Abstract
Aim We studied the role of mini-TyrRS and mini-TrpRS in angiogenesis by using small interfering RNA-mediated mini-TyrRS/mini-TrpRS knockout in hypoxic culture of human umbilical vein endothelial cells. Methods SiRNA was used as the main method to inhibited the gene function. Silencing efficiency was assayed by real-time reverse transcription-polymerase chain reaction and western blotting. The angiogenic activity in vitro was evaluated by transwell migration assay and Matrigel-induced capillary tube formation in hypoxic culture. Cell proliferation was determined by crystal violet staining. Results The results showed that levels of the mini-TyrRS/mini-TrpRS gene and protein in mock transfection group and negative control group were higher, but noticeably decreased in experimental group. However, no significant difference was detected between mock transfection group and negative control group, but there was a statistically significant difference compared with experimental group. For mini-TyrRS-siRNA group, the cell migration, tube formation and the rate of cell proliferation were respectively inhibited by (47.4, 56.3, 65.4, 73.7%), (60.5, 69.1, 75.9, 83.6%) and (40.4, 56.2, 61.2, 68.0%). For mini-TrpRS-siRNA, were respectively increased by (18.0, 33.8, 45.1, 56.4%), (18.3, 31.2, 40.3, 45.7%) and (8.4, 26.4, 38.2, 46.6%). Conclusion These results indicated that angiogenesis is either stimulated by mini-TyrRS or inhibited by mini-TrpRS in matrigel models in hypoxic culture, raising the possibility that mini-TyrRS stimulates a common downstream signaling event. Thus, naturally occurring fragments of two proteins involved in translation, TyrRS and TrpRS, have opposing activity on endothelial cell angiogenesis in the matrigel assays. The opposing activities of the two tRNA synthetases suggest tight regulation of the balance between pro- and anti-angiogenic stimuli.
Keywords: Mini-TyrRS, Mini-TrpRS, Hypoxia, Human umbilical vein endothelial cells, siRNA, Angiogenesis
Introduction
Coronary atherosclerotic heart disease (CAHD) is the most common cause of cardiovascular disability and death in many Western countries. In China, the incidence and prevalence of this disease is also increasing year by year. Although treatments are being developed rapidly, such as percutaneous transluminal coronary angioplasty (PTCA) and coronary artery bypass grafting (CABG), many patients who have diffuse coronary pathological changes cannot obtain good therapeutic efficacy, and their prognoses are very bad. Research on therapies for these patients will be of central importance to CAHD treatment. Remedial angiogenesis is a new concept for healing the advanced stage patients. It means using some cytokines, genes or cells to promote angiogenesis and rebuild the myocardial blood transportation.
The process of angiogenesis, the formation of a new blood vessels, is central to physiological and pathophysiological conditions including placental development, wound healing, diabetic retinopathy, coronary heart disease, and tumor growth (Folkman 1985; Risau 1997; Yancopoulos et al. 1998). Because of the great potential for treatment of human disease through manipulation of angiogenesis, fundamental information regarding the molecular pathways that regulate endothelial cell differentiation is of central importance. It is well known that endothelial cells are capable of aggregating in vitro to form capillary-like structures (Folkman and Haudenschild 1980; Maciag et al. 1982; Madri and Williams 1983; Montesano 1983). This process, which is thought to mimic the process by which endothelial cells form capillaries in vivo, requires specialized culture conditions. To study the cellular and molecular basis of angiogenesis, matrigel, as an in vitro model of vascular formation, has been shown to induce human umbilical vein endothelial cells (HUVECs) to form a three-dimensional capillary network (Grant et al. 1989; Kubota et al. 1988).
Aminoacyl-tRNA synthetases catalyze the first step of protein synthesis that consists of the aminoacylation of tRNAs. But they have a broad repertoire of functions beyond protein synthesis, including transcriptional and translational regulation as well as cell signaling (Martinis et al. 1999). Recently, it has been demonstrated that two of the tRNA synthetases, human tyrosyl-tRNA synthetase (TyrRS) and human tryptophanyl-tRNA synthetase (TrpRS), have novel cytokine functions (Wakasugi and Schimmel 1999a). This demonstration established a link between protein synthesis and signal transduction. At the same time, mammalian TyrRS and TrpRS have also been shown to regulate angiogenesis (Wakasugi and Schimmel 1999b; Wakasugi et al. 2002a, b; Otani et al. 2002; Ewalt and Schimmel 2002; Ibba 2000).
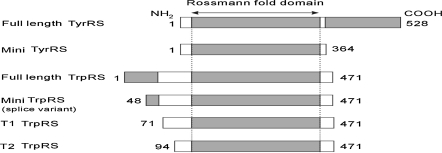
Under apoptotic conditions in culture, full length TyrRS is secreted, and two distinct cytokines can then be generated by an extracellular protease such as leukocyte elastase (Wakasugi and Schimmel 1999a). The NH2-terminal catalytic fragment, mini-TyrRS, binds strongly to the CXC-chemokine receptor CXCR1 and, like IL-8, functions as a chemoattractant for polymorphonuclear leukocytes (PMNs), to promote angiogenesis (Wakasugi and Schimmel 1999a), whereas the full-length enzyme lacks cytokine activity. The catalytic core domain of TrpRS is a close homologue of the catalytic domain of TyrRS (Ribas de Pouplana et al. 1996; Lee et al. 2004; Fleckner et al. 1995). In normal cells, human TrpRS exists as two forms. The major form is the full-length protein, and the other is a truncated TrpRS (mini-TrpRS) in which most of the extra NH2-terminal domain is deleted because of alternative splicing of the pre-mRNA (Kise et al. 2004; Jorgensen et al. 2000), with Met-48 being deduced as the NH2-terminal residue of mini-TrpRS (Kise et al. 2004). PMN elastase digestion of recombinant full-length TrpRS produced two fragments designated T1-TrpRS and T2-TrpRS, respectively. These fragments were similar in size to mini-TrpRS. In addition, mini-TrpRS and T2-TrpRS blocked vascular endothelial growth factor-stimulated angiogenesis in both chick cell adhesion molecule and mouse matrigel assays in vivo (Wakasugi et al. 2002b; Otani et al. 2002). The full-length enzyme lacks cytokine activities. The construction and relationship of TyrRS, TrpRS, and their variants is shown in Fig. 1.
Fig. 1.
Schematic representation of human TyrRS, TrpRS and their variant constructs used in this study. Shaded regions of full-length TyrRS and TrpRS represent COOH- and NH2-terminal appended domains, respectively. Numbers on the left and right correspond to the NH2- and COOH-terminal residues relative to the human full-length enzymes, respectively
Because all the previous studies mentioned above were based on standard cell culture, what the results would be if performed in a hypoxic atmosphere was still a problem. Why did we choose the hypoxia as our research target? The reason is that some ischemia diseases (like CAHD), are actually pathophysiologically caused by hypoxia. Many organs lose their function due to lack of oxygen. So, we designed this research simulating the conditions of ischemia in order to prove whether mini-TyrRS and mini-TrpRS could be expressed in hypoxic culture, and to begin to comprehend their angiogenesis mechanism. All of this may be helpful for healing ischemia diseases, such as CAHD.
Methods
Materials
HUVECs (HUV-EC-C, ATCC® Number: CRL-1730™) were obtained from the Laboratory of Peptides Related with Human Diseases, West China Hospital, Sichuan University (Sichuan, China). pGPU6/GFP/Neo siRNA expression vector kit and RNAi-Mate transfection reagent were purchased from Genepharma (Shanghai, China). Dulbecco’s modified Eagle’s medium (DMEM) low-glucose medium was from Invitrogen (Carlsbad, CA, USA). The ECL chemiluminescence kit and BCA protein quantitative kit was from Pierce (Rockford, IL, USA). The plasmid extraction kit (Promega, Madison, WI, USA) and Matrigel (Becton Dickinson, San Jose, CA, USA) were purchased. Goat monoclonal anti-human mini-TyrRS or mini-TrpRS IgG antibodies, and horseradish peroxidase (HRP) conjugated rabbit anti-goat IgG were all from Santa Cruz Biotechnology (Santa Cruz, CA, USA). HRP-conjugated GAPDH was from Kang Chen Biotechnology Company (China). Revert Aid™ First Strand cDNA Synthesis Kit and Protein Marker were purchased from MBI Company (Lithuania). Cytoplasm proteins extraction kit was from Keygen Biotechnology (Nanjing, China).
SiRNA target site selection
Beginning with the AUG start codon, the transcript was scanned for AA dinucleotide sequences. Each AA and the 3′ adjacent 19 nucleotides were considered as potential siRNA target sites when the G/C ratio was 30–50% as measured with Ambion siRNA analysis software (http://www.ambion.com/techlib/misc/siRNA_finder.html). Since some regions of mRNA may be either highly structured or bound by regulatory proteins, we selected each of the three siRNA target sites at different positions along the length of the gene sequence (the procedure is described in detail as follows). We did not see a correlation between the position of target sites on the mRNA and siRNA potency. Then we compared the potential target sites to the appropriate genome database and eliminated from consideration any target sequences with more than 16–17 contiguous base pairs of homology to other coding sequences. We used BLAST, which can be found on the NCBI server at: http://www.ncbi.nlm.nih.gov/BLAST. A complete siRNA experiment should include a negative control siRNA with the same nucleotide composition as siRNA but which lacks significant sequence homology to the genome. In order to design a negative control siRNA, we scrambled the nucleotide sequence of each of our prospective siRNAs and conducted a search to make sure that it lacked homology to any other gene. We then transfected the siRNAs to choose the best target site position as follows. One day before transfection, cells were plated in 2 mL of growth medium without antibiotics such that they were 30–50% confluent at the time of transfection. To obtain the highest transfection and low non-specific effects, we optimized the transfection conditions by varying RNA and Lipofectamine™ 2000 concentrations. 0.5 mL siRNA oligomer—Lipofectamine™ 2000 complexes, and 1.5 mL DMEM without serum and antibiotics were added to each well for the final volume 2 mL.
Hairpin siRNA template oligonucleotide design and preparation
Two complementary oligonucleotide strands were designed with the following sequences: mini-TyrRS (Top strand: 5′-CACCGCCTGCACTTGGCTATTCAATTTCAAGAGAAATTGAATAGCCAAGTGCAGGTTTTTTG-3′; and bottom strand: 5′-GATCCAAAAAACCTGCACTTGGCTATTCAATTTCTCTTGAAATTGAATAGCCAAGTGCAGGC-3′); mini-TrpRS(Top strand: 5′-CACCGAGATGTTGGTGTCATTAAATTTCAAGAGAAATTTAATGACACCAACATCTTTTTTTG-3′; and bottom strand: 5′-GATCCAAAAAAAGATGTTGGTGTCATTAAATTTCTCTTGAAATTTAATGACACCAACATCTC-3′). A 5 μL of sense siRNA template oligonucleotide strand (100 μM), 5 μL of antisense siRNA template oligonucleotide strand (100 μM), and 5 μl of 10× siRNA annealing buffer solution were mixed, and 35 μL sterilized deionized H2O was added to a final volume 50 μL. The reaction mixture was denatured at 95 °C for 3 min, then annealed at room temperature for 20 min until it formed double-stranded oligonucleotides (ds-oligo).
Ligation of annealed siRNA template insert into pGPU6/GFP/Neo expression vector
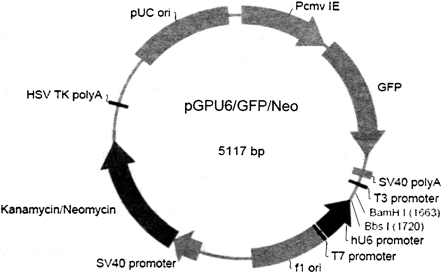
pGPU6/GFP/Neo expression vector containing an RNA polymerase III expression element (Fig. 2) was used as the vector in this research. The annealed siRNA template insert was diluted with nuclease-free water to a final concentration of 100 nM. Two 10 μL ligation reactions were set up. The first was a plus-insert ligation: 1 μL of diluted annealed siRNA insert, 6.5 μL of nuclease-free water, 1 μL of 10 × T4 DNA ligase buffer, 1 μL of pGPU6/GFP/Neo expression vector and 0.5 μL of T4 DNA ligase (5 U/μL). The second was the minus-insert negative control: 1 μL of 1 × siDNA annealing solution, 6.5 μL of nuclease-free water, 1 μL of 10 × T4 DNA ligase buffer, 1 μL of pGPU6/GFP/Neo expression vector and 0.5 μL of T4 DNA ligase (5 U/μL). T4 DNA ligase was used to clone the ds-oligo into the linear pGPU6/GFP/Neo expression vector. The ligation reactions were incubated overnight at 16 °C for high ligation efficiency. TOPO 10 competent cells of Escherichia coli were transformed with the plus-ligation products and the minus-ligation products at room temperature for 10 min, on ice for 35 min, then, spread on Luria Broth (LB) solid medium with Kanamycin (100 μg/mL) and grown overnight at 37 °C. Monoclonal colonies were selected 1 day later, and the plasmid DNA was isolated and digested with BamHI and EcoRI. The plasmids were confirmed by sequencing by Genepharma (Shanghai, China). Finally, pGPU6/GFP/Neo expression vector was transfected into HUVECs, following the instructions of the RNAi-Mate in a 24-well format. We used 0.8 μg DNA and 3.2 μg RNAi-Mate in our experiment.
Fig. 2.
pGPU6/GFP/Neo expression vector containing a RNA polymerase III expression element. Silencing efficiency was assayed by real-time fluorescent quantitation PCR by expression of Green Fluorescence Protein (GFP)
Cell culture and experimental groups
HUVECs were cultivated in DMEM low-glucose medium supplemented with 10% inactivated calf serum and 100 U/mL penicillin and 100 μg/mL streptomycin in an atmosphere of 5% CO2 in air at 37 °C according to the instructions of the incubator, before being used in experiments. In our study, cells were cultivated in hypoxic culture for 3, 6, 12, or 24 h. The hypoxic atmosphere consisted of 1% O2, 5% CO2 and 94% N2 at 37 °C according to the instructions of the hypoxia incubator. Three experimental groups were constructed: (1) untransfected HUVECs (mock transfection); (2) pGPU6/GFP/Neo negative control; (3) Mini-TyrRS or mini-TrpRS pGPU6/GFP/Neo expression vector transfected HUVECs (experimental group). Purity of EC cultureswas verified by immunostaining with an antibody against CD31. For all data shown, each individual experiment was performed with an independent preparation of HUVECs.
Cell proliferation
A cell proliferation assay was performed as previously described. In brief, HUVECs (5 × 103 cells per well) were plated onto 0. 1% glutin-coated 96-well plates in culture medium. The next day, the cells were pretreated with or without siRNA. The cells were cultured in DMEM containing 1% FBS to avoid cell detachment. After 24 hours, cell proliferation was determined by crystal violet staining. Counts were performed on three wells, and experiments were performed in triplicate. Results were normalized by arbitrarily setting the OD490 nm of control to 100%.
Matrigel-induced capillary tube formation
This assay was performed by the method of Yeh et al. as described (Yeh et al. 1998). Matrigel, a basement membrane, was diluted to 4 mg/mLwith cold D-hank’s and added to 24-well plates to a total volume of 400 μL in each well. Plates were stored at 37 °C for 30 min to form a gel layer. After gel formation, 1 mL HUVECs (5 × 105 cells) in a medium containing 20% fetal bovine serum was applied onto each well. They were incubated in the hypoxic atmosphere for 3, 6, 12, or 24 h. After incubation, the cells were observed with an inverted phase-contrast microscope (Olympus, Japan) and photographed. The total tube area were analyzed in 3 different fields at 40× magnification using Image-proplus 5.1 software. Results were normalized by arbitrarily setting the total tube area of control to 100%.
Cell migration assays
Transwell migration in vitro was performed as described previously (Grant et al. 1989). Briefly, cell migration was assayed with 8 μm pore size Transwell migration chambers (Costar). The undersides of membraneswere coated at room temperature over night with 0. 1% glutin (BD Biosciences). Cells (1 × 105) were added to the chambers in DMEM containing 1% fetal bovine serum. Cell migration was allowed to proceed for 4 hours at 37 °C in a hypoxia culture incubator.cells then were removed from the upper surface of the membranes with a cotton swab. Cells that migrated to the lower surface were stained withWright’s & Giemsa staining for 15 minutes and washed with water. Dried membranes were cut out and mounted on glass slides in immersion oil. At least 5 random high-power fields from each of trip licate membranes were counted for each experimental condition. Allmigration assayswere repeated three times. Results were normalized by arbitrarily setting the migration cell number of control to 100%.
Western blot analysis
Cytoplasmic protein extracts were prepared according to the instructions of the cytoplasm proteins extraction kit. The extracted proteins (35 μg) were subjected to 10% SDS-PAGE. After electrophoresis, proteins were transferred onto PVDF membranes (Millipore, Billerica, MA, USA). The membranes were then placed into blocking buffer containing nonfat dry milk. Goat monoclonal anti-human mini-TyrRS or mini-TrpRS IgG antibodies (final dilution 1:500) were used as the primary antibody, and horseradish peroxidase (HRP) conjugated rabbit anti-goat IgG (final dilution 1:10000) was used as the secondary reagent.An internal control, HRP-conjugated GAPDH and Protein Marker were also used in this experiment. Detection was performed using an ECL chemiluminescence kit. The data were scanned and analyzed by the Gel Doc 1000 gel imaging system (Bio-Rad Company, Hercules, CA, USA). The same test was repeated 3 times.
Real time fluorescent quantitation PCR (RT-PCR)
After hypoxic cultivation, HUVECs (1 × 106 cells) were harvested at the times indicated, washed twice with ice-cold phosphate-buffered saline (PBS) and collected by centrifugation. Total RNA was isolated from HUVECs using the Trizol reagent (MRC, USA) according to the manufacturer’s instructions. Total RNA (5 μL) was converted to complementary DNA (cDNA) using Revert Aid™ First Strand cDNA Synthesis Kit. A 5μL aliquot of the resulting cDNA was used as template for PCR amplification with the following primers: human TrpRS, P1 (forward, 5′-CCC TGC TGC ACT CCA CCT T-3′), P2 (reverse, 5′-ACG CAT GCT TAT TGA CCT TG-3′); human TyrRS, P3 (forward, 5′-CAT CTG ATG AAT CCT ATG GTT-3′), P4 (reverse, 5′-GGA TCA CAA ACT CGG ACT TA-3′); human β-actin, P5 (forward, 5′-GCC AAC ACA GTG CTG TCT-3′), P6 (reverse, 5′-AGG AGC AAT GAT CTT GAT CTT-3′). The amplifications were performed by an initial denaturation (94 °C for 2 min), followed by 45 cycles of denaturation, annealing and extension (94 °C for 20 s, 54 °C for 20 s, 72 °C for 30 s), and a final extension (72 °C for 5 min). The transcript of β-actin was also amplified by RT-PCR from the same cDNA template and was used as an internal control. All the primers were designed and synthesized by Genepharma (Shanghai, China). The identity of each PCR product was confirmed by DNA sequencing. The pictures were scanned and analyzed by the Gel Doc 1000 gel imaging system.
Statistical analysis
Results are expressed as mean ± standard deviation. Comparison of means was performed by means of the analysis of variance procedure (Student–Newman–keuls test, SPSS 13.0 for Windows). p < 0. 05 was considered statistically significant.
Results
Mini-TyrRS and mini-TrpRS RNAi induced gene silencing
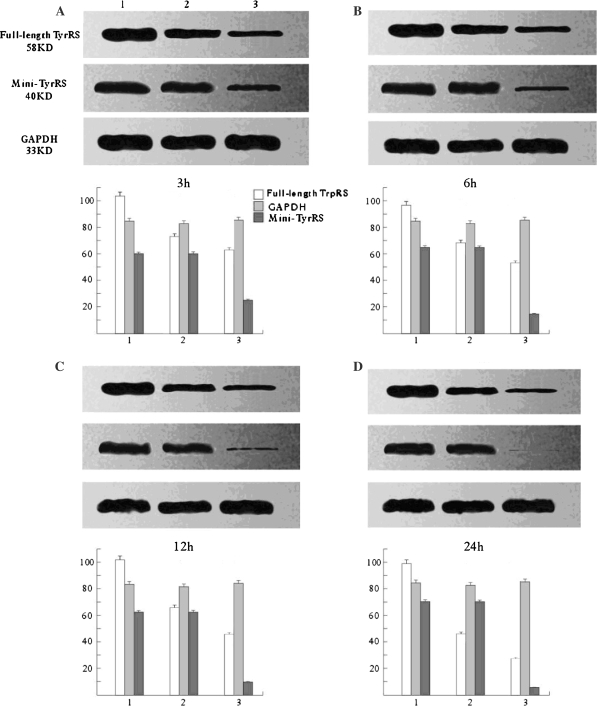
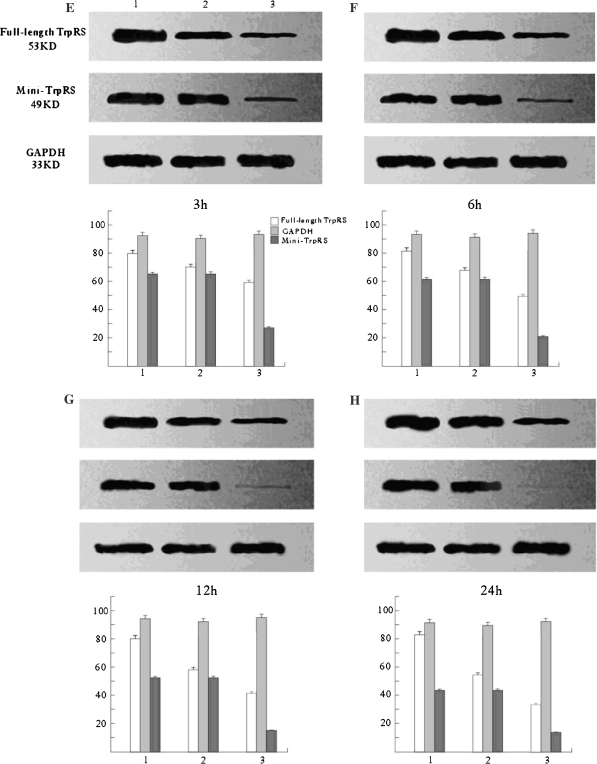
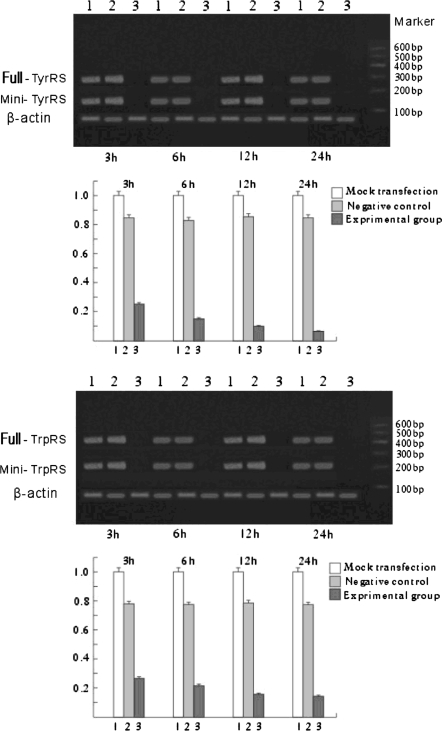
Mini-TyrRS and mini-TrpRS RNAi induced gene silencing Western blotting (Figs. 3, 4), and Real-time fluorescence quantitative PCR (Fig. 5) were used to compare the levels of Mini-TyrRS and mini-TrpRS gene and protein among three groups of HUVECs: (1) untransfected HUVECs (mock transfection); (2) pGPU6/GFP/Neo negative control; (3) Mini-TyrRS or mini-TrpRS pGPU6/GFP/Neo expression vector transfected HUVECs (experimental group). The results showed that levels of the mini-TyrRS/mini-TrpRS gene and protein in groups 1 and 2 were higher, but noticeably decreased in group 3. However, no significant difference was detected between group 1 and 2, while there was a significant statistical difference compared with group 3. Expression of the mini-TyrRS gene and protein were increased in hypoxia culture, after transfection with mini-TyrRS-siRNA at 3, 6, 12, and 24 h, were respectively decreased by 75, 85, 90, and 94% compared with group 1 and 2 (Figs. 3, 5), and for mini-TrpRS-siRNA, the expression were specifically decreased by 73, 79, 85, 86% (Figs. 4, 5).While the expression of full length TyrRS and TrpRS were also found inhibited, the same level with their variant constructs.
Fig. 3.
Assay for mini-TrpRS gene silencing efficiency by Western blotting. Only the mini-TyrRS-specific siRNA inhibited the expression of mini-TyrRS protein. The silencing efficiency had no significant difference detected between the mock and untransfected groups. (1) Untransfected HUVECs (mock transfection); (2) negative control; (3) mini-TyrRS pGPU6/GFP/Neo expression vector transfected HUVECs (experimental group). Compared with GAPDH, protein expression of the full-length TyrRS maintained the same level from 3 to 24 h, but the mini-TyrRS protein expression increased during that time
Fig. 4.
Assay for mini-TyrRS gene silencing efficiency by Western blotting. Only the mini-TrpRS-specific siRNA inhibited the expression of mini-TrpRS protein. The silencing efficiency had no significant difference detected between the mock and untransfected groups. (1) Untransfected HUVECs (mock transfection); (2) negative control; (3) mini-TrpRS pGPU6/GFP/Neo expression vector transfected HUVECs (experimental group). Compared with GAPDH, protein expression of the full-length TrpRS maintained the same level from 3 to 24 h, but the Mini-TrpRS protein expression decreased during that time
Fig. 5.
Assay for mini-TyrRS/mini-TrpRS gene silencing efficiency by real-time fluorescence quantitative reverse transcription-polymerase chain reaction (RT-PCR). Only the mini-TyrRS/mini-TrpRS specific siRNA inhibited expression of the mini-TyrRS/mini-TrpRS gene. No mini-TyrRS/mini-TrpRS knockdown was observed with the negative group. (1) Untransfected HUVECs (mock transfection); (2) negative control; (3) mini-TrpRS or mini-TyrRS pGPU6/GFP/Neo expression vector transfected HUVECs (experimental group)
Cell migration assay and tube formation
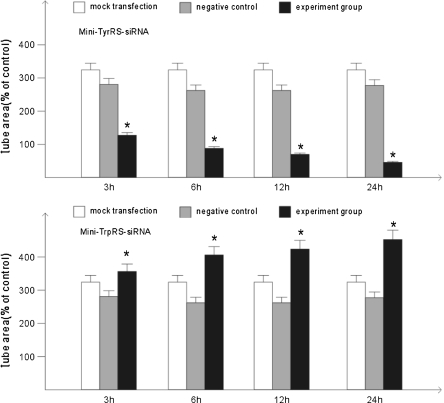
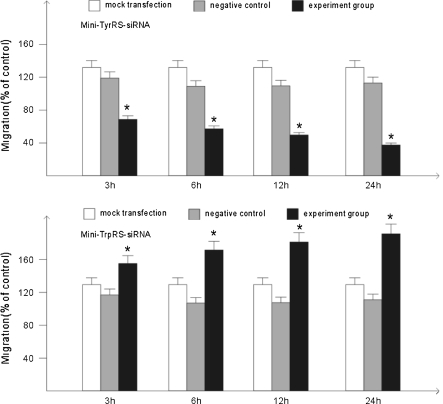
The cell migration assay and tubule formation assay are two crucial assays to evaluate the angiogenic ability of endothelial cells. We examined the effects of the mini-TyrRS or mini-TrpRS-induced tube formation and migration. The data in Fig. 3 showed that tube formation of HUVECs induced by mini-TyrRS-siRNA at 3, 6, 12, and 24 h were significantly inhibited by 60.5, 69.1, 75.9, 83.6%, and for mini-TrpRS-siRNA, were rapidly increased by 18.3, 31.2, 40.3, 45.7%, respectively. At the same time, using migration assasy, mini-TyrRS–siRNA group were also inhibited by 47.4, 56.3, 65.4, 73.7%, and mini-TrpRS-siRNA group were increased by 18.0, 33.8, 45.1, 56.4%.
Cell proliferation assay
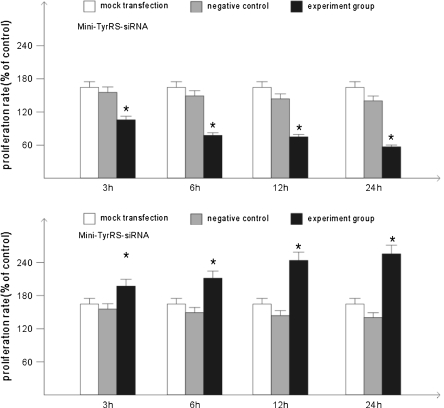
Next, we explored whether the ability of mini-TyrRS and mini-TrpRS affected endothelial cell proliferation. The data in Fig. 6 showed that mini-TrpRS-siRNA at 3, 6, 12, and 24 h significantly increased 8.4, 26.4, 38.2, 46.6%, HUVECs proliferation. But, proliferation in HUVECs by mini-TyrRS-siRNA significantly reduced by 40.4, 56.2, 61.2, 68.0% (Figs. 7, 8).
Fig. 6.
Mini-TyrRS/mini-TrpRS-siRNA induced tube formation assay. *p < 0.05, compared with the mock transfection group and negative control group. There were no statistic difference between the mock transfection group and the negative control group
Fig. 7.
Assay of cell migration induced by mini-TyrRS/mini-TrpRS-siRNA. *p < 0.05, compared with the mock transfection group and negative control group. There were no statistic difference between the mock transfection group and the negative control group
Fig. 8.
Rate of cell proliferation induced by mini-TyrRS/mini-TrpRS-siRNA. *p < 0.05, compared with the mock transfection group and negative control group. There were no statistic difference between the mock transfection group and the negative control group
Discussion
RNA interference (RNAi) by double stranded RNA (dsRNAs) molecules of approximately 20–25 nucleotides termed short interfering (siRNAs) is a powerful method for preventing the expression of a particular gene. The dsRNA molecules can target mRNAs with complementary sequence for degradation via a cellular process (Elbashir et al. 2001). This technique was first developed in Caenorhabditis elegans, and was rapidly applied to a wide range of organisms. Methods for expressing siRNAs in cells in culture and in vivo using viral vectors, and for transfecting cells with synthetic siRNAs, have been developed and are being used to establish the functions of specific proteins in various cell types and organisms (Sui et al. 2002; Lee et al. 2001; Paul et al. 2002). For example, chemically synthesized or in vitro transcribed siRNAs can be transfected into cells, injected into mice, or introduced into plants (Brummelkamp et al. 2002). siRNAs can also be expressed endogenously from siRNA expression vectors or PCR products in cells or in transgenic animals (Paddison et al. 2002). Using siRNA expression vectors has the advantage that the expression of a target gene can be reduced for weeks or even months (Byrom et al. 2002), eclipsing the 6–10 days typically observed with in vitro prepared siRNA used for transient transfection (Byrom et al. 2002).
In normal cells, human TrpRS exists as a full length form and as a truncated form designated mini-TrpRS, which is produced by alternative splicing (Yeh et al. 1998). Expression of mini-TrpRS is highly stimulated in human cells by the addition of IFN-γ (Tolstrup et al. 1995; Fleckner et al. 1991; Shaw et al. 1999). Although both human full-length TrpRS and mini-TrpRS are enzymatically active in aminoacylation, they differ in angiostatic activity (Lee et al. 2004; Tzima et al. 2005). The same phenomenon is also present between full-length TyrRS and mini-TyrRS. Researches have found a kind of angiogenesis regulatory factor, CXC chemotactic factor, but their function was dependent on whether or not they have the Glu-Leu-Arg (ELR) motif (Clark et al. 1993). The CXC chemotactic factors which have the ELR motif could promote angiogenesis, while CXC chemotactic factors which do not have the ELR motif would be the antagonistic factors of angiogenisis. The situation is analogous to human mini-TyrRS and mini-TrpRS. Mini-TyrRS has an ELR motif and promotes angiogenesis, but mini-TrpRS does not have the ELR motif. Therefore, their function was totally opposite (Jeong et al. 2000).
BD Matrigel™ Matrix is a solubilized basement membrane preparation extracted from EHS mouse sarcoma, a tumor rich in ECM proteins. Its major component is laminin, followed by collagen IV, heparan sulfate proteoglycans, and entactin. At room temperature, BD Matrigel Matrix polymerizes to produce biologically active matrix material resembling the mammalian cellular basement membrane. Cells behave as they do in vivo when they are cultured on BD Matrigel Matrix. It provides a physiologically relevant environment for studies of cell morphology, biochemical function, migration or invasion, and gene expression. BD Matrigel Matrix serves as a substrate for in vitro endothelial cell invasion and tube formation assays. It can also be used to assess in vivo angiogenic activity of different compounds via the BD Matrigel Plug Assay.
We were aware that angiogenic and angiostatic factors may work together to regulate angiogenesis. To test our hypothesis that mini-TyrRS (containing a natural ELR sequence) is an angiogenic factor as well as a PMN cell chemoattractant, and that mini-TrpRS (containing a natural ELQ sequence) is an angiostatic factor, we evaluated whether mini-TyrRS/mini-TyrRS induced endothelial cells to format capillary-like structures using BD Matrigel Matrix. We found out that the expression of mini-TrpRS was decreased, but that the mini-TyrRS was overexpressed in the hypoxic culture. Using mini-TyrRS siRNA knockdown, the formation speed of capillary-like structures was slowed, but for mini-TrpRS, the result was the opposite (Figs. 4, 5). In contrast, angiogenesis was not observed with full-length TyrRS and TrpRS. Interestingly, angiogenesis is stimulated by either mini-TyrRS or is inhibited by mini-TrpRS in matrigel models during hypoxic culture, raising the possibility that mini-TyrRS stimulates a common downstream signaling event. Thus, naturally occurring fragments of the two proteins involved in translation, TyrRS and TrpRS, have opposing activity on endothelial cell angiogenesis in the matrigel assays. The opposing activities of the two tRNA synthetases suggest tight regulation of the balance between pro- and anti-angiogenic stimuli, but the exact mechanism and relationship between them has not been clearly demonstrated. More research should be done to further explore the mechanism.
References
- Brummelkamp TR, Bernards R, Agami R (2002) A system for stable expression of short interfering RNAs in mammalian cells. Sciences (New York) 296:550–553 [DOI] [PubMed]
- Byrom M, Pallotta V, Brown D et al (2002) Visualizing SiRNA in mammalian cells: fluorescence analysis of the RNAi effect. Ambion Technotes 9(3):6–8
- Clark LI, Dewald B, Geiser T et al (1993) Platelet factor 4 binds to interleukin 8 receptors and activates neutrophils when its N terminus is modified with Glu-Leu-Arg. Proc Natl Acad Sci USA 90:3574–3577. doi:10.1073/pnas.90.8.3574 [DOI] [PMC free article] [PubMed]
- Elbashir SM, Harborth J, Lendeckel W et al (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in Drosophila melanogaster embryo lysate. Nature 411:494–498. doi:10.1038/35078107 [DOI] [PubMed]
- Ewalt KL, Schimmel P (2002) Activation of angiogenic signaling pathways by two human tRNA synthetases. Biochemistry 41:13344–13349. doi:10.1021/bi020537k [DOI] [PubMed]
- Fleckner J, Rasmussen HH, Justesen J (1991) Human interferon gamma potently induces the synthesis of a 552 kDa protein highly homologous to rabbit peptide chain release factor and bovine tryptophan. Proc Natl Acad Sci USA 88:11520–11524. doi:10.1073/pnas.88.24.11520 [DOI] [PMC free article] [PubMed]
- Fleckner J et al (1995) Differential regulation of the human interferon inducible tryptophanyl-tRNA synthetase by various cytokines in cell lines. Cytokine 7:70–77. doi:10.1006/cyto.1995.1009 [DOI] [PubMed]
- Folkman J (1985) Toward an understanding of angiogenesis: search and discovery. Perspect Biol Med 29:10–36 [DOI] [PubMed]
- Folkman J, Haudenschild C (1980) Angiogenesis in vitro. Nature 288:551–556. doi:10.1038/288551a0 [DOI] [PubMed]
- Grant DS, Tashiro K, Segui-Real B et al (1989) Two different laminin domains mediate the differentiation of human endothelial cells into capillary-like structures in vitro. Cell 58:933–943. doi:10.1016/0092-8674(89)90945-8 [DOI] [PubMed]
- Ibba M (2000) SÊ ll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem 69:617–650. doi:10.1146/annurev.biochem.69.1.617 [DOI] [PubMed]
- Jeong EJ, Hwang GS, Kim KH et al (2000) Structural analysis of multi-functional peptide motifs present in human bifunctional tRNA synthetase: identification of RNA-binding residues and functional implications for tandem repeats. Biochemistry 39:15775–15782. doi:10.1021/bi001393h [DOI] [PubMed]
- Jorgensen R, Sogaard TM, Rossing AB, Martensen PM, Justesen J (2000) Identification and characterization of human mitochondrial tryptophanyl-tRNA synthetase. J Biol Chem 275:16820–16826. doi:10.1074/jbc.275.22.16820 [DOI] [PubMed]
- Kise Y et al (2004) A short peptide insertion crucial for angiostatic activity of human tryptophanyl-tRNA synthetase. Nat Struct Mol Biol 11:149–156. doi:10.1038/nsmb722 [DOI] [PubMed]
- Kubota Y, Kleinman HK, Martin GR et al (1988) Roles of laminin and basement membrane in the morphological differentiation of human endothelial cells into capillary-like structures. J Cell Biol 107:1589–1598. doi:10.1083/jcb.107.4.1589 [DOI] [PMC free article] [PubMed]
- Lee NS, Dohjima T, Bauer G et al (2001) Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat Biotechnol 19:500–505 [DOI] [PubMed]
- Lee SW et al (2004) Aminoacyl-tRNA synthetase complexes: beyond translation. J Cell Sci 117:3725–3734. doi:10.1242/jcs.01342 [DOI] [PubMed]
- Maciag T, Kadish J, Wilkins L et al (1982) Organizational behavior of human umbilical vein endothelial cells. J Cell Biol 94:511–520. doi:10.1083/jcb.94.3.511 [DOI] [PMC free article] [PubMed]
- Madri JA, Williams SK (1983) Capilary endothelial cell culture phenotypic modulation by matrix components. J Cell Biol 97:153–165. doi:10.1083/jcb.97.1.153 [DOI] [PMC free article] [PubMed]
- Martinis SA, Plateau P, Cavarelli J et al (1999) Aminoacyl-tRNA synthetases: a family of expanding functions. EMBO J 18:4591–4596. doi:10.1093/emboj/18.17.4591 [DOI] [PMC free article] [PubMed]
- Montesano RL, Orci L, Vassalli P (1983) In vitro rapid organization of endothelial cells into capillary-like networks is promoted by collagen matrices. J Cell Biol 97:1648–1652. doi:10.1083/jcb.97.5.1648 [DOI] [PMC free article] [PubMed]
- Otani A, Slike BM, Dorrell M et al (2002) A fragment of human TrpRS as a potent antagonist of ocular angiogenesis. Proc Natl Acad Sci USA 99:178–183. doi:10.1073/pnas.012601899 [DOI] [PMC free article] [PubMed]
- Paddison PJ, Caudy AA, Berstein E et al (2002) Short hairpin RNAs induces sequences-specific silencing in mammalian cells. Genes Dev 16:948–958. doi:10.1101/gad.981002 [DOI] [PMC free article] [PubMed]
- Paul CP, Good PD, Winer I (2002) Effective expression of small interfering RNA in human cells. Nat Biotechnol 20:505–508. doi:10.1038/nbt0502-505 [DOI] [PubMed]
- Ribas de Pouplana L, Frugier M, Quinn CL, Schimmel P (1996) Evidence that two present-day components needed for the genetic code appeared after nucleated cells separated from eubacteria. Proc Natl Acad Sci USA 93:166–170. doi:10.1073/pnas.93.1.166 [DOI] [PMC free article] [PubMed]
- Risau W (1997) Mechanism of angiogenesis. Nature 386:671–674. doi:10.1038/386671a0 [DOI] [PubMed]
- Shaw AC, Rossel LM, Roepstorff P et al (1999) Mapping and identification of interferon gamma-regulated HeLa cell proteins separated by immobilized pH gradient two-dimensional gel electrophoresis. Electrophoresis 20:984–993. doi :10.1002/(SICI)1522-2683(19990101)20:4/5≤984::AID-ELPS984≥3.0.CO;2-R [DOI] [PubMed]
- Sui G, Soohoo C, Affar EB et al (2002) A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci USA 99:5515–5520. doi:10.1073/pnas.082117599 [DOI] [PMC free article] [PubMed]
- Tolstrup AB, Bejder A, Fleckner J et al (1995) Transcriptional regulation of the interferon-γ inducible tryptophanyl-tRNA synthetase includes alternative splicing. J Biol Chem 270:397–403. doi:10.1074/jbc.270.1.397 [DOI] [PubMed]
- Tzima E et al (2005) VE-cadherin links tRNA synthetase cytokine to anti-angiogenic function. J Biol Chem 280:2405–2408. doi:10.1074/jbc.C400431200 [DOI] [PubMed]
- Wakasugi K, Schimmel P (1999a) Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science 284:147–151. doi:10.1126/science.284.5411.147 [DOI] [PubMed]
- Wakasugi K, Schimmel P (1999b) Highly differentiated motifs responsible for two cytokine activities of a split human tRNA synthetase. J Biol Chem 274:23155–23159. doi:10.1074/jbc.274.33.23155 [DOI] [PubMed]
- Wakasugi K, Slike BM, Hood J et al (2002a) Introduction of angiogenesis by a fragment of human tyrosyl-tRNA synthetase. J Biol Chem 277:20124–20126. doi:10.1074/jbc.C200126200 [DOI] [PubMed]
- Wakasugi K, Slike BM, Hood J et al (2002b) A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc Natl Acad Sci USA 99:173–177. doi:10.1073/pnas.012602099 [DOI] [PMC free article] [PubMed]
- Yancopoulos GD, Klagsbrum M, Folkman J (1998) Vasculogenesis, angiogenesis and growth factors: ephrins enter the fray at the border. Cell 93:661–664. doi:10.1016/S0092-8674(00)81426-9 [DOI] [PubMed]
- Yeh CH, Peng HC, Huang TF (1998) Accutin, a new disintegrin, inhibits angiogenesis in vitro and in vivo by acting as integrin alphavbeta-antagonist and inducing apoptosisl. J Blood 92:3268–3276 [PubMed]