed] [Google Scholar]
- Hong, F., Breitling, R., McEntee, C.W., Wittner, B.S., Nemhauser, J.L., and Chory, J. (2006). RankProd: A bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 22 2825–2827. [DOI] [PubMed] [Google Scholar]
- Hu, H., Dai, M., Yao, J., Xiao, B., Li, X., Zhang, Q., and Xiong, L. (2006). Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 103 12987–12992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, J.D., Estep, P.W., Tavazoie, S., and Church, G.M. (2000). Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 296 1205–1214. [DOI] [PubMed] [Google Scholar]
- Ingram, J., and Bartels, D. (1996). The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 377–403. [DOI] [PubMed] [Google Scholar]
- Irizarry, R.A., Bolstad, B.M., Collin, F., Cope, L.M., Hobbs, B., and Speed, T.P. (2003). Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen, K.R., Gilmour, S.J., Zarka, D.G., Schabenberger, O., and Thomashow, M.F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280 104–106. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5 387–405. [Google Scholar]
- Jones-Rhoades, M.J., and Bartel, D.P. (2004). Computational identification of plant microRNAs and their targets, including a stress induced miRNA. Mol. Cell 14 787–799. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades, M.W., Bartel, D.P., and Bartel, B. (2006). MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 57 19–53. [DOI] [PubMed] [Google Scholar]
- Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17 287–291. [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal, S., Morgan, R., Dahlbeck, D., Borsani, O., Villegas, A.J., Zhu, J.-K., Staskawicz, B.J., and Jin, H. (2006). A pathogen-inducible endogenous siRNA in plant immunity. Proc. Natl. Acad. Sci. USA 103 18002–18007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, J.H., Yang, S.H., and Han, K.H. (2006). Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J. 47 343–355. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Leon-Kloosterziel, K.M., Schwartz, S.H., and Zeevaart, J.A.D. (1998). The genetic and molecular dissection of abscisic acid biosynthesis and signal transduction in Arabidopsis. Plant Physiol. Biochem. 36 83–89. [Google Scholar]
- Lee, H.S., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (2003). Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc. Natl. Acad. Sci. USA 100 2152–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemichez, E., Wu, Y., Sanchez, J.P., Mettouchi, A., Mathur, J., and Chua, N.H. (2001). Inactivation of AtRac1 by abscisic acid is essential for stomatal closure. Genes Dev. 15 1808–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C., Xie, Z., Kasschau, K.D., and Carrington, J.C. (2002). Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297 2053–2056. [DOI] [PubMed] [Google Scholar]
- Lotan, T., Ohto, M., Yee, K.M., West, M.A., Lo, R., Kwong, R.W., Yamagishi, K., Fischer, R.L., Goldberg, R.B., and Harada, J.J. (1998). Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93 1195–1205. [DOI] [PubMed] [Google Scholar]
- Mantovani, R. (1999). The molecular biology of the CCAAT-binding factor NF-Y. Gene 239 15–27. [DOI] [PubMed] [Google Scholar]
- Nelson, D.E., et al. (2007). Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. USA 104 16450–16455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei, Z.M., Ghassemian, M., Kwak, C.M., McCourt, P., and Schroeder, J.I. (1998). Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science 282 287–290. [DOI] [PubMed] [Google Scholar]
- Rabino, R., and Mancinelli, A.L. (1986). Light, temperature, and anthocyanin production. Plant Physiol. 81 922–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romier, C., Cocchiarella, F., Mantovani, R., and Moras, D. (2003). The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 278 1336–1345. [DOI] [PubMed] [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (1996). Molecular responses to drought and cold stress. Curr. Opin. Biotechnol. 7 161–167. [DOI] [PubMed] [Google Scholar]
- Shinozaki, K., Yamaguchi-Shinozaki, K., and Seki, M. (2003). Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6 410–417. [DOI] [PubMed] [Google Scholar]
- Sieber, P., Wellmer, F., Gheyselinc, J., Riechmann, J.L., and Meyerowitz, E.M. (2007). Redundancy and specialization among plant microRNAs: Role of the MIR164 family in developmental robustness. Development 134 1051–1060. [DOI] [PubMed] [Google Scholar]
- Singh, K.B., Foley, R.C., and Oñate-Sánchez, L. (2002). Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 5 430–436. [DOI] [PubMed] [Google Scholar]
- Sunkar, R., Chinnusamy, V., Zhu, J., and Zhu, J.-K. (2007). Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 12 301–309. [DOI] [PubMed] [Google Scholar]
- Sunkar, R., Girke, T., Jain, P.K., and Zhu, J.-K. (2005). Cloning and characterization of microRNAs from rice. Plant Cell 17 1397–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar, R., Kapoor, A., and Zhu, J.-K. (2006). Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18 2051–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar, R., and Zhu, J.-K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16 2001–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar, R., and Zhu, J.-K. (2007). MicroRNAs and short-interfering RNAs in plants. J Integr Plant Biol. 49 817–826. [Google Scholar]
- Tang, G. (2005). siRNA and miRNA: an insight into RISCs. Trends Biochem. Sci. 30 106–114. [DOI] [PubMed] [Google Scholar]
- Verslues, P.E., Agarwal, M., Katiyar-Agarwal, S., Zhu, J., and Zhu, J.-K. (2006). Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 45 523–539. [DOI] [PubMed] [Google Scholar]
- Wenkel, S., Turck, F., Singer, K., Gissot, L., Gourrierec, J.L., Samach, A., and Couplanda, G. (2006). CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell 18 2971–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong, L., Schumaker, K.S., and Zhu, J.-K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14(suppl.): S165–S183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57 781–803. [DOI] [PubMed] [Google Scholar]
- Zhang, J.Z., Creelman, R.A., and Zhu, J.-K. (2004). From laboratory to field. using information from Arabidopsis to engineer salt, cold, and drought tolerance in crops. Plant Physiol. 135 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.-K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53 247–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.