Abstract
Several accessory proteins are required for the maturation of two nickel-containing enzymes in the gastric pathogen Helicobacter pylori. These two enzymes are hydrogenase and urease. Among the accessory/maturation proteins, the nickel-binding HypA protein has been previously shown to be required for the full activity of both the hydrogenase and the urease enzymes, while another nickel-binding protein, UreE, is known to be solely involved in the urease maturation process. In this study, UreE was shown to be required under all nickel levels for full activation of the apourease. By use of cross-linking studies, an interaction between purified HypA and UreE proteins was identified, leading to the formation of a 34 kDa heterodimer complex. The cross-linked adduct was detected by immunoblotting with either anti-HypA or anti-UreE antiserum. By using a two-plasmid system in Escherichia coli, the highest urease activity was achieved under low nickel conditions only when the UreE protein was expressed along with the wild-type HypA protein, but not with its nickel-binding-deficient variant HypA H2A. Addition of only 1 µM NiCl2 into minimal medium abolished the need for HypA to activate the urease. Although various attempts to show direct nickel transfer from HypA to UreE failed, these results suggest that interactions between the nickel-binding accessory proteins HypA and UreE are required to allow nickel transfer from HypA eventually to the apourease in H. pylori.
INTRODUCTION
Helicobacter pylori is a spiral, Gram-negative, microaerophilic bacterium that colonizes the gastric epithelium (Marshall & Warren, 1984) in about 50% of the world’s population (Covacci et al., 1999). H. pylori has been shown to be the aetiological agent of peptic ulcer disease or chronic atrophic gastritis (Blaser, 1995), which can subsequently develop into gastric cancer (Sipponen et al., 1998). Urease and hydrogenase are two nickel-containing enzymes that have been shown to be of particular importance for this bacterium. The urease enzyme raises the pH in the microenvironment surrounding the cell by producing ammonia (Mobley et al., 1991). In addition, it has been shown to be a key component of pH homeostasis within the cell (Sachs et al., 2002; Stingl et al., 2002) and it is the most abundant protein in H. pylori [10–15% of the total protein content of the cell (Bauerfeind et al., 1997)]. Therefore, it is essential for both early colonization events and for virulence (Eaton et al., 1991; Wirth et al., 1998). In contrast, the membrane-bound hydrogen-uptake Ni-Fe hydrogenase is important for efficient respiratory-based energy generation, aiding the in vivo maintenance of the pathogen (Maier et al., 1996; Olson & Maier, 2002).
Each nickel enzyme requires a battery of accessory proteins to become catalytically active. For instance, coexpression of the ureI, ureE, ureF, ureG and ureH genes has been shown to be required along with structural genes ureA and ureB to yield fully active urease in Escherichia coli (Cussac et al., 1992). In agreement with these results, stepwise addition of ureE and ureI to ureFGH significantly increased urease activity in E. coli (Park et al., 2005). Furthermore, Voland et al. (2003) have shown that H. pylori ureE, ureF, ureG and ureH mutants are severely deficient in urease activity. However, these proteins do not appear to be involved in hydrogenase maturation, since the ureE or ureG mutants have hydrogenase activities comparable to those of wild-type cells (Benoit et al., 2004; Mehta et al., 2003a). Interactions among urease accessory proteins have been reported, including UreF-UreH and UreG-UreE interactions, as well as interactions between the urease catalytic subunits UreA/B and the acid-gated urea channel, UreI (Voland et al., 2003). Among these proteins, UreE and UreG have been well characterized: the UreE protein has been shown to form homodimeric forms in solution, with a capability of binding 1 mol nickel (mol dimer)−1, while the UreG protein has been shown to possess a conserved nucleotide-binding-domain (GSGKT) (Benoit & Maier, 2003; Mehta et al., 2003a). Site-directed mutagenesis of the Lys14 residue in this domain abolished the urease activity (Mehta et al., 2003a).
Similarly, the hyp genes are required to obtain full hydrogenase activity. Indeed, mutants in the hypA, hypB, hypC, hypD, hypE or hypF genes are deficient for hydrogenase activity (Benoit et al., 2004; Olson et al., 2001). Among these genes, hypA and hypB are of particular interest because hypA or hypB mutants lack not only hydrogenase but also urease activity (Olson et al., 2001). Both proteins have been extensively studied; HypA is found as a dimer in solution, able to bind up to two Ni2+ ions (per dimer), with one histidine residue (His2) shown to be critical for nickel binding (Mehta et al., 2003b); HypB, also found as a dimer in solution, is a GTPase-like protein which relies on a lysine residue (Lys59) to be fully efficient (Mehta et al., 2003a). Both proteins have been shown to interact with each other in a 1:1 molar ratio, by using cross-linking and immunoblotting (Mehta et al., 2003b). The protein-protein interactions that specify the particular nickel-dependent maturation pathway (urease vs hydrogenase) are of interest and would be expected to involve Hyp-Ure accessory protein interactions. In this study, the role of UreE was investigated further by determining the urease activity of a ureE mutant grown in the presence of various concentrations of nickel in the medium; the nickel content of cells was also analysed in this mutant and compared to that in a hypA mutant and in a ΔureAB mutant (devoid of structural urease subunits). In addition, both nickel-binding proteins, UreE and HypA, were shown to interact intimately with each another. Evidence for a direct physical interaction was provided by immunoblotting using either anti-HypA or anti-UreE antiserum after cross-linking of both purified proteins. This interaction was further studied by using a two-plasmid system in E. coli. When cells were grown in M9 minimal medium with no nickel added, the expression of HypA (but not its nickel-deficient derivative HypA H2A) along with UreE significantly enhanced urease activity in this organism. This effect was not seen when 1 µM NiCl2 was added to the medium, suggesting that HypA and UreE are required predominantly under low nickel conditions.
METHODS
Bacterial strains and plasmids
The E. coli and H. pylori strains and the plasmids used in this study are listed in Table 1. The H. pylori ΔureAB::cat mutant was constructed by a PCR method, using the primers and protocol described by (Tan & Berg 2004). The deletion of the ureAB genes was confirmed by PCR and urease assays (data not shown). To construct plasmid pET-HypC, a 250 bp sequence containing H. pylori hypC was amplified by PCR using primers NdehypC (5'-GCATTCGTTCATATGTGTCTAGCGATCCCC-3') and SalhypC (5'-GCTTCAGTCGACTTATTGCGTTTCGTT-3'), designed to incorporate an NdeI and a SalI restriction site, respectively. The PCR product was digested with both restriction enzymes, gel-purified and ligated with similarly cut pET21b vector to generate plasmid pET-HypC. This plasmid was sequenced at the Sequencing and Synthesis Facility, University of Georgia.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics * | Source or reference |
|---|---|---|
| E. coli | ||
| TOP10 | Cloning strain | Invitrogen |
| BL21DE3 | Host strain for two-plasmid system | Stratagene |
| BL21Rosetta/pLysS | Host strain for overexpressing H. pylori proteins; Cmr | Novagen |
| H. pylori | ||
| 43504 | Parental strain | ATCC† |
| hypA mutant | hypA::aphA3; Kmr | Olson et al. (2001) |
| hypA H2A mutant | Codon for His2 replaced by Ala within hypA | Mehta et al. (2003b) |
| ureE mutant | ureE::aphA3; Kmr | Voland et al. (2003) |
| ΔureAB mutant | ΔureAB::cat; Cmr | This study |
| Plasmid | ||
| pHP8080 | pACYC184 derivative with H. pylori ureABIEFGH and nixA; Cmr | McGee et al. (1999) |
| pET21b | Cloning and expression vector; Apr | Novagen |
| pET-HypA | pET derivative plasmid for overexpression of HypA | Mehta et al. (2003b) |
| pET-HypA-H2A | pET-HypA with His2 replaced by Ala | Mehta et al. (2003b) |
| pET-HypB | pET derivative plasmid for overexpression of HypB | Mehta et al. (2003b) |
| pET-HypC | pET derivative plasmid for overexpression of HypC | This study |
| pET-UreE | pET derivative plasmid for overexpression of UreE | Benoit & Maier (2003) |
| pET-UreG | pET derivative plasmid for overexpression of UreG | Mehta et al. (2003a) |
| pET-HypA+UreE | pET derivative plasmid for overexpression of HypA and UreE | This study |
Ap, Ampicillin; Cm, chloramphenicol; Km, kanamycin.
American Type Culture Collecion, Manassas, VA, USA.
The two-plasmid system included plasmid pHP8080 (McGee et al., 1999). The second plasmid was either plasmid pET21b (Novagen) or one of its derivatives, pET-HypA (Mehta et al., 2003b), pET-HypA-H2A (Mehta et al., 2003b), pET-UreE (formerly pET-HP; Benoit & Maier, 2003) or pET-HypA+UreE. Plasmid pET-HypA+UreE was constructed as follows: plasmid pET-HypA was digested with EcoRI, blunt-ended with T4 polymerase, gel-purified and then digested with HindIII before being gel-purified again; plasmid pET-UreE was digested with XbaI, blunt-ended with T4 polymerase, gel-purified and digested with HindIII to release a 575 bp DNA fragment including the ureE gene and the T7 promoter region. This fragment was ligated into the partially blunt-ended, HindIII-cut pET-HypA to generate pET-HypA+UreE. This plasmid allows co-expression of both hypA and ureE genes when introduced in any E. coli strain expressing the T7 RNA polymerase.
Growth conditions
E. coli cells were grown aerobically in Luria-Bertani (LB) medium or plates at 37°C. Ampicillin (100 µg ml−1), chloramphenicol (30 µg ml−1) and IPTG (0.2 mM) were added as needed. For preparation of urease extracts, E. coli cells were grown overnight in M9 minimal medium, consisting of 1 × M9 salts (1−1: 6 g Na2HPO4, 3 g KH2PO4, 0.5 g NaCl, 1 g NH4Cl), 0.4% glycerol, 1 mM MgSO4, 0.1 mM CaCl2, 100 µg ampicillin ml−1 and 30 µg chloramphenicol ml−1, with or without 1 µM NiCl2. H. pylori was routinely grown on Brucella agar plates supplemented with 10% defibrinated sheep blood at 37°C under microaerophilic conditions with 5% O2, 5% CO2 and 90% N2. For 63Ni studies, cells from the wild-type strain, or the hypA, hypA H2A, ureE or ΔureAB mutants were grown for 24 h on BA plates before harvesting and resuspension into PBS. Cells were transferred to a defined medium supplemented with 63NiCl2 (100 nM). This defined medium was adapted from Reynolds & Penn (1994) with previously described modifications (Bruggraber et al., 2004; Chevalier et al., 1998). In each case, cultures were grown in 5 ml medium in 70 ml sealed glass bottles aseptically flushed with anaerobic gas mixture (5% CO2/10% H2/85% N2). O2 was added aseptically after flushing to a final headspace concentration of 6%. Liquid cultures were inoculated to an initial OD600 of ~0.05 and shaken at 37°C. After 24 h, the final OD600 measurement was taken, then cells were harvested, washed three times with PBS, broken by three freeze-thaw cycles and spun down to remove unbroken cells. The 63Ni content of these cell-free extracts was measured on a Beckman LS6000TA scintillation counter and related to the amount of protein.
Urease assays
Overnight-grown E. coli cotransformants were harvested and washed three times with 50 mM HEPES (pH 7.5) before being resuspended in the same buffer. Lysates of freshly sonicated cells were incubated for 20 min with the same buffer plus 25 mM urea, and the amount of ammonia released was assayed by using the phenol-hypochlorite assay (Weatherburn, 1967). A standard ammonium chloride concentration curve was used to convert the absorbance at 625 nm to nmol ammonia. Data are presented as urease specific activity, defined as nmol ammonia produced min-1 (mg total protein)−1. As previously reported, urease assays are reproducible within the same experiment, but there is a high variability between experiments (independent cultures) (McGee et al., 1999). In this study, we report urease activities and standard deviations obtained from one experiment to minimize this inter-assay variability. However, the overall trend and the differences between strains were the same for each of the three to five independent growth experiments.
Purification of accessory proteins from E. coli
HypA, HypB, UreE or UreG proteins were purified from E. coli BL21 DE3 Rosetta strain expressing each protein from a pET derivative plasmid, as described previously (Benoit & Maier, 2003; Mehta et al., 2003a, b). For overproduction and purification of HypC, BL21 Rosetta (pLysS) cells harbouring plasmid pET-HypC were grown at 37°C to an OD600 of 0.4 in 500 ml LB supplemented with chloramphenicol and ampicillin. Expression of HypC was induced by adding 0.2 mM IPTG into the medium, and cells were grown for an additional 3 hat 37°C and harvested by centrifugation (5000 g, 15 min, 4°C). All subsequent steps were performed at 4°C. Cells were washed with 200 ml 20 mM Tris/HCl (pH 7.5)/20 mM NaCl (buffer A) and resuspended in 5 ml of the same buffer. PMSF was added to a final concentration of 0.5 mM. Bacteria were lysed by three passages through a cold French pressure cell at 18 000 p.s.i., cell debris were removed by centrifugation at 20 000 g and the supernatant was subjected to ultracentrifugation at 100 000 g for 2 h. Solid (NH4)2SO4 was added stepwise to the membrane-free supernatant, with centrifugation steps (20 000 g, 15 min) between each addition of (NH4)2SO4. Finally, the pellet obtained after the addition of 3.2 M (NH4)2SO4 was resupended in 5 ml buffer A and dialysed against the same buffer. The sample (5 ml) was applied to a Q-Sepharose anion exchange column (GE Healthcare) and the protein was eluted with a linear gradient of 0.02–1 M NaCl in buffer A. HypC (pI=4.3) was eluted from the resin at approximately 0.5 M NaCl. Fractions of interest were pooled, concentrated to a final volume of 0.5 ml and applied to a Sephacryl S-200 10/60 gel filtration column. Fractions containing apparently pure HypC (Ve=16–20 ml) were pooled and concentrated.
Cross-linking assay
An equimolar mixture of purified wild-type proteins (HypA, HypB, HypC, UreE or UreG; 15 µM final concn) was incubated for 30 min at 25°C in the presence of 5 mM dimethyl suberimidate (DMS; Pierce). The reaction was quenched by adding 1 M Tris/HCl (pH 7.5) followed by incubation for 15 min at 25°C. After quenching, samples were subjected to SDS-PAGE (12.5 %) with a Mini-Protean II apparatus (Bio-Rad) according to the method of Laemmli (1970) and electrophoretically transferred to a nitrocellulose membrane (0.45 µM pore size; Osmonics) (Towbin et al., 1979).
Immunoblotting
The membrane was blocked by incubation with either 5% non-fat dry milk or 3% gelatin prepared in 20 mM Tris/ HCl (pH 7.6) plus 137 mM NaCl buffer (Tris buffer saline, TBS). This was followed by a 1–3 h incubation along with a 1 : 1000 dilution of either anti-HypA or anti-UreE antiserum (Benoit & Maier, 2003; Mehta et al., 2003b). The membrane was washed five times with TBS buffer plus 0.1% Tween 20 (TTBS) and then incubated for 1 h with a 1:2000 dilution of the secondary antibody (goat anti-rabbit immunoglobulin G coupled with alkaline phosphatase). The membrane was again washed five times with TTBS buffer. Bound antibodies were detected following addition of the chromogenic reagents nitro blue tetrazolium (0.25 mg ml−1) and 5-bromo-4-chloro-3-indolyl phosphate (0.125 mg ml-1) (Sigma).
RESULTS
UreE is required for nickel activation of urease
Although H. pylori hypA mutants are severely deficient in both hydrogenase and urease activities, both enzyme activities can be restored to wild-type levels by addition of nickel into the medium (Olson et al., 2001). In a ureE mutant, the urease activity is severely affected (Benoit & Maier, 2003; Voland et al., 2003) and it was important to see whether addition of nickel would complement this phenotype. Therefore H. pylori wild-type and ureE mutant cells were grown on BA plates supplemented with 0, 1, 2, 5, 10, 20 or 50 µM NiCl2 and subsequent urease activity (four replicates) was determined on cell-free extracts. Although increased nickel concentration induced the urease activity in the ureE mutant (specific activity increased from 2±1 units for non-supplemented medium to 332±20 units for BA with 50 µM nickel), these activities remained negligible when compared to those of wild-type cells (specific activity of 10 068±1781 units for non-supplemented medium to 62 632±235 for BA with 50 µM). For any given nickel concentration, the urease activity in the ureE mutant never exceeded 0.5% of the urease activity in the wild-type. Therefore, in contrast with the HypA accessory protein, UreE is required for full nickel maturation of the apourease, regardless of the nickel level supplied.
Further evidence for the accessory protein requirement was provided by measuring the 63Ni content of the H. pylori hypA, hypA H2A and ureE mutants or the wild-type strain grown in a defined medium supplemented with 63NiCl2 (100 nM). After 24 h at 37°C, there was no significant difference in cell growth (as measured by OD600) between strains (data not shown). Compared to the wild-type [237 c.p.m. (µg protein)-1=100%], there was 40, 63 or 6% labelled nickel in the cell-free extracts of the hypA, hypA H2A or ureE mutants, respectively. Because of its nickel-binding ability [24 atoms of nickel bound per (UreAB)12 complex (Hawtin et al., 1991; Pinkse et al., 2003)] and its abundance (10–15% of the total protein content of the cell; Bauerfeind et al., 1997), urease is likely to be the major nickel sink in the cell and has to account for most of the 63Ni detected in cell-free extracts. In light of this, the 63Ni results confirm the need for HypA and the requirement for UreE to deliver nickel to the apourease. As a control, an H. pylori ureAB deletion strain was constructed, grown and processed as for the other strains. This mutant contained only 26% of the nickel content of the wild-type, confirming that UreAB is responsible for most of the nickel detected in the cell. These experiments were repeated twice with similar results. The marked decrease of labelled nickel detected in the hypA and hypA H2A mutants suggest that the 63Ni concentration used in this experiment (100 nM) is not saturating. While both nickel-binding proteins HypA and UreE are important for activation of the urease apoenzyme, the strict dependency on UreE suggests that this protein is involved in later or more stringent stages of the nickel maturation steps.
Direct interaction between UreE and HypA is revealed by cross-linking and immunoblotting
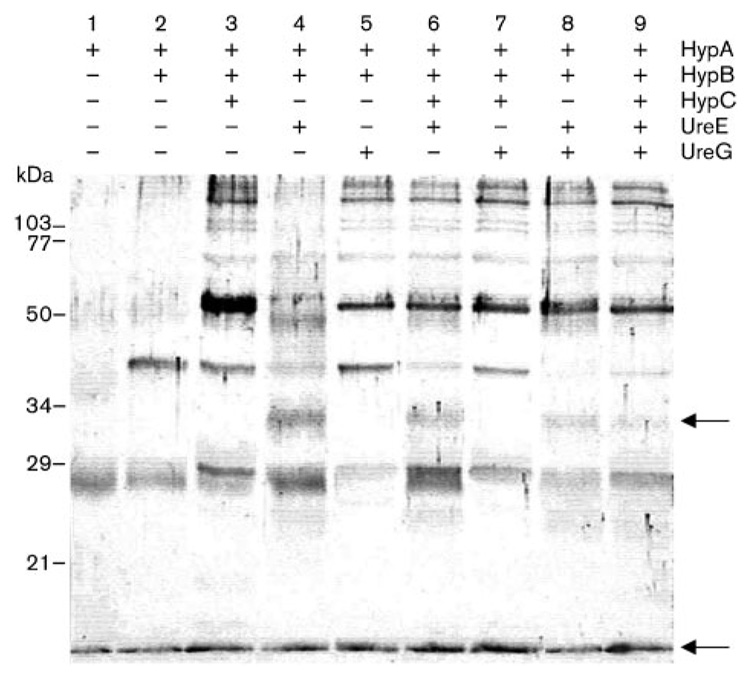
Previous findings revealed a direct interaction between the nickel-binding protein HypA and the GTPase-like protein HypB (Mehta et al., 2003b). As both of these proteins have been shown to be required for urease as well as hydrogenase maturation, possible interactions of these two proteins with other proteins involved in either the hydrogenase maturation process or the urease maturation pathway only were investigated. Equimolar concentrations of purified HypA (predicted mass, 13.2 kDa) were incubated with purified HypB (27.5 kDa), with or without the following purified accessory proteins: HypC (8.7 kDa), shown to be needed only for hydrogenase activity (Benoit et al., 2004); UreE (19.4 kDa) or UreG (22 kDa), both known to be required for urease maturation/activity only (Benoit & Maier, 2003; Mehta et al., 2003a; Voland et al., 2003). The homobifunctional cross-linker DMS was added to the protein mixture, and the protein complexes were subjected to SDS-PAGE and immunoblotting using anti-HypA antiserum (Fig. 1). In each lane, a protein with an apparent mass of 13–15 kDa was observed, corresponding to the HypA monomer; in addition, an estimated 27–28 kDa product was also observed, corresponding to the HypA homodimer. The HypA protein has already been shown to form dimers in solution (Mehta et al., 2003b). Additionally, when HypB was present in the reaction (lanes 2–9), a product with an apparent mass of approximately 40 kDa was seen; this corresponded to the previously described heterodimeric HypA-HypB complex (Mehta et al., 2003b). Interestingly, an additional protein complex with an apparent molecular mass of about 33 kDa could be seen in lanes 4, 6, 8 and 9, only when purified UreE was added to the reaction. This size corresponds to a 1:1 molar ratio of HypA to UreE. Since the presence of this band correlates with the fading of the 40 kDa HypA-HypB band, it is possible that HypB and UreE compete for HypA under these conditions. This finding would make the formation of a higher molecular mass complex of HypA, HypB, UreE and UreG unlikely, although HypA-HypB, HypA-UreE or UreE-UreG heterodimeric complexes have been reported (Mehta et al., 2003b; Voland et al., 2003). Nevertheless, other higher molecular mass complexes, including one with an approximate mass of 52 kDa could also be seen when HypC (lane 3) or UreG (lane 5) was added to the HypA+HypB mixture; however, the HypA-HypB complex remained unaffected under these circumstances. In an independent experiment, interactions between HypB and UreG were revealed by immunoblotting using anti-HypB antiserum (S. L. Benoit & R. J. Maier, unpublished data).
Fig. 1.
Immunoblotting analysis of cross-linked products arising from a mixture of HypA, HypB, HypC, UreE and UreG. The membrane was probed with anti-HypA antiserum. The cross-linking was carried out in the presence (+) or absence (−) of each protein, as indicated above the panel. The positions of the HypA monomer and the putative HypA–UreE complex are shown with arrows. Sizes of molecular mass standards are indicated on the left.
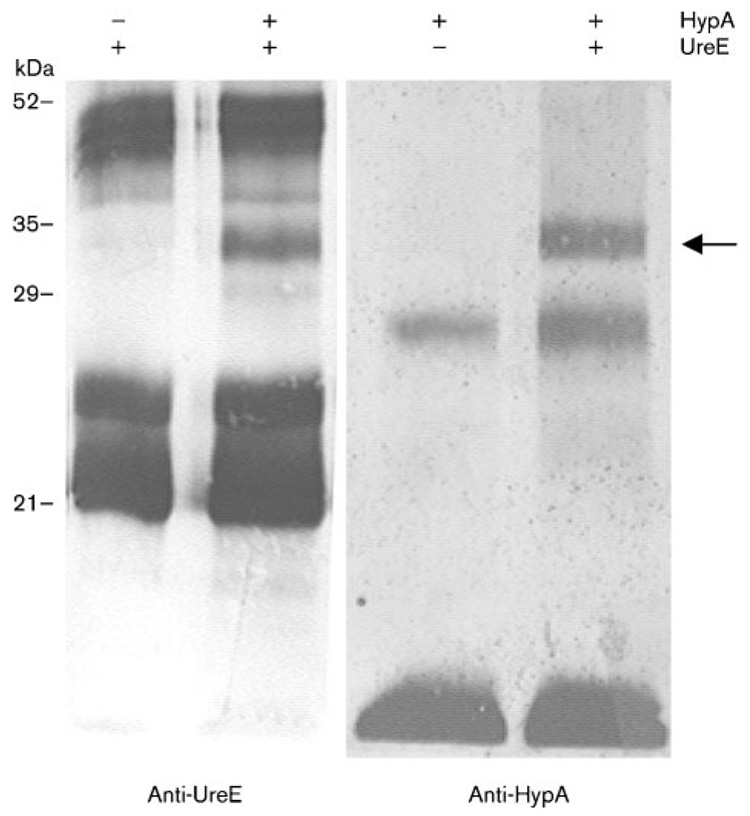
Confirmation of the HypA-UreE interaction was sought by performing a second cross-linking experiment, with only HypA and UreE being mixed together. The complex was then analysed by using either anti-HypA or anti-UreE antiserum (Fig. 2). In both cases, incubation of HypA along with UreE in the presence of DMS resulted in the formation of a unique 33 kDa heterodimeric adduct, which was not seen when either of these proteins was tested individually. Moreover, this intimate interaction between HypA and UreE was captured regardless of nickel concentration, as the same complex was seen in the absence or presence of 50 µM NiCl2 in the reaction mixture (data not shown). It therefore appears that HypA and UreE, both of them nickel-binding proteins, intimately interact to form a complex involved in the nickel enzyme maturation process; still, this interaction does not seem to be nickel-dependent.
Fig. 2.
Immunoblotting analysis of cross-linked products arising from a mixture of HypA and UreE. The cross-linking was carried out in presence (+) or absence (−) of HypA or UreE, as indicated above the panels. The membrane was probed with either anti-HypA or anti-UreE antiserum, as shown below each panel. The position of the HypA–UreE adduct is shown with the arrow. Sizes of molecular mass standards are indicated on the left.
Coexpression of UreE and HypA activates urease activity in E. coli cells under low nickel conditions only
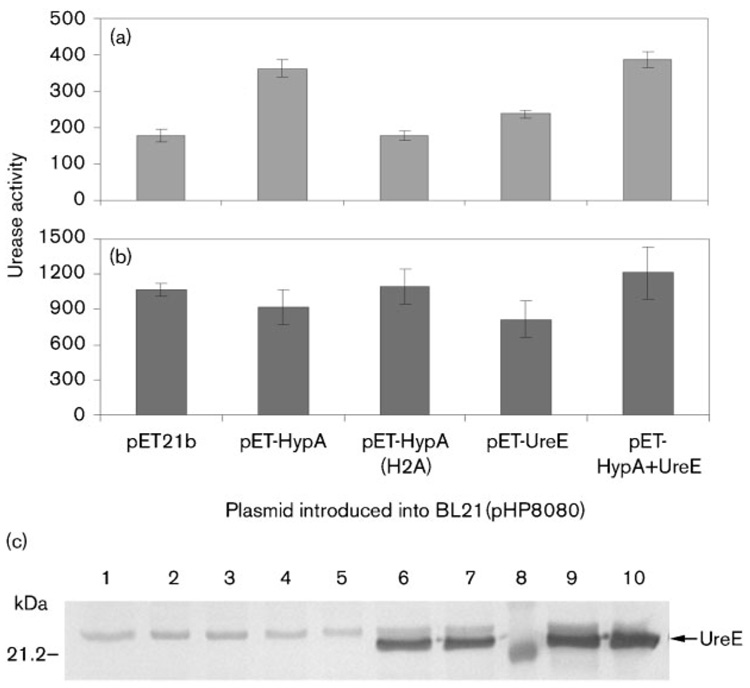
We investigated the physiological relevance of the HypA-UreE interaction. As an alternate way to study the role of a given gene on urease activity in H. pylori, a two-plasmid system in E. coli can be used. This system features a plasmid which harbours the ureA and ureB urease structural genes, along with the accessory ureEFGH genes, the ureI gene involved in urea import and the nixA gene encoding the NixA nickel transporter (McGee et al., 1999). Expression of this plasmid (pHP8080) alone confers urease activity to E. coli, and genes acting as urease-decreasing factors or urease-enhancing factors can be isolated when expressed from a second plasmid (McGee et al., 1999). Plasmids expressing HypA (pET-HypA), its nickel-binding-deficient derivative HypA-H2A (pET-HypA H2A) or UreE (pET-UreE) were introduced into E. coli strain BL21 DE3 along with pHP8080 to determine the effect of the expressed proteins on urease activity. In addition, a plasmid expressing both HypA and UreE was constructed to determine whether coexpression of both proteins from the high-copy-number pET21b plasmid would affect urease activity in E. coli. Cells were grown overnight in M9 minimal medium, with or without 1 µM NiCl2 added, and urease activity was determined in cell-free extracts (Fig. 3a, b). At the same time, these extracts were separated by SDS-PAGE, transferred to nitrocellulose and probed against either UreE (Fig. 3c) or HypA (data not shown) to determine the respective amount of H. pylori accessory proteins synthesized from pET21b in E. coli in each case. Although at first the BL21(pET) expression system was selected for the convenience of expressing the proteins at will by adding IPTG to the medium, we could not take full advantage of this feature because addition of even small amounts of IPTG (5 µM) was detrimental to the cells and induced lysis. However, according to the manufacturers (Novagen, Invitrogen), and from our previous experiences with similar vectors, the system has been shown to be leaky in this particular host strain, which means that expression of genes cloned under the control of the T7 promoter usually happens even without the addition of IPTG. Indeed, expression of UreE from either pET-UreE or pET-HypA+ UreE with no IPTG added in the medium was confirmed by immunoblotting using anti-UreE antiserum (Fig. 3c). The specific amount of HypA synthesized in each condition could not be determined, as non-specific bands with the same molecular mass (from E. coli proteins) were present in each lane (data not shown). Nevertheless, all growth experiments were carried out in the absence of IPTG.
Fig. 3.
(a) and (b) Urease activity in E. coli cells harbouring plasmid pHP8080 and a pET-derivative plasmid. Results shown are representative of one experiment and three replicates. Cells were grown overnight in M9 minimal medium with (a) no nickel added to the medium or (b) 1 µM NiCl2 added to the medium. (a) The urease activities obtained with pET-HypA, pET-UreE or pET-HypA+UreE were significantly greater (P<0.05) than the pET21b or pET-HypA(H2A) results. Urease activities are expressed as nmol ammonia produced min−1 (mg total protein)−1. (c) Immunoblotting of UreE. Ten micrograms of total protein from E. coli BL21(pHP8080) grown without nickel (lanes 2, 4, 6 and 9) or with 1 µM NiCl2 (lanes 1, 3, 5, 7 and 10) was loaded onto each lane. The second plasmid present in the cell was either pET21b (lane 1), pET-HypA (lanes 2 and 3), pET-HypA(H2A) (lanes 4 and 5), pET-UreE (lanes 6 and 7) or pET-HypA+UreE (lanes 9 and 10). A molecular mass marker was loaded on lane 8, with the size of one of the markers indicated on the left. The position of UreE is indicated by an arrow. A non-specific band can be seen in all lanes.
When no nickel was added to the medium, the presence of plasmid pET-HypA in BL21(pHP8080) increased the urease activity two- to threefold compared to the control which was BL21(pHP8080) with vector pET21b (Fig. 3a).This result suggested that the HypA protein expressed from plasmid pET-HypA was capable of activating urease in E. coli, probably through interaction with UreE encoded from plasmid pHP8080. When plasmid pET-HypAH2A was introduced into strain BL21(pHP8080) the urease activity was comparable to the activity observed for the control. This result confirmed the critical role of the His2 residue of HypA in the binding of nickel and the activation of urease, as proposed previously (Mehta et al., 2003b). It also indicated that HypA was not likely to play any role besides nickel transfer to UreE. Although the ureE gene was already present in the cell (plasmid pHP8080), expression of additional UreE from plasmid pET-UreE also increased the urease activity, probably by increasing the overall nickel-binding capacity of the cell (Fig. 3a). This result was in good agreement with previous results from our lab, in which expression of additional UreE protein in H. pylori hypA or hypB mutant backgrounds was found to increase urease activity (Benoit & Maier, 2003). Coexpression of both HypA and UreE from plasmid pET-HypA+UreE resulted in the highest urease activity under these conditions, suggesting that the combination (i.e. interaction) of both proteins resulted in the most efficient transfer of the metal to the urease. However, this level was not statistically significantly greater than the pET-HypA urease results. Nevertheless, the highest urease activities were always observed when a functional HypA was present, as opposed to when no HypA (pET21b) or a nickel-deficient HypA (HypA H2A) was present.
Addition of only 1 µMNiCl2 to the medium had a dramatic enhancing effect on all urease activities and dramatically reduced differences between strains (Fig. 3b). This effect was not reported by McGee et al. (1999) who used the same nickel concentration. This might be because we used a different E. coli strain (BL21 DE3 in this study, DH5α or SE5000 in their case), and their results showed an important strain-to-strain variability. Another probable reason lies in the medium used to grow the cells; McGee and coworkers used an M9 medium supplemented with glucose, Casamino acids and 1% LB, while we grew the cells under more stringent conditions, with M9 medium being supplemented only with glycerol. Although the assays were necessarily carried out in E. coli, these results suggest that HypA is probably required only when the nickel concentration encountered (by H. pylori) is very low. The presence of as little as 1 µM nickel in the medium bypassed the need for the HypA accessory protein. This is in agreement with previous findings from our lab where the urease activity of an H. pylori hypA mutant was restored when 5 µM NiCl2 was added to the medium (Olson et al., 2001). The addition of nickel into the medium did not seem to have any effect on the expression of H. pylori proteins from pET derivatives, since the amount of UreE synthesized in BL21 from either pET-UreE or pET-HypA+UreE was the same regardless of the level of nickel supplied in the medium (Fig. 3c).
Nickel transfer between HypA and UreE
Finally, we sought to investigate the relationship between HypA and UreE and the possibility of a direct nickel transfer between both partners. Three different approaches were followed. First, purified HypA protein which had been previously EDTA-treated and then 63Ni-labelled was incubated with EDTA-treated UreE, along with protein extracts and/or other purified accessory proteins and with GTP, with the expectation that UreE may become 63Niloaded. After incubation, the UreE protein was repurified and subjected to scintillation spectrometry. Unfortunately, the results from this in vitro approach turned out to be ambiguous; although 63Ni-UreE was detected, this nickel could not be attributed to nickel donation from HypA as apo-UreE readily bound the label even in the absence of 63Ni-HypA. Next, we tried an in vivo approach, by assessing the nickel content of UreE purified from four different background strains: wild-type strain 43504 and the hypA, hypA H2A or ureE mutant strains, with the latter being our negative control. We hypothesized that if the UreE protein relied preferentially on the HypA protein as its preferred nickel donor, it would probably have a higher nickel content in a strain where the HypA protein is fully functional, as compared to strains in which HypA is absent or unable to bind nickel. H. pylori cells were grown under low nickel conditions, and the UreE accessory protein was purified following the same protocol used for recombinant UreE overexpressed in E. coli (Benoit & Maier, 2003), but with an additional size exclusion chromatography step. After the final step, no protein could be seen by Coomassie or SYPRO ruby staining, but UreE could be detected by immunoblotting (data not shown). However, the nickel content (determined by graphite furnace atomic absorption spectrophotometry) was statistically the same in all purified UreE preparations, even in the control fraction obtained from a ureE mutant. Finally, the last approach involved purification of a T7-tagged UreE from different background strains grown in a defined medium in presence of 63Ni. Although T7-UreE was indeed isolated in each strain, other nickel-binding proteins also bound to the anti-T7 affinity column and it was therefore impossible to discriminate the UreE-specific nickel from the background.
DISCUSSION
Our results showed that the UreE accessory protein, known to be involved in nickel maturation of urease only, was capable of interacting in vitro with the accessory protein HypA, known to be required for the nickel activation of both the hydrogenase and the urease. This interaction was shown by immunoblotting with either anti-HypA or anti-UreE antiserum. In addition, the UreE accessory protein seems to compete with the GTPase HypB protein for interaction with HypA, since formation of the HypA-UreE complex correlated with the disappearance of the HypA-HypB adduct. When other proteins (HypC, UreG) were added to the protein mix, higher molecular mass adducts could be seen using anti-HypA antiserum, suggesting that these proteins can also interact with HypA or other proteins bound to HypA, like UreE or HypB. Indeed, a HypA/HypB heterodimeric interaction has been shown by Mehta et al.(2003b), while Voland et al. (2003) reported a UreE-UreG dimeric complex; in addition, preliminary data from our lab suggest the existence of a HypB/UreG interaction, as shown by immunoblotting using anti-HypB antiserum (S. L. Benoit & R. J. Maier, unpublished data). Although HypC has been shown to be involved specifically in the maturation of hydrogenase (but not urease), its putative role as a chaperone might imply it interacts with HypA or HypB, since both the latter accessory proteins are also involved in the hydrogenase maturation process. Therefore, multi-protein complexes involving HypA, HypB, HypC, UreE and UreG may be expected in vitro. However, whenever the heterologous proteins were added to HypA, the amount of the immunologically detectable 13 kDa band corresponding to the HypA monomer was not altered, which raises the question of the degree to which each protein can challenge the identified HypA-containing complexes. Further studies will include the use of antiserum raised against proteins other than HypA or UreE.
Expression of HypA significantly enhanced urease activity in E. coli cells co-expressing H. pylori UreE, suggesting that the intimate interaction captured by cross-linking happens in vivo. While it is also possible that HypA interacts with other H. pylori proteins synthesized from plasmid pHP8080 and that were not part of our in vitro screening (such as UreF, UreH or UreI), such interactions remain to be demonstrated. Since both HypA and UreE proteins have been shown to bind nickel (Benoit & Maier, 2003; Mehta et al., 2003b), this interaction is likely to trigger nickel transfer from HypA to UreE, although such a direct transfer remains to be demonstrated. Nevertheless, results obtained with the two-plasmid system in E. coli strongly suggest such a transfer: indeed when the nickel-deficient version of HypA (HypA H2A) was expressed (along with UreE), urease activity was not increased. The HypA-UreE interaction seems to be required only when nickel concentrations are low in the cell, since addition of only 1 µM of NiCl2 in the medium abolished differences between strains, bypassing the dependency on HypA. A plausible explanation lies in the respective nickel-binding capacity and affinity of each accessory protein; while HypA can bind 2Ni2+ per dimer with an estimated Kd of 13 µM (Mehta et al., 2003b), UreE can only bind 1Ni2+ per dimer with an estimated Kd of 1 µM (Benoit & Maier, 2003). Therefore, HypA is probably better than UreE at chelating the metal, which suggests that under low nickel concentrations the apo-urease may rely on the HypA-UreE interaction in order to be activated. It seems likely that H. pylori may have recruited the hydrogenase accessory (nickel-binding) protein HypA to aid its nickel sequestering capacity for urease maturation, as its UreE is relatively poor in total nickel-binding capacity. It is notable that other UreE proteins with histidine-rich regions, such as UreE from Proteus mirabilis (Jones & Mobley, 1989) or from Klebsiella aerogenes (Mulrooney & Hausinger, 1990) have a much greater nickel-binding capacity than H. pylori UreE. A comparison between the respective H. pylori and K. aerogenes urease pools may aid our understanding of why H. pylori would need additional nickel-binding accessory proteins: while the gastric pathogen has to provide enough nickel for a urease apoenzyme which can account for as much as 10–15% of the total protein content of the cell (Bauerfeind et al., 1997), K. aerogenes can rely on a UreE protein able to bind up to 6Ni2+ ions per dimer to fulfil the demand of a urease protein which represents only 0.1% of the total protein content (Maier et al., 2007). In support of this, when engineered versions of UreE with increased nickel-binding capacity (achieved by addition of histidine residues) were expressed in H. pylori hypA or hypB strains, the urease activity was enhanced five- to tenfold over strains lacking the engineered versions of UreE.
Finally, regardless of the nickel concentration in the cell, UreE is required to activate the urease, since urease activity is adversely affected when this accessory protein is absent. The acute urease maturation dependency on UreE was con- firmed by growing H. pylori cells in a defined medium in presence of 100 nM 63Ni. The amount of 63Ni in the ureE mutant strain was only 6% of the level in the wild-type strain. Because urease is the most abundant protein in H. pylori, the difference in the accumulated cellular nickel pool is probably due to the amount of nickel in the urease. Since ?urease expression has been shown to be affected by growth phase (Thompson et al., 2003), growth of the mutants and the wild-type was monitored by OD600 after 24 h (before harvesting the cells). There was no significant growth curve difference between the strains. Therefore the difference in 63Ni concentration between strains is not due to a difference in growth phase or yields. Finally, in contrast with H. pylori hypA or hypA H2A mutants, in which the HypA protein is either absent or unable to bind nickel, supplementation of the medium with nickel could not restore urease activity in the ureE mutant. Combined with a previous study (Benoit & Maier, 2003), the new results show the key role played by UreE in the overall urease maturation process.
ACKNOWLEDGEMENTS
This work was supported by the Georgia Research Foundation and by National Institute of Health Grant RO1DK062852. We would like to thank Dr David McGee, Department of Microbiology & Immunology, Louisiana State University Health Sciences Center, Shreveport, LA, who kindly provided us with plasmid pHP8080, and Dr David Scott, Department of Physiology, VA Greater Los Angeles Healthcare System, Los Angeles, CA, for the gift of the H. pylori ureE:: aphA3 mutant strain.
Abbreviations
- DMS
dimethyl suberimidate
REFERENCES
- Bauerfeind P, Garner R, Dunn BE, Mobley HL. Synthesis and activity of Helicobacter pylori urease and catalase at low pH. Gut. 1997;40:25–30. doi: 10.1136/gut.40.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit S, Maier RJ. Dependence of Helicobacter pylori urease activity on the nickel-sequestering ability of the UreE accessory protein. J Bacteriol. 2003;185:4787–4795. doi: 10.1128/JB.185.16.4787-4795.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit S, Mehta N, Wang G, Gatlin M, Maier RJ. Requirement of hydD, hydE, hypC and hypE genes for hydrogenase activity in Helicobacter pylori. Microb Pathog. 2004;36:153–157. doi: 10.1016/j.micpath.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Blaser MJ. The role of Helicobacter pylori in gastritis and its progression to peptic ulcer disease. Aliment Pharmacol Ther. 1995;9(suppl 1):27–30. doi: 10.1111/j.1365-2036.1995.tb00780.x. [DOI] [PubMed] [Google Scholar]
- Bruggraber SF, French G, Thompson RP, Powell JJ. Selective and effective bactericidal activity of the cobalt (II) cation against Helicobacter pylori. Helicobacter. 2004;9:422–428. doi: 10.1111/j.1083-4389.2004.00264.x. [DOI] [PubMed] [Google Scholar]
- Chevalier SFA, Davidson I, King A, French GL, Thompson RPH, Powell JJ. A fully chemically defined broth medium for Helicobacter pylori growth at variable pHs. Gut. 1998;43:A7. [Google Scholar]
- Covacci A, Telford JL, Del Giudice G, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- Cussac V, Ferrero RL, Labigne A. Expression of Helicobacter pylori urease genes in Escherichia coli grown under nitrogen-limiting conditions. J Bacteriol. 1992;174:2466–2473. doi: 10.1128/jb.174.8.2466-2473.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton KA, Brooks CL, Morgan DR, Krakowka S. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect Immun. 1991;59:2470–2475. doi: 10.1128/iai.59.7.2470-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawtin PR, Delves HT, Newell DG. The demonstration of nickel in the urease of Helicobacter pylori by atomic absorption spectroscopy. FEMS Microbiol Lett. 1991;61:51–54. doi: 10.1016/0378-1097(91)90012-y. [DOI] [PubMed] [Google Scholar]
- Jones BD, Mobley HL. Proteus mirabilis urease: nucleotide sequence determination and comparison with jack bean urease. J Bacteriol. 1989;171:6414–6422. doi: 10.1128/jb.171.12.6414-6422.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maier RJ, Fu C, Gilbert J, Moshiri F, Olson J, Plaut AG. Hydrogen uptake hydrogenase in Helicobacter pylori. FEMS Microbiol Lett. 1996;141:71–76. doi: 10.1111/j.1574-6968.1996.tb08365.x. [DOI] [PubMed] [Google Scholar]
- Maier RJ, Benoit SL, Seshadri S. Biometals. 2007. Nickel-binding accessory proteins facilitating urease and hydrogenase maturation in Helicobacter pylori. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- McGee DJ, May CA, Garner RM, Himpsl JM, Mobley HL. Isolation of Helicobacter pylori genes that modulate urease activity. J Bacteriol. 1999;181:2477–2484. doi: 10.1128/jb.181.8.2477-2484.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta N, Benoit S, Maier RJ. Roles of conserved nucleotide-binding domains in accessory proteins, HypB and UreG, in the maturation of nickel-enzymes required for efficient Helicobacter pylori colonization. Microb Pathog. 2003a;35:229–234. doi: 10.1016/s0882-4010(03)00151-7. [DOI] [PubMed] [Google Scholar]
- Mehta N, Olson JW, Maier RJ. Characterization of Helicobacter pylori nickel metabolism accessory proteins needed for maturation of both urease and hydrogenase. J Bacteriol. 2003b;185:726–734. doi: 10.1128/JB.185.3.726-734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobley HL, Hu LT, Foxal PA. Helicobacter pylori urease: properties and role in pathogenesis. Scand J Gastroenterol. 1991;187(Suppl):39–46. [PubMed] [Google Scholar]
- Mulrooney SB, Hausinger RP. Sequence of the Klebsiella aerogenes urease genes and evidence for accessory proteins facilitating nickel incorporation. J Bacteriol. 1990;172:5837–5843. doi: 10.1128/jb.172.10.5837-5843.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson JW, Maier RJ. Molecular hydrogen as an energy source for Helicobacter pylori. Science. 2002;298:1788–1790. doi: 10.1126/science.1077123. [DOI] [PubMed] [Google Scholar]
- Olson JW, Mehta NS, Maier RJ. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol Microbiol. 2001;39:176–182. doi: 10.1046/j.1365-2958.2001.02244.x. [DOI] [PubMed] [Google Scholar]
- Park JU, Song JY, Kwon YC, Chung MJ, Jun JS, Park JW, Park SG, Hwang HR, Choi SH, et al. Effect of the urease accessory genes on activation of the Helicobacter pylori urease apoprotein. Mol Cells. 2005;20:371–377. [PubMed] [Google Scholar]
- Pinkse MW, Maier CS, Kim JI, Oh BH, Heck AJ. Macromolecular assembly of Helicobacter pylori urease investigated by mass spectrometry. J Mass Spectrom. 2003;38:315–320. doi: 10.1002/jms.443. [DOI] [PubMed] [Google Scholar]
- Reynolds DJ, Penn CW. Characteristics of Helicobacter pylori growth in a defined medium and determination of its amino acid requirements. Microbiology. 1994;140:2649–2656. doi: 10.1099/00221287-140-10-2649. [DOI] [PubMed] [Google Scholar]
- Sachs G, Scott D, Weeks D, Melchers K. The compartment buffered by the urease of Helicobacter pylori: cytoplasm or periplasm? Trends Microbiol. 2002;10:217–218. doi: 10.1016/s0966-842x(02)02346-6. author reply 218–219. [DOI] [PubMed] [Google Scholar]
- Sipponen P, Hyvarinen H, Seppala K, Blaser MJ. Review article: pathogenesis of the transformation from gastritis to malignancy. Aliment Pharmacol Ther. 1998;12(Suppl. 1):61–71. doi: 10.1111/j.1365-2036.1998.00005.x. [DOI] [PubMed] [Google Scholar]
- Stingl K, Altendorf K, Bakker EP. Acid survival of Helicobacter pylori: how does urease activity trigger cytoplasmic pH homeostasis? Trends Microbiol. 2002;10:70–74. doi: 10.1016/s0966-842x(01)02287-9. [DOI] [PubMed] [Google Scholar]
- Tan S, Berg DE. Motility of urease-deficient derivatives of Helicobacter pylori. J Bacteriol. 2004;186:885–888. doi: 10.1128/JB.186.3.885-888.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LJ, Merrell DS, Neilan BA, Mitchell H, Lee A, Falkow S. Gene expression profiling of Helicobacter pylori reveals a growth-phase-dependent switch in virulence gene expression. Infect Immun. 2003;71:2643–2655. doi: 10.1128/IAI.71.5.2643-2655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voland P, Weeks DL, Marcus EA, Prinz C, Sachs G, Scott D. Interactions among the seven Helicobacter pylori proteins encoded by the urease gene cluster. Am J Physiol Gastrointest Liver Physiol. 2003;284:G96–G106. doi: 10.1152/ajpgi.00160.2002. [DOI] [PubMed] [Google Scholar]
- Weatherburn MW. Phenol-hypochlorite reaction for determination of ammonia. Anal Chem. 1967;39:971–974. [Google Scholar]
- Wirth HP, Beins MH, Yang M, Tham KT, Blaser MJ. Experimental infection of Mongolian gerbils with wild-type and mutant Helicobacter pylori strains. Infect Immun. 1998;66:4856–4866. doi: 10.1128/iai.66.10.4856-4866.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]