Abstract
The Guard Model for disease resistance postulates that plant resistance proteins act by monitoring (guarding) the target of their corresponding pathogen effector. We posit, however, that guarded effector targets are evolutionarily unstable in plant populations polymorphic for resistance (R) genes. Depending on the absence or presence of the R gene, guarded effector targets are subject to opposing selection forces (1) to evade manipulation by effectors (weaker interaction) and (2) to improve perception of effectors (stronger interaction). Duplication of the effector target gene or independent evolution of a target mimic could relax evolutionary constraints and result in a decoy that would be solely involved in effector perception. There is growing support for this Decoy Model from four diverse cases of effector perception involving Pto, Bs3, RCR3, and RIN4. We discuss the differences between the Guard and Decoy Models and their variants, hypothesize how decoys might have evolved, and suggest ways to challenge the Decoy Model.
Plants have evolved sophisticated mechanisms to perceive pathogen attack and trigger an effective innate immune response. An important and well-characterized perception mechanism is based on resistance (R) genes in plants whose products confer recognition of cognate avirulence (Avr) proteins in the pathogen. This gene-for-gene hypothesis was introduced by Flor in the 1940s, and dozens of R-Avr gene combinations have since been characterized (Dangl and Jones, 2001).
Although the gene-for-gene hypothesis is now firmly supported by the characterization of many R-Avr gene pairs, the underlying perception mechanism has been subject to debate for more than a decade. Initially it was widely thought that products of R genes act as receptors that directly interact with the products of Avr genes (Keen, 1990). This ligand-receptor model was supported by the fact that some Avr gene products are small and colocalize with R gene products, most of which encode receptor-like proteins carrying Leu-rich repeats (LRRs). Indeed, direct binding of a few R-Avr combinations was found, consistent with a receptor-ligand mode of action (e.g., Jia et al., 2000; Deslandes et al., 2003; Dodds et al., 2006; Ueda et al., 2006). However, for a number of R-Avr combinations, physical interactions have not been observed, and perception is thought to be indirect.
Meanwhile, it has become evident that many Avr proteins contribute to pathogen virulence on plants lacking the cognate R gene. Avr proteins are now considered to be part of a larger repertoire of pathogen-secreted proteins that are called effectors to stress their presumed intrinsic virulence function. Avr recognition by plants has been coined effector-triggered immunity to contrast it with pathogen-associated molecular pattern (PAMP)–triggered immunity (Chisholm et al., 2006; Jones and Dangl, 2006). Effectors are pathogen-secreted proteins that manipulate host cell functions, whereas PAMPs define molecular motifs common to many pathogens. The concept that effectors have specific targets in the host is an essential component of a popular model that explains indirect perception mechanism of effectors by R proteins. This Guard Model predicts that R proteins act by monitoring (guarding) the effector target and that modification of this target by the effector results in the activation of the R protein, which triggers disease resistance in the host (Van der Biezen and Jones, 1998; Dangl and Jones, 2001).
The Guard Model was originally proposed to explain the mechanism of Pseudomonas syringae AvrPto perception by the tomato proteins Pto and Prf (Van der Biezen and Jones, 1998) and was later generalized to perception of other effector proteins (Dangl and Jones, 2001). The indirect effector perception mechanism postulated by the Guard Model explains how multiple effectors could be perceived by a single R protein, thus enabling a relatively small R gene repertoire to target the broad diversity of pathogens that attack plants (Dangl and Jones, 2001). Implicit in the Guard Model is the notion that the guarded effector target (also called the guardee) is indispensible for the virulence function of the effector protein in the absence of the cognate R protein. Support for the Guard Model has accumulated over the past decade with the description of guarded effector targets. Classical examples of these presumed guardees are Arabidopsis RIN4 and PBS1 and tomato RCR3 and Pto (Jones and Dangl, 2006).
Over the past few years, new data on indirectly recognized effectors have emerged that are inconsistent with the original description of the Guard Model. It is now well documented that many pathogen effectors have multiple targets in the host and that classical guardee proteins are often dispensable for the virulence activities of effectors in plants lacking the R protein. New data on additional targets of AvrPto and AvrBs3 prompted proposals of the concept that some host targets of effectors act as decoys to detect pathogen effectors via R proteins (Zhou and Chai, 2008; Zipfel and Rathjen, 2008). Here, we further develop these ideas into a Decoy Model that is consistent with most of the data described so far and is coherent with the current knowledge of evolution in plant–pathogen interactions. Also, we discuss the experiments required to discriminate between the Guard and Decoy Models as well as the possible implications of the Decoy Model. For clarity, Table 1 lists the terms used throughout this essay.
Table 1.
Definitions of Terms Used in This Manuscript
| Term | Definition |
|---|---|
| Avr protein | Pathogen effector that triggers resistance via activation of specific cognate host R proteins. |
| Decoy | Effector target required for R protein function but with no function in host defense or susceptibility in the absence of its cognate R protein; effector alteration of the decoy does not result in enhanced pathogen fitness in plants that lack the R protein and triggers innate immunity in plants that carry the R protein. |
| Effector | Secreted pathogen protein that manipulates host cell functions. |
| Guardee | Effector target required for R protein function and with a function in host defense or susceptibility in the absence of its cognate R protein; effector alteration of the guardee results in enhanced pathogen fitness in plants that lack the R protein and triggers innate immunity in plants that carry the R protein. |
| Operative target | Host target that when manipulated by a pathogen effector results in enhanced pathogen fitness. |
| R protein | Protein that confers resistance by mediating direct or indirect recognition of a pathogen Avr protein. This is often but not always an NB-LRR protein. |
THE DECOY MODEL
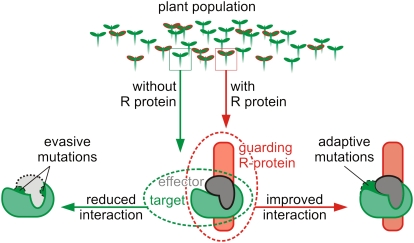
From an evolutionary point of view, the guarded effector target is in an unstable situation since it is subject to two opposing natural selection forces in plant populations where R genes are polymorphic. In this case, R gene polymorphism means the presence/absence of functional R genes in different individuals in a plant population (Figure 1). In the absence of a functional R gene, natural selection is expected to drive the guardee to decrease its binding affinity with the effector and thereby evade detection and modification by the effector. However, in the presence of a functional R gene, natural selection is expected to favor guardees with improved interaction with an effector to enhance pathogen perception. These two conflicting selection pressures on the same effector interaction surface of the guardee results in an evolutionarily unstable situation that could be relaxed upon the evolution of a host protein, termed here “decoy,” that specializes in perception of the effector by the R protein but itself has no function either in the development of disease or resistance. Thus, the decoy mimics effector targets to trap the pathogen into a recognition event. Decoys might evolve from effector targets by gene duplication followed by subsequent evolution or evolve independently by mimicking effector targets (target mimicry). In any case, the Decoy Model implies that the effector target monitored by the R protein is a decoy that mimics the operative effector target but only functions in perception of pathogen effectors without contributing pathogen fitness in the absence of its cognate R protein. This Decoy Model is distinct from the classical and refined Guard Models that imply that the manipulation of the guarded effector target by the effector benefits pathogen fitness in the absence of the R protein (Figure 2).
Figure 1.
Opposing Selection Forces on Guarded Effector Targets in a Plant Population Polymorphic for R Genes.
Opposing selection forces are expected to operate on guarded effector targets in plants with or without the associated R protein. In the absence of the R protein (green arrows), targets will be under selective pressure to reduce the interaction and evade manipulation (left). In the presence of the R protein (red arrows), the guarded effector target will be under selective pressure to improve the interaction with the effector and enhance pathogen perception (right). The figure represents protein complexes, but similar models can be drawn for nonprotein effector targets. A gene duplication of the effector target or the independent evolution of a target mimic would reduce the evolutionary constraints imposed on the guarded effector target, allowing it to specialize as a coreceptor (decoy) that regulates the activation of the R protein.
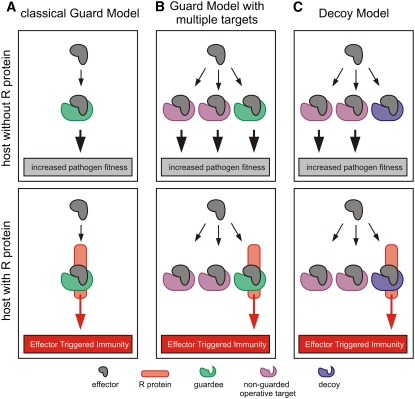
Figure 2.
Comparisons of the Guard and Decoy Models.
The classical Guard Model (A) is contrasted with a modified Guard Model in which the effector targets multiple plant proteins (B) and the Decoy Model (C). Effectors are depicted in gray, operative effector targets in purple, guardee in green, decoy in blue, and the R protein in orange.
This concept of a decoy is also distinct from animal decoy receptors that are defined as inactive receptors that act as sinks that deplete ligands, thereby preventing them from binding their operative receptor (Ashkenazi and Dixit, 1999; Montovani et al., 2001). These competing decoys, however, act in the absence of monitoring R proteins and are therefore different from the decoys that are enslaved in effector perception mechanisms. However, it remains possible that plant decoys that act in perception also compete with operative targets for effector target binding. In the absence of the R protein, these competing decoys would then limit rather than promote pathogen fitness.
The key assumptions behind the Decoy Model are inferred from our current understanding of plant–microbe interactions. First, R genes are typically polymorphic in natural plant populations. This has been observed repeatedly in both single gene studies (e.g., Arabidopsis RPM1 [Stahl et al., 1999] and tomato Cf-9 [Van der Hoorn et al., 2001]) and genome-wide analyses (Bakker et al., 2006; Clark et al., 2007). Second, effector targets are under selection for decreased binding affinity to effectors. Examples include the recessive resistance mutations in rice xa13 that evolved to evade transcriptional activation by Xanthomonas oryzae pv oryzae effectors (Iyer-Pascuzzi and McCouch, 2007; Yang et al., 2007). Furthermore, recessive mutations in transcription factor IIA and elongation factor elF4E were found to evade manipulation by bacterial blight and potyviral VPg, respectively (Iyer-Pascuzzi and McCouch, 2006; Charron et al., 2008). Also, enhanced patterns of diversifying selection have been described in inhibitor binding residues of several plant enzymes that operate at the plant pathogen interface (reviewed in Misas-Villamil and Van der Hoorn, 2008). These include soybean endo-β-1,3-glucanase that is targeted by the glucanase-inhibitor protein-1 from Phytophthora sojae (Bishop et al., 2005; Damasceno et al., 2008) and the tomato proteases targeted by Cladosporium fulvum Avr2 (Shabab et al., 2008). Third, in the presence of the R protein, there is selection on the guarded effector target to maintain or improve its interaction with the effector. This has not been shown directly, but this process should not be different from the adaptation of R proteins that physically interact with effectors. Flax rust resistance alleles L5, L6, and L7, for example, have probably been selected for enhanced interactions with the different alleles of AvrL567 (Ellis et al., 2007).
It is interesting to note that each of the four players in this antagonistic molecular interaction are under selection forces to adapt: (1) the operative target is under selection to evade manipulation by the effector; (2) the effector is under selection to target the adjusted operative targets while preventing interactions with the decoy, which would trigger defense responses in the presence of the R protein; (3) the decoy is under selection to adapt to adjusted effectors and is under additional selection to prevent autoimmune responses; and (4) the R protein is under selection to adapt to novel decoy-effector complexes while preventing autoimmune responses. As a result, each component is part of a molecular arms race in which each player is a target of the next.
SUPPORT FOR THE DECOY MODEL
The Decoy Model is consistent with recent findings on effector activities and perception by plants and is supported by four cases of well-studied effector perception mechanisms. These cases are discussed below and are summarized in Table 2. Overall, there is a striking diversity in the perception mechanisms, R protein structure, and pathosystems, indicating that decoys have evolved frequently and independently in antagonistic plant–pathogen interactions.
Table 2.
Four Cases Supporting the Decoy Model
| Case | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Plant species | Tomato | Pepper | Tomato | Arabidopsis |
| Pathogen | P. syringae pv tomato (bacterium) | X. campestris pv vesicatoria (bacterium) | C. fulvum (fungus) | P. syringae (bacterium) |
| Site of perception | Cytoplasm | Nucleus | Apoplast | Cytoplasm |
| R protein | Prf | Bs3 | Cf-2 | RPS2 |
| Biochemical function of R protein | NB-LRR | Flavin monooxygenase | Receptor-like protein | NB-LRR |
| Decoy | Pto | pBs3 | RCR3 | RIN4 |
| Biochemical function of decoy | Kinase | upa box in promoter of Bs3 gene | Cys protease | Negative regulator of basal defense (Kim et al., 2005) |
| Operative target | Le FLS2? | pUpa20 | PIP1 | Not yet identified |
| Structure and function of operative target | Receptor-like kinase required for basal resistance | upa box in promoter of cell size regulator Upa20 and other genes | Cys protease secreted abundantly during defense | Unknown |
| Effector | AvrPto | AvrBs3 | Avr2 | AvrRpt2 |
| Biochemical function of effector | Kinase inhibitor | Transcription factor | Protease inhibitor | Cys protease |
| Presumed perception mechanism | Pto inhibition by AvrPto activates Prf (Mucyn et al., 2006; Xing et al., 2007) | AvrBs3 binds and activates promoter of Bs3 (Römer et al., 2007) | Avr2 inhibits Rcr3, and Avr2-Rcr3 complex probably activates Cf-2 (Rooney et al., 2005) | AvrRpt2 cleaves RIN4 from the RIN4-RPS2 complex, activating RPS2 (Axtell and Staskawicz, 2003; Mackey et al., 2003) |
| Virulence role of the effector? | Yes: AvrPto contributes to virulence on tomato (Chang et al., 2000) and Arabidopsis (Xiang et al., 2008) | Yes: AvrBs3 contributes to virulence on pepper under field conditions (Wichmann and Bergelson, 2004) | Yes: Avr2 contributes to virulence on tomato (van Esse et al., 2008) | Yes: AvrRpt2 contributes to virulence on Arabidopsis (Guttman and Greenberg, 2001) |
| Does pathogen benefit from manipulating decoy? | No?: no enhanced virulence on pto/Prf compared with Pto/prf tomato lines (Chang et al., 2000) | No: not anticipated | No?: No enhanced virulence on MM-Cf2/rcr3 compared with MM-Cf0 tomato lines (Dixon et al., 2000) | No?: No enhanced virulence on rin4/rps2 compared with RIN4/rps2 Arabidopsis lines (Belkhadir et al., 2004; Lim and Kunkel, 2004) |
| Does pathogen benefit from manipulating operative target? | Yes: AvrPto inhibits FLS2 kinase domain and no longer contributes to virulence on fls2 mutants (Xiang et al., 2008) | Yes: AvrBs3 activates the promoter of Upa20, resulting in enhanced cell size, a phenotype that is thought to be beneficial for the bacteria (Kay et al., 2007) | Yes?: Avr2 inhibits the abundant, defense-related protease PIP1 (Shabab et al., 2008). However, a role of PIP1 in defense has not yet been demonstrated. | Not investigated: operative targets are not yet known. |
Case 1: Pto
P. syringae AvrPto is a kinase inhibitor that blocks the function of FLS2 and EFR, two receptor-like kinases involved in PAMP-triggered immunity (Xing et al., 2007; Xiang et al., 2008). Tomato Pto encodes a Ser/Thr kinase that confers resistance to P. syringae strains carrying avrPto, an interaction that also requires the nucleotide binding (NB)–LRR R protein Prf. AvrPto contributes to virulence on tomato and Arabidopsis but not on Arabidopsis lacking FLS2, indicating that FLS2 is an operative virulence target of AvrPto (Xiang et al., 2008). Considering that AvrPto contributes to virulence on tomato, even in the absence of Pto (Chang et al., 2000), it is possible that the tomato ortholog of FLS2 and other receptor-like kinases are the operative targets of AvrPto and that Pto itself is a decoy. That Pto is a decoy confined to the regulation of Prf is further supported by the observation that Pto associates with Prf in vivo and that Pto accumulation is dependent of Prf accumulation (Mucyn et al., 2006). As an interesting variation of the Decoy Model, it has been proposed that Pto competes with FLS2 for AvrPto binding (Zhou and Chai, 2008; Zipfel and Rathjen, 2008). This competing decoy model is similar to that of animal decoy receptors and implies that Pto would restrict rather than promote pathogen fitness in the absence of Prf.
Case 2: pBS3
Xanthomonas campestris pv vesicatoria AvrBs3 is a type-III effector that functions as a transcription factor by binding and activating promoters in the nucleus of host pepper cells. An important role for AvrBs3 is to induce host cell size expansion (hypertrophy) by binding and activating the promoter of Upa20 (pUpa20), a master regulator of cell size (Kay et al., 2007). In resistant plants, AvrBs3 also activates the promoter of the pepper Bs3 gene (pBs3), an unusual R gene in that it encodes a flavin monooxygenase (Römer et al., 2007). Expression of Bs3 has not been detected in the absence of AvrBs3, suggesting that this gene may not have any obvious function in the absence of avirulent bacteria and does not contribute to defense to bacteria that lack AvrBs3. These data are consistent with a model in which pBs3 is a decoy and pUpa20 is one of the operative targets of AvrBs3 (Zhou and Chai, 2008). In addition, AvrHah1, an AvrBs3-homologous effector of Xanthomonas gardneri that functions as a transcriptional activator and induces water-soaking in susceptible pepper plants, also activates the Bs3 promoter resulting in hypersensitive cell death in Bs3 pepper plants (Schornack et al., 2008). This indicates that pBs3 is a decoy that traps at least two distinct effectors from different species of Xanthomonas (Schornack et al., 2008).
Case 3: RCR3
The effector protein Avr2 of the fungus C. fulvum is a secreted protein that inhibits RCR3 and PIP1, two secreted, defense-induced Cys proteases of tomato that are under diversifying selection (Rooney et al., 2005; Shabab et al., 2008). PIP1 is a pathogenesis-related protein that accumulates to high levels during infection by diverse pathogens and by salicylic acid (Tian et al., 2007; Shabab et al., 2008). RCR3, on the other hand, also accumulates as a pathogenesis-related protein but to much lower levels compared with PIP1 (Shabab et al., 2008). The role of PIP1 and RCR3 in the apoplastic defense response remains to be determined. However, rcr3 mutants (MM-Cf2/rcr3 lines) are similarly susceptible to C. fulvum as tomato lacking the Cf-2 gene cluster (MM-Cf0 line), indicating that RCR3 inhibition does not contribute to virulence (Dixon et al., 2000). Together, these data suggest that PIP1 is an operative target of Avr2 and that RCR3 acts as a decoy to trap the fungus into a recognition event in plants carrying Cf-2 (Shabab et al., 2008).
Case 4: RIN4
Arabidopsis RIN4 is a negative regulator of basal defense that is targeted by multiple P. syringae effectors (AvrRpm1, AvrRpt2, and AvrB) and monitored by at least two R proteins (RPM1 and RPS2) (Kim et al., 2005). Basal defense responses are suppressed in RIN4 overexpression lines and slightly enhanced in rin4 mutant lines (Kim et al., 2005). RIN4 is targeted by AvrRpm1 and AvrRpt2 for phosphorylation and degradation, respectively (Mackey et al., 2002; Axtell and Staskawicz, 2003), but it is unknown if and how these modifications benefit pathogen virulence. One hypothesis is that RIN4 cleavage by AvrRpt2 releases RIN4 fragments that suppress basal defense responses. Although these data are consistent with the Guard Model, RIN4 could also be a decoy as long as the definite link that RIN4 manipulation promotes pathogen virulence has not been demonstrated. For example, AvrRpm1 and AvrRpt2 promote virulence to a similar extent in rin4 knockout plants as in wild-type plants, and AvrRpt2 mutants that do not cleave RIN4 still contribute to virulence (Belkhadir et al., 2004; Lim and Kunkel, 2004). Thus, both AvrRpm1 and AvrRpt2 appear to have operative targets other than RIN4. AvrRpt2 is a protease that cleaves several Arabidopsis proteins besides RIN4, but the role of these AvrRpt2 targets in defense remains to be examined (Chisholm et al., 2005; Takemoto and Jones, 2005). Thus, the key information to define RIN4 as a guardee or a decoy is whether or not RIN4 modification by the effectors benefits the pathogen.
Other cases for which the Guard Model has been proposed also fit the Decoy Model, although there is no evidence to distinguish between the Guard and Decoy Models at this stage. P. syringae AvrPphB, for example, cleaves the PBS1 kinase in the host Arabidopsis, resulting in recognition by the RPS5 R protein (Shao et al., 2003; Ade et al., 2007). However, the virulence effect of AvrPphB in the presence and absence of PBS1 remains to be investigated. Also, perception of X. oryzae pv oryzae AvrXa27, C. fulvum Avr9, Tobacco mosaic virus p50, Turnip crinkle virus CP, and Potato virus X CP by their cognate R proteins appears to be indirect, but the effector targets and/or perception mechanisms remain to be elucidated (Kooman-Gersmann et al., 1998; Ren et al., 2000, 2005; Gu et al., 2005; Sacco et al., 2007; Tameling and Baulcombe, 2007; Caplan et al., 2008).
EVOLUTION OF DECOYS
How do decoys evolve? Conceptually, decoys can be evolutionarily related to operative targets or may evolve independently by target mimicry. Either one of these scenarios might apply for Pto. AvrPto inhibits multiple defense-related kinases, indicating that Pto could have directly evolved from one of these targets. For instance, Pto may have evolved from a receptor-like kinase that lost the extracellular domains that are not required for AvrPto perception. This is consistent with the observation that Pto is most closely related to the kinase domains of receptor-like kinases (Hardie, 1999). Alternatively, Pto may have functioned in a kinase pathway that was not originally targeted by AvrPto but was then recruited to function in effector perception by mimicking the operative targets of Pto (target mimicry). In the case of RCR3, this decoy has most likely evolved by target duplication since it is phylogenetically closely related to the presumptive operative target PIP1 and the Rcr3 and Pip1 genes reside at the same locus in the tomato genome, suggesting that they were generated by gene duplication and divergent evolution (Tian et al., 2007). On the other hand, target mimicry might best explain the evolution of the AvrBs3 binding box in the Bs3 promoter though it also could have originated from a recombination event between the promoter of an operative target with a flavin monooxygenase gene.
Although the various examples illustrated above are plausible, there is at least one observation that is not explained by the Decoy Model. The Decoy Model predicts that features that are not relevant to effector perception will be lost during decoy evolution. However, if Pto and Rcr3 are specialized decoy proteins, why are they active enzymes? Our current knowledge is insufficient to provide a satisfactory answer, but three scenarios might apply. First, these decoys may have evolved only recently and have not yet lost their enzymatic activity. This explanation contradicts the observation that both the Pto and Rcr3 genes are ancient and accumulated sequence variation in regions without affecting their activity (Rose et al., 2005; Shabab et al., 2008). Second, these decoys may have additional functions unrelated to pathogen perception. It is common for proteins to have multiple functions. Pto, for example, could act in a signaling pathway that includes Pti phosphorylation (Zhou et al., 1995). Similarly, Rcr3 could function in processes unrelated to defense. However, this explanation contradicts the observation that Rcr3 expression is defense related (Shabab et al., 2008) and that the absence of Pto is common in plant populations (Rose et al., 2005). Third, catalytic activity of these decoys could be required for effector perception. Indeed, kinase activity of Pto is essential for AvrPto perception (Rathjen et al., 1999), and AvrPto phosphorylation is required for its recognition, though this phosphorylation occurs in the absence of Pto (Anderson et al., 2006). It has not been investigated whether or not Rcr3 activity is required for Avr2 perception. Avr2 itself is probably not cleaved by Rcr3, and Rcr3 inhibition by other inhibitors is insufficient to activate Cf-2 signaling (Rooney et al., 2005). However, Rcr3 activity might be required for autocatalytic removal of its prodomain to create the binding site for Avr2. In conclusion, the observation that decoys can be active enzymes is not fully understood. Future work will shed light on these issues and help to clarify how decoys evolve in plant pathosystems.
GENERATING EXPERIMENTAL EVIDENCE FOR GUARD AND DECOY MODELS
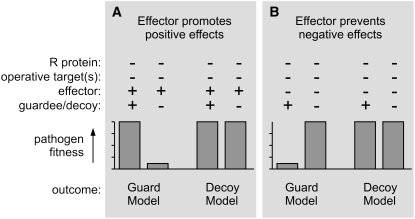
Providing experimental evidence to discriminate between the Guard and Decoy Models is challenging for several reasons. First, the two models are not necessarily mutually exclusive since intermediate stages may occur while a guardee evolves into a decoy. As a result, many of the predictions of the Guard Model also hold for the Decoy Model. Second, the issue of redundancy of effector targets can confound genetic analyses. For example, if effectors have several operative targets, then removing one of the targets may not alter the phenotype. Third, the definition of decoys is based on a lack of evidence for a role in defense, which is always difficult to establish with certainty. Despite these limitations, it remains possible to devise genetic experiments to discriminate between the Guard and Decoy Models (Figure 3). These assays involve comparing the effect of the presence or absence of the guardee/decoy on pathogen fitness in genetic backgrounds that lack the corresponding R protein and other effector targets. For example, pathogen fitness can be compared between Pto and pto tomato plants lacking both tomato FLS2 and Prf to determine whether Pto contributes to pathogen fitness and thus discriminate between the Decoy and Guard Models. The type of test depends on the presumed action of the effector: Does the effector promote host processes that are positive for the pathogen (e.g., release of nutrients) or prevent responses that are negative for the pathogen (e.g., suppression of defense responses)? For instance, the contribution of RIN4 to host defense needs to be assessed in the presence of the Cys protease AvrRpt2 to reveal contributions by the cleaved RIN4. On the other hand, the contribution of Pto to host defense should be assessed in the absence of the kinase inhibitor AvrPto. In summary, although such experiments can be difficult to set up since they require the identification of all operative targets, the generation of the appropriate genetic material, and the development of quantitative assays for pathogen fitness, these assays offer a direct test to exclude or support the Decoy Model.
Figure 3.
Genetic Tests to Discriminate between the Guard and Decoy Models.
Plants lacking both the R protein and the presumed operative target(s) should be challenged with pathogens in the absence or presence of the guardee/decoy. A differential pathogen growth supports the Guard Model, whereas an unaffected pathogen growth supports the Decoy Model. The test of choice depends on the nature of the effector.
(A) Effectors that promote positive effects on pathogen growth by manipulating their target should be present during the test to reveal target contributions.
(B) Effectors that prevent negative effects on pathogen growth should be omitted to avoid them from suppressing a possible phenotype.
CONCLUSION
The Decoy Model remains to be experimentally demonstrated, but it is consistent with a number of recent observations and provides a challenging platform for future experiments. We hope that new data and experiments will challenge the Decoy Model and generate a basis for a deeper understanding of effector perception in plants, ultimately leading to novel approaches to manipulate innate immunity and improve pathogen resistance.
Acknowledgments
We thank Brande Wulff, David Mackey, Edgar Huitema, Frank Takken, Jane Parker, John Rathjen, Paul Schulze-Lefert, Ralph Panstruga, Réka Tóth, Sebastian Schornack, Silke Robatzek, and the three anonymous reviewers for useful discussions, suggestions, and/or critical reading. R.A.L.V. is supported by the Max Planck Society and S.K. is supported by The Gatsby Charitable Foundation.
References
- Ade, J., DeYoung, D.J., Golstein, C., and Innes, R.W. (2007). Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc. Natl. Acad. Sci. USA 104 2531–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J.C., Pascuzzi, P.E., Xiao, F., Sessa, G., and Martin, G.B. (2006). Host-mediated phosphorylation of type III effector AvrPto promotes Pseudomonas virulence and avirulence in tomato. Plant Cell 18 502–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazi, A., and Dixit, V.M. (1999). Apoptosis control by death and decoy receptors. Curr. Opin. Cell Biol. 11 225–260. [DOI] [PubMed] [Google Scholar]
- Axtell, M.J., and Staskawicz, B.J. (2003). Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112 369–377. [DOI] [PubMed] [Google Scholar]
- Bakker, E.G., Toomajian, C., Kreitman, M., and Bergelson, J. (2006). A genome-wide survey of R gene polymorphisms in Arabidopsis. Plant Cell 18 1803–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir, Y., Nimchuk, Z., Hubert, D.A., Mackey, D.A., and Dangl, J.L. (2004). Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell 16 2822–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, J.G., Ripoll, D.R., Bashir, S., Damasceno, C.M.B., Seeds, J.D., and Rose, J.K.C. (2005). Selection on Glycine β-1,3-Endoglucanase genes differentially inhibited by a Phytophthora glucanase inhibitor protein. Genetics 169 1009–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan, J.L., Mamillapalli, P., Burch-Smith, T.M., Czymmek, K., and Dinesh-Kumar, S.P. (2008). Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell 132 449–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, J.H., Rathjen, J.P., Bernal, A.J., Staskawicz, B.J., and Michelmore, R.W. (2000). avrPto enhances growth and necrosis caused by Pseudomonas syringae pv. tomato in tomato lines lacking either Pto or Prf. Mol. Plant Microbe Interact. 13 568–571. [DOI] [PubMed] [Google Scholar]
- Charron, C., Nicolai, M., Gallois, J.-L., Robaglia, C., Moury, B., Palloix, A., and Caranta, C. (2008). Natural variation and functional analyses provide evidence for co-evolution between plant elF4E and potyviral VPg. Plant J. 54 56–68. [DOI] [PubMed] [Google Scholar]
- Chisholm, S.T., Coaker, G., Day, B., and Staskawicz, B.J. (2006). Host-microbe interactions: Shaping the evolution of the plant immune response. Cell 124 803–814. [DOI] [PubMed] [Google Scholar]
- Chisholm, S.T., Dahlbeck, D., Krishnamurthy, N., Day, B., Sjolander, K., and Staskawicz, B.J. (2005). Molecular characterization of proteolytic cleavage sites of the Pseudomonas syringae effector AvrRpt2. Proc. Natl. Acad. Sci. USA 102 2087–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, R.M., et al. (2007). Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317 338–342. [DOI] [PubMed] [Google Scholar]
- Damasceno, C.M.B., Bishop, J.G., Ripoll, D.R., Win, J., Kamoun, S., and Rose, J.K.C. (2008). Structure of the glucanase inhibitor protein (GIP) family from Phytophthora species suggests coevolution with plant endo-beta-1,3-glucanases. Mol. Plant Microbe Interact. 21 820–830. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D.G. (2001). Plant pathogens and integrated defence responses to infection. Nature 411 826–833. [DOI] [PubMed] [Google Scholar]
- Deslandes, L., Olivier, J., Peeters, N., Feng, D.X., Khounlotham, M., Boucher, C., Somssich, I., Genin, S., and Marco, Y. (2003). Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc. Natl. Acad. Sci. USA 100 8024–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, M.S., Golstein, C., Thomas, C.M., Van der Biezen, E.A., and Jones, J.D.G. (2000). Genetic complexity of pathogen perception by plants: The example of Rcr3, a tomato gene required specifically by Cf-2. Proc. Natl. Acad. Sci. USA 97 8807–8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N., Lawrence, G.J., Catanzariti, A.M., Teh, T., Wang, C.I., Ayliffe, M.A., Kobe, B., and Ellis, J.G. (2006). Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. USA 103 8888–8893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J.G., Dodds, P.N., and Lawrence, G.J. (2007). Flax rust resistance gene specificity is based on direct resistance-avirulence protein interactions. Annu. Rev. Phytopathol. 45 289–306. [DOI] [PubMed] [Google Scholar]
- Gu, K., Yang, B., Tian, D., Wu, L., Wang, D., Sreekala, C., Yang, F., Chu, Z., Wang, G.-L., White, F.F., and Yin, Z. (2005). R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435 1122–1125. [DOI] [PubMed] [Google Scholar]
- Guttman, D.S., and Greenberg, J.T. (2001). Functional analysis of the type III effectors AvrRpt2 and AvrRpm1 of Pseudomonas syringae with the use of a single-copy genomic integration system. Mol. Plant Microbe Interact. 14 145–155. [DOI] [PubMed] [Google Scholar]
- Hardie, D.G. (1999). Plant protein serine/thereonine kinases: Classification and functions. Annu. Rev. Plant Biol. 50 97–131. [DOI] [PubMed] [Google Scholar]
- Iyer-Pascuzzi, A.S., and McCouch, S.R. (2007). Recessive resistance genes and the Oryza sativa-Xanthomonas oryzae pv. oryzae pathosystem. Mol. Plant Microbe Interact. 20 731–739. [DOI] [PubMed] [Google Scholar]
- Jia, Y., McAdams, S.A., Bryan, G.T., Hershey, H.P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D.G., and Dangl, J.L. (2006). The plant immune system. Nature 444 323–329. [DOI] [PubMed] [Google Scholar]
- Kay, S., Hahn, S., Marois, E., Hause, G., and Bonas, U. (2007). A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 318 648–651. [DOI] [PubMed] [Google Scholar]
- Keen, N.T. (1990). Gene-for-gene complementarity in plant-pathogen interactions. Annu. Rev. Genet. 24 447–473. [DOI] [PubMed] [Google Scholar]
- Kim, M.G., da Cunha, L., McFall, A.J., Belkhadir, Y., DebRoy, S., Dangl, J.L., and Mackey, D. (2005). Two Pseudomonas syringae type III effectors inhibit RIN4-regulated basal defense in Arabidopsis. Cell 121 749–759. [DOI] [PubMed] [Google Scholar]
- Kooman-Gersmann, M., Vogelsang, R., Vossen, P., van den Hooven, H.W., Mahé, E., Honee, G., and de Wit, P.J.G.M. (1998). Correlation between binding affinity and necrosis-inducing activity of mutant AVR9 peptide elicitors. Plant Physiol. 117 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, M.T.S., and Kunkel, B.N. (2004). The Pseudomonas syringae type III effector AvrRpt2 promotes virulence independently of RIN4, a predicted virulence target in Arabidopsis thaliana. Plant J. 40 790–798. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Belkhadir, Y., Alfonso, J.M., Ecker, J.R., and Dangl, J.L. (2003). Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112 379–389. [DOI] [PubMed] [Google Scholar]
- Mackey, D., Holt, B.F., Wiig, A., and Dangl, J.L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistence in Arabidopsis. Cell 108 743–754. [DOI] [PubMed] [Google Scholar]
- Misas-Villamil, J.C., and Van der Hoorn, R.A.L. (2008). Enzyme-inhibitor interactions at the plant-pathogen interface. Curr. Opin. Plant Biol. 11 380–388. [DOI] [PubMed] [Google Scholar]
- Montovani, A., Locati, M., Vecchi, A., Sozzani, S., and Allavena, P. (2001). Decoy receptors: A strategy to regulate inflammatory cytokines and chemokines. Trends Immunol. 22 328–336. [DOI] [PubMed] [Google Scholar]
- Mucyn, T.S., Clemente, A., Andriotis, V.M.E., Balmuth, A.L., Oldroyd, G.E.D., Staskawicz, B.J., and Rathjen, J.P. (2006). The tomato NBARC-LRR protein Prf interacts with Pto kinase in vivo to regulate specific plant immunity. Plant Cell 18 2792–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathjen, J.P., Chang, J.H., Staskawicz, B.J., and Michelmore, R.W. (1999). Constitutively active Pto induces Prf-dependent hypersensitive response in the absence of avrPto. EMBO J. 12 3232–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, T., Qu, F., and Morris, T.J. (2005). The nuclear localisation of the Arabidopsis transcription factor TIP is blocked by its interaction with the coat protein of Turnip crincle virus. Virology 331 316–324. [DOI] [PubMed] [Google Scholar]
- Ren, T., Qu, F., and Morris, T.J. (2000). HRT gene function requires interaction between a NAC protein and viral capsid protein to confer resistance to turnip crinkle virus. Plant Cell 12 1917–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römer, P., Hahn, S., Jordan, T., Strauβ, T., Bonas, U., and Lahaye, T. (2007). Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science 318 645–648. [DOI] [PubMed] [Google Scholar]
- Rooney, H.C.E., van't Klooster, J.W., Van der Hoorn, R.A.L., Joosten, M.H.A.J., Jones, J.D.G., and De Wit, P.J.G.M. (2005). Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 308 1783–1786. [DOI] [PubMed] [Google Scholar]
- Rose, L.E., Langley, C.H., Bernal, A.J., and Michelmore, R.W. (2005). Natural variation in the Pto pathogen resistance gene within species of wild tomato (Lycopersicon). I. Functional analysis of Pto alleles. Genetics 171 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacco, M.A., Mansoor, S., and Moffett, P. (2007). A RanGAP protein physically interacts with the NB-LRR protein Rx, and is required for Rx-mediated viral resistance. Plant J. 52 82–93. [DOI] [PubMed] [Google Scholar]
- Schornack, S., Minsavage, G.V., Stall, R.E., Jones, J.B., and Lahaye, T. (2008). Characterization of AvrHah1, a novel AvrBs3-like effector from Xanthomonas gardneri with virulence and avirulence activity. New Phytol., in press. [DOI] [PubMed]
- Shabab, M., Shindo, T., Gu, C., Kaschani, F., Pansuriya, T., Chintha, R., Harzen, A., Colby, T., Kamoun, S., and Van der Hoorn, R.A.L. (2008). Fungal effector protein Avr2 targets diversifying defence-related Cys proteases of tomato. Plant Cell 20 1169–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, F., Golstein, C., Ade, J., Stoutemyer, M., Dixon, J.E., and Innes, R.W. (2003). Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301 1230–1233. [DOI] [PubMed] [Google Scholar]
- Stahl, E.A., Dwyer, G., Mauricio, R., Kreitman, M., and Bergelson, J. (1999). Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 400 667–671. [DOI] [PubMed] [Google Scholar]
- Tameling, W.I.L., and Baulcombe, D.C. (2007). Physical association of the NB-LRR resistance protein Rx with a Ran GTPase-activating protein is required for extreme resistance to Potato virus X. Plant Cell 19 1682–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto, D., and Jones, D.A. (2005). Membrane release and destabilisation of Arabidopsis RIN4 following cleavage by Pseudomonas syringae AvrRpt2. Mol. Plant Microbe Interact. 18 1258–1268. [DOI] [PubMed] [Google Scholar]
- Tian, M., Win, J., Song, J., Van der Hoorn, R.A.L., Van der Knaap, E., and Kamoun, S. (2007). A Phytophthora infestans cystatin-like protein interacts with and inhibits a tomato papain-like apoplastic protease. Plant Physiol. 143 364–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, H., Yamaguchi, Y., and Sano, H. (2006). Direct interaction between the tobacco mosaic virus helicase domain and the ATP-bound resistance protein, N factor, during the hypersensitive response in tobacco plants. Plant Mol. Biol. 61 31–45. [DOI] [PubMed] [Google Scholar]
- Van der Biezen, E.A., and Jones, J.D.G. (1998). Plant disease-resistance proteins and the gene-for-gene concept. Trends Plant Sci. 23 454–456. [DOI] [PubMed] [Google Scholar]
- Van der Hoorn, R.A.L., Kruijt, M., Roth, R., Brandwagt, B.F., Joosten, M.H.A.J., and De Wit, P.J.G.M. (2001). Intragenic recombination generated two distinct Cf genes that mediate AVR9 regognition in the natural population of Lycopersicon pimpinellifolium. Proc. Natl. Acad. Sci. USA 98 10493–10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Esse, H.P., van't Klooster, J.W., Bolton, M.D., Yadeta, K.A., van Baarlen, P., Boeren, S., Vervoort, J., de Wit, P.J.G.M., and Thomma, B.P.H.J. (2008). The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell 20 1948–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann, G., and Bergelson, J. (2004). Effector genes of Xanthomonas axonopodis pv. vesicatoria promote transmission and enhance other fitness traits in the field. Genetics 166 693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, T., Zhong, N., Zou, Y., Wu, Y., Zhang, J., Xing, W., Li, Y., Tang, X., Zhu, L., Chai, J., and Zhou, J.-M. (2008). Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr. Biol. 18 74–80. [DOI] [PubMed] [Google Scholar]
- Xing, W., et al. (2007). The structural basis for activation of plant immunity by bacterial effector protein AvrPto. Nature 449 243–247. [DOI] [PubMed] [Google Scholar]
- Yang, B., Sugio, A., and White, F.F. (2007). Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. USA 103 10503–10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J.M., and Chai, J. (2008). Plant pathogenic bacterial type III effectors subdue host responses. Curr. Opin. Microbiol. 11 179–185. [DOI] [PubMed] [Google Scholar]
- Zhou, J.M., Loh, Y.T., Bressan, R.A., and Martin, G.B. (1995). The tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in hte hypersensitive response. Cell 83 925–935. [DOI] [PubMed] [Google Scholar]
- Zipfel, C., and Rathjen, J.P. (2008). Plant Immunity: AvrPto targets the frontline. Curr. Biol. 18 R218–R220. [DOI] [PubMed] [Google Scholar]