Abstract
Epigenetic modifications of histone play important roles for regulation of cell activity, such as cell division, cell death, and cell differentiation. A SET domain consisting of about 130 amino acids has lysine methyltransferase activity in the presence of the cosubstrate S-adenosyl-methionine. More than 60 SET domain-containing proteins have been predicted in various organisms. One of them, the SMYD family genes which contain a SET domain and a zinc-finger MYND domain are reported to regulate cell cycle and muscle formation. Here we examined the expression and function of smyd1 and 2 in Xenopus. smyd1 and 2 were expressed in various muscle tissues. While smyd1 expression was observed mainly in cardiac muscle and skeletal muscle, smyd2 expression was done abundantly in skeletal muscle and face region. Moreover, by loss-of-function experiments using antisense morpholino oligonucleotides, it was suggested that smyd1 and 2 related to muscle cells differentiation.
Keywords: Heart, Muscle, MYND, Myogenesis, SET, smyd, Xenopus laevis
Introduction
Epigenetic modifications of histone regulate chromatin structure and gene expression in various cellular functions, such as cell cycle, cell death, maintenance of cell state, and cell differentiation. One of epigenetic modifications, methylation of histone tail is thought to play an important role for cell fate determination (Zhang and Reinberg 2001; Jenuwein and Allis 2001; Kouzarides 2002). In recent studies, it is reported that Polycomb group (PcG) and trithorax group (TrxG) regulate differentiation state of embryonic cells by methylating histone H3 lysine 27 (H3K27), H3K4 and H3K9 of differentiation factor genes locus, such as homeobox (Hox), Distal-less homeobox (Dlx) paired box (Pax) and sine-oculis-related homeobox (Six) gene locus (Caretti et al. 2004; Szutorisz et al. 2005; Azuara et al. 2006; Boyer et al. 2006; O’Neill et al. 2006; Rinn et al. 2007; Reik 2007). The SET domain is a catalytic domain conserved among lysine methyltransferases proteins and more than 60 SET protein genes are identified in various organisms (Jenuwein et al. 1998; Trievel et al. 2002; Qian and Zhou 2006). For examples, Ezh2 which is a member of PcG regulates myogenesis by histone H3-K27 methylation (O’Carroll et al. 2001; Czermin et al. 2002; Caretti et al; 2004). Mll and Mll2 involved in TrxG sustain Hox genes regulation in normal development (Yu et al. 1995; Yu et al. 1998; Hanson et al. 1999; Glaser et al. 2006). Furthermore, Set9 and Smyd2 can methylate not only histone but also p53 and regulate the cell cycle (Chuikov et al. 2004; Huang et al. 2006). However, functions of many of SET proteins have not resolved yet.
Among SET proteins, SMYD proteins which contain MYND (named from myeloid translocation protein 8, Nervy, and DEAF-1) and SET domains. The MYND domain is a conserved zinc binding domain and defined by seven conserved cysteine residues and a single histidine residue that are arranged in a C4-C2HC consensus (Spadaccini et al. 2006). The 5 varieties of smyd genes in M. musculus and H. sapiens have been cloned. It is well established that SMYD1 plays a role in normal development of heart in mouse (Hwang and Gottlieb 1997; Gottlieb et al. 2002; Phan et al. 2005) and muscle in zebrafish (Du et al. 2006; Tan et al. 2006). Also, SMYD2 and SMYD3 are reported to might be involved in repression and activation of cell cycle (Brown et al. 2006; Tsuge et al. 2005; Hamamoto et al. 2004).
In this study, we have characterized the expression patterns of smyd1 and 2 in Xenopus laevis embryos. smyd1 and 2 were expressed in various muscle tissues. While smyd1 expression was observed mainly in cardiac muscle and skeletal muscle, smyd2 expression was done abundantly in skeletal muscle and face region. Taken together with the loss-of-function experiments, it was suggested that smyd1 and 2 are related to muscle tissue development.
Materials and methods
Embryos manipulation
Xenopus laevis embryos were obtained and cultured by standard methods (Sive et al. 2000). Embryos were allowed to develop in 1× Steinberg’s solution and staged according to the normal table of Nieuwkoop and Faber (1967). Animal cap cells were dissected from embryos at stage 9 and cultured in 1× modified Barth’s saline (MBS) supplemented with 0.1% BSA. Subsequently, these were treated with 5 ng/ml activin A.
smyd1 and 2 antisense morphlono oligonucleotides (Smyd1MO and Smyd2MO) were designed and supplied by Gene Tools LCC as follows, Smyd1MO: 5′-catggctgttccgcttctaccctgt-3′, and Smyd2MO: 5′-gttccagaccctcgggctgtcccat-3′. Microinjection was performed in 1× MBSH containing 5% ficoll solution. Morpholino oligonucleotide were microinjected into the animal side of stage 1 embryos.
RNA extraction and RT-PCR
RNAs of each animal cap and embryo were extracted using TRIzol reagent (Invitrogen). cDNA was synthesized using M-MLV reverse transcriptase (Invitrogen). Oligo-dT primed reverse-transcription was performed using 1 μg total RNA as a template. Each cDNA was amplified by PCR using the following primer pairs and under the following cycling conditions; pax3 (NM_001095524), forward: 5’-tttacccgttactcatggatagtgt-3′, reverse: 5′-aatgtcacataaaatccaaaaagga-3′ for 30 cycles, myoD (NM_203641), forward: 5′-tctctccagcatcgtcgagc-3′, reverse: 5′-ggaattcattgtccgtttgg-3′ for 30 cycles, myf-5 (X56738), forward: 5′-ccatgagagaacggagaagg-3′, reverse: 5′-cggggtgatagagtctggaa-3′ for 30 cycles, smyd1 (MGC80131), forward: 5′-cgtgttgtgaaagaggtg-3′, reverse: 5′-gggttcatccatgacttg-3′ for 30 cycles, smyd2 (MGC82991), forward: 5′-tgatgcacctccctttg-3′, reverse: 5′-caaaccgtaaaccaagctc-3′ for 30 cycles and an internal control, ornithine decarboxylase (odc, NM_001086698), forward: 5′-gcgggcaaaggagcttaatg-3′, reverse: 5′-taacgccagaatctgctggg-3′ for 25 cycles. Five micro liters of each PCR product was resolved by 1% agarose gel electrophoresis and observed with ethidium bromide.
Whole mount in situ hybridization
smyd1 and 2 PCR products were inserted into pBluescript II (SK-) (TOYOBO) and then these were prepared to construct probes for whole mount in situ hybridization. Digoxigenin-labeled antisense RNA probes were in vitro transcribed with T7 or T3 polymerase (Roche Molecular Biochemicals) from template cDNA, for smyd1 and 2. Whole mount in situ hybridization was performed according to the method of Harland (Sive et al. 2000), except that the chromogenic reaction was done using BM purple as the substrate (Roche Molecular Biochemicals).
Results and discussion
Sequence comparison
We found that AAH72803 and AAH73650 correspond to SMYD1 and 2, respectively, by BLAST search and domain analysis (Fig. 1a). The post-SET domain shown in Fig. 1b is not essential for methylation activity, but it associates with the SET domain and exerts histone methyltransferase activity (Rea et al. 2000).
Fig. 1.
Comparison of SMYD proteins sequence among H. sapiens, SMYD1 (HsSMYD1, NM_198274), SMYD2 (HsSMYD2, NM_020197), SMYD3 (HsSMYD3, NM_022743), SMYD4 (HsSMYD4, NM_052928) and SMYD5 (HsSMYD5, NM_006062), and X. laevis, SMYD1 (XlSMYD1) and SMYD2 (XlSMYD2). (a) MYND domain sequences. (b) SET domain and post-SET domain sequences. Identical amino acids are indicated by asterisks. Conserved domain residues are boxed
Expression pattern of smyd1 and 2 during early development
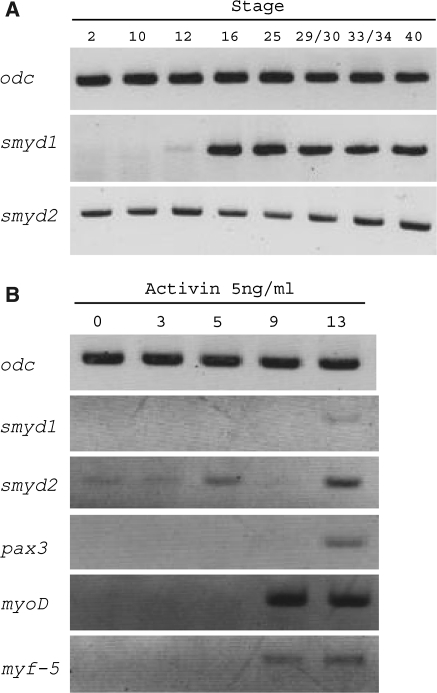
We examined the expression pattern of smyd1 and 2 in X. laevis early development by using RT-PCR (Fig. 2a). We used embryos at stage 2, 10, 12, 16, 25, 29/30, 33/34 and 40 according to the normal table of Nieuwkoop and Faber (1967). smyd2 expression was identified at stage 2, indicating that smyd2 mRNA existed maternally, and was persistent through stage 40 (Fig. 2a). On the other hand, smyd1 expression was not identified maternally, but began to increase after the gastrula stage.
Fig. 2.
Gene expression patterns of smyd1 and 2. (a) RT-PCR analysis from whole embryo RNA at the indicated stage according to Nieuwkoop and Faber (1967). Ornithine decarboxylase (odc) was used as control. (b) RT-PCR analysis from animal cap assay. myoD, myf-5 and pax3 were used as control of myogenesis
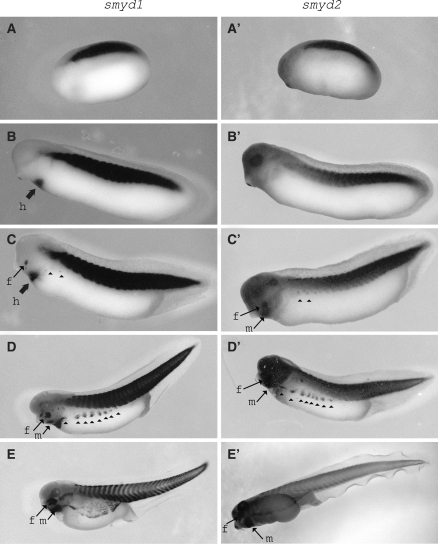
Next, we analyzed the spatial expression patterns of smyd1 and 2 by using whole mount in situ hybridization. The expression of smyd1 and 2 was observed distinctly in the prospective muscle tissue regions, somite, at stage 22 (Fig. 3a and a′). Subsequently, expression of both smyd1 and 2 was observed also at the prospective ventral body wall, which began to migrate from the somite at later stage (Fig. 3c–e and c′–e′). These migrating cells correspond to dermomyotome.
Fig. 3.
Spatial expression patterns of smyd1 and 2 in early development. smyd1 and 2 mRNA localized in somite at stage 23 (a and a′). At stage 28, smyd1 expression was observed in the presumptive heart (b) but smyd2 was not (b’). After that, when the ventral body wall cells began to migrate from the somite at stage 33-34 (c and c′) and expanded completely by stage 41(e and e′), smyd1 and 2 expression was observed in these migrating cells (c–d and c′–d′, arrow heads). h; heart, f; face and m; mandibular
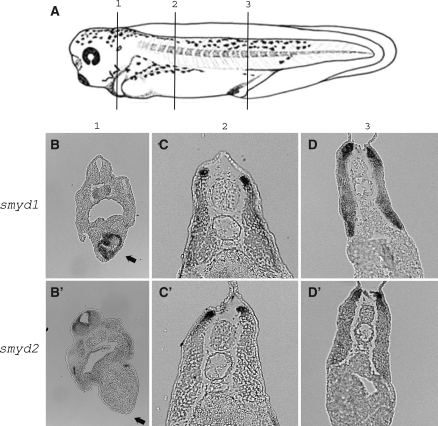
Furthermore, we identified the differences between smyd1 and 2 expression. At first, only smyd1 expression was detected in the heart (Figs. 3 and 4). This expression is coincident with the fact that smyd1 expression in heart is important for beating cardiac muscle in zebrafish (Du et al. 2006; Tan et al. 2006). Secondly, in the section at the anterior somite, while smyd1 was observed within the entire region of the dermomyotome (Fig. 4c), smyd2 expression was concentrated more within the dorsomedial lip (Fig. 4c′). Finally, in the face region, smyd2 expression was more widely than smyd1 (Fig. 3c, d, c′ and d′). Also, in mandibular regions, following the smyd2 expression at stage 33/34, smyd1 expression began at stage 37/38 (Fig. 3d, e c′, and e′).
Fig. 4.
smyd1 and 2 expression in somite region. (a) Position of the partial sections (stage 37/38). smyd1 expression was observed in the heart (b, arrow) but smyd2 was not (b′). smyd1 was observed within dermomyotome (c). smyd2 was observed within dorsomedial lip (c′). In middle of somite, both smyd1 and 2 mRNA existed in dermomyotome (d, d′)
These expression patterns in somite migration cell, heart and face regions are similar to those of myoD and myf-5, which are homeobox domain transcription factors and are referred to myogenic regulatory factors (Martin and Harland 2001). Taken together, we suggest that the expression of smyd1 and 2 may correlate with muscle tissue formation during X. laevis early development.
Expression during muscle differentiation
To examine the expression smyd1 and 2 during myogenesis, we carried out animal cap assays. Animal caps cells are multipotent cells and are able to differentiate into various types of cells by responding to differentiation inducing factors. When animal cap cells are treated with activin A, they become mesodermal tissues including muscle cells (Tamai et al. 1999). The animal cap cells were dissected from blastula embryo (stage 9), treated with 5 ng/ml activin A, and then used for RT-PCR. As myoD, myf-5 and pax3, which are known as myogenic regulatory factor, were expressed by treatment of activin A (Fig. 2b), the animal cap cells differentiated into muscle lineage cells. smyd1 and 2 expression also increased after the treatment with activin A (Fig. 2b 13 h). The existence of smyd2 mRNA from 0 to 3 hrs after treatment with activin A (Fig. 2b) may be due to the occurrence of its mRNA as a maternal substance (Fig. 2a). This indicated that smyd1 and 2 are related to myogenesis process.
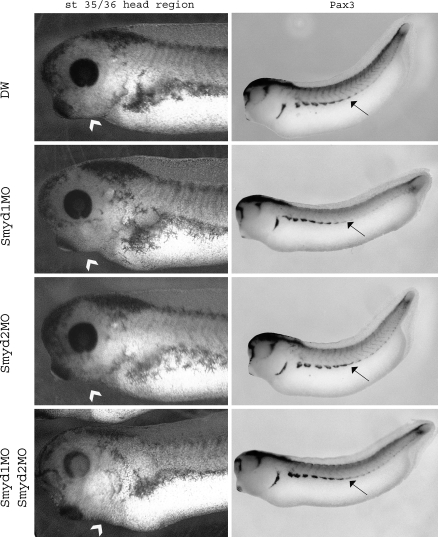
Next, we examined the smyd1 and 2 importance in embryogenesis by loss-of-function analysis using the antisense morpholino oligonucleotide (MO). The embryos injected with Smyd1MO had a immature somite (Fig. 5) and could not swim (data not shown). Moreover, injecting with both Smyd1MO and Smyd2MO, the embryos possess not only an abnormal somite, but also an abnormal mandibular tissue (Fig. 5) and the extent of malformations became to be more intensive. When Smyd2MO was injected into embryos, the abnormalities were not observed. Because the difference of smyd2 expression from smyd1 was only in face region, it is possible that we could not detect the malformation of face in this assay. From these results, smyd1 is essential for the heart and skeletal muscle development.
Fig. 5.
The loss-of-function analysis of smyd1 and 2. DW, Smyd1MO, Smyd2MO and Smyd1MO-Smyd2MO were injected to stage 1 embryos. Right panels: whole mount in situ hybridization by using pax3 as a probe. The arrow heads show the mandibular and arrow show the somite
Conclusion
From the results described here, it is revealed that expression of smyd1 and 2 plays a role during muscle development. And we suggested that at least smyd1 is necessary for muscle cell formations. However, there are temporal and anatomical differences between smyd1 and 2 expression. To clarify the mechanism of muscle tissue differentiation and muscle tissue formation, we have to analyze the roles of smyd1 and 2 at molecular level.
Acknowledgements
We thank Dr. William Watt for critical reading of manuscripts and Dr. Kaori Kubota for technical advices. This research was supported by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
References
- Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG (2006) Chromatin signatures of pluripotent cell lines. Nat Cell Biol 8:532–538 [DOI] [PubMed]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441:349–353 [DOI] [PubMed]
- Brown MA, Sims RJ 3rd, Gottlieb PD, Tucker PW (2006) Identification and characterization of Smyd2: a split SET/MYND domain-containing histone H3 lysine 36-specific methyltransferase that interacts with the Sin3 histone deacetylase complex. Mol Cancer 5:26–37 [DOI] [PMC free article] [PubMed]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V (2004) The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Gene Dev 18:2627–2638 [DOI] [PMC free article] [PubMed]
- Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, Barlev NA, Reinberg D (2004) Regulation of p53 activity through lysine methylation. Nature 432:353–360 [DOI] [PubMed]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111:185–196 [DOI] [PubMed]
- Du SJ, Rotllant J, Tan X (2006) Muscle-specific expression of the smyd1 gene is controlled by its 5.3-kb promoter and 5′-flanking sequence in zebrafish embryos. Dev Dyn 235:3306–3315 [DOI] [PubMed]
- Glaser S, Schaft J, Lubitz S, Vintersten K, van der Hoeven F, Tufteland KR, Aasland R, Anastassiadis K, Ang SL, Stewart AF (2006) Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development 133:1423–1432 [DOI] [PubMed]
- Gottlieb PD, Pierce SA, Sims RJ, Yamagishi H, Weihe EK, Harriss JV, Maika SD, Kuziel WA, King HL, Olson EN, Nakagawa O, Srivastava D (2002) Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat Genet 31:25–32 [DOI] [PubMed]
- Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, Yagyu R, Nakamura Y (2004) SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol 6:731–740 [DOI] [PubMed]
- Hanson RD, Hess JL, Yu BD, Ernst P, van Lohuizen M, Berns A, van der Lugt NM, Shashikant CS, Ruddle FH, Seto M, Korsmeyer SJ (1999) Mammalian Trithorax and polycomb-group homologues are antagonistic regulators of homeotic development. Proc Natl Acad Sci USA 96:14372–14377 [DOI] [PMC free article] [PubMed]
- Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T, Berger SL (2006) Repression of p53 activity by Smyd2-mediated methylation. Nature 444:629–632 [DOI] [PubMed]
- Hwang I, Gottlieb PD (1997) The Bop gene adjacent to the mouse CD8b gene encodes distinct zinc-finger proteins expressed in CTLs and in muscle. J Immunol 158:1165–1174 [PubMed]
- Jenuwein T, Allis CD (2001) Translating the Histone code. Science 293:1074–1080 [DOI] [PubMed]
- Jenuwein T, Laible G, Dorn R, Reuter G (1998) SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell Mol Life Sci 54:80–93 [DOI] [PMC free article] [PubMed]
- Kouzarides T (2002) Histone methylation in transcriptional control. Curr Opin Genet Dev 12:198–209 [DOI] [PubMed]
- Martin BL, Harland RM (2001) Hypaxial muscle migration during primary myogenesis in Xenopus laevis. Dev Biol 239:270–280 [DOI] [PubMed]
- Nieuwkoop PD, Faber J (1967) Normal Table of Xenopus laevis. North Holland, Daudin, Amsterdam
- O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T (2001) The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol 21:4330–4336 [DOI] [PMC free article] [PubMed]
- O’Neill LP, VerMilyea MD, Turner BM (2006) Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nat Genet 38:835–841 [DOI] [PubMed]
- Phan D, Rasmussen TL, Nakagawa O, McAnally J, Gottlieb PD, Tucker PW, Richardson JA, Bassel-Duby R, Olson EN (2005) BOP, a regulator of right ventricular heart development, is a direct transcriptional target of MEF2C in the developing heart. Development 132:2669–2678 [DOI] [PubMed]
- Qian C, Zhou MM (2006) SET domain protein lysine methyltransferases: Structure, specificity and catalysis. Cell Mol Life Sci 63:2755–2763 [DOI] [PMC free article] [PubMed]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593–599 [DOI] [PubMed]
- Reik W (2007) Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447:425–432 [DOI] [PubMed]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129:1311–1323 [DOI] [PMC free article] [PubMed]
- Sive HL, Grainger RM, Harland RM (2000) Early development of Xenopus laevis: A labpratory manual. Cold Spring Harbor Laboratory Press, New York
- Spadaccini R, Perrin H, Bottomley MJ, Ansieau S, Sattler M (2006) Structure and functional analysis of the MYND domain. J Mol Biol 358:498–508 [DOI] [PubMed]
- Szutorisz H, Canzonetta C, Georgiou A, Chow CM, Tora L, Dillon N (2005) Formation of an active tissue-specific chromatin domain initiated by epigenetic marking at the embryonic stem cell stage. Mol Cell Biol 25:1804–1820 [DOI] [PMC free article] [PubMed]
- Tamai K, Yokota C, Ariizumi T, Asashima M (1999) Cytochalasin B inhibits morphogenetic movement and muscle differentiation of activin-treated ectoderm in Xenopus. Dev Growth Differ 41:41–49 [DOI] [PubMed]
- Tan X, Rotllant J, Li H, De Deyne P, Du SJ (2006) SmyD1, a histone methyltransferase, is required for myofibril organization and muscle contraction in zebrafish embryos. Proc Natl Acad Sci USA 103:2713–2718 [DOI] [PMC free article] [PubMed]
- Trievel RC, Beach BM, Dirk LM, Houtz RL, Hurley JH (2002) Structure and catalytic mechanism of a SET domain protein methyltransferase. Cell 111:91–103 [DOI] [PubMed]
- Tsuge M, Hamamoto R, Silva FP, Ohnishi Y, Chayama K, Kamatani N, Furukawa Y, Nakamura Y (2005) A variable number of tandem repeats polymorphism in an E2F-1 binding element in the 5′ flanking region of SMYD3 is a risk factor for human cancers. Nat Genet 37:1104–1107 [DOI] [PubMed]
- Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y (2001) Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell 8:1207–1217 [DOI] [PubMed]
- Xiao T, Hall H, Kizer KO, Shibata Y, Hall MC, Borchers CH, Strahl BD (2003) Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev 17:654–663 [DOI] [PMC free article] [PubMed]
- Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ (1995) Korsmeyer, Altered Hox expression and segmental identity in Mll-mutant mice. Nature 378:505–508 [DOI] [PubMed]
- Yu BD, Hanson RD, Hess JL, Horning SE, Korsmeyer SJ (1998) MLL, a mammalian trithorax-group gene, functions as a transcriptional maintenance factor in morphogenesis. Proc Natl Acad Sci USA 95:10632–10636 [DOI] [PMC free article] [PubMed]
- Zhang Y, Reinberg D (2001) Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev 15:2343–2360 [DOI] [PubMed]