Abstract
Leaf primordia initiate from the shoot apical meristem with inherent polarity; the adaxial side faces the meristem, while the abaxial side faces away from the meristem. Adaxial/abaxial polarity is thought to be necessary for laminar growth of leaves, as mutants lacking either adaxial or abaxial cell types often develop radially symmetric lateral organs. The milkweed pod1 (mwp1) mutant of maize (Zea mays) has adaxialized sectors in the sheath, the proximal part of the leaf. Ectopic leaf flaps develop where adaxial and abaxial cell types juxtapose. Ectopic expression of the HD-ZIPIII gene rolled leaf1 (rld1) correlates with the adaxialized regions. Cloning of mwp1 showed that it encodes a KANADI transcription factor. Double mutants of mwp1-R with a microRNA-resistant allele of rld1, Rld1-N1990, show a synergistic phenotype with polarity defects in sheath and blade and a failure to differentiate vascular and photosynthetic cell types in the adaxialized sectors. The sectored phenotype and timing of the defect suggest that mwp1 is required late in leaf development to maintain abaxial cell fate. The phenotype of mwp1; Rld1 double mutants shows that both genes are also required early in leaf development to delineate leaf margins as well as to initiate vascular and photosynthetic tissues.
INTRODUCTION
A fundamental question in developmental biology is how polarity is established and maintained during organogenesis. Plants are an attractive system to address this question because of the repetitive nature of leaf initiation. Leaves develop from a population of founder cells at the periphery of the meristem and exhibit polarity from the onset of initiation. The side of the leaf adjacent to the meristem, the adaxial side, has histological characteristics distinct from the abaxial side, the side furthest from the meristem (Kaplan, 1975; Steeves and Sussex, 1989). Proximal/distal polarity is also established upon leaf initiation, with the distal leaf tip growing away from the meristem. Finally, medial/lateral polarity is established at initiation by the presence of a midvein in the center of the leaf (Sharman, 1942; Esau, 1960). Although an inherent polarity exists in leaf primordia due to their relationship to the meristem, how polarity is maintained during leaf development as the leaf grows away from the meristem is not known.
A developmental genetic approach has provided insight into how abaxial/adaxial polarity is established and maintained in leaves. Phenotypic analyses in phantastica mutants of Antirrhinum majus provided the basis for an elegant model in which the juxtaposition of abaxial and adaxial cell fates leads to lamina growth (Waites and Hudson, 1995; Waites et al., 1998). Additional genes have been described that are specifically expressed in the abaxial or adaxial domains of the leaf (Kidner and Timmermans, 2007). Arabidopsis thaliana kanadi1 (kan1) mutants were first identified in screens for the presence of trichomes in the abaxial epidermis of the first two rosette leaves, which normally lacks trichomes (Kerstetter et al., 2001), and as enhancers of mutations in CRABS CLAW, a YABBY gene that is required for abaxial identity in carpels (Eshed et al., 1999). Like the YABBY genes, KAN1 and the other three KAN genes in Arabidopsis are expressed in lateral organs and encode transcription factors that redundantly specify abaxial fate. kan1 kan2 and kan1 kan2 kan3 loss-of-function mutants develop radial leaf primordia, abaxial outgrowths in the blade, and vascular bundles with xylem surrounding phloem (Eshed et al., 2001, 2004; Emery et al., 2003).
In contrast with the abaxial expression patterns of KAN genes, the transcripts of HD-ZIPIII genes are confined to the adaxial domain of lateral organs in wild-type plants (Zhong and Ye, 1999; Eshed et al., 2001; McConnell et al., 2001; Otsuga et al., 2001; Emery et al., 2003; Juarez et al., 2004b; Prigge et al., 2005; Williams et al., 2005). Several regulatory mechanisms contribute to this adaxial localization pattern. First, HD-ZIPIII transcripts are negatively regulated by two related microRNAs, miR165 and miR166 (Juarez et al., 2004b; Kidner and Martienssen, 2004; Williams et al., 2005). Mutations that disrupt the complementarity between HD-ZIPIII transcripts and these microRNAs behave as dominant, gain-of-function alleles that expand HD-ZIPIII expression outside of the adaxial domain (Zhong et al., 1999; McConnell et al., 2001; Emery et al., 2003; Zhong and Ye, 2004; Ochando et al., 2006). Such dominant mutations not only prevent transcript cleavage but have also been correlated with changes in the methylation levels of the corresponding chromosomal loci, suggesting that chromatin modifications also participate in the transcriptional silencing of HD-ZIPIII genes (Bao et al., 2004). Second, KAN genes function as genetic repressors of HD-ZIPIII transcripts, as inferred from the ectopic localization of PHABULOSA transcripts in kan1 kan2 kan3 triple mutants and their downregulation by ectopic KAN2 expression (Eshed et al., 2004). This antagonism between KAN and HD-ZIPIII genes provides a molecular mechanism for the generation of two discrete cell populations with mutually exclusive cell fates, as predicted by the juxtaposition model. Finally, the recent discovery of a family of small leucine zipper proteins, named LITTLE ZIPPER (ZPR), has added further complexity to the abaxial/adaxial patterning regulatory network (Wenkel et al., 2007; Kim et al., 2008). ZPR proteins are upregulated by HD-ZIPIII proteins and, in turn, negatively regulate HD-ZIPIII function by forming ZPR/HD-ZIPIII heterodimers that fail to bind DNA. Consistent with this model, overexpression of ZPR proteins yields an abaxialized leaf phenotype.
The maize (Zea mays) leaf has distinct proximal/distal and adaxial/abaxial domains, each with characteristic epidermal features and internal cellular organization (Sylvester et al., 1990). The proximal domain of the maize leaf is the sheath, which wraps around the culm and provides mechanical support to the leaf. The distal domain is the blade, which lays flat to optimize light capture and photosynthesis. The boundary between the sheath and blade is occupied by the ligule fringe and the auricles. The ligule extends across the leaf on the adaxial surface, sealing the space between the sheath and the stem to exclude potentially damaging external agents. The auricles, located on either side of the midrib, act as a hinge between sheath and blade. In maize, adaxial identity can easily be assessed by the presence of a ligule at the auricle-sheath boundary and by specific features, such as bulliform cells and macrohairs, in the blade domain. Combined with the large size of maize leaves, this rich diversity of anatomical and histological markers has facilitated the interpretation of mutant phenotypes resulting from local changes in polarity.
Maize leaf mutants that affect each of the three axes of polarity have been described. Rolled leaf1 (Rld1) (Nelson et al., 2002; Juarez et al., 2004b), leafbladeless1 (lbl1) (Timmermans et al., 1998; Juarez et al., 2004a; Nogueira et al., 2007), ragged seedling2 (Henderson et al., 2005, 2006), indeterminate gametophyte1 (ig1) (Evans, 2007), and required to maintain repression6 (Parkinson et al., 2007) mutants affect abaxial/adaxial patterning in leaves. Rld1 mutants are adaxialized and carry nucleotide substitutions at the miR166 complementarity site of the maize ortholog of the HD-ZIPIII gene REVOLUTA (Juarez et al., 2004b), indicating a regulatory interaction between HD-ZIPIII transcripts and miR166 that is conserved in Arabidopsis and maize. lbl1 encodes a key component in the transacting small interfering RNA pathway, revealing that transacting small interfering RNAs are also important in leaf polarity (Nogueira et al., 2007). The ig1 mutant, which identifies a role in abaxial/adaxial patterning for a LOB domain protein, illustrates the potential of maize for identifying subtle abaxial/adaxial polarity phenotypes (Evans, 2007).
Here, we describe the isolation and cloning of milkweed pod1 (mwp1), a gene that is required for abaxial/adaxial patterning in the sheath domain. We show that mwp1 encodes a member of the KAN family of GARP transcription factors. GARP is an acronym for the founding members in which the GARP DNA binding domain was originally found: maize Golden2, ARR B-class (Arabidopsis response regulators), and Psr1 (phosphorous stress response1 from Chlamydomonas) (Riechmann et al., 2000). The abaxial side is adaxialized in the sheath of mwp1 leaves. The late onset of the phenotype, the sectored distribution of the phenotype, and the expression pattern in the epidermis are consistent with mwp1 functioning in the maintenance of abaxial/adaxial polarity. Furthermore, adaxialized sectors in mwp1 plants correlate with the ectopic expression of rld1 mRNA. The synergistic phenotype of mwp1; Rld1 double mutants shows that expression of mwp1 and rld1 is also needed early in development to pattern vasculature and photosynthetic tissues.
RESULTS
Recessive mwp1 Mutations Alter the Abaxial/Adaxial Polarity of Sheath Tissues
The reference allele of mwp1, mwp1-R, is a spontaneous recessive mutation named after the resemblance of the husk leaf-enclosed ears to the seedpods of milkweed (Asclepias syriaca) plants. mwp1-R was introgressed into the W23, Mo17, A188, and B73 inbred backgrounds for more than three generations before characterizing the phenotype. Additional alleles were obtained by transposon tagging, as described below. The phenotype of mwp1-R is representative of the other alleles.
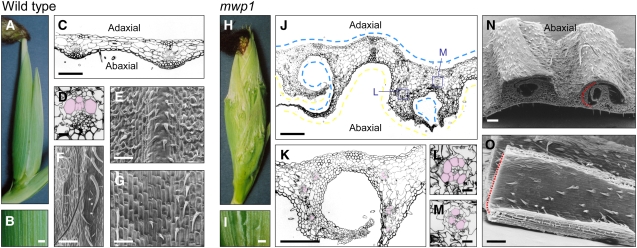
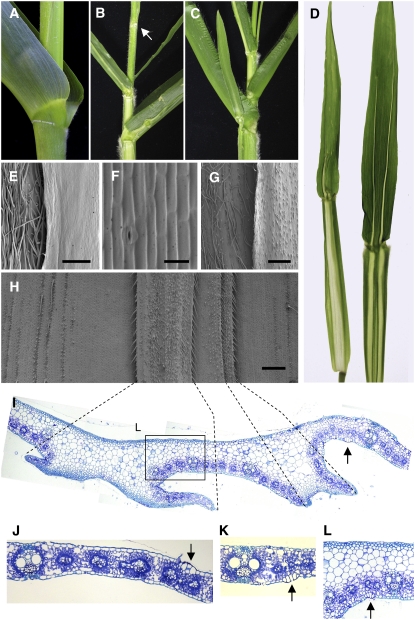
The mwp1 phenotype is most pronounced on the outer, abaxial surface of husk leaves. Whereas wild-type husk leaves have smooth surfaces (Figures 1A and 1B), mwp1-R husk leaves are rough, with outgrowths that run longitudinally, in parallel to the proximal-distal axis (Figures 1H and 1I). To examine these outgrowths, we performed scanning electron microscopy and light microscopy analyses for both wild-type and mutant husk leaves. The adaxial epidermis of wild-type husk leaves is relatively smooth, with brick-shaped cells and short prickle-like hairs (Figure 1G). The abaxial epidermis has rounded cells and more hairs (Figure 1E) with especially long hairs near the margin (Figure 1F). Scanning electron microscopy revealed that the outgrowths on mwp1-R husk leaves occur in pairs (Figures 1J, 1K, and 1N). The outer surfaces of these flaps are hairy with rounded cells (Figure 1N), therefore resembling abaxial margin tissue. The inner surface of the ectopic flaps (Figure 1O) is smooth with brick-like cells and only short prickle-like hairs, similar to those in the adaxial epidermis (Figure 1G). In transverse sections, wild-type husk leaves have a slight ribbed appearance (Figure 1C) with widely spaced vascular bundles that have xylem at the adaxial pole and phloem at the abaxial pole (Figure 1D). In the ectopic flaps of mwp1 husks, vascular bundles are oriented with xylem facing the adaxialized, inner side of the flaps (Figure 1K). Vascular bundles at junctions between flaps and the main lamina are often partially or fully radialized (Figure 1J, boxes), with xylem surrounding the phloem (Figures 1L and 1M). These phenotypes suggest that the ectopic flaps result from localized adaxialization of abaxial tissues and are consistent with the juxtaposition model (Waites and Hudson, 1995).
Figure 1.
Phenotype of Wild-Type and mwp1-R Husk Leaves.
(A) to (G) Wild-type husk leaves.
(A) Wild-type ear covered by smooth husk leaves.
(B) Magnified view of the surface of a wild-type husk leaf.
(C) Transverse section through a wild-type husk leaf.
(D) Close-up view of a vascular bundle, showing a collateral distribution of adaxial xylem (pseudocolored pink) and abaxial phloem.
(E) Scanning electron micrograph of the abaxial epidermis.
(F) Scanning electron micrograph of the abaxial epidermis near the margin, showing characteristically long hairs.
(G) Scanning electron micrograph of the adaxial epidermis.
(H) to (O) mwp1-R husk leaves.
(H) mwp1-R ears are covered by rough husk leaves with outgrowths.
(I) Magnified view of the surface of a mwp1-R husk leaf.
(J) Transverse section through a mwp1-R husk leaf, showing the development of pairs of flaps flanking sectors of adaxialized epidermis on the abaxial side. Parts of the epidermis with adaxial and abaxial characteristics are indicated by blue and yellow lines, respectively.
(K) Magnified view of (J), showing that the vascular bundles in the flaps have xylem oriented toward the inner, adaxialized side.
(L) and (M) Examples of radialized bundles from mwp1-R leaf flaps, with xylem at the periphery (pseudocolored pink).
(N) Scanning electron micrograph of a piece of mwp1-R husk leaf, showing the hairy outer surface of the flaps, characteristic of abaxial margin identity.
(O) Scanning electron micrograph of the inner surface of a flap, showing adaxial sheath characteristics.
Bars = 1 mm in (B) and (I), 20 μm in (D), (L), and (M), and 200 μm in all others.
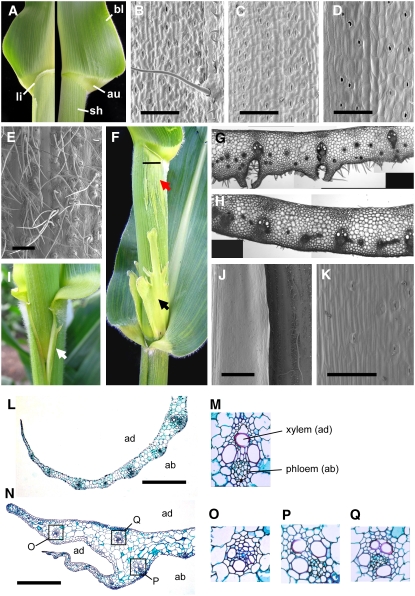
Given that the mwp1 defect was first identified in husk leaves, which consist almost entirely of sheath tissue, we next examined the sheath of leaves on the main shoot for signs of adaxialization. Most mwp1-R leaves have an ectopic ligule on the abaxial surface at the auricle-sheath boundary (Figure 2F), at a location on the proximal/distal axis matching the normal, adaxial ligule (Figure 2A). Depending on the inbred background, the ectopic ligule may extend the entire width of the leaf or only a portion of the leaf (Figure 2F). Ectopic ligules were often associated with sectors of lighter green color in the sheath tissue immediately below. Transverse sections through normal sheaths revealed large vascular bundles alternating with intermediate bundles, both of which are connected to the epidermis through hypodermal sclerenchyma, and a few small bundles that are separated from the epidermis by several cell layers (Figure 2H) (Russell and Evert, 1985). Sections through the light-green sectors of mwp1-R sheaths revealed large, correctly patterned bundles alternating with groups of small bundles that are separated from the abaxial surface by proliferating ground tissue (Figure 2G). This defect may account for the lighter pigmentation and rough surface of the sectors. Because vascular patterning is a continuous, hierarchical process (Nelson and Langdale, 1992), this alternating pattern suggests that mwp1 mutations do not disrupt the abaxial/adaxial polarity at the time of leaf initiation but rather that they fail to maintain organ polarity at a relatively late developmental stage.
Figure 2.
The mwp1 Phenotype in Vegetative Leaves.
(A) Wild-type leaves consist of three domains along the proximal/distal axis: sheath (sh), auricle (au), and blade (bl). The ligule (li) normally develops at the sheath-auricle boundary on the adaxial side (left) and is absent from the abaxial side (right).
(B) to (E) Different domains of the wild-type leaf are distinguishable based on epidermal characteristics.
(B) Adaxial blade, showing macrohairs.
(C) Abaxial blade, lacking macrohairs.
(D) Adaxial sheath, which is not hairy.
(E) Abaxial sheath, which is hairy.
(F) mwp1-R mutants have an ectopic ligule at the sheath-auricle boundary as well as adaxialized sectors (red arrow) that extend below the ligule and above the node, sometimes with development of large laminar flaps (black arrow). The horizontal line marks the position of the section shown in (G).
(G) Transverse section through a mwp1 mutant sheath, showing ectopic cell proliferation and extra vascular bundles lacking the abaxial hypodermal sclerenchyma.
(H) Transverse section through the wild-type sheath.
(I) Single flaps (white arrow) often develop at the sheath margins.
(J) Scanning electron micrograph of the margin region, including a nonhairy margin flap (left) that develops adjacent to normal-looking hairy sheath (right).
(K) Close-up view of the flap epidermis showing absence of hairs.
(L) Transverse section across the tapered sheath margin of a wild-type leaf.
(M) Close-up view of a wild-type vascular bundle, showing adaxial (ad) xylem and abaxial (ab) phloem.
(N) Transverse section through the margin region of a mwp1-R mutant leaf, showing the true margin (blunt) and the ectopic margin (tapered).
(O) to (Q) Close-up views of mwp1-R vascular bundles, with xylem at a peripheral location, as in (O) and (Q), or correctly oriented, as in (P).
Bars = 500 μm in (E), 1 mm in (J), and 200 μm in all others.
Sectors of adaxialized sheath were also found at the base and margins of the sheath (Figures 2F and 2I), but no adaxialization defects were found in the blade (data not shown). The adaxialization at the base of the sheath tends to occur in bilaterally symmetric sectors that occasionally grow into irregularly shaped laminar projections (black arrow in Figure 2F). The adaxialization at the sheath margin, revealed by vertical patches of shiny, pale-green tissue on both sides of the sheath (Figure 2I), is accompanied by a blunt true margin and development of an ectopic flap on the abaxial side (Figure 2N). The margin of the ectopic flap was tapered, similar to normal sheath margins (Figure 2L). The epidermis between the true and ectopic margins was smooth and similar to normal adaxial epidermis, whereas the rest of the abaxial epidermis was hairy (Figure 2I). The vascular bundles near the true margin of mwp1-R leaves had xylem on both poles and phloem at the center (Figures 2O and 2Q), in contrast with the collateral arrangement of the vascular bundles in normal-looking parts of mwp1 mutant and wild-type leaves (Figures 2M and 2P). The blunt shape of the true margin (Figure 2N), which is adaxialized, and the tapered shape of the ectopic margin, which has normal adaxial/abaxial polarity, may reflect a requirement for adaxial and abaxial cell types for proper margin development. Because the sheath margins are the last part of the leaf to differentiate (Sylvester et al., 1990; Scanlon, 2003), the distribution of adaxialized sectors in the sheath of mwp1 mutants is likely to reflect a requirement for mwp1 activity late in leaf development.
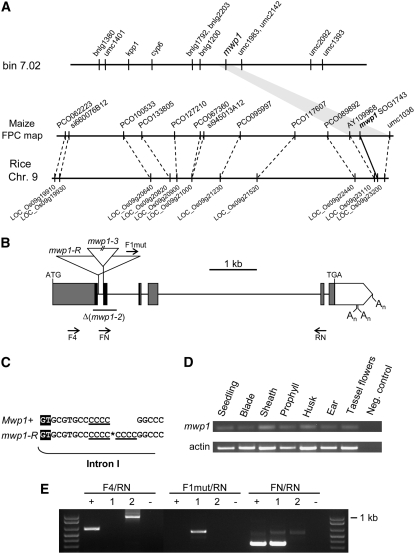
Positional Cloning of mwp1
Using a mapping population of 600 individuals, we placed mwp1 in bin 7.02, within a 0.65-centimorgan interval flanked by two polymorphic markers (AY109968 and umc1036) that reside in contig 309 of the agarose FPC physical map (7/19/05 release) (Figure 3A). Twelve markers in contig 309 detect sequences located on rice chromosome 9 in the same relative order (Figure 3A; see Supplemental Table 1 online). One of these markers was a sorghum genomic clone (SOG1743; accession number BH245825) with sequence similarity to members of the KAN gene family in Arabidopsis (Eshed et al., 2001; Kerstetter et al., 2001), suggesting that the mwp1 interval contained a KAN gene. The two partial genomic sequences that most likely corresponded to the maize ortholog of the sorghum gene, AZM4_25886 and AZM4_123646, were found to be absolutely linked to the mwp1 phenotype and could be amplified from BAC clones detected by the sorghum probe. Given the similarity between the mwp1 and kan phenotypes, we pursued this gene as a candidate.
Figure 3.
Molecular Cloning and Characterization of mwp1 Alleles.
(A) Mapping strategy used to clone mwp1. mwp1 was first mapped to bin 7.02 using molecular markers and subsequently assigned to an interval defined by markers AY109968 and umc1036 on a contig of the physical map with synteny to rice chromosome 9.
(B) Structure of the mwp1 transcriptional unit, which consists of six exons (boxes) that encode an open reading frame (shaded boxes) with similarity to members of the KAN protein family. The sequence encoding the GARP domain characteristic of these proteins is in black. Triangles mark the insertions in the mwp1-R and mwp1-3 alleles. The region deleted in the mwp1-2 allele is indicated by a horizontal line. Arrows (not to scale) indicate the approximate position of some of the oligonucleotides used. An, polyadenylation sites.
(C) Duplication of four nucleotides at the insertion site of a retrotransposon in mwp1-R. A retrotransposon was found inserted at the site marked by an asterisk, 13 nucleotides downstream of the exon/intron boundary, and was flanked by a direct repeat of four nucleotides (underlined). The first two nucleotides (GT) of intron I are highlighted.
(D) RT-PCR analysis of mwp1 expression in assorted tissues.
(E) RT-PCR analysis of mwp1 expression in wild-type and mutant seedlings. Primers F4 and RN fail to amplify a band using cDNA from mwp1-R mutant seedlings but amplify larger products in mwp1-2 due to mis-splicing. Primers F1-mut (specific to the mwp1-R insertion) and RN amplify a processed chimeric transcript only in mwp1-R, consisting of retrotransposon and gene sequences. Primers FN (which is specific to the region deleted in the mwp1-2 allele) and RN amplify bands of the expected size using cDNA of wild-type and mwp1-R seedlings. Lanes correspond to the wild type (+), mwp1-R (1), mwp1-2 (2), and water control (−). Molecular weight marker is 1 kb Plus (Invitrogen).
mwp1-R is thought to have originated spontaneously in crosses involving inbred lines and tropical land races. PCR amplification of the KAN candidate gene in mwp1-R plants yielded a band 1.8 kb larger than the one obtained from several wild-type strains tested, including the A188, B73, Mo17, W22, and A632 inbred lines. We cloned and sequenced this fragment and found an insertion within the small first intron, 13 bp downstream of the splice donor site (Figures 3B and 3C). Consistent with the spontaneous origin of mwp1-R, the inserted sequence seems to correspond to a retrotransposon, as it is flanked by identical 479-bp-long terminal repeats in direct orientation, each with the canonical TG-CA terminal nucleotides. The insertion caused the tandem duplication of four nucleotides at the insertion site (Figure 3C).
To isolate additional mwp1 mutant alleles, we crossed mwp1-R to lines carrying Mutator (Mu) transposons. Two additional alleles, mwp1-2 and mwp1-3, were identified in noncomplementation screens among ∼28,000 F1 plants. To identify the insertion sites in these new alleles, we searched for Mu insertions using PCR, by combining mwp1-specific primers with a Mu-out primer. No insertion was present in the mwp1-2 allele, but instead we found a 443-bp deletion that was absent from the progenitors (Figure 3B). The deletion in mwp1-2 affects the first two exons; therefore, mwp1-2 transcripts are expected to be mis-spliced. The resulting sequence is predicted to encode a nonfunctional protein lacking most of the conserved GARP domain and truncated by a premature stop codon. The mwp1-3 allele was found to carry a Mu element inserted at the splice acceptor site of the first intron and thus is also expected to interfere with normal intron splicing. Combined, the lesions identified in three independently isolated alleles provide strong evidence that mwp1 corresponds to this KAN gene.
mwp1 Encodes a Conserved KAN Transcription Factor
The mwp1 transcriptional unit consists of six exons that encode a predicted protein of 477 amino acids with an estimated molecular mass of 48.36 kD (Figure 3B). Several polyadenylation sites were identified downstream of the stop codon using 3′ rapid amplification of cDNA ends (RACE). Although we failed to identify the 5′ untranslated region of mwp1 by means of 5′ RACE, the position of the translation start site is supported by our RT-PCR results and by the high sequence similarity to barley (Hordeum vulgare) and switchgrass (Panicum virgatum) ESTs (accession numbers BI777239 and FE615893) and the rice (Oryza sativa) genome. The most similar sequence in rice belongs to a predicted gene, LOC_Os09g23200, which can be considered the ortholog of mwp1 based on sequence identity and chromosomal synteny. The GARP domains of mwp1 and LOC_Os09g23200 are identical and differ only in a single amino acid residue from those of the Arabidopsis KAN1 and KAN2 proteins.
Using RT-PCR, we found evidence for the transcriptional activity of mwp1 in all tissues and organs tested, including seedlings, vegetative leaves, prophylls, husk leaves, ears, and immature tassel flowers (Figure 3D). BLAST searches detected three ESTs matching the 3′ untranslated region (BM500926, DW902148, and DW890724), two of which are from a cDNA library of laser capture microdissected shoot apical meristems (Emrich et al., 2007). No transcripts were detected by RT-PCR in mwp1-R mutant seedlings using primers flanking the insertion, but we managed to amplify chimeric transcripts in mwp1-R, originating within the insertion and extending into the 3′ part of the gene, by combining gene-specific primers with primers for the retrotransposon (Figure 3E). Mis-spliced transcripts, with various combinations of intronic and exonic sequences, were amplified in mwp1-2 using primers flanking the deletion, as a result of the use of cryptic and normal splice sites (Figure 3E).
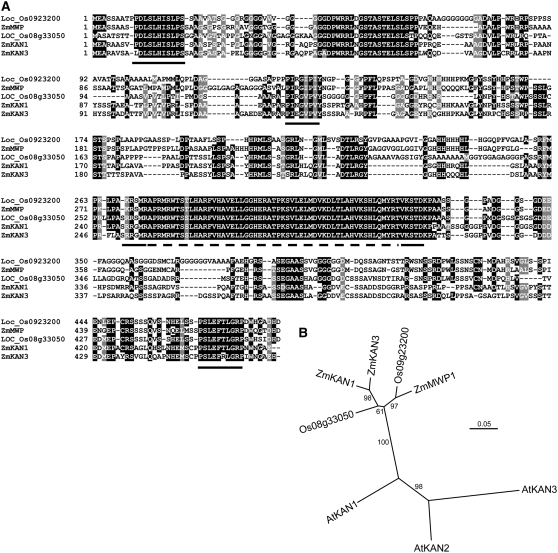
The rice genome encodes at least six members of the KAN family; the most similar paralog of LOC_Os09g23200 is LOC_Os08g33050. Our search for KAN genes among available B73 genomic and cDNA sequences indicates that the maize genome encodes at least three genes closely related to LOC_Os09g23200 and LOC_Os08g33050, indicating that the family has expanded during maize evolution. To clarify the phylogenetic relationship between mwp1 and its closest homologs, we performed a phylogenetic analysis using the aligned nucleotide sequences encoding the GARP domains of LOC_Os09g23200 and LOC_Os08g33050 with those of mwp1 and the two most similar genes in the maize genome (named Zm KAN1 and Zm KAN3), which were identified in the sequence of the c0112F01 (chromosome 1) and c0072N10 (chromosome 4) BACs, respectively. Our analysis further supported that mwp1 is an ortholog of LOC_Os09g23200 and suggests that mwp1 represents a single-copy gene in the maize genome. By contrast, the two maize genes closest to mwp1 are more similar to each other than to mwp1 and LOC_Os08g33050, possibly representing a duplicate pair of genes (Figure 4; see Supplemental Figure 1 online). Trees with the same topology were obtained when the predicted full-length sequences of the proteins were used (data not shown). Previous authors have pinpointed additional regions in KAN proteins that are highly conserved across the dicotyledonous plants Arabidopsis and Ipomoea (Iwasaki and Nitasaka, 2006). These domains were also uncovered by our alignment of maize and rice sequences, suggesting that they are also important for KAN function in monocots (Figure 4A). Based on sequence comparisons of these domains, mwp1 is most similar to KAN1.
Figure 4.
Sequence Alignment and Phylogenetic Analysis of mwp1 and Related KAN Genes.
(A) Alignment of the amino acid sequences of mwp1 and the four most similar proteins in maize and rice. A dashed line indicates the GARP domain. Black lines indicate additional conserved regions in KAN proteins, first identified by Iwasaki and Nitasaka (2006), which are also conserved in KAN proteins from grasses.
(B) Neighbor-joining phylogenetic tree based on an alignment of the nucleotide sequence coding for the GARP domain. Numbers at the branches are percentages based on 10,000 bootstrap repetitions.
Expression Analysis of mwp1 and rld1
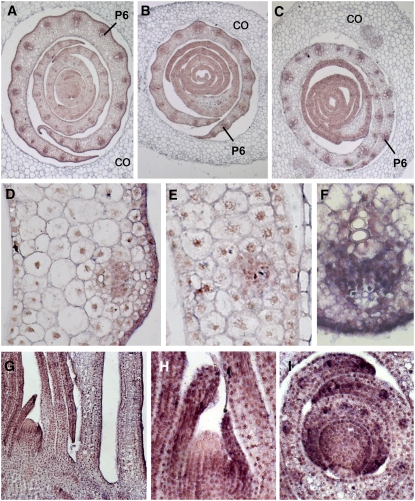
In situ hybridization was performed to determine the tissue localization of mwp1. In wild-type seedlings, mRNA expression of mwp1 was most intense in the abaxial epidermis of the sheath, clearly visible at plastochron 6 (P6) (Figures 5A and 5D). A similar trend was also observed in leaves that initiated later in development. In young leaf primordia (P1-3), expression was seen throughout the primordia, but more pronounced at the margins and in the vasculature (Figures 5F to 5I). The expression at the margin may reflect the requirement for mwp1 activity inferred from the observed margin phenotypes (Figure 2I). Longitudinal sections showed that mwp1 expression was absent from the apex of the meristem (Figures 5G and 5H), a region where rld1 expression is high (Juarez et al., 2004b). Consistent with the expression patterns of KAN genes in Arabidopsis (Kerstetter et al., 2001), transverse sections showed reduced expression in the center of the meristem (Figure 5I). Expression was reduced, but not eliminated in mwp1-2 (Figure 5B), which has a deletion but is not an RNA null (Figure 3E).
Figure 5.
In Situ Hybridization Analysis of mwp1 Expression.
(A) to (C) Transverse sections of young seedlings 2 d after germination, showing several developing leaves covered by the coleoptile (CO). The oldest (most external) leaf primordia are at plastochron 6 (P6) stage and were sectioned at the level of the sheath.
(A) Wild-type seedling.
(B) mwp1-2 seedling.
(C) Rld1-N1990 seedling.
(D) Magnified view of the sheath of a wild-type developing leaf, showing higher expression in the abaxial epidermis.
(E) Magnified view of (C) showing reduced expression in the abaxial epidermis.
(F) Close-up view of a wild-type vascular bundle showing mwp1 expression between the xylem and phloem.
(G) Longitudinal section of a wild-type shoot apical meristem and the surrounding leaf primordia.
(H) Close-up view of the meristem in (G). Expression is absent from the apex of the meristem.
(I) Transverse section of a wild-type shoot apical meristem showing strong expression in the margins of P2 and P3 leaf primordia.
We also studied the expression of mwp1 in dominant Rld1-N1990 mutants, which carry a point mutation that disrupts the complementarity between rld1 transcripts and the miR166 microRNA (Juarez et al., 2004b). In contrast with wild-type plants, mwp1 transcripts were absent from the abaxial epidermis of Rld1-N1990 mutants (Figures 5C and 5E), as expected if rld1 is a direct or indirect negative regulator of mwp1.
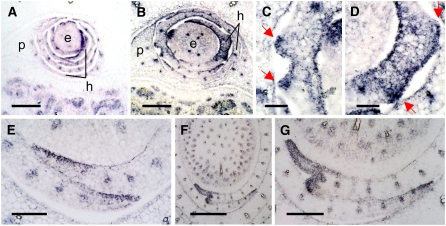
The expression pattern of rld1 (Juarez et al., 2004a, 2004b) was examined to determine if genes that are normally expressed adaxially are misexpressed in mwp1 mutants. rld1 was expressed on the adaxial side of wild-type husk leaf primordia and persisted at the margins and in vascular bundles (Figure 6A) in a pattern similar to that seen in vegetative leaves (Figure 6E). Ectopic outgrowths on the abaxial side of developing mwp1 husk leaves were associated with rld1 misexpression (Figures 6B to 6D). In the region delimited by the outgrowths, rld1 was expressed both adaxially and abaxially, with strong expression in the abaxial epidermis (Figures 6C and 6D). rld1 expression was also examined in mwp1-R leaf primordia with developing sheath margin outgrowths. In the wild type, rld1 was expressed on the adaxial side of young leaf primordia. Expression persisted at the margins and in developing vascular bundles of older primordia (Figure 6E). However, in mwp1-R leaves, rld1 expression was seen throughout the sheath margin (Figures 6F and 6G). The boundary between rld1-expressing and nonexpressing tissues corresponded to the position of the developing outgrowth. These results suggest that the expression of mwp1 at the abaxial margin is required to keep rld1 restricted to the adaxial margin, the loss of mwp1 function leading to a pronounced defect in this location (Figures 2I and 2N).
Figure 6.
In Situ Hybridization Analysis of rld1 Expression in Wild-Type and mwp1 Plants.
(A) and (B) Transverse sections of female axillary shoots, showing the developing ear (e) covered by several husk leaves (h) and a prophyll (p).
(A) The wild type.
(B) mwp1-R showing ectopic abaxial expression of rld1 in the husk leaves associated with developing pairs of leaf flaps.
(C) and (D) Magnified views of the developing flap pairs in (B). The pairs of flaps in are indicated by red arrows.
(E) Wild-type vegetative leaf margins showing adaxial rld1 expression.
(F) mwp1-R vegetative leaves showing ectopic rld1 expression and a developing margin flap.
(G) Magnified view of (F).
Bars = 50 μm in (C) and (D), 500 μm in (F), and 200 μm in all others.
The Phenotype of Rld1/rld+; mwp1 Double Mutants Is Synergistic
To further investigate the functional relationship between mwp1 and rld1, we analyzed the double mutant. Rld1 mutants are characterized by upwardly curving leaf blades and by the presence of patches of ligule (Figure 7A), macrohairs, and bulliform cells (arrow in Figure 7K) on the abaxial side of the leaf (Nelson et al., 2002; Juarez et al., 2004b). In addition, small sectors of pale green tissue lacking the minor vasculature have been reported in the blade and the sheath (Nelson et al., 2002).
Figure 7.
The Synergistic Phenotype of mwp1; Rld1 Double Mmutants.
(A) Rld1-N1990/rld1+ mutant.
(B) mwp1-R; Rld1-N1990/rld1+ double mutant showing longitudinal sectors that extend the length of sheath and blade. A leafless node is indicated by a white arrow.
(C) Same genotype as in (B) but with an ectopic blade fused to and developing from a sector in the main leaf.
(D) mwp1-R; Rld1-N1990/rld1+ double mutant leaves extended to show the shape and propagation of the sectors from sheath to blade.
(E) Scanning electron micrograph of the sheath of a mwp1-R; Rld1-N1990/rld1+ mutant leaf, as seen from the abaxial side, showing normal-looking sheath (left side) and the adaxialized, hairless epidermis of a sector (right side).
(F) Magnified view of the adaxialized epidermis of the sector shown in (E).
(G) Sheath of mwp1-R; Rld1-N1990/rld1+ mutant leaf showing an adaxialized sector with small hairs (right side), in contrast with the hairy, normal-looking sheath of adjacent tissue.
(H) Scanning electron micrograph of the sectors and flaps observed on the abaxial surface of mwp1-R; Rld1-N1990/rld1+ leaf blades.
(I) Transverse section corresponding to the region shown in (H). Dashed lines connect equivalent positions in (H) and (I), corresponding to developing ectopic blade margins.
(J) Transverse section of an mwp1-R leaf that, like the wild type, shows bulliform cells only on the adaxial epidermis (arrow).
(K) Transverse section of a Rld1-N1990/rld1+ leaf, showing bulliform cells (arrow) on the abaxial side but not on the adaxial.
(L) Detailed view of (I), showing abaxial bulliform cells (arrow) as in (K).
The Rld1-N1990/rld+; mwp1-R double mutants had husk leaves with outgrowths similar to those of mwp1-R single mutants (data not shown). In addition, a new phenotype consisting of broad longitudinal sectors of nonphotosynthetic tissue flanked by laminar flaps was consistently observed on the abaxial side of the uppermost vegetative leaves (Figures 7B to 7D). These pale sectors were often positioned at paired, bilaterally symmetric locations on both sides of the midrib and extended along the entire sheath and most of the blade, where they were narrower but still flanked by flaps (Figure 7D). The sectors continued from leaf to leaf, suggesting that they originated in the meristem and maintained their fate through subsequent cell divisions. Sectors in consecutive leaves were connected by internode sectors with slightly lighter pigmentation, in which the vasculature failed to appress to the stem surface (see Supplemental Figure 2 online). Occasionally, laminar growth associated with these sectors gave rise to leaf-like organs fused at the sheath of the leaf from which they originated, detaching distally and differentiating into a blade and an auricle (Figure 7C). In addition, one or two nodes immediately below the lowest tassel branch were occasionally leafless, their leaves being replaced by membranous ligule-like outgrowths (Figure 7B).
To understand the nature of these sectors, cell types in the double mutant were analyzed and compared with those of the wild type. The abaxial side of the sheath was covered by hairs, as in the wild type, except at the sector where the epidermis was hairless and consisted of brick-shaped, rectangular cells and rows of stomatal complexes, resembling the epidermis of the adaxial side (Figures 7E and 7F). Occasionally, small prickle-like hairs, similar to those seen in adaxial husk leaves, were visible in the sectors (right-hand side of Figure 7G). These differed from the longer hairs in the flanking abaxial sheath (left-hand side of Figure 7G). In general, the phenotype of the Rld1-N1990/rld+ mwp1-R sheath was similar to that of the mwp1-R husk leaves. By contrast, most of the blade epidermis resembled that of Rld1-N1990/rld+ single mutants; broad areas of the epidermis showed reversed polarity, with macrohairs and bulliform cells on the abaxial surface but not on the adaxial surface (Figures 7H and 7L). Regions with reversed polarity were often intermixed with regions of normal polarity. Transverse sections through the blade sectors revealed thickened areas with symmetrically organized internal tissues and a complete absence of photosynthetic and vascular tissues (Figure 7I). Bulliform cells could be seen on the abaxial side immediately adjacent to the flaps (Figure 7L, arrow), suggesting that these cells were adaxialized. However, adjacent to these bulliform cells were areas in which macrohairs were correctly patterned: absent from the abaxial side (see hairless area surrounding the flaps in Figure 7H) and present on the adaxial side (data not shown). The abaxial surface of blade sectors themselves was covered by small hairs and, sometimes, macrohairs, suggesting that they in fact originated from adaxialization of tissues within the sector. The presence of adaxial features within the sector suggests that the juxtaposition of this localized adaxialization within a region of abaxial identity led to formation of flaps. In summary, the double mutant phenotype appears to be synergistic as the sectors of adaxialization extend the entire length of the leaf and go from leaf to leaf as if they originated in the meristem. Within the sectors, certain tissues are not simply adaxialized, but fail to differentiate at all.
DISCUSSION
mwp1-R is a recessive mutant characterized by sectors of adaxialized tissue in the sheath domain of vegetative and husk leaves. The mutant phenotype suggests a role for mwp1 in the initiation and/or maintenance of abaxial identity. We cloned mwp1 by following a positional approach and found that it encodes a member of the KAN family of GARP transcription factors. In Arabidopsis, KAN gene expression marks the abaxial side of leaves and the peripheral region of the meristem (Kerstetter et al., 2001). Similarly, we found mwp1 expressed in the abaxial epidermis, consistent with a function in promoting abaxial identity and repressing adaxial identity. Although mwp1 transcripts were found in sheath and blade, we only observed adaxialization in the sheath of mwp1 mutant leaves. In Arabidopsis, similar severe phenotypes are only observed in double or triple kan mutant combinations. The observation of a mutant phenotype in mwp1 single mutants suggests that some KAN genes have evolved unique functions in grass leaves and therefore are not as functionally redundant as their Arabidopsis counterparts.
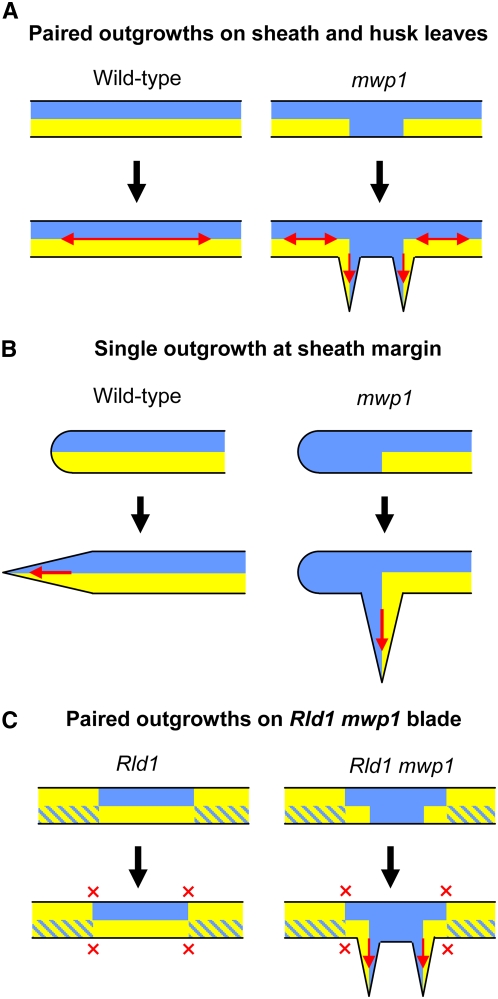
The mwp1 phenotype was particularly evident in husk leaves, with pairs of flaps that often extended the entire sheath length. In Arabidopsis, disorganized growth, expansion of the blade in various planes, or development of ectopic leaf-like organs have been reported for kan1 kan2 (Eshed et al., 2001) and kan1 kan2 kan4 triple mutants (Izhaki and Bowman, 2007), but their compromised development made interpretation of the phenotypes difficult in terms of local changes in abaxial/adaxial polarity. Our observations of mwp1 husk leaves allow a clearer interpretation of the phenotype and demonstrate that new planes of laminar growth develop at the sites where the adaxialized sectors and the surrounding abaxial tissue juxtapose. According to the model of Waites and Hudson (1995), a pair of ectopic leaf flaps develops when sectors with adaxial identity are flanked on both sides by cells with abaxial identity (Figure 8A). A similar model can be applied to the single leaf flaps at the sheath margin. The loss of abaxial identity at the margin generates only one new boundary between abaxial and adaxial cells and thus only one ectopic flap develops (Figure 8B). Despite the absence of morphological markers in Arabidopsis, the ectopic expression of a leaf margin reporter in the abaxial outgrowths of kan1 kan2 double mutants suggests that a similar mechanism operates in Arabidopsis (Eshed et al., 2004).
Figure 8.
Model for the Development of Laminar Outgrowths in mwp1 Mutants and in mwp1; Rld1 Double Mutants.
(A) In wild-type husk leaves, the juxtaposition of adaxial (blue) and abaxial (yellow) cell types promotes laminar growth (red arrows). In mwp1 leaves, a failure to initiate or maintain abaxial identity in husk leaves results in ectopic adaxial cell types on the adaxial side, creating new boundaries of juxtaposed abaxial and adaxial identity. As predicted by the juxtaposition model, growth then occurs at the planes defined by these new boundaries.
(B) When ectopic adaxial cell types are present at the sheath margin, a single ectopic boundary is created, resulting in the development of a single outgrowth.
(C) Rld1 mutations often cause a complete switch in the abaxial/adaxial polarity of the blade. Sectors with normal polarity are often located adjacent to sectors with reversed polarity, but this juxtaposition does not normally lead to laminar outgrowths (red crosses). Development of outgrowths in mwp1; Rld1 blades may occur in response to adaxialization in sectors of the blade that otherwise retain abaxial identity on the abaxial side.
The sectored nature of the mwp1 phenotype shows that the shift from abaxial to adaxial identity occurs locally, rather than in the sheath as a whole. The polar (along the proximal/distal axis) orientation of the mwp1 defects raises the possibility that they originate from a local failure in the maintenance or initiation of abaxial fate that is subsequently propagated clonally as leaf cells proliferate. A failure in maintaining stable transcriptional states through mitosis was also hypothesized as the cause for the mosaicism seen in other mutants, as in the incurvata2 (icu2) mutants of Arabidopsis, which ectopically express the floral identity gene AGAMOUS in leaf sectors (Serrano-Cartagena et al., 2000). Cloning of ICU2 has shown that it encodes a DNA polymerase α-subunit that physically interacts with other proteins involved in chromatin-mediated gene repression (Barrero et al., 2007). Interestingly, the abaxial outgrowths in kan1 kan2 double mutants are suppressed by loss-of-function alleles of the SWI/SNF gene SPLAYED in Arabidopsis (Eshed et al., 2004), possibly reflecting the participation of a mechanism of cellular memory that involves chromatin remodeling.
In vegetative leaves, the sheath responds to loss of mwp1 function in a different way. The adaxializing effect of mwp1 mutations is clearly shown by the development of ectopic ligules on the abaxial side. The location of such ectopic ligules at the sheath-auricle boundary indicates that the site of ligule development is specified by a combination of positional cues from the proximal/distal and the abaxial/adaxial axes. In addition, leaf flaps appeared at various locations, including the proximal end of the sheath, below the ectopic ligules, and along the sheath margin. In many cases, however, the shape, internal organization, and distribution of these sectors seem incompatible with their origin from a single, clonally propagated event. The possible involvement of a mechanism of epigenetic silencing, however, is supported by the observation of adaxialized phenotypes strikingly similar to those of mwp1-R in mutants such as required to maintain repression6 (Parkinson et al., 2007) and mediator of paramutation1 (D. Lisch, personal communication), both of which fail to maintain stable epigenetic states during paramutation. A mosaic analysis previously showed that non-cell autonomous signals from Rld1 mutant sectors trigger a reversal of polarity in the underlying wild-type cell layers (Nelson et al., 2002). Further clonal and mosaic analyses will be required to understand whether the sectoring in mwp1 mutants is clonal or involves cell-to-cell communication.
Given that maize leaves elaborate and differentiate from the tip to the base and from the midrib to the margin (Sharman, 1942; Sylvester et al., 1990; Scanlon, 2003), it appears that the mwp1 defect becomes more pronounced in tissues that differentiate late. In tissues that differentiate early, either no defect is seen (in blade and at midrib) or minor defects are seen (at the top of the sheath). Consistent with a role for mwp1 in tissues that differentiate late in leaf development, we also see a similar trend within the plant as a whole. The mwp1 mutant phenotype is not seen in juvenile leaves and becomes more severe in leaves that form late in the life of the maize plant, which includes the husk leaves that are initiated during the reproductive state.
Despite the fact that mwp1-R and Rld1 mutations both result in adaxialized leaf phenotypes, they differ in the specificity of their effects on sheath and blade, respectively. The anatomy of the sheath remains unaffected in Rld1 mutants, but the blade exhibits a distinct upward curling and an altered distribution of hypodermal sclerenchyma, bulliform cells, and macrohairs, all of which are characteristically patterned along the abaxial-adaxial axis in normal leaves (Nelson et al., 2002; Juarez et al., 2004b). Although the Rld1 epidermis has juxtaposed abaxial and adaxial cell identities, as determined by the presence of macrohairs, no flaps occur. In contrast with Rld1 mutants, we did not observe changes in the anatomy of the blade in mwp1 mutants, despite the range of mutant phenotypes present in the sheath. Outgrowths were consistently observed in the sheath of mwp1 mutants at the sites where abaxial and adaxial identities juxtapose but have not been reported in the sheath or the blade of Rld1 mutants or flanking the boundaries of rld1+ sectors in Rld1 mosaic plants (Nelson et al., 2002). Thus, Rld1 and mwp1 mutants also differ in their ability to initiate new axes of growth in leaves.
Double mutants of mwp1-R and Rld1-N1990 showed a synergistic phenotype in which the sectors and paired flaps characteristic of mwp1, normally confined to the sheath domain, extend outside the sheath and into the blade. These flaps occur in a tissue that is already adaxialized. A plausible explanation for these flaps is that the mwp1 mutation affects the abaxial identity in regions of the blade that have maintained normal polarity (Figure 8C). The sectors observed in mwp1; Rld1 double mutants have adaxial features on both surfaces but appear to be flanked on the abaxial surface by normal abaxial cell types.
The expansion of the mwp1 defect into the blade in an Rld1 mutant background may be due to a redundant KAN gene functioning in the blade in mwp1-R mutants. At least two other KAN genes, Zm KAN1 and Zm KAN3, are expressed in blade tissue (see Supplemental Figure 3 online). With such a scenario, both KAN genes would be repressed by ectopic HD-ZIPIII expression in Rld1 mutants. Rld1 single mutants have clearings observed above and below the blade-sheath boundary, which are pale green and fail to develop intermediate and transverse veins (Nelson et al., 2002). The sectors observed in mwp1; Rld1 double mutants most likely represent an enhancement of these clearings as a consequence of increased rld1 expression or a change in the timing or domain of expression beyond what is seen in Rld1 single mutants. Although previous results have suggested that Rld1 mutations behave as antimorphic or dominant-negative alleles (Nelson et al., 2002), molecular data suggest that dominant Rld1 alleles are best described as hypermorphic or neomorphic. If this is actually the case, the molecular basis of the observed enhancement may lie at the titration of ZPR proteins (Wenkel et al., 2007; Kim et al., 2008) by increased RLD1 levels, causing increased adaxialization in double mutants.
The timing of polarity establishment is likely to be critical for vascular development. The adaxialized sectors in mwp1; Rld1 double mutants demonstrate that initiation of vasculature and differentiation of photosynthetic mesophyll cells cannot proceed without abaxial identity or without the formation of adaxial-abaxial boundaries. These sectors are likely to originate early in leaf initiation, since they continue for the length of the leaf and appear to go from leaf to leaf. Their position in the leaf between the midrib and the margin may indicate a particular domain of the leaf, such as that defined by narrow sheath (Scanlon, 2000) or Wavy auricle in blade1 (Hay and Hake, 2004). By contrast, the sectors of mwp1 single mutants are confined to the sheath and do not go from leaf to leaf. Thus, correct polarity regulated by MWP1 and RLD1 is needed early in leaf development to create vasculature and later in leaf development to maintain cell identity and prevent new lamina from forming.
Other authors have proposed a dual abaxializing and adaxializing function for KAN genes based on the abaxialized vasculature seen in the petioles of some kan1 kan2 double mutants (Ha et al., 2007). One attractive possibility is that normal development in distinct leaf domains (petiole/blade in Arabidopsis or sheath/blade in maize) involves the same developmental pathways but a different interpretation of positional information. In line with this idea, it is remarkable that sheath and blade of normal maize leaves exhibit reciprocal patterns of hairiness on their adaxial and abaxial surfaces. One might also speculate as to whether the absence of laminar growth in the petioles of Arabidopsis rosette leaves is attained by regulating abaxial-adaxial patterning. Further research on the adaxial-abaxial patterning mechanisms might therefore contribute to advance our understanding of the morphogenesis of differently shaped leaf domains.
METHODS
Plant Materials
The mwp1-R allele was obtained from the Maize Genetics Cooperation Stock Center (accession number 5804F; http://maizecoop.cropsci.uiuc.edu/) and backcrossed at least four times to several inbreds, including B73, A632, W23, and A188. An Rld1-N1990 mutant line that had been backcrossed twice to W23 was also obtained from the stock center (accession number 927K). Double mutants were identified in the F2 generation of a cross of this line and mwp1-R. The Rld1-N1990/rld1+; mwp1-R double mutant has since been maintained by repeated backcrossing to mwp1-R mutant introgressed in W23. Plants were grown in our Gill Tract summer nursery or in the greenhouse for phenotypic and molecular studies in Albany, CA.
Isolation of mwp1-2 and mwp1-3
Two additional alleles were identified in the F1 progeny of crosses of females carrying active Mu elements and mwp1-R males. F1 plants were field grown and screened either in the Gill Tract or at the University of Illinois at Urbana-Champaign. Putative mutants were crossed to one or more inbred lines, and their F1 progenies self-pollinated to establish F2 families segregating either mwp1-R or the newly induced allele. mwp1-2 was outcrossed to several inbred backgrounds for four generations. No families segregating the mwp1-3 allele have been found, suggesting a transmission problem or that the insertion in the original mutant was somatic. However, we managed to amplify a Mu insertion in mwp1-3 using DNA purified from mutant tissue collected at the time of the screen.
Mapping and Characterization of Alleles
Genetic linkage was first detected between mwp1-R and the visible Rough sheath1 mutant, which maps to chromosome 7, in a family segregating both traits. Linkage to chromosome 7 was then confirmed using bulked segregant analysis (Michelmore et al., 1991) of simple sequence repeat markers and subsequently tested on individual plants. Our mapping population was the progeny of a testcross segregating mwp1 and wild-type plants in a 1:1 ratio. Information on the simple sequence repeat markers used (shown in Figure 3) is available from the MaizeGDB website (http://www.maizegdb.org). We identified additional single nucleotide polymorphisms in genes previously localized in the maize FPC physical map (http://www.genome.arizona.edu/fpc/WebAGCoL/maize/WebFPC/) or in genes selected based on the synteny with the rice genome, which were likely to map to the candidate interval. Such single nucleotide polymorphic markers were scored by direct sequencing of PCR fragments spanning the polymorphism, using an ABI 3100 genome analyzer and BigDye Terminator v3.1 cycle sequencing chemistry (Applied Biosystems).
For characterizing the alleles, primers were designed to amplify the genomic region defined by the AZM4_25886 and AZM4_123646 contigs and to bridge the gap between them. Two such primers, mwp-F4 (5′-CTGCCAATCCGAGGGATAC-3′) and mwp-R4 (5′-TTTCATTCACCGTCTGGAGTT-3′), amplified a bigger fragment when genomic DNA of mwp1-R was used as the template. The same primers detected a smaller fragment in mwp1-2. The Mu insertion of the mwp1-3 allele was detected by PCR combining gene-specific primers with Pioneer's 9242 Mu-specific degenerate primer (5′-AGAGAAGCCAACGCCAWCGCCTCYATTTCGTC-3′) (Blauth et al., 2001).
Light Microscopy
For plastic sections, the tissue was fixed overnight in FAA (50% ethanol, 5% acetic acid, and 3.7% formaldehyde), dehydrated through a series of increasing ethanol concentration, and embedded in Technovit 7100 according to the manufacturer's instructions (Heraeus Kulzer). Alternatively, the tissue was fixed in a solution containing 2% glutaraldehyde and 2% paraformaldehyde in 0.1 M phosphate buffer and embedded in Procure 812 (ProSciTech). Sections of 1 to 4 μm thickness were cut using an HM340 microtome (Microm), stained with 0.05% (w/v) Toluidine blue O, rinsed with water, allowed to dry, mounted on slides with Permount (Fisher Scientific), and photographed in a bright-field microscope.
Scanning Electron Microscopy
For scanning electron microscopy, plant tissue was fixed overnight as described above, dehydrated with ethanol, and critical-point dried with liquid CO2. Samples were then coated with gold palladium using a Desk II sputter coater (Denton Vacuum) for 45 s and examined on a Hitachi S-4700 scanning electron microscope. Alternatively, samples were coated with 25-nm gold using a Polaron E 5400 sputter coater (SCD-050; Bal-Tec) and examined on a Cambridge 250 Mark III scanning electron microscope (Cambridge InstrumentsK).
Gene Expression Analysis
Total RNA was purified from the aboveground, green parts of 2-week-old seedlings of the B73 inbred line using Trizol reagent according to the manufacturer's instructions (Invitrogen). For 3′ RACE, RNA was treated with the GeneRacer RACE Ready cDNA kit and reverse transcribed with SuperScriptIII reverse transcriptase and the kit's oligo(dT) primer according to the manufacturer's instructions (Invitrogen). The resulting cDNA was used in nested PCR reactions with the gene-specific 3RACE-1F (5′-GGCTGTCGTCGAATTCATGCAACA-3′) and 3RACE-2F (5′-GCAGCCCGAGCCTGGAGTTCAC-3′) primers, which were combined with the GeneRacer 3′ and GeneRacer 3′ nested primers from the kit. The resulting bands were extracted from the gel, cloned into pGEM-T Easy (Promega), and sequenced to determine the position of the polyadenylation sites. For RT-PCR amplification of wild-type and mutant transcripts, the mwp1-F4 (see above), mwp1-RT-FN (5′-CATGGACGTGAAGGATCTGACC-3′), and mwp1-Fmut (5′-GGGGAATACAAGGGTCCAGT-3′) forward primers were combined with the mwp1-RT-RN (5′-GATGAGCATCCATGTTGCATGA-3′) reverse primer.
Phylogenetic Analysis
Alignments of predicted full-length amino acid sequences were done automatically using MUSCLE software (Edgar, 2004) and subsequently refined manually. Alignments were also done with the segment of the nucleotide sequence that encodes the conserved GARP domain. Consensus phylogenetic trees were constructed for the aligned sequences with MEGA version 3.1 (Kumar et al., 2004) using the neighbor-joining and minimum evolution methods. Bootstrap values in the bootstrap consensus tree were produced with 10,000 replications.
In Situ Hybridization
In situ hybridization was performed using the method of Jackson (1991), as modified by Bortiri et al. (2006). To generate a probe, a fragment of the first exon upstream of the GARP domain was amplified with mwp-probe-F (5′-GAGGGTGTCGGTGAGCAT-3′) and mwp-probe-R (5′-GAACGGGAGGAGGAGGAG-3′) primers and cloned into pGEM-T Easy (Promega).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: BAC clone c0112F01 (AC199826), BAC clone c0072N10 (AC184836), mwp1 cDNA (EU925398), KAN1 cDNA (EU935003), and KAN3 (EU925399).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of Nucleotide Sequences.
Supplemental Figure 2. Cross Section of Rld1; mwp1 Double Mutant Stem.
Supplemental Figure 3. RT-PCR Analysis of KAN1 and KAN3 Expression in Maize.
Supplemental Table 1. Maize-Rice Synteny for Selected Markers in Contig 309.
Supplemental Data Set 1. Alignment of Protein Sequences (from Figure 4A).
Supplemental Data Set 2. Alignment of Nucleotide Sequences (Used for the Tree in Figure 4B).
Acknowledgments
This work was supported by grant NSF IOS 0445387 and USDA–Agricultural Research Service funding to S.H. and a Marsden grant to T.F. (HRT0501) from the Royal Society of New Zealand. H.C. was a recipient of a postdoctoral fellowship of the Ministerio de Educación y Ciencia of Spain and R.J. of a FRST Bright Futures Scholarship. We thank Torbert Rocheford for field space, Marty Sachs and the Maize Genetics Cooperation Stock Center for maize strains, David Hantz and Julie Calfas for greenhouse support, Delilah F. Wood and Tina Williams for the use of the electron microscopy facility, Doug Hopcroft and Raymond Bennett for assistance with scanning electron microscopy, and Marja Timmermans for providing the rld1 probe. We also wish to acknowledge the helpful discussions and editing from lab members.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Sarah Hake (maizesh@nature.berkeley.edu).
Online version contains Web-only data.
References
- Bao, N., Lye, K.W., and Barton, M.K. (2004). MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev. Cell 7 653–662. [DOI] [PubMed] [Google Scholar]
- Barrero, J.M., Gonzalez-Bayon, R., del Pozo, J.C., Ponce, M.R., and Micol, J.L. (2007). INCURVATA2 encodes the catalytic subunit of DNA polymerase alpha and interacts with genes involved in chromatin-mediated cellular memory in Arabidopsis thaliana. Plant Cell 19 2822–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blauth, S.L., Yao, Y., Klucinec, J.D., Shannon, J.C., Thompson, D.B., and Guilitinan, M.J. (2001). Identification of Mutator insertional mutants of starch-branching enzyme 2a in corn. Plant Physiol. 125 1396–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortiri, E., Chuck, G., Vollbrecht, E., Rocheford, T., Martienssen, R., and Hake, S. (2006). ramosa2 encodes a LATERAL ORGAN BOUNDARY domain protein that determines the fate of stem cells in branch meristems of maize. Plant Cell 18 574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R.C. (2004). MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery, J.F., Floyd, S.K., Alvarez, J., Eshed, Y., Hawker, N.P., Izhaki, A., Baum, S.F., and Bowman, J.L. (2003). Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 13 1768–1774. [DOI] [PubMed] [Google Scholar]
- Emrich, S.J., Barbazuk, W.B., Li, L., and Schnable, P.S. (2007). Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res. 17 69–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau, K. (1960). Anatomy of Seed Plants. (New York: Wiley).
- Eshed, Y., Baum, S.F., and Bowman, J.L. (1999). Distinct mechanisms promote polarity establishment in carpels of Arabidopsis. Cell 99 199–209. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Baum, S.F., Perea, J.V., and Bowman, J.L. (2001). Establishment of polarity in lateral organs of plants. Curr. Biol. 11 1251–1260. [DOI] [PubMed] [Google Scholar]
- Eshed, Y., Izhaki, A., Baum, S.F., Floyd, S.K., and Bowman, J.L. (2004). Asymmetric leaf development and blade expansion in Arabidopsis are mediated by KANADI and YABBY activities. Development 131 2997–3006. [DOI] [PubMed] [Google Scholar]
- Evans, M.M. (2007). The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo Sac and leaf development. Plant Cell 19 46–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha, C.M., Jun, J.H., Nam, H.G., and Fletcher, J.C. (2007). BLADE-ON-PETIOLE 1 and 2 control Arabidopsis lateral organ fate through regulation of LOB domain and adaxial-abaxial polarity genes. Plant Cell 19 1809–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay, A., and Hake, S. (2004). The dominant mutant Wavy auricle in blade1 disrupts patterning in a lateral domain of the maize leaf. Plant Physiol. 135 300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson, D.C., Muehlbauer, G.J., and Scanlon, M.J. (2005). Radial leaves of the maize mutant ragged seedling2 retain dorsiventral anatomy. Dev. Biol. 282 455–466. [DOI] [PubMed] [Google Scholar]
- Henderson, D.C., Zhang, X.L., Brooks, L., and Scanlon, M.J. (2006). RAGGED SEEDLING2 is required for expression of KANADI2 and REVOLUTA homologues in the maize shoot apex. Genesis 44 372–382. [DOI] [PubMed] [Google Scholar]
- Iwasaki, M., and Nitasaka, E. (2006). The FEATHERED gene is required for polarity establishment in lateral organs especially flowers of the Japanese morning glory (Ipomoea nil). Plant Mol. Biol. 62 913–925. [DOI] [PubMed] [Google Scholar]
- Izhaki, A., and Bowman, J.L. (2007). KANADI and class III HD-Zip gene families regulate embryo patterning and modulate auxin flow during embryogenesis in Arabidopsis. Plant Cell 19 495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, D. (1991). In situ hybridization in plants. In Molecular Plant Pathology: A Practical Approach, D.J. Bowles, S.J. Gurr, and M. McPherson, eds (Oxford, UK: Oxford University Press), pp. 163–174.
- Juarez, M.T., Kui, J.S., Thomas, J., Heller, B.A., and Timmermans, M.C.P. (2004. b). microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428 84–88. [DOI] [PubMed] [Google Scholar]
- Juarez, M.T., Twigg, R.W., and Timmermans, M.C.P. (2004. a). Specification of adaxial cell fate during maize leaf development. Development 131 4533–4544. [DOI] [PubMed] [Google Scholar]
- Kaplan, D.R. (1975). Comparative developmental evaluation of the morphology of unifacial leaves in the monocotyledons. Bot. Jahrb. 95 1–105. [Google Scholar]
- Kerstetter, R.A., Bollman, K., Taylor, R.A., Bomblies, K., and Poethig, R.S. (2001). KANADI regulates organ polarity in Arabidopsis. Nature 411 706–709. [DOI] [PubMed] [Google Scholar]
- Kidner, C.A., and Martienssen, R.A. (2004). Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428 81–84. [DOI] [PubMed] [Google Scholar]
- Kidner, C.A., and Timmermans, M.C.P. (2007). Mixing and matching pathways in leaf polarity. Curr. Opin. Plant Biol. 10 13–20. [DOI] [PubMed] [Google Scholar]
- Kim, Y.S., Kim, S.G., Lee, M., Lee, I., Park, H.Y., Seo, P.J., Jung, J.H., Kwon, E.J., Suh, S.W., Paek, K.H., and Park, C.M. (2008). HD-ZIP III activity is modulated by competitive inhibitors via a feedback loop in Arabidopsis shoot apical meristem development. Plant Cell 20 920–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., and Nei, M. (2004). MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5 150–163. [DOI] [PubMed] [Google Scholar]
- McConnell, J.R., Emery, J., Eshed, Y., Bao, N., Bowman, J., and Barton, M.K. (2001). Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411 709–713. [DOI] [PubMed] [Google Scholar]
- Michelmore, R.W., Paran, I., and Kesseli, R.V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis - A rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88 9828–9832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, J.M., Lane, B., and Freeling, M. (2002). Expression of a mutant maize gene in the ventral leaf epidermis is sufficient to signal a switch of the leaf's dorsoventral axis. Development 129 4581–4589. [DOI] [PubMed] [Google Scholar]
- Nelson, T., and Langdale, J.A. (1992). Developmental genetics of C-4 photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43 25–47. [Google Scholar]
- Nogueira, F.T.S., Madi, S., Chitwood, D.H., Juarez, M.T., and Timmermans, M.C.P. (2007). Two small regulatory RNAs establish opposing fates of a developmental axis. Genes Dev. 21 750–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochando, I., Jover-Gil, S., Ripoll, J.J., Candela, H., Vera, A., Ponce, M.R., Martinez-Laborda, A., and Micol, J.L. (2006). Mutations in the microRNA complementarity site of the INCURVATA4 gene perturb meristem function and adaxialize lateral organs in arabidopsis. Plant Physiol. 141 607–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuga, D., DeGuzman, B., Prigge, M.J., Drews, G.N., and Clark, S.E. (2001). REVOLUTA regulates meristem initiation at lateral positions. Plant J. 25 223–236. [DOI] [PubMed] [Google Scholar]
- Parkinson, S.E., Gross, S.M., and Hollick, J.B. (2007). Maize sex determination and abaxial leaf fates are canalized by a factor that maintains repressed epigenetic states. Dev. Biol. 308 462–473. [DOI] [PubMed] [Google Scholar]
- Prigge, M.J., Otsuga, D., Alonso, J.M., Ecker, J.R., Drews, G.N., and Clark, S.E. (2005). Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell 17 61–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann, J.L., et al. (2000). Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 290 2105–2110. [DOI] [PubMed] [Google Scholar]
- Russell, S.H., and Evert, R.F. (1985). Leaf vasculature in Zea mays L. Planta 164 448–458. [DOI] [PubMed] [Google Scholar]
- Scanlon, M.J. (2000). NARROW SHEATH1 functions from two meristematic foci during founder-cell recruitment in maize leaf development. Development 127 4573–4584. [DOI] [PubMed] [Google Scholar]
- Scanlon, M.J. (2003). The polar auxin transport inhibitor N-1-naphthylphthalamic acid disrupts leaf initiation, KNOX protein regulation, and formation of leaf margins in maize. Plant Physiol. 133 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Cartagena, J., Candela, H., Robles, P., Ponce, M.R., Perez-Perez, J.M., Piqueras, P., and Micol, J.L. (2000). Genetic analysis of incurvata mutants reveals three independent genetic operations at work in Arabidopsis leaf morphogenesis. Genetics 156 1363–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharman, B.C. (1942). Developmental anatomy of the shoot of Zea mays L. Ann. Bot. (Lond.) 6 245–282. [Google Scholar]
- Steeves, T.A., and Sussex, I.M. (1989). Patterns in Plant Development. (Cambridge, UK: Cambridge University Press).
- Sylvester, A.W., Cande, W.Z., and Freeling, M. (1990). Division and differentiation during normal and liguleless-1 maize leaf development. Development 110 985–1000. [DOI] [PubMed] [Google Scholar]
- Timmermans, M.C.P., Schultes, N.P., Jankovsky, J.P., and Nelson, T. (1998). Leafbladeless1 is required for dorsoventrality of lateral organs in maize. Development 125 2813–2823. [DOI] [PubMed] [Google Scholar]
- Waites, R., and Hudson, A. (1995). phantastica - A gene required for dorsoventrality of leaves in Antirrhinum majus. Development 121 2143–2154. [Google Scholar]
- Waites, R., Selvadurai, H.R.N., Oliver, I.R., and Hudson, A. (1998). The PHANTASTICA gene encodes a MYB transcription factor involved in growth and dorsoventrality of lateral organs in Antirrhinum. Cell 93 779–789. [DOI] [PubMed] [Google Scholar]
- Wenkel, S., Emery, J., Hou, B.H., Evans, M.M., and Barton, M.K. (2007). A feedback regulatory module formed by LITTLE ZIPPER and HD-ZIPIII genes. Plant Cell 19 3379–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, L., Grigg, S.P., Xie, M., Christensen, S., and Fletcher, J.C. (2005). Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 132 3657–3668. [DOI] [PubMed] [Google Scholar]
- Zhong, R., and Ye, Z.H. (1999). IFL1, a gene regulating interfascicular fiber differentiation in Arabidopsis, encodes a homeodomain-leucine zipper protein. Plant Cell 11 2139–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R.Q., Taylor, J.J., and Ye, Z.H. (1999). Transformation of the collateral vascular bundles into amphivasal vascular bundles in an Arabidopsis mutant. Plant Physiol. 120 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R.Q., and Ye, Z.H. (2004). amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant Cell Physiol. 45 369–385. [DOI] [PubMed] [Google Scholar]