Abstract
Despite growing evidence that HIV-1-specific CD4+ T helper (Th) cells may play a role in the control of viremia, discrete Th cell epitopes remain poorly defined. Furthermore, it is not known whether Th cell responses generated using vaccines based on clade B virus sequences will elicit immune responses that are effective in regions of the world where non-clade B viruses predominate. To address these issues we isolated CD4+ T cell clones from individuals with vigorous HIV-1-specific Th cell responses and identified the minimum epitopes recognized. The minimum peptide length required for induction of CD4+ T cell proliferation, IFN-γ secretion, and cytolytic activity ranged from 9 to 16 amino acids in the five epitopes studied. Cross-clade recognition of the defined epitopes was examined for variant peptides from clades A, B, C, D, and AE. Over half the variant epitopes (17 of 32) exhibited impaired recognition, defined as less than 50% of the IFN-γ secretion elicited by B clade consensus sequence. There was no evidence for antagonistic activity mediated by the variant peptides, and despite strong responses there was no escape of autologous virus from Th responses in the epitopes we studied. Abrogated recognition of variant CD4+ T cell epitopes presents a potential obstacle to vaccine development.
INTRODUCTION
While the relative contribution of Th cell responses in HIV-1 infection is becoming increasingly understood, the precise targets of the Th cell response are not well characterized. Well over 100 CD8+ CTL epitopes have been defined, whereas the number of reported HIV-1-specific CD4+ Th epitopes is comparatively small 1-3. CTL epitopes are often defined as optimal, with the optimal length peptide recognized at concentrations logs lower than peptides one or two amino acid shorter or longer. Conflicting results have been generated in murine studies as to whether analogous optimal length epitopes exist for Th cells 4-14. While there may be no universal rule governing the optimum length of Th epitopes, few studies have addressed the issue in humans, and the data are not consistent as to whether longer peptides are more effective than minimum length peptides in stimulating Th responses 15, 16.
The bulk of HIV-1 vaccines in development or clinical trials utilize sequences based on clade B virus (see http://www.hvtn.org/trials/). However, the majority of individuals infected with HIV-1 are infected with non-clade B virus 17. A major concern is that vaccines developed using the clade B sequence might not be effective in preventing or attenuating infection with non-clade B HIV-1. Multiple studies have addressed cross-clade recognition of CTL epitopes and found varying degrees of cross-clade reactivity. Many studies showed common cross-clade recognition after stimulation of CTL with whole HIV-1 protein constructs 18-22. Analyses of individual CTL epitopes have shown more variable recognition of cross-clade epitopes 19, 23-26.
In the present study we evaluated five epitopes in Gag-p24 protein. In contrast to described CTL epitopes, there was no readily identifiable optimum length of peptide for the HIV-1-specific Th cell clones examined here. Cross-clade studies revealed that many of the naturally occurring HIV-1 epitope variants from clades A, B, C, D and AE were poorly recognized at the clonal and polyclonal level but did not antagonize the response of clones to the clade B virus sequence.
MATERIALS AND METHODS
Study subjects
Four persons with vigorous p24-specific proliferative responses were studied by cloning, including two long-term nonprogressors (LTNP), 161J and CTS-01, and two subjects with treated primary HIV-1 infection. CD4+ T cell clones were isolated from acute infection subjects AC-01 and AC-25 eleven and eighteen months after initiation of therapy, respectively. Results are also shown from stimulation of T cell lines from three additional LTNP, LT-04, LT-09, and LT-10. LTNP were defined as being HIV-1 infected for at least ten years and maintaining virus load less than 2000 RNA copies/ml. The only LTNP to receive antiretroviral therapy was LT-04 for four months in 1992, ten years prior to the current study. Study subject characteristics are summarized in Table 1. HLA typing was performed by PCR using sequence-specific primes at the Massachusetts General Hospital Histocompatibility Lab.
Table 1.
Study subject characteristics. CD4 counts and virus load are listed at the time of study
| Subject | Status | Infection date | CD4 count (cells/ml) | Virus load (RNA copies/ml) | ART | DR | DRw | DQ |
|---|---|---|---|---|---|---|---|---|
| AC-01 | Acute | 1997 | 875 | <400 | yes | 11, 14 | 52 | 3, 5 |
| AC-25 | Acute | 1998 | 471 | 150 | yes | 1, 11 | 52 | 5, 7 |
| CTS-01 | LTNP | late 1980s | 571 | 545 | never | 11, 15 | 51, 52 | 6, 7 |
| 161J | LTNP | mid 1980s | 842 | <50 | never | 4, 15 | 51, 53 | 3, 6 |
| LT-04 | LTNP | 1992 | 903 | <50 | never | 13 | 4, 5 | |
| LT-09 | LTNP | 1988 | 2103 | <50 | 4 months | 7, 11 | 52, 53 | 2, 3 |
| LT-10 | LTNP | 1992 | 435 | 5280 | never | 4, 13 | 52, 53 | 3 |
Peptides and Antibodies
Recombinant HIV-1 p24 protein (amino acids 133 to 373) derived from the NY-5 strain of HIV-1 was produced in a baculovirus expression system (Protein Science, Meriden, CT). Shorter p24 peptides were generated as free acids using an Advanced ChemTech (Texas) 396Ω peptide synthesizer 27. DR blocking antibody was a gift from Kai Wucherpfennig and DQ blocking antibody was from Immunotech (Fullerton, CA). Bispecific anti-CD3-anti-CD8 antibody was produced by Johnson Wong at the Massachusetts General Hospital using described methods 28. Briefly, a hybrid-hybridoma was produced by fusion of 12F6 and OKT8 hybridomas and purification of the antibody was accomplished by preparative isoelectric focusing.
T cell clones and lines
T cell clones were generated by limiting dilution as previously described 29. T cell clones and lines were maintained in media consisting of RPMI 1640 (Sigma, St. Louis, MO) with penicillin/streptomycin (Mediatech, Herndon, VA), HEPES (Mediatech), L-glutamine (Mediatech) (R+), plus 10% heat inactivated human AB serum (R10H). T cell lines were generated from 5×106 PBMC suspended in 100 μl of a mixture of 37 peptides each at 50 μg/ml in PBS. The 37 peptides represented the clade B consensus sequence epitopes plus naturally occurring variant sequences described in Table 3. After 1h the cells were washed twice with media and resuspended at 106 cells/ml in R10H containing 50 U/ml recombinant IL-2 (Hoffman La Roche). After 10-14 days the cells were restimulated nonspecifically with bispecific anti-CD3-anti-CD8 antibody (obtained from Johnson Wong 28, 0.5 μg/ml), 1×106 irradiated (30 Gy) feeder cells/ml, and 50 U/ml recombinant IL-2. The anti-CD3-anti-CD8 antibody depletes CD3+CD8+ cells and expands CD3+ CD4+ cells; 10-14 days after treatment cell lines were used for experiments and were typically >90% CD3+CD4+ (data not shown).
Table 3. Prevalence of epitope variants across clades.
Th epitopes are listed in each column with clade variants listed in the rows below. The percentage frequency of each variant peptide is listed in the shaded column to the right of the peptide. These frequencies are calculated by dividing the number of sequences for each listed in the HIV database Epilign section by the total number of viruses sequenced in each clade. The total number of sequences for each clade consisted of 11 for clade A, 37 for clade B, 17 for clade C, 5 for clade D, and 11 for clade AE. The epitope EPRGSDIAGT was lengthened by one amino acid at the N-terminus as the longer epitope was recognized more frequently by subjects whose Gag-specific Th responses were mapped (L. Cosimi, unpublished data)
 |
Proliferation assays
Antigen was presented by irradiated (120 Gy) autologous or partially HLA matched B lymphoblastoid cell lines (B-LCL). T cell clones and B-LCL were plated in triplicate wells at 50,000 cells/well each in 96 well plates in R10H. After 48 hours, 1 μCi of 3H-thymidine (Dupont Nen, Boston, MA) in 50 μl R10H was added per well. Plates were harvested onto glass fiber filters after 18 hours. Results were expressed as net CPM, the difference between the counts in the presence of antigen and the counts without antigen. Significant responses were considered to be net CPM greater than 1000 based on multiple assays with negative control stimulation 29.
Interferon-gamma ELISPOT assays
ELISPOT plates were coated with anti-IFN-γ antibody (Endogen, Woburn, MA) and incubated overnight at 4°C. The following day plates were washed 6X with phosphate buffered saline (PBS, Mediatech), then blocked with PBS plus 1% human serum for one hour. B-LCL (5×104) and antigen (1 μg/ml) were added in 100 μl R10H. T cell clones were added at 100 cells/well and T cell lines were added at 50,000 cells/well in 100 μl R10H. After overnight incubation the cells were discarded and plates were washed 6X with PBS, and biotinylated anti-IFN-γ (Endogen) was added for 1.5 hours at 25°C. The plates were then washed 6X with PBS. Streptavidin (Bio-Rad, Hercules, CA, 100 μl/well) was added and plates were incubated 45 minutes at 25°C. After 6 more PBS washes 100 μl/well coloring reagent (NBT/BCIP, Bio-Rad) was added. Once dots appeared the reaction was stopped with 3 water washes and plates were inactivated 10 minutes with PBS 10% Tween. Background responses to wells with no antigen or irrelevant antigen ranged from 0.5 to 2.5 spots per well, which translated to 20 to 80 SFC/million for the PBMC and T cell line experiments. Responses greater than 5 spots per well and 5 times maximum background were considered significant.
Cytotoxicity assays
Target cells (B-LCL) were incubated overnight with peptides and Na251CrO4 (60 μCi/ml, Dupont NEN). The following day the target cells were washed twice with refrigerated R+ before effector cells were added. Effector and target cells were incubated together in triplicate wells for 4 hours at 37°C, then the plates were centrifuged at 1000 rpm for 5 minutes at 4°C. Supernatant (25 μl) was assayed for 51Cr release. Maximal release was obtained by mixing the targets with 100 μl 1% Triton X-100. The percentage specific 51Cr release was calculated as the ratio 100 X (cpm experimental release - cpm spontaneous release)/(cpm maximal release - cpm spontaneous release). Spontaneous lysis was generally less than 20% and was noted if greater than 30%. For the purpose of interpretation specific lysis of greater than 10% was considered significant as lysis of targets pulsed with control peptides was always less than 3%.
Sequencing of autologous virus
Viral DNA was isolated from PBMC (5×106 cells) and viral RNA was extracted from plasma samples and reverse-transcribed as described previously 30. PCR cycling conditions were as follows: 94 °C for 2 minutes, 35-50 cycles of 30 s at 94 °C, 30 s at 56 °C, 2 min at 72 °C and a final extension of 68 °C for 20 min. RT-PCR cycling conditions were as follows: 50 °C for 60 min, 95 °C for 15 min and cycling conditions as noted above. External Gag-specific PCR primers used were 737-F (5′GCGACTGGTGAGTACGCC3′) and 2095-R (5′TTCCCTAAAAAATTAGCCTG3′). Nested primers utilized were 988-F (5′CCCTTCAGACAGGATCAGAAG3′) and 1754-R (5′CAACAAGGTTTCTGTCATCC3′). PCR fragments were then gel purified and cloned (TOPO TA, Invitrogen, Carlsbad, CA). Plasmid DNA was isolated using the QiaPrep Turbo Miniprep kit (Qiagen, Valencia, CA) and sequenced bi-directionally on an ABI 3100 PRISM automated sequencer. Sequencher (Gene Codes Corp., Ann Arbor, MI) and MacVector 4.1 (Oxford Molecular) were used to edit and align sequences. Blast searches and phylogenetic tree analyses were performed to rule out contamination of samples (MacVector 4.1). At least four clones were sequenced per sample.
RESULTS
Characterization of minimum and optimum epitopes of p24-specific clones
The minimum peptide able to induce proliferation, IFN-γ secretion, and cytolysis was defined, allowing assessment of whether different effector functions of CD4+ T cells were similarly influenced by peptide length (Fig 1). The minimum epitope for clone two was well defined, with complete abrogation of responses when one amino acid was removed from the N- or C-terminus of peptide EPRGSDIAGT (Fig. 1B). Clone five had an equally sharply defined minimum epitope based on peptide titration (data not shown). For clones one, three and four the minimum epitopes were less sharply delineated. Truncations of the minimum by one C- or N-terminal residue still activated the clones, though at a concentration ten-fold greater than the minimum epitope for at least one of the three functions assayed.
FIG 1, A-D.
Optimum epitopes for the Th cell clones, with parts A to D corresponding to clones one to four respectively. Peptides were diluted in log increments from 10 μg/ml to 0.01 μg/ml. Negative control peptides in each assay include peptide truncations shorter than the minimum epitope.
Top panel: Proliferation data, results are expressed as Net CPM, the difference between proliferation of the clone in the presence of peptide pulsed and unpulsed B-LCL. Peptides were added to irradiated B-LCL at the same time T cell clones were added.
Middle panel: IFN-γ secretion quantitated by ELISPOT assay. Fifty to one hundred clone cells were added per well. Resulting spots were multiplied by 10,000 or 20,000 to give results in SFC per million. Peptides were added to B-LCL at the same time T cell clones were added.
Bottom panel: Cytolytic activity by 51Cr release assay. Peptides and 51Cr were incubated with B-LCL overnight and washed twice the day of the assay. Spontaneous lysis was 36% for clone 2, peptide EPRGSDIAG at 01. μg/ml and 45% for clone 2, peptide EPRGSDIAGTT at 1 μg/ml.
We questioned whether there would be hierarchical induction of Th responses based on peptide concentration. Clones one and two both maintained cytolytic activity at a peptide concentration one log lower than that required for induction of proliferation and IFN-γ secretion. In contrast, clone three was able to proliferate and secrete IFN-γ at a peptide concentration one log lower than that required for cytolytic activity. Clone five secreted IFN-γ at peptide concentrations insufficient to induce proliferation or cytolysis (data not shown). Given the heterogeneous patterns seen, no clear hierarchy of Th functions was present in the group of clones we studied. A summary of the minimum epitopes for each clone is given in Table 2. The minimum epitope and HLA restriction for clone five was published previously 29.
Table 2.
Summary of clone HLA restriction and minimum epitopes. The abbreviation and corresponding numerical amino acid positions within Gag are listed to the right of each epitope. Of note, the clones’ response is not completely abrogated when the C-terminal proline is removed from the minimum epitope in clone 1, the N-terminal proline for clone 3 and the N-terminal glutamate for clone 4 (See Fig. 1).
| Clone | HLA restriction | Minimum epitope | ||
|---|---|---|---|---|
| AC-01, Clone 1 | DQ5 | EEKAFSPEVIP | (EP11) | (161-171) |
| AC-01, Clone 2 | DQ7 | EPRGSDIAGT | (ET10) | (231-240) |
| AC-25, Clone 3 | DR1 | PEVIPMFSALSEGATP | (PP16) | (167-182) |
| 161J, Clone 4 | DR4 | EVIPMFSALS | (ES10) | (168-177) |
| CTS01, Clone 5 | DQ7 | VHAGPIAPG | (VG9) | (219-227) |
We defined the HLA restriction of the clonal responses using blocking antibodies and partially HLA matched B-LCL. The first step in determining the HLA restricting allele was performed with blocking antibodies directed against DR or DQ alleles. Subsequently, partially HLA matched B-LCL were used to identify the precise DR or DQ allele presenting antigen; only B-LCL sharing the restricting class II allele with the clone were able to present the identified epitope (data not shown). A summary of the HLA restriction for the clones is presented in Table 2 29.
While isolation of minimum epitopes and definition of HLA restriction are important steps in characterizing the HIV-1-specific immune response, we evaluated whether an optimal Th epitope length existed, similar to optimal CTL epitopes 31. We found that the 22 amino acid peptide and many variations slightly longer than the minimum were often as potent a stimulus as the minimum epitope (Fig. 1). As the B-LCL antigen presenting cells were not fixed, some processing of the longer peptides may have occurred. Up to six additional peptides longer than the minimum for each clone were screened (data not shown). The only clone that recognized an optimum epitope was clone two (Fig. 1B). The peptide REPRGSDIAGTT (two amino acids longer than the minimum epitope) elicited a slightly better immune response at each peptide concentration. In general, however, no single length of epitope was significantly more potent than all others for these CD4+ T cell clones, in marked contrast to the case for CD8+ T cell clones31.
Degree of variation in defined Th epitopes
T cell recognition of epitopes could be affected by HIV-1 sequence variation, so we searched for variants in the Los Alamos Laboratory HIV sequence database (http://hiv-web.lanl.gov). We utilized the Epilign function as it contains only full-length protein sequences and no duplicate sequences from single individuals, to better reflect the prevalence of reported virus sequence variation around the globe (Bette Korber, personal communication). The majority of non-clade B consensus sequences were identical to the clade B consensus sequence for the epitopes we studied (Table 3). Exceptions included the clade AE consensus for EP11, clade C consensus for PP16 and ES10, and the clade D and AE consensus for VG9 (Table 3). For each of the epitopes studied, variation from the clade B sequence was seen in only a minority of reported samples, reflecting reasonable conservation of Gag across clades. Finally, to assess the variability of the subjects’ virus sequence within Gag, we sequenced Gag in each of the subjects studied (Table 4). Only minor variant sequences were seen in the epitopes from subject AC-01, but they did not persist over time, indicating that the patients’ autologous virus had not escaped Th responses in the epitopes we studied.
Table 4. Autologous virus sequences for study subjects.
The epitope sequences recognized by each individual are boxed. Virus was sequenced from subject AC-01 at two timepoints nineteen months apart. Minor variation was seen in the two epitopes from subject AC-01 at the earlier timepoint, but did not persist. One of the four sequences analyzed for subject 161J has been previously published 62
 |
Cross-clade recognition of variant peptides
To test whether or not Th responses raised against clade B virus sequence would recognized non-clade B HIV-1 sequences, we stimulated the clones with the consensus B sequence and variant peptides (Table 3) and measured IFN-γ production in an ELISPOT assay. Only variant sequences present in five percent or more of isolates in the database were tested. The clone that showed widest cross-clade reactivity was directed against the epitope ET11 (Fig. 2B, upper left panel). All of the other clones exhibited decreased sensitivity for the majority of the variant peptides, with less than 50% of the IFN-γ secretion elicited by B clade consensus sequence (Fig. 2A and C-D, upper left panels). A number of variant peptides were recognized equally as well as the B consensus peptides, EDRGSDIAGT, EDRGSDIAGA, and EPRGSAIAGT for the epitope ET11, EVIPMFPALS for the epitope ES9, and IHAGPIAPG for the epitope VG9. In spite of poor cross-clade recognition for most of the epitopes studied, 15 of 32 variants induced what we defined as a partial response, at least 50% of maximal IFN-γ secretion to clade B epitopes at high peptide concentrations (10 μg/ml).
FIG 2, A-E.

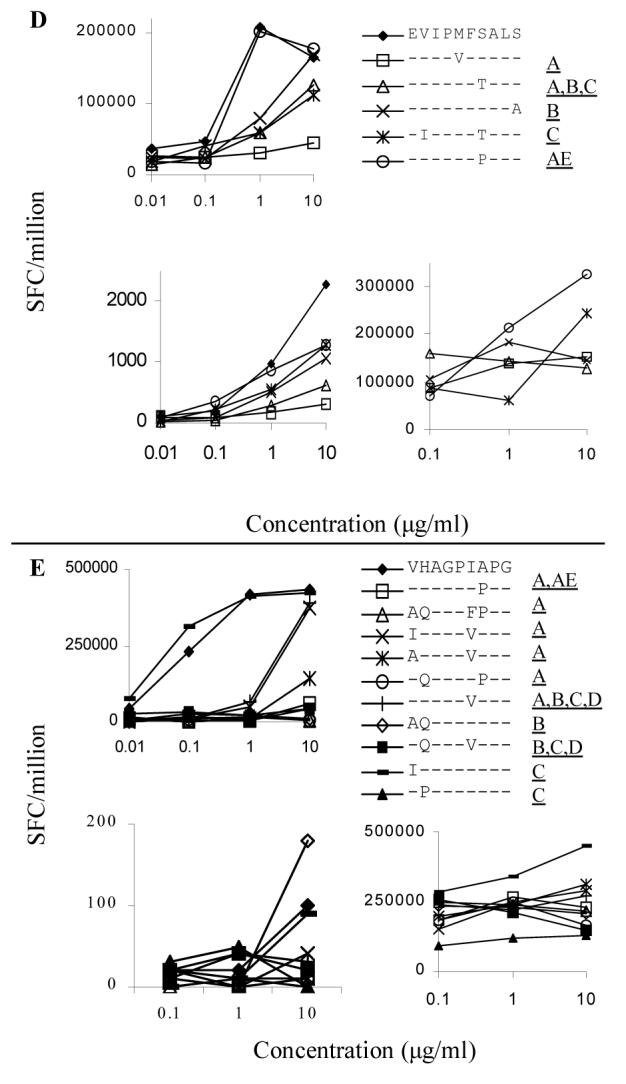
Upper left panels: Clonal response to cross-clade peptides. IFN-γ secretion was measured by ELISPOT over a range of peptide concentrations from 0.1 to 10 μg/ml. The clone used to analyze each epitope is listed in Table 1. The clade B consensus peptide is represented by the filled diamond (◆) for each of the epitopes. Peptides that elicit more than 50% less IFN-γ than the clade B epitope are marked in the legend with a dashed line. Clones were assayed at 100 cells per well and responses are listed as SFC/million. Each condition was performed in duplicate and these results represent the average of at least two assays performed on separate occasions.
Lower left panels: Polyclonal response to cross-clade peptides. PBMC and p24-specific T cell lines were stimulated with the same panel of peptides studied at the clonal level. Fig. 1D shows data generated with fresh PBMC, all other graphs were derived using T cell lines. The subject from whom each T cell line was derived is listed above the graph. PBMC responses were too low to consistently detect in most individuals so T cell lines were generated. Lines were restimulated 10-14 days after initial antigen-specific stimulation and were assayed 10 days later. PBMC and T cell lines were assayed at 50,000 cells/well and results are listed as SFC/million. IFN-γ secretion was about 100-fold less in the polyclonal than in the clonal response. Each condition was performed in duplicate. Background ranged from 1 to 2.5 spots per well and was not subtracted in the figures.
Lower right panels: Antagonism of clonal responses. Cross-clade peptides were unable to antagonize the clonal response to the clade B consensus peptide. The clone used to analyze each epitope is listed in Table 1. All wells included 1 μg/ml of clade B consensus peptide, except for epitope ET10 that required 10 μg/ml for activation of the clone. The variant peptides were added at increasing concentration to the wells and each condition was performed in duplicate. Clones were assayed at 100 cells/well and results represent the average of two separate experiments. No consistent inhibition of the clonal response to the clade B peptide was seen.
Targeting an epitope with multiple clones has been proposed as a means of preventing viral escape from CTL responses 32. We therefore tested whether polyclonal responses would improve recognition of variant peptides. 161J was the only subject whose responses were strong enough to consistently measure in PBMC prior to in vitro expansion. All other responses were measured with p24-specific CD4+ T cell lines derived by stimulating freshly isolated PBMC with a mixture of all the variant peptides, to avoid the bias of initially stimulating with only clade B sequence. T cell lines were derived from different individuals from whom the clones were originally isolated, as many of the original individuals were not available for follow up study. A total of eight additional subjects were studied, and the results from T cell lines from three individuals with the best cross-clade recognition are shown (Fig. 2A-E, lower left panels). Recognition of some cross-clade epitopes was improved at the polyclonal level, while other epitopes showed decreased recognition compared to clonal responses. There was no net increase in recognition of variant peptides at the polyclonal compared to the clonal level, as 15 of 32 variants were again partially recognized as defined above. In summary, cross-clade variation resulted in partial to complete abrogation of Th responses for most of the epitopes studied here.
Antagonism mediated by altered peptide ligands
Virus sequence variation can lead not only to escape from recognition by T cells, but also to antagonism of immune responses to the index viral sequence. Antagonism has been described for both HIV-1-specific CTL 33 and Th responses 34, and antagonism of Th epitopes has been proposed as a potential mechanism for HIV-1 vaccine failure 35. To test for antagonism, clones were incubated with clade B peptide at a submaximal concentration and variant peptides were titrated into the ELISPOT assay. Peptides with antagonist properties would cause a dose dependent decrease in Th cell responses. While many of the cross-clade peptides were poorly recognized (Fig.2, left panels), none of the variant peptides proved antagonistic (Fig. 2A-E, lower right panels).
DISCUSSION
We studied five HIV-1 Gag-p24 epitopes and found that naturally occurring variant peptides were poorly recognized, but none of the altered peptide ligands caused antagonism of IFN-γ secretion. We defined four HIV-1 Th epitopes and found no discernable hierarchy in the induction of proliferation, IFN-γ secretion or cytolytic capability based on peptide concentration or length.
A number of reports have shown a dependence of T cell activation on regions flanking the core of amino acids that contact the HLA molecule 36-39. We found one clone with enhanced responses to a peptide extended beyond the minimum 10 amino acids by one amino acid at the N- and C-termini (clone 2). The sequence was recognized at one to two logs lower concentration than the minimum epitope, but the overall effect was small. It does not appear that Th epitopes show the same strict length dependence as optimal CTL epitopes. One of the epitopes we identified is promiscuously recognized. While we show the epitope EVIPMFSALS to be DR4 restricted, a separate study showed it to be restricted by DR1. The same group defined the epitope EEKAFSPEV restricted by DQ5 40. We also tested the hierarchy of induction of Th functions, as studies have shown that Th cell cytolytic activity can be induced by shorter peptides 41 or at lower peptide concentration 42 than proliferation or IFN-γ secretion. We did not find any consistent hierarchical pattern based on peptide concentration or length.
In addition to defining a number of minimum Th epitopes in HIV-1, we tested their worldwide significance across HIV-1 clades. Cross-clade recognition by both clones and polyclonal T cell lines was found to be limited. It is possible that stimulation of the T cell lines led to a restricted repertoire of T cells and biased toward a narrow response with poor cross clade recognition. However, recent work has shown that stimulation with anti-CD3/CD4 antibody does not perturb the V-β repertoire of expanded CD8+ T cells 43, and that peptide stimulation of CD4+ T cells results in a diverse repertoire of T cell receptors 44. The poor recognition of variant Th epitopes is consistent with abrogated CTL recognition of variant peptides and decline in responses to the original infecting strain of virus seen in the setting of HIV-1 superinfection 45. Antagonism of Th responses to the cognate peptide by altered peptide ligands has been documented 46-49. Additionally, selective blockade of Th cell responses has been demonstrated, such as induction of IL-4 secretion but not proliferation by altered peptide ligands 50. We found no evidence for antagonistic properties mediated by the variant peptides in our study. However, we only measured IFN-γ secretion and could have missed antagonism of other Th cell functions, such as proliferation or IL-2 secretion.
The epitope VG9 showed the poorest cross-clade recognition (Fig. 2E), as well as the highest degree of variability seen within the epitopes studied (Table 3). Not only were cross-clade variant peptides poorly recognized for epitope VG9, but clade B variants also showed abrogated recognition. One possible explanation for the greater variability in VG9 is evolution due to immune pressure. The Th epitope is very close to the B7 restricted CTL epitope HPVHAGPIA (aa 84-92) 51, so this region might fall under immune pressure from both CD4+ and CD8+ T cell responses. The clade B consensus sequence for VG9 matches the ancestral sequence (http://hiv-web.lanl.gov/content/hiv-db/CONSENSUS/M_GROUP/M_GROUP_ancestral_sequence), but there is significant variation in the epitope even among clade B isolates. The high mutation rate in the region VG9 is also surprising as this region of the virus lies within the cyclophilin A binding region 52. Cyclophilin A is a host protein that is a member of a ubiquitous family of proteins catalyzing protein folding and is required for an early step in the life cycle of HIV-1 53. Mutations in the region VG9 have been described that allow replication of HIV-1 without incorporation of cyclophilin A 54. It is possible that interaction between HIV-1 and cyclophilin A has driven diversity in this region of Gag to optimize the interaction of the virus with cyclophilin A.
The current study suggests that vaccines utilizing one clade of virus may not generate Th cell responses that would be effective against variants found in other clades. The limitation of single clade vaccines would be ameliorated by the fact that the majority of non-clade B consensus sequences for the epitopes analyzed in our study were identical to the clade B consensus sequence (Table 3). Decreased recognition of Th epitope variant sequences has been described in HIV-1 and malaria 35, 55-59. Limited Th cell recognition of clade variants of whole Gag and shorter Env proteins has been described 60, 61, but variation across clades has never been examined for individual HIV-1 Th epitopes. Since variant virus sequences are less common than consensus sequence virus (Table 3), 60-100% of reported clade B,C, and D virus sequences would be at least partially recognized at the highest peptide concentration tested. Partial recognition lends hope that highly immunogenic vaccines can induce responses that will recognize cross-clade variant viruses. However, other regions of the virus such as Env are more variable than Gag and might present even greater limitations to cross-clade recognition.
In summary, we characterized in detail a number of antigenic regions of Gag p24 protein in HIV-1 infected individuals with strong Th cell responses. In our search for optimum antigenic peptide length we found the addition of amino acids to the minimum peptide length had little effect on Th cell activation. While recognition of cross-clade variant peptides was usually abrogated, variation within most of the Gag epitopes studied here was relatively rare. It appears that vaccination with a clade B vaccine would elicit Th responses cross-reactive with most circulating virus strains in the Gag epitopes we defined in spite of poor recognition of variant peptides.
Acknowledgments
This work was supported by the National Institutes of Health (Grants AI01698 and AI40873). PJN, ESR, and BDW are supported by the Doris Duke Charitable Foundation. BDW is a Doris Duke Distinguished Clinical Science Professor.
Sequence Data
The GenBank accession numbers for subject AC-01’s first timepoint are AF281722 to AF281733 and for the second are AY317076 to AY317085. Sequences for AC-25 are listed as AY317086 to AY317097. Previously unpublished sequences from 161J are listed as AF073420-AF073423. Sequences for CTS-01 are listed as AY319949 to AY319964.
References
- 1.Cosimi LA, Rosenberg ES. The Characterization of HIV-1 Specific CD4+ T Helper Epitopes. In: Korber BTM, Brander C, Haynes BF, Koup R, Kuiken C, Moore JP, Walker BD, Watkins DI, editors. HIV Molecular Immunology 2000. Los Alamos National Laboratory, Theoretical Biology and Biophysics; Los Alamos, NM: 2000. pp. I-55–62. [Google Scholar]
- 2.Korber B, Moore J, Brander C, Walker B, Haynes B, Koup R. HIV Molecular Immunology Database. Los Alamos National Laboratory; 1999. [Google Scholar]
- 3.Wilson CC, Palmer B, Southwood S, et al. Identification and antigenicity of broadly cross-reactive and conserved human immunodeficiency virus type 1-derived helper T-lymphocyte epitopes. J Virol. 2001;75:4195–4207. doi: 10.1128/JVI.75.9.4195-4207.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhayani H, Carbone FR, Paterson Y. The activation of pigeon cytochrome c-specific T cell hybridomas by antigenic peptides is influenced by non-native sequences at the amino terminus of the determinant. J Immunol. 1988;141:377–382. [PubMed] [Google Scholar]
- 5.Kim JE, Kojima M, Houghten R, et al. Characterization of a helper T cell epitope recognized by mice of a low responder major histocompatibility type. Mol Immunol. 1990;27:941–946. doi: 10.1016/0161-5890(90)90116-h. [DOI] [PubMed] [Google Scholar]
- 6.Ertl HC, Dietzschold B, Otvos L., Jr. T helper cell epitope of rabies virus nucleoprotein defined by tri- and tetrapeptides. Eur J Immunol. 1991;21:1–10. doi: 10.1002/eji.1830210102. [DOI] [PubMed] [Google Scholar]
- 7.Srinivasan M, Domanico SZ, Kaumaya PT, Pierce SK. Peptides of 23 residues or greater are required to stimulate a high affinity class II-restricted T cell response. Eur J Immunol. 1993;23:1011–1016. doi: 10.1002/eji.1830230504. [DOI] [PubMed] [Google Scholar]
- 8.Shimizu T, Uenishi H, Teramura Y, et al. Fine structure of a virus-encoded helper T-cell epitope expressed on FBL-3 tumor cells. J Virol. 1994;68:7704–7708. doi: 10.1128/jvi.68.12.7704-7708.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vacchio MS, Berzofsky JA, Krzych U, Smith JA, Hodes RJ, Finnegan A. Sequences outside a minimal immunodominant site exert negative effects on recognition by staphylococcal nuclease-specific T cell clones. J Immunol. 1989;143:2814–2819. [PubMed] [Google Scholar]
- 10.Gammon G, Geysen HM, Apple RJ, et al. T cell determinant structure: cores and determinant envelopes in three mouse major histocompatibility complex haplotypes. J Exp Med. 1991;173:609–617. doi: 10.1084/jem.173.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vignali DA, Strominger JL. Amino acid residues that flank core peptide epitopes and the extracellular domains of CD4 modulate differential signaling through the T cell receptor. J Exp Med. 1994;179:1945–1956. doi: 10.1084/jem.179.6.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noll A, Autenrieth IB. Yersinia-hsp60-reactive T cells are efficiently stimulated by peptides of 12 and 13 amino acid residues in a MHC class II (I-Ab)-restricted manner. Clin Exp Immunol. 1996;105:231–237. doi: 10.1046/j.1365-2249.1996.d01-758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y, Boros DL. Identification of the immunodominant T cell epitope of p38, a major egg antigen, and characterization of the epitope-specific Th responsiveness during murine schistosomiasis mansoni. J Immunol. 1998;160:5420–5427. [PubMed] [Google Scholar]
- 14.Zaliauskiene L, Kang S, Sparks K, et al. Enhancement of MHC class II-restricted responses by receptor-mediated uptake of peptide antigens. J Immunol. 2002;169:2337–2345. doi: 10.4049/jimmunol.169.5.2337. [DOI] [PubMed] [Google Scholar]
- 15.Honorati MC, Dolzani P, Mariani E, et al. Epitope specificity of Th0/Th2 CD4+ T-lymphocyte clones induced by vaccination with rHBsAg vaccine. Gastroenterology. 1997;112:2017–2027. doi: 10.1053/gast.1997.v112.pm9178695. [DOI] [PubMed] [Google Scholar]
- 16.Bitmansour AD, Douek DC, Maino VC, Picker LJ. Direct ex vivo analysis of human CD4(+) memory T cell activation requirements at the single clonotype level. J Immunol. 2002;169:1207–1218. doi: 10.4049/jimmunol.169.3.1207. [DOI] [PubMed] [Google Scholar]
- 17.Janssens W, Buve A, Nkengasong JN. The puzzle of HIV-1 subtypes in Africa. Aids. 1997;11:705–712. doi: 10.1097/00002030-199706000-00002. [DOI] [PubMed] [Google Scholar]
- 18.Betts MR, Krowka J, Santamaria C, et al. Cross-clade human immunodeficiency virus (HIV)-specific cytotoxic T-lymphocyte responses in HIV-infected Zambians. J Virol. 1997;71:8908–8911. doi: 10.1128/jvi.71.11.8908-8911.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao H, Mani I, Vincent R, et al. Cellular immunity to human immunodeficiency virus type 1 (HIV-1) clades: relevance to HIV-1 vaccine trials in Uganda. J Infect Dis. 2000;182:1350–1356. doi: 10.1086/315868. [DOI] [PubMed] [Google Scholar]
- 20.Buseyne F, Chaix ML, Fleury B, et al. Cross-clade-specific cytotoxic T lymphocytes in HIV-1-infected children. Virology. 1998;250:316–324. doi: 10.1006/viro.1998.9373. [DOI] [PubMed] [Google Scholar]
- 21.Ferrari G, Humphrey W, McElrath MJ, et al. Clade B-based HIV-1 vaccines elicit cross-clade cytotoxic T lymphocyte reactivities in uninfected volunteers. Proc Natl Acad Sci U S A. 1997;94:1396–1401. doi: 10.1073/pnas.94.4.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lynch JA, deSouza M, Robb MD, et al. Cross-clade cytotoxic T cell response to human immunodeficiency virus type 1 proteins among HLA disparate North Americans and Thais. J Infect Dis. 1998;178:1040–1046. doi: 10.1086/515652. [DOI] [PubMed] [Google Scholar]
- 23.Cao H, Kanki P, Sankale JL, et al. Cytotoxic T-lymphocyte cross-reactivity among different human immunodeficiency virus type 1 clades: implications for vaccine development. J Virol. 1997;71:8615–8623. doi: 10.1128/jvi.71.11.8615-8623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dorrell L, Willcox BE, Jones EY, et al. Cytotoxic T lymphocytes recognize structurally diverse, clade-specific and cross-reactive peptides in human immunodeficiency virus type-1 gag through HLA-B53. Eur J Immunol. 2001;31:1747–1756. doi: 10.1002/1521-4141(200106)31:6<1747::aid-immu1747>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 25.Dorrell L, Dong T, Ogg GS, et al. Distinct recognition of non-clade B human immunodeficiency virus type 1 epitopes by cytotoxic T lymphocytes generated from donors infected in Africa. J Virol. 1999;73:1708–1714. doi: 10.1128/jvi.73.2.1708-1714.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McAdam S, Kaleebu P, Krausa P, et al. Cross-clade recognition of p55 by cytotoxic T lymphocytes in HIV-1 infection. Aids. 1998;12:571–579. doi: 10.1097/00002030-199806000-00005. [DOI] [PubMed] [Google Scholar]
- 27.Rosenberg ES, Billingsley JM, Caliendo AM, et al. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science. 1997;278:1447–1450. doi: 10.1126/science.278.5342.1447. see comments. [DOI] [PubMed] [Google Scholar]
- 28.Wong JT, Colvin RB. Bi-specific monoclonal antibodies: selective binding and complement fixation to cells that express two different surface antigens. J Immunol. 1987;139:1369–1374. [PubMed] [Google Scholar]
- 29.Norris PJ, Sumaroka M, Brander C, et al. Multiple effector functions mediated by human immunodeficiency virus-specific cd4(+) t-cell clones. J Virol. 2001;75:9771–9779. doi: 10.1128/JVI.75.20.9771-9779.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen TM, O’Connor DH, Jing P, et al. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407:386–390. doi: 10.1038/35030124. see comments. [DOI] [PubMed] [Google Scholar]
- 31.Altfeld MA, Trocha A, Eldridge RL, et al. Identification of dominant optimal HLA-B60- and HLA-B61-restricted cytotoxic T-lymphocyte (CTL) epitopes: rapid characterization of CTL responses by enzyme-linked immunospot assay. J Virol. 2000;74:8541–8549. doi: 10.1128/jvi.74.18.8541-8549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Douek DC, Betts MR, Brenchley JM, et al. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J Immunol. 2002;168:3099–3104. doi: 10.4049/jimmunol.168.6.3099. [DOI] [PubMed] [Google Scholar]
- 33.Johnson RP, Trocha A, Yang L, et al. HIV-1 gag-specific cytotoxic T lymphocytes recognize multiple highly conserved epitopes. Fine specificity of the gag-specific response defined by using unstimulated peripheral blood mononuclear cells and cloned effector cells. J Immunol. 1991;147:1512–1521. [PubMed] [Google Scholar]
- 34.Fenoglio D, Li Pira G, Lozzi L, et al. Natural analogue peptides of an HIV-1 GP120 T-helper epitope antagonize response of GP120-specific human CD4 T-cell clones. J Acquir Immune Defic Syndr. 2000;23:1–7. doi: 10.1097/00126334-200001010-00001. [DOI] [PubMed] [Google Scholar]
- 35.Kent SJ, Greenberg PD, Hoffman MC, Akridge RE, McElrath MJ. Antagonism of vaccine-induced HIV-1-specific CD4+ T cells by primary HIV-1 infection: potential mechanism of vaccine failure. Journal of Immunology. 1997;158:807–815. [PubMed] [Google Scholar]
- 36.Carson RT, Vignali KM, Woodland DL, Vignali DA. T cell receptor recognition of MHC class II-bound peptide flanking residues enhances immunogenicity and results in altered TCR V region usage. Immunity. 1997;7:387–399. doi: 10.1016/s1074-7613(00)80360-x. [DOI] [PubMed] [Google Scholar]
- 37.Carson RT, Desai DD, Vignali KM, Vignali DA. Immunoregulation of Th cells by naturally processed peptide antagonists. J Immunol. 1999;162:1–4. [PubMed] [Google Scholar]
- 38.Godkin AJ, Smith KJ, Willis A, et al. Naturally processed HLA class II peptides reveal highly conserved immunogenic flanking region sequence preferences that reflect antigen processing rather than peptide-MHC interactions. J Immunol. 2001;166:6720–6727. doi: 10.4049/jimmunol.166.11.6720. [DOI] [PubMed] [Google Scholar]
- 39.Arnold PY, La Gruta NL, Miller T, et al. The majority of immunogenic epitopes generate CD4+ T cells that are dependent on MHC class II-bound peptide-flanking residues. J Immunol. 2002;169:739–749. doi: 10.4049/jimmunol.169.2.739. [DOI] [PubMed] [Google Scholar]
- 40.Boritz E, Palmer BE, Livingston B, Sette A, Wilson CC. Diverse Repertoire of HIV-1 p24-Specific, IFN-gamma-Producing CD4(+) T Cell Clones Following Immune Reconstitution on Highly Active Antiretroviral Therapy. J Immunol. 2003;170:1106–1116. doi: 10.4049/jimmunol.170.2.1106. [DOI] [PubMed] [Google Scholar]
- 41.Heemskerk MH, Schoemaker HM, De Jong I, Schijns VE, Spaan WJ, Boog CJ. Differential activation of mouse hepatitis virus-specific CD4+ cytotoxic T cells is defined by peptide length. Immunology. 1995;85:517–522. [PMC free article] [PubMed] [Google Scholar]
- 42.Hemmer B, Stefanova I, Vergelli M, Germain RN, Martin R. Relationships among TCR ligand potency, thresholds for effector function elicitation, and the quality of early signaling events in human T cells. J Immunol. 1998;160:5807–5814. [PubMed] [Google Scholar]
- 43.Jones N, Agrawal D, Elrefaei M, et al. Evaluation of antigen-specific responses using in vitro enriched T cells. J Immunol Methods. 2003;274:139–147. doi: 10.1016/s0022-1759(02)00510-0. [DOI] [PubMed] [Google Scholar]
- 44.Cameron TO, Cohen GB, Islam SA, Stern LJ. Examination of the highly diverse CD4(+) T-cell repertoire directed against an influenza peptide: a step towards TCR proteomics. Immunogenetics. 2002;54:611–620. doi: 10.1007/s00251-002-0508-y. [DOI] [PubMed] [Google Scholar]
- 45.Altfeld M, Allen TM, Yu XG, et al. HIV-1 superinfection despite broad CD8(+) T-cell responses containing replication of the primary virus. Nature. 2002;420:434–439. doi: 10.1038/nature01200. [DOI] [PubMed] [Google Scholar]
- 46.Ostrov D, Krieger J, Sidney J, Sette A, Concannon P. T cell receptor antagonism mediated by interaction between T cell receptor junctional residues and peptide antigen analogues. J Immunol. 1993;150:4277–4283. [PubMed] [Google Scholar]
- 47.De Magistris MT, Alexander J, Coggeshall M, et al. Antigen analog-major histocompatibility complexes act as antagonists of the T cell receptor. Cell. 1992;68:625–634. doi: 10.1016/0092-8674(92)90139-4. [DOI] [PubMed] [Google Scholar]
- 48.Alexander J, Snoke K, Ruppert J, et al. Functional consequences of engagement of the T cell receptor by low affinity ligands. J Immunol. 1993;150:1–7. [PubMed] [Google Scholar]
- 49.Racioppi L, Ronchese F, Matis LA, Germain RN. Peptide-major histocompatibility complex class II complexes with mixed agonist/antagonist properties provide evidence for ligand-related differences in T cell receptor-dependent intracellular signaling. J Exp Med. 1993;177:1047–1060. doi: 10.1084/jem.177.4.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evavold BD, Allen PM. Separation of IL-4 production from Th cell proliferation by an altered T cell receptor ligand. Science. 1991;252:1308–1310. doi: 10.1126/science.1833816. [DOI] [PubMed] [Google Scholar]
- 51.Brander C, Goulder PJR. The Identification of Optimal HIV-Derived CTL Epitopes in Diverse Populations Using HIV Clade-Specific Consensus. In: Korber BTM, Brander C, Haynes BF, Koup R, Kuiken C, Moore JP, Walker BD, Watkins DI, editors. HIV Molecular Immunology 2001. Los Alamos National Laboratory, Theoretical Biology and Biophysics; Los Alamos, NM: 2001. pp. I-1–20. [Google Scholar]
- 52.Franke EK, Yuan HE, Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. see comments. [DOI] [PubMed] [Google Scholar]
- 53.Braaten D, Franke EK, Luban J. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. Journal of Virology. 1996;70:3551–3560. doi: 10.1128/jvi.70.6.3551-3560.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Braaten D, Aberham C, Franke EK, Yin L, Phares W, Luban J. Cyclosporine A-resistant human immunodeficiency virus type 1 mutants demonstrate that Gag encodes the functional target of cyclophilin A. Journal of Virology. 1996;70:5170–5176. doi: 10.1128/jvi.70.8.5170-5176.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ratto S, Sitz KV, Scherer AM, et al. CD4+ T-lymphocyte lines developed from HIV-1-seropositive patients recognize different epitopes within the V3 loop. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;11:128–136. doi: 10.1097/00042560-199602010-00003. [DOI] [PubMed] [Google Scholar]
- 56.Siliciano RF, Lawton T, Knall C, et al. Analysis of host-virus interactions in AIDS with anti-gp120 T cell clones: effect of HIV sequence variation and a mechanism for CD4+ cell depletion. Cell. 1988;54:561–575. doi: 10.1016/0092-8674(88)90078-5. [DOI] [PubMed] [Google Scholar]
- 57.Harcourt GC, Garrard S, Davenport MP, Edwards A, Phillips RE. HIV-1 variation diminishes CD4 T lymphocyte recognition. J Exp Med. 1998;188:1785–1793. doi: 10.1084/jem.188.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Plebanski M, Flanagan KL, Lee EA, et al. Interleukin 10-mediated immunosuppression by a variant CD4 T cell epitope of Plasmodium falciparum. Immunity. 1999;10:651–660. doi: 10.1016/s1074-7613(00)80064-3. [DOI] [PubMed] [Google Scholar]
- 59.Zevering Y, Khamboonruang C, Good MF. Natural amino acid polymorphisms of the circumsporozoite protein of Plasmodium falciparum abrogate specific human CD4+ T cell responsiveness. Eur J Immunol. 1994;24:1418–1425. doi: 10.1002/eji.1830240627. [DOI] [PubMed] [Google Scholar]
- 60.Fernandez MH, Fidler SJ, Pitman RJ, Weber JN, Rees AD. CD4+ T-cell recognition of diverse clade B HIV-1 isolates. Aids. 1997;11:281–288. doi: 10.1097/00002030-199703110-00004. [DOI] [PubMed] [Google Scholar]
- 61.Moss RB, Giermakowska W, Wallace MR, Savary J, Jensen F, Carlo DJ. T-helper-cell proliferative responses to whole-killed human immunodeficiency virus type 1 (HIV-1) and p24 antigens of different clades in HIV-1-infected subjects vaccinated with HIV-1 immunogen (Remune) Clin Diagn Lab Immunol. 2000;7:724–727. doi: 10.1128/cdli.7.5.724-727.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alexander L, Weiskopf E, Greenough TC, et al. Unusual polymorphisms in human immunodeficiency virus type 1 associated with nonprogressive infection. J Virol. 2000;74:4361–4376. doi: 10.1128/jvi.74.9.4361-4376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]



