Abstract
BACKGROUND
Brain-derived neurotrophic factor (BDNF) has been found to be important in energy homeostasis in animal models, but little is known about its role in energy balance in humans. Heterozygous, variably sized, contiguous gene deletions causing haploinsufficiency of the WT1 and PAX6 genes on chromosome 11p13, approximately 4 Mb centromeric to BDNF (11p14.1), result in the Wilms’ tumor, aniridia, genitourinary anomalies, and mental retardation (WAGR) syndrome. Hyperphagia and obesity were observed in a subgroup of patients with the WAGR syndrome. We hypothesized that the subphenotype of obesity in the WAGR syndrome is attributable to deletions that induce haploinsufficiency of BDNF.
METHODS
We studied the relationship between genotype and body-mass index (BMI) in 33 patients with the WAGR syndrome who were recruited through the International WAGR Syndrome Association. The extent of each deletion was determined with the use of oligonucleotide comparative genomic hybridization.
RESULTS
Deletions of chromosome 11p in the patients studied ranged from 1.0 to 26.5 Mb; 58% of the patients had heterozygous BDNF deletions. These patients had significantly higher BMI z scores throughout childhood than did patients with intact BDNF (mean [±SD] z score at 8 to 10 years of age, 2.08±0.45 in patients with heterozygous BDNF deletions vs. 0.88±1.28 in patients without BDNF deletions; P = 0.03). By 10 years of age, 100% of the patients with heterozygous BDNF deletions (95% confidence interval [CI], 77 to 100) were obese (BMI ≥95th percentile for age and sex) as compared with 20% of persons without BDNF deletions (95% CI, 3 to 56; P<0.001). The critical region for childhood-onset obesity in the WAGR syndrome was located within 80 kb of exon 1 of BDNF. Serum BDNF concentrations were approximately 50% lower among the patients with heterozygous BDNF deletions (P = 0.001).
CONCLUSIONS
Among persons with the WAGR syndrome, BDNF haploinsufficiency is associated with lower levels of serum BDNF and with childhood-onset obesity; thus, BDNF may be important for energy homeostasis in humans. (ClinicalTrials.gov number, NCT00006073.)
Studies in animal models suggest that brain-derived neurotrophic factor (BDNF) plays a key role in energy homeostasis.1–6 BDNF is believed to act primarily within the ventromedial hypothalamus to regulate energy intake1,2 downstream of the leptin–proopiomelanocortin signaling pathway.3,5 In mice, genetic BDNF haploinsufficiency leads to obesity.7–10 Mice that are heterozygous for inactivated BDNF have a 50% reduction in hypothalamic expression of BDNF, and they have hyperphagia and obesity, which are reversed by intracerebroventricular infusions of BDNF.8–10
Although studies in animals provide support for a role of BDNF in energy homeostasis, data in humans are relatively limited. Some studies have shown an inverse association between the peripheral BDNF concentration and the body-mass index (BMI) (the weight in kilograms divided by the square of the height in meters) in children and adults.11–14 A common BDNF polymorphism, Val66Met, has been inconclusively associated with altered body weight.14–17 The most relevant data are from two case reports. One described an obese child with hyperphagia and a heterozygous 11p13p15.3 inversion that resulted in what has been termed “functional” BDNF haploinsufficiency (as determined from measurements performed in a lymphoblastoid cell line)18; the other, more convincingly, described an obese child with hyperphagia and a heterozygous missense substitution resulting in impaired signaling of the cognate receptor of BDNF, TrkB.19 Thus, the available data suggest the importance of BDNF in energy homeostasis in humans, but the evidence is not definitive.
We therefore systematically investigated a naturally occurring model of genetic BDNF haploinsufficiency in humans. This rare disorder (estimated prevalence, 1 case per 500,000 to 1,000,000 persons)20,21 is characterized by Wilms’ tumor, aniridia, genitourinary anomalies, and mental retardation, and it is thus called the WAGR syndrome. It is caused by heterozygous contiguous gene deletions that involve at least two genes, WT1 and PAX6, which are present in the 11p13 region. These genes are positioned approximately 4 Mb centromeric to the BDNF locus at 11p14.1 (Fig. 1A). Haploinsufficiency for WT1 and PAX6 has been observed in all patients with the WAGR syndrome and accounts for the common oncogenic, ocular, and genitourinary features of the syndrome. Although persons with the WAGR syndrome typically have low-normal birth weight,21 marked obesity subsequently develops in a substantial subgroup.20 Case reports involving single patients22–28 have described severe hyperphagia and obesity in a few persons with deletions that included the 11p14 BDNF locus. Such reports have led to the hypothesis that BDNF haploinsufficiency may be responsible for the obesity subphenotype of the WAGR syndrome.8,18 To test this hypothesis, we examined the relationship between BDNF haploinsufficiency and childhood BMI in children and adults with the WAGR syndrome.
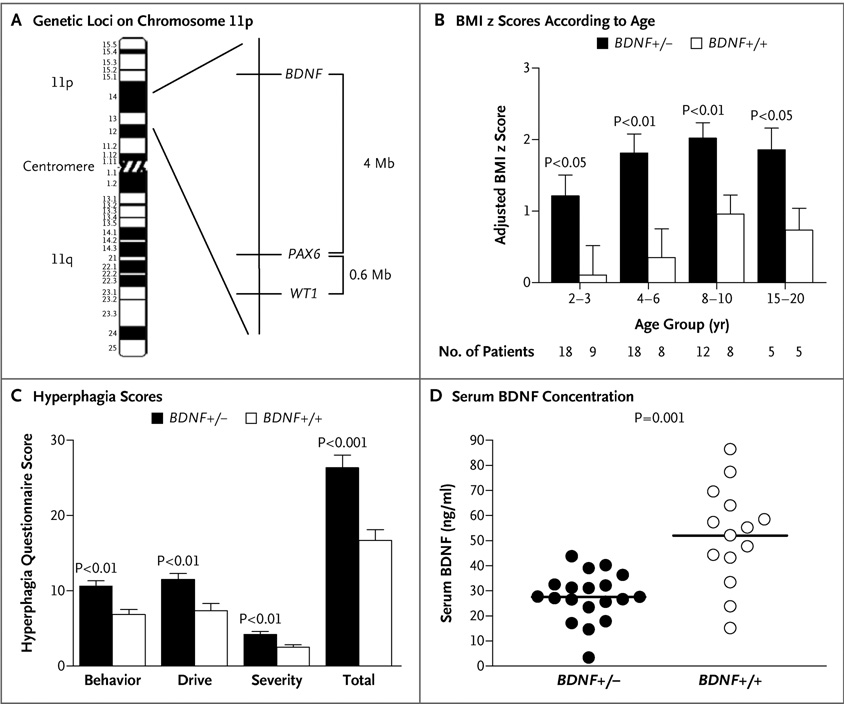
Figure 1. Genetic Loci on Chromosome 11, Body-Mass Index, Hyperphagia, and Concentrations of Serum BDNF in Patients with the WAGR Syndrome.
As shown in Panel A, the WAGR syndrome is caused by deletions on chromosome 11p that result in haploinsufficiency for the PAX6 and WT1 genes. BDNF is located approximately 4 Mb telomeric to PAX6. Panel B shows body-mass index (BMI) z scores in patients 20 years of age or younger, adjusted for the BMI of the mother and father, in patients with BDNF haploinsufficiency (BDNF+/−) and patients with intact BDNF (BDNF+/+). Anthropometric data were obtained from medical records. The sample size for each age range is shown. Values are means ±SE. Nominal P values are shown. Panel C shows the scores for types of behavior, the drive to eat, and severity of symptoms from the hyperphagia questionnaire in patients with BDNF haploinsufficiency (BDNF+/−) and patients with intact BDNF (BDNF+/+). The minimum total score for this questionnaire is 11, and the maximum total score is 55 with higher scores indicating greater hyperphagia. Subtotal and total scores are shown as means ±SE. Panel D shows that the concentration of serum BDNF was significantly lower in patients with BDNF haploinsufficiency (BDNF+/−) than in patients with intact BDNF (BDNF+/+). Individual values are shown, and horizontal bars indicate group means.
METHODS
PATIENTS AND PROCEDURES
Patients were recruited through the International WAGR Syndrome Association. The study was approved by the institutional review board of the National Institute of Child Health and Human Development. Written informed consent was obtained from adults who were competent to provide consent and from the parents or legal guardians of children and adults with cognitive impairment. Assent was obtained from patients who were unable to give consent. Patients were categorized as having the WAGR syndrome and were eligible to participate in the study if they had karyotypes with 11p13 deletions or fluorescence in situ hybridization analyses showing WT1 and PAX6 deletion or if they had a clinical history of aniridia plus genitourinary anomalies, Wilms’ tumor, or both. Patients were excluded if they had serious chronic medical conditions during childhood that affected energy balance, such as cyanotic congenital heart disease, or a physical impediment influencing oral intake.
The height and weight of probands with the WAGR syndrome and their parents were measured in triplicate. Previous anthropometric data for probands were also obtained through a review of medical records collected by the patients’ primary care and subspecialty providers. To assess hyperphagia, parents completed a validated hyperphagia questionnaire developed for studies of the Prader–Willi syndrome.29 Because case reports18,19 and studies in animals30 suggest that BDNF may also play a role in modulating pain sensation, we assessed pain perception using a questionnaire31 completed by the parents of the patients. This questionnaire evaluated behavioral responses to typically painful stimuli (for details, see Appendix 1 in the Supplementary Appendix, available with the full text of this article at www.nejm.org). Venous blood samples were obtained from probands and their parents for genetic analysis and measurement of serum BDNF concentrations (BDNF Emax ImmunoAssay System, Promega) (for details see Appendix 2 in the Supplementary Appendix). Platelet counts were also obtained so that concentrations of serum BDNF could be adjusted for the number of platelets, since BDNF is stored within platelets.32 Data were collected between June 2006 and September 2007.
DELETION MAPPING
The extent of the deletion in each patient was determined by means of comparative genomic hybridization, as described in Appendix 3 in the Supplementary Appendix. We custom-designed a microarray platform (Agilent Technologies) containing 57,925 probes for chromosome 11p spaced at approximately 400-bp intervals (excluding repeat regions); 121 probes were located within BDNF. Comparative genomic hybridization of labeled proband and reference DNA was performed according to Agilent’s standard protocol (version 4). With DNA obtained from the probands and their parents, microsatellite-length polymorphism analysis was used to confirm the extent of each deletion (for details, see Appendix 4 in the Supplementary Appendix). For patients with deletion boundaries detected within 250 kb of BDNF, we performed polymerase-chain-reaction amplification and sequencing of the region containing the deletion splice site (for details, see Appendix 5 in the Supplementary Appendix).
STATISTICAL ANALYSIS
Patients with deletions involving all or a portion of the BDNF gene were categorized as having heterozygous BDNF deletions; patients with both BDNF genes intact were categorized as having no BDNF deletions. Our primary hypothesis was that BMI z scores would be higher in patients with heterozygous BDNF deletions. BMI z scores, which were normalized for age and sex, were determined with the use of the modified LMS method33 in the Centers for Disease Control and Prevention 2000 growth charts,34 which are based on normative standards of the U.S. population in the 1970s. Normative data for persons 20 years of age were used to calculate BMI z scores from the measured weight and height of five adults who were 22 to 25 years of age.
The primary analyses used to compare BMI z scores in patients with and those without BDNF deletions were analyses of covariance conducted at the ages of 2 to 3 years, 4 to 6 years, 8 to 10 years, and 15 to 20 years. For BMI z-score comparisons, the BMIs of both parents were used as covariates. The measured height and weight were not available for the fathers of three patients; thus, the heights and weights of these fathers were based on the estimates reported by the mothers. Each patient was represented by one data point (if available) for each age-category analysis. Therefore, multiple observations were used for each patient, but only one observation per age category was used. A secondary analysis compared the prevalence of childhood-onset obesity, defined as a BMI z score of 1.64 or more (the 95th percentile) by 10 years of age in patients with and those without BDNF deletions. For this analysis, patients 10 years of age or older who had a BMI z score below the 95th percentile were considered to be of normal weight; data for patients who were younger than 10 years of age but who had a BMI z score of less than 1.64 were categorized as indeterminate. A third analysis compared concentrations of serum BDNF in patients with and those without BDNF deletions, with the use of sex, current age, BMI, and platelet count as covariates.11,32
Data were analyzed with the use of SPSS software (version 12.0). The Kolmogorov–Smirnov test for normality was performed before analysis. Data that were not normally distributed were log-transformed. Independent-sample t-tests and Fisher’s exact tests were used to compare unadjusted differences in demographic and clinical characteristics between patients with and those without BDNF deletions. Scores for hyperphagia and pain questionnaires were compared with the use of the nonparametric Mann–Whitney U test. Results are reported as means ±SD unless otherwise noted, and nominal P values are reported.
RESULTS
ASSOCIATION BETWEEN BDNF HAPLOINSUFFICIENCY AND BODY WEIGHT
A total of 33 patients with the WAGR syndrome were recruited (Table 1). Patients had heterozygous 11p deletions ranging in size from 1.0 to 26.5 Mb (deletion boundaries are shown in Appendix 6 in the Supplementary Appendix). Each deletion caused haploinsufficiency for WT1 and PAX6, confirming the WAGR syndrome. At 10 years of age, 16 patients had become obese, 8 patients had normal weight, and the weights of 9 patients were indeterminate because they were not yet 10 years of age.
Table 1.
Characteristics of the Study Participants.*
| Characteristic | Patients with BDNF Deletions (N = 19) | Patients without BDNF Deletions (N = 14) | P Value† |
|---|---|---|---|
| Age — yr | 0.65 | ||
| Mean | 11.4±6.5 | 12.6±8.7 | |
| Range | 3.8–25.8 | 2.0–24.9 | |
| Female sex — % (95 % CI) | 42 (20–66) | 64 (35–87) | 0.30 |
| Race or ethnic group — % (95% CI)‡ | 0.56 | ||
| Non-Hispanic white | 95 (74–100) | 86 (57–98) | |
| Other | 5 (<1–26) | 14 (2–43) | |
| Deletion — % (95% CI) | |||
| PAX6 | 100 (82–100) | 100 (77–100) | 1.00 |
| WT1 | 100 (82–100) | 100 (77–100) | 1.00 |
| Wilms’ tumor — % (95% CI) | 63 (38–84) | 50 (23–77) | 0.50 |
| Age at diagnosis of Wilms’ tumor — yr | 1.9±1.3 | 3.2±2.7 | 0.29 |
| Platelet count per mm3 | 295,000±97,000 | 305,000±84,000 | 0.75 |
| Obesity — % (95% CI) | |||
| In father | 32 (13–57) | 36 (13–65) | 1.00 |
| In mother | 26 (9–51) | 29 (8–58) | 1.00 |
| In both parents | 10 (1–33) | 14 (2–43) | 1.00 |
| Serum BDNF in parents — ng/ml§ | 33.74±12.32 | 31.86±9.90 | 0.53 |
| Weight | |||
| At birth — kg | 3.1±0.4 | 3.2±0.5 | 0.42 |
| At birth — z score | −0.71±0.77 | −0.42±0.94 | 0.35 |
| At 1 yr — kg | 8.9±1.8 | 8.4±1.6 | 0.50 |
| At 1 yr — z score | −1.15±1.54 | −1.46±1.82 | 0.66 |
| BMI z score¶ | |||
| At 2–3 yr∥ | 1.22±1.36 | 0.09±0.95 | 0.03 |
| At 4–6 yr** | 1.80±1.17 | 0.38±1.70 | 0.02 |
| At 8–10 yr†† | 2.08±0.45 | 0.88±1.28 | 0.03 |
| At 15–20 yr‡‡ | 1.88±0.46 | 0.70±1.06 | 0.05 |
| Current§§ | 1.61±0.75 | 0.66±1.29 | 0.01 |
| Current adjusted§§ | 1.63±0.86 | 0.64±0.86 | 0.003 |
Plus–minus values are means ±SD. BDNF denotes brain-derived neurotrophic factor, and BMI body-mass index.
Nominal P values are shown.
Race or ethnic group was self-reported.
Data are for 33 parents of persons with heterozygous BDNF deletions and 27 parents of persons without BDNF deletions.
There were differences in the sample size for BMI z scores because some participants had incomplete growth records and others were of young age. Multiple observations were obtained from each patient, but only one data point (if available) for each age-category analysis was used.
Data are for 18 patients with heterozygous BDNF deletions and 9 patients without BDNF deletions.
Data are for 18 patients with heterozygous BDNF deletions and 8 patients without BDNF deletions.
Data are for 12 patients with heterozygous BDNF deletions and 8 patients without BDNF deletions.
Data are for 5 patients with heterozygous BDNF deletions and 5 patients without BDNF deletions.
Data are for 19 patients with heterozygous BDNF deletions and 14 patients without BDNF deletions. Covariates for the adjusted current BMI z score were the BMI values of the parents.
Nineteen of 33 patients (58%) had BDNF haploinsufficiency: 17 had heterozygous complete deletion of BDNF, and 2 had heterozygous deletion of a portion of BDNF. The patients with and those without BDNF deletions did not differ significantly with regard to age, sex, race, the prevalence of Wilms’ tumor, the age at which Wilms’ tumor was diagnosed, the prevalence of obesity among their parents, or weight at birth (Table 1). However, patients with heterozygous BDNF deletions had significantly higher measured BMI z scores at the time of enrollment (Table 1) and higher BMI z scores throughout childhood than patients without BDNF deletions, with respect to both unadjusted values and values adjusted for the BMI of their parents. These differences were evident by 2 years of age (Table 1 and Fig. 1B). Results of questionnaires completed by the parents of the patients suggested significantly more symptoms of hyperphagia in patients with heterozygous BDNF deletions than in patients without BDNF deletions (Fig. 1C).
The mean serum BDNF concentration was 47% lower in patients with heterozygous BDNF deletions than in patients without these deletions (27.6±9.6 vs. 52.0±19.6 ng per milliliter, P = 0.001) (Fig. 1D). This result remained significant after adjustment for sex, current age, BMI, and platelet count (P = 0.004).
The typical BMI values according to age in patients with and those without heterozygous BDNF deletions are shown in Figure 2. Among the 24 participants with known weight status, the prevalence of childhood-onset obesity was significantly greater among patients with heterozygous BDNF deletions (100%; 95% confidence interval [CI], 77 to 100) than in patients without BDNF deletions (20%; 95% CI, 3 to 56, P<0.001).
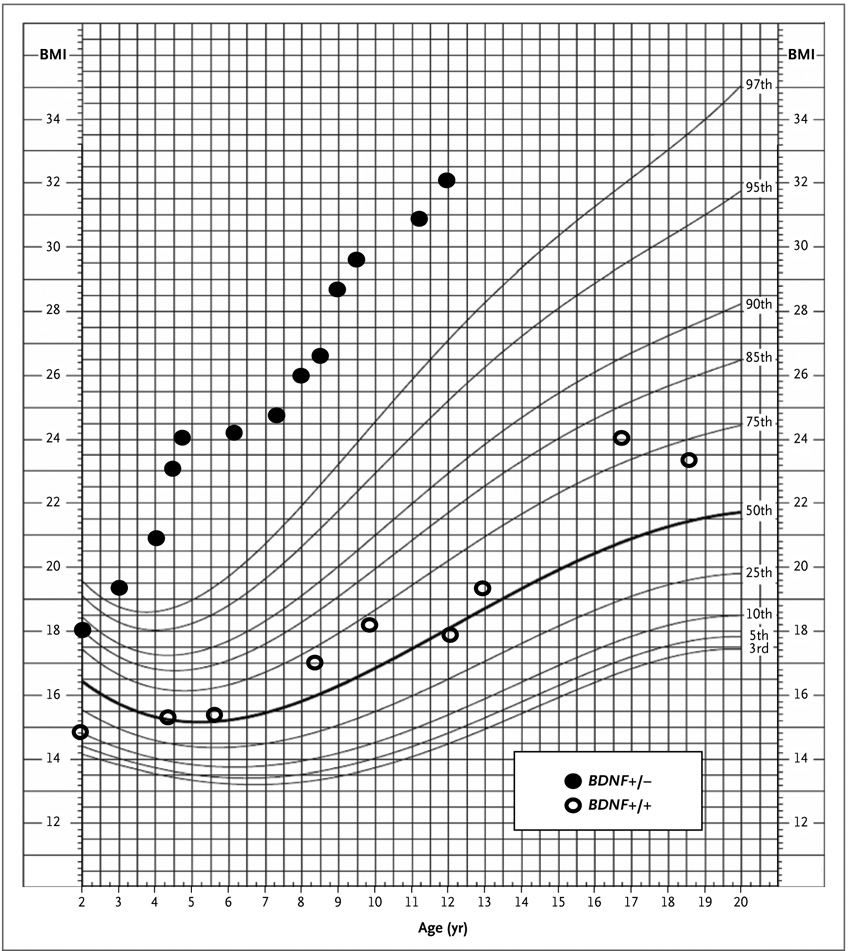
Figure 2. Body-Mass Index According to Age in Two Subjects.
The percentiles for body-mass index (BMI) according to age in a female subject with BDNF haploinsufficiency (Subject 10 in Appendix 6 in the Supplementary Appendix; BDNF+/−) and a female subject with intact BDNF (Subject 31 in Appendix 6 in the Supplementary Appendix; BDNF+/+) are shown in this standard growth chart for girls, from the Centers for Disease Control and Prevention. These patients had deletions of similar size (approximately 13 Mb) but divergent BMI values.
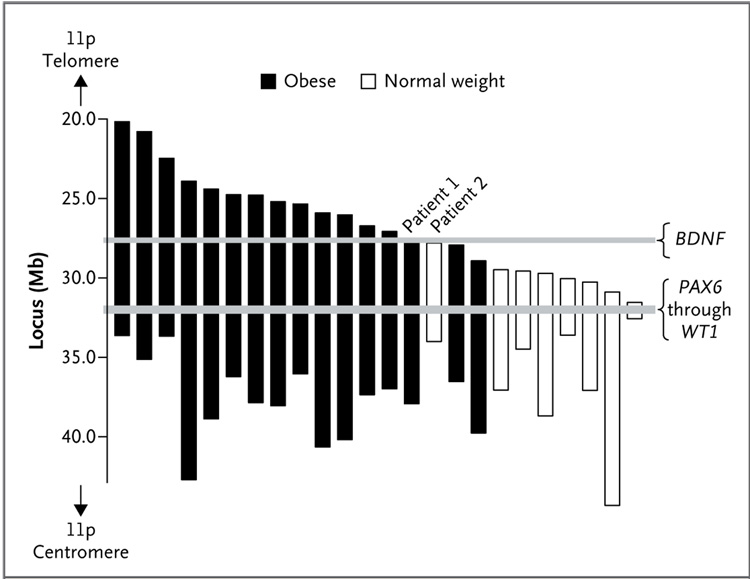
To explore the gene region associated with obesity further, the centromeric and telomeric boundaries of each patient’s deletion were examined. There was no association between the extent of the patients’ centromeric deletions and childhood obesity (Fig. 3). However, an analysis of the telomeric deletion boundaries indicated the presence of a critical region for childhood-onset obesity that started within 80 kb of BDNF exon 1 (Fig. 3). All patients with deletions that involved any portion of the BDNF gene became obese by 10 years of age. Deletion of BDNF exons 1 through 3, but preservation of the subsequent exons of BDNF, occurred in one obese patient (Patient 1 in Fig. 3). In contrast, in each patient who had normal weight during childhood, BDNF was intact. In the telomeric direction, the largest deletion associated with normal weight (Patient 2 in Fig. 3) ended 72.5 kb before BDNF exon 1. We analyzed the genomic DNA sequence within the 72.5-kb region upstream of BDNF exon 1, but we did not identify any protein-encoding sequences in this region.
Figure 3. Regions of Deletion on Chromosome 11p.
The region of deletion on chromosome 11p is shown for each of the 24 patients in whom the presence or absence of childhood obesity (body-mass index [BMI] ≥95th percentile by 10 years of age) could be determined. No association between the centromeric deletion boundary and childhood obesity was observed. However, for the telomeric deletion boundary, all patients with heterozygous deletion of all or a portion of BDNF had childhood obesity, whereas no deletions involved BDNF in the patients who were of normal weight. In Patient 1, who was obese, there was a heterozygous deletion of BDNF exons 1 through 3. In Patient 2, who had a normal weight (BMI, approximately 20th percentile at 10 years of age), the deletion region ended 72.5 kb upstream of BDNF. Only 20% of the patients without BDNF deletions were obese; this rate is similar to the prevalence of childhood obesity in the general U.S. population.35
ANALYSIS OF HUMAN HYPOTHALAMIC BDNF EXPRESSION
The observation of obesity in a child with deletion of only exons 1 through 3 of BDNF but preservation of all subsequent exons, including exon 9, which encodes the entire mature BDNF protein, prompted us to investigate whether exons 1 through 3 are expressed in the human hypothalamus; this would corroborate the potential role of these early exons in energy homeostasis. Transcripts involving BDNF exons 1 through 3 accounted for 64% of normal human hypothalamic expression. The methods and results of this study are provided in Appendix 7 in the Supplementary Appendix.
ASSOCIATION BETWEEN BDNF HAPLOINSUFFICIENCY AND IMPAIRED NOCICEPTION
The parents of 31 patients completed the pain questionnaire (see Appendix 1 in the Supplementary Appendix). Patients with BDNF haploinsufficiency had lower pain scores than patients with intact BDNF (P = 0.03), suggesting impaired nociception similar to that described anecdotally in the case reports of a child with a chromosomal inversion of the BDNF region18 and of a child with a TrkB-inactivating mutation.19 These data are consistent with results of studies in mice that suggest that BDNF plays a role in the modulation of pain sensation.30
DISCUSSION
A recent national health survey reported that approximately 16% of children in the United States were considered to be obese.35 Known single-gene mutations36 or syndromes37 may explain a small fraction of cases of childhood-onset obesity, but in the majority of cases, obesity is attributed to the interaction between a permissive environment and multiple genetic factors. Because data from studies in animals and limited studies in humans implicated insufficient BDNF signaling as a potential cause of hyperphagia and obesity, we studied body weight in a sample of patients with the WAGR syndrome that was expected to include people with haploinsufficiency for BDNF.
Among the patients in our study who did not have BDNF deletions, the prevalence of obesity was 20%; this rate was similar to that reported in the general pediatric population.35 However, the patients with BDNF haploinsufficiency had significantly higher BMIs during childhood, with a 100% prevalence of childhood-onset obesity. These observations suggest that BDNF may play an important role in energy homeostasis in humans.
The present results are derived from a hypothesis-driven clinical study in which we used a systematic approach with a sample size providing adequate power to examine whether BDNF haploinsufficiency is associated with obesity. Experiments involving inactivation of BDNF in mice, which can provide direct evidence of gene function, are not possible in humans; therefore, we undertook the most direct approach possible: an examination of a naturally occurring deletion syndrome. In patients with the WAGR syndrome, we were able to study intrinsic haploinsufficiency (i.e., the presence of a single intact allele), which provided an opportunity to observe the effects of gene loss directly. Analysis of the chromosomal deletion boundaries in patients with other genetic conditions that are caused by genetic haploinsufficiency and that have an obesity subphenotype may help to identify genes that are important for energy balance in humans.
In animals, BDNF primarily regulates energy intake.1,2,8–10 The birth weights of mice with heterozygous BDNF inactivation are similar to those of mice with intact BDNF.9,10 However, the mice with BDNF inactivation subsequently have hyperphagia,10 and excessive adiposity develops.9 We found that obesity after infancy also developed in patients with the WAGR syndrome who had heterozygous BDNF deletions. Food diaries underestimate intake,38 and they are particularly inaccurate in the case of children who, because of hyperphagia, seek food without parental knowledge. Thus, we assessed hyperphagia with a questionnaire29 that was applicable to patients with the WAGR syndrome because it was validated in patients with the Prader–Willi syndrome, another disorder that causes hyperphagia, food-seeking behavior, and variable cognitive impairment. Studies that use direct observation and objective measures of energy intake are required to confirm parents’ reports of hyperphagia in children with BDNF haploinsufficiency.
A possible limitation of our study is the fact that all patients with the WAGR syndrome and BDNF haploinsufficiency have large heterozygous 11p deletions. Thus, in this cohort, we could not examine patients who had isolated BDNF haploinsufficiency due to copy-number variations involving only the BDNF gene. At least 12 protein-coding genes, 6 pseudogenes, and 1 miscellaneous RNA gene lie in the 4-Mb region that separates PAX6 from BDNF. No phenotypic abnormalities have been reported to be associated with heterozygous mutations of these genes in animals or humans. Two patients in the study had telomeric deletion boundaries at particularly revealing locations. One, which was located in the intron between exons 3 and 4 of BDNF, was associated with obesity; the other, which was located 72.5 kb upstream of BDNF exon 1, was observed in a lean person. Human BDNF exons 1 through 8h are variably spliced with exon 9 (which encodes the entire mature BDNF protein) to form tissue-specific BDNF messenger RNA (mRNA) isoforms.39 Since, to our knowledge, BDNF isoform expression has not been characterized in the human hypothalamus, we studied pooled samples obtained from healthy adults and identified high expression of isoforms containing exons 1 and 2. These findings, along with the obesity observed in all the patients in our study who had deletions of the first three BDNF exons, suggest that exons 1 and 2 may be important for the hypothalamic function of BDNF as a regulator of energy balance.
The concentration of serum BDNF in the group of patients with heterozygous BDNF deletions was approximately half that in patients without BDNF deletions. This finding is consistent with the hypothesis that there is no compensatory increase in transcription from the intact gene in persons with BDNF haploinsufficiency. Serum is BDNF-enriched because of platelet degranulation during the clotting process.32 Mega-karyocytes, from which platelets derive, contain no detectable BDNF mRNA. Platelets are thus believed to bind and store BDNF from other, as yet unidentified, sources.32 Brain-derived enrichment of plasma BDNF in humans has recently been reported.14 Reduced concentrations of serum BDNF thus appear to reflect the haploinsufficient state and could well parallel decreased BDNF expression within the central nervous system.8
Since the discovery of leptin was reported in 1994,40 important strides have been made in our understanding of the regulation of energy homeostasis. Key molecules along the leptin signaling pathway have been elucidated in studies involving patients with rare monogenetic causes of obesity (e.g., mutations in the genes encoding leptin, leptin receptor, proopiomelanocortin, and prohormone convertase 1) as well as those with more common function-altering mutations in the gene encoding the melanocortin 4 receptor.41 In the present study, we investigated BDNF, one of the major signaling systems believed to function downstream of the melanocortin 4 receptor. Our results suggest that BDNF may play an important role in energy homeostasis in humans. Hyperleptinemia is a hallmark of almost all forms of obesity; thus, understanding the role of downstream mediators of leptin action such as BDNF holds promise for laying the groundwork for the development of new pharmaceutical approaches to the global obesity epidemic. Further studies are needed to assess the potential therapeutic role of BDNF replacement in persons in whom insufficient BDNF protein is produced and the potential contributions of more common allelic variants at this locus for susceptibility to obesity among persons in the general population.
Supplementary Material
Acknowledgments
Supported by grants from the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development of the National Institutes of Health (NIH) (Z01-HD-00641) and from the NIH Office of Rare Diseases.
We thank the members of International WAGR Syndrome Association for assistance in identifying families who were willing to participate in this study, Dr. Jack W. Tsao for his assistance with evaluation of nociception in our patients and review of an earlier version of the manuscript, and Chanelle Wijensinghe for administrative assistance.
Footnotes
Dr. Uhl reports holding patents for OPRM1 and DAT (SLC6A3), which might play roles in obesity, but having no patents or other financial interests that are related to brain-derived neurotrophic factor (BDNF); and Dr. Yanovski, receiving research support for an obesity-related clinical trial from Obecure but having no financial interests that are related to BDNF. Drs. Han and Yanovski are commissioned officers in the U.S. Public Health Service. No other potential conflict of interest relevant to this article was reported.
REFERENCES
- 1.Unger TJ, Calderon GA, Bradley LC, Sena-Esteves M, Rios M. Selective deletion of Bdnf in the ventromedial and dorsomedial hypothalamus of adult mice results in hyperphagic behavior and obesity. J Neurosci. 2007;27:14265–14274. doi: 10.1523/JNEUROSCI.3308-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang C, Bomberg E, Levine A, Billington C, Kotz CM. Brain-derived neurotrophic factor in the ventromedial nucleus of the hypothalamus reduces energy intake. Am J Physiol Regul Integr Comp Physiol. 2007;293:R1037–R1045. doi: 10.1152/ajpregu.00125.2007. [DOI] [PubMed] [Google Scholar]
- 3.Xu B, Goulding EH, Zang K, et al. Brain-derived neurotrophic factor regulates energy balance downstream of melanocortin-4 receptor. Nat Neurosci. 2003;6:736–742. doi: 10.1038/nn1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelleymounter MA, Cullen MJ, Wellman CL. Characteristics of BDNF-induced weight loss. Exp Neurol. 1995;131:229–238. doi: 10.1016/0014-4886(95)90045-4. [DOI] [PubMed] [Google Scholar]
- 5.Levin BE. Neurotrophism and energy homeostasis: perfect together. Am J Physiol Regul Integr Comp Physiol. 2007;293:R988–R991. doi: 10.1152/ajpregu.00434.2007. [DOI] [PubMed] [Google Scholar]
- 6.Nakagawa T, Tsuchida A, Itakura Y, et al. Brain-derived neurotrophic factor regulates glucose metabolism by modulating energy balance in diabetic mice. Diabetes. 2000;49:436–444. doi: 10.2337/diabetes.49.3.436. [DOI] [PubMed] [Google Scholar]
- 7.Rios M, Fan G, Fekete C, et al. Conditional deletion of brain-derived neurotrophic factor in the postnatal brain leads to obesity and hyperactivity. Mol Endocrinol. 2001;15:1748–1757. doi: 10.1210/mend.15.10.0706. [DOI] [PubMed] [Google Scholar]
- 8.Kernie SG, Liebl DJ, Parada LF. BDNF regulates eating behavior and locomotor activity in mice. EMBO J. 2000;19:1290–1300. doi: 10.1093/emboj/19.6.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lyons WE, Mamounas LA, Ricaurte GA, et al. Brain-derived neurotrophic factor-deficient mice develop aggressiveness and hyperphagia in conjunction with brain serotonergic abnormalities. Proc Natl Acad Sci U S A. 1999;96:15239–15244. doi: 10.1073/pnas.96.26.15239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fox EA, Byerly MS. A mechanism underlying mature-onset obesity: evidence from the hyperphagic phenotype of brain-derived neurotrophic factor mutants. Am J Physiol Regul Integr Comp Physiol. 2004;286:R994–R1004. doi: 10.1152/ajpregu.00727.2003. [DOI] [PubMed] [Google Scholar]
- 11.El-Gharbawy AH, Adler-Wailes DC, Mirch MC, et al. Serum brain-derived neurotrophic factor concentrations in lean and overweight children and adolescents. J Clin Endocrinol Metab. 2006;91:3548–3552. doi: 10.1210/jc.2006-0658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lommatzsch M, Zingler D, Schuhbaeck K, et al. The impact of age, weight and gender on BDNF levels in human platelets and plasma. Neurobiol Aging. 2005;26:115–123. doi: 10.1016/j.neurobiolaging.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 13.Chaldakov GN, Fiore M, Stankulov IS, et al. Neurotrophin presence in human coronary atherosclerosis and metabolic syndrome: a role for NGF and BDNF in cardiovascular disease? Prog Brain Res. 2004;146:279–289. doi: 10.1016/S0079-6123(03)46018-4. [DOI] [PubMed] [Google Scholar]
- 14.Krabbe KS, Nielsen AR, Krogh-Madsen R, et al. Brain-derived neurotrophic factor (BDNF) and type 2 diabetes. Diabetologia. 2007;50:431–438. doi: 10.1007/s00125-006-0537-4. [DOI] [PubMed] [Google Scholar]
- 15.Friedel S, Horro FF, Wermter AK, et al. Mutation screen of the brain derived neurotrophic factor gene (BDNF): identification of several genetic variants and association studies in patients with obesity, eating disorders, and attention-deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet. 2005;132B:96–99. doi: 10.1002/ajmg.b.30090. [DOI] [PubMed] [Google Scholar]
- 16.Ribasés M, Gratacòs M, Fernández-Aranda F, et al. Association of BDNF with restricting anorexia nervosa and minimum body mass index: a family-based association study of eight European populations. Eur J Hum Genet. 2005;13:428–434. doi: 10.1038/sj.ejhg.5201351. [DOI] [PubMed] [Google Scholar]
- 17.Gunstad J, Schofield P, Paul RH, et al. BDNF Val66Met polymorphism is associated with body mass index in healthy adults. Neuropsychobiology. 2006;53:153–156. doi: 10.1159/000093341. [DOI] [PubMed] [Google Scholar]
- 18.Gray J, Yeo GS, Cox JJ, et al. Hyperphagia, severe obesity, impaired cognitive function, and hyperactivity associated with functional loss of one copy of the brain-derived neurotrophic factor (BDNF) gene. Diabetes. 2006;55:3366–3371. doi: 10.2337/db06-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeo GS, Connie Hung CC, Rochford J, et al. A de novo mutation affecting human TrkB associated with severe obesity and developmental delay. Nat Neurosci. 2004;7:1187–1189. doi: 10.1038/nn1336. [DOI] [PubMed] [Google Scholar]
- 20.Fischbach BV, Trout KL, Lewis J, Luis CA, Sika M. WAGR syndrome: a clinical review of 54 cases. Pediatrics. 2005;116:984–988. doi: 10.1542/peds.2004-0467. [DOI] [PubMed] [Google Scholar]
- 21.Breslow NE, Norris R, Norkool PA, et al. Characteristics and outcomes of children with the Wilms tumor-aniridia syndrome: a report from the National Wilms Tumor Study Group. J Clin Oncol. 2003;21:4579–4585. doi: 10.1200/JCO.2003.06.096. [DOI] [PubMed] [Google Scholar]
- 22.Gül D, Oğur G, Tunca Y, Ozcan O. Third case of WAGR syndrome with severe obesity and constitutional deletion of chromosome 11)(p12p14) Am J Med Genet. 2002;107:70–71. doi: 10.1002/ajmg.10013. [DOI] [PubMed] [Google Scholar]
- 23.Marlin S, Couet D, Lacombe D, Cessans C, Bonneau D. Obesity: a new feature of WAGR (del 11p) syndrome. Clin Dysmorphol. 1994;3:255–257. [PubMed] [Google Scholar]
- 24.Tiberio G, Digilio MC, Giannotti A. Obesity and WAGR syndrome. Clin Dysmorphol. 2000;9:63–64. doi: 10.1097/00019605-200009010-00014. [DOI] [PubMed] [Google Scholar]
- 25.Amor DJ. Morbid obesity and hyperphagia in the WAGR syndrome. Clin Dysmorphol. 2002;11:73–74. doi: 10.1097/00019605-200201000-00016. [DOI] [PubMed] [Google Scholar]
- 26.Brémond-Gignac D, Crolla JA, Copin H, et al. Combination of WAGR and Potocki Shaffer contiguous deletion syndromes in a patient with an 11p11.2-p14 deletion. Eur J Hum Genet. 2005;13:409–413. doi: 10.1038/sj.ejhg.5201358. [DOI] [PubMed] [Google Scholar]
- 27.Jung R, Rauch A, Salomons GS, et al. Clinical, cytogenetic and molecular characterization of a patient with combined succinic semialdehyde dehydrogenase deficiency and incomplete WAGR syndrome with obesity. Mol Genet Metab. 2006;88:256–260. doi: 10.1016/j.ymgme.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 28.McGaughran JM, Ward HB, Evans DG. WAGR syndrome and multiple exostoses in a patient with del(11)(p11.2p14.2) J Med Genet. 1995;32:823–824. doi: 10.1136/jmg.32.10.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dykens EM, Maxwell MA, Pantino E, Kossler R, Roof E. Assessment of hyperphagia in Prader-Willi syndrome. Obesity (Silver Spring) 2007;15:1816–1826. doi: 10.1038/oby.2007.216. [DOI] [PubMed] [Google Scholar]
- 30.Allen SJ, Dawbarn D. Clinical relevance of the neurotrophins and their receptors. Clin Sci (Lond) 2006;110:175–191. doi: 10.1042/CS20050161. [DOI] [PubMed] [Google Scholar]
- 31.Breau LM, McGrath PJ, Camfield CS, Finley GA. Psychometric properties of the non-communicating children’s pain checklist-revised. Pain. 2002;99:349–357. doi: 10.1016/s0304-3959(02)00179-3. [DOI] [PubMed] [Google Scholar]
- 32.Fujimura H, Altar CA, Chen R, et al. Brain-derived neurotrophic factor is stored in human platelets and released by agonist stimulation. Thromb Haemost. 2002;87:728–734. [PubMed] [Google Scholar]
- 33.Cole TJ. The LMS method for constructing normalized growth standards. Eur J Clin Nutr. 1990;44:45–60. [PubMed] [Google Scholar]
- 34.Kuczmarski RJ, Ogden CL, Grummer-Strawn LM, et al. Advance data from vital and health statistics. No. 314. Hyattsville, MD: National Center for Health Statistics; 2000. CDC growth charts: United States; pp. 1–27. [PubMed] [Google Scholar]
- 35.Ogden CL, Carroll MD, Flegal KM. High body mass index for age among US children and adolescents, 2003–2006. JAMA. 2008;299:2401–2405. doi: 10.1001/jama.299.20.2401. [DOI] [PubMed] [Google Scholar]
- 36.Bell CG, Walley AJ, Froguel P. The genetics of human obesity. Nat Rev Genet. 2005;6:221–234. doi: 10.1038/nrg1556. [DOI] [PubMed] [Google Scholar]
- 37.Chung WK, Leibel RL. Molecular physiology of syndromic obesities in humans. Trends Endocrinol Metab. 2005;16:267–272. doi: 10.1016/j.tem.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 38.De Castro JM. Methodology, correlational analysis, and interpretation of diet diary records of the food and fluid intake of free-living humans. Appetite. 1994;23:179–192. doi: 10.1006/appe.1994.1045. [DOI] [PubMed] [Google Scholar]
- 39.Pruunsild P, Kazantseva A, Aid T, Palm K, Timmusk T. Dissecting the human BDNF locus: bidirectional transcription, complex splicing, and multiple promoters. Genomics. 2007;90:397–406. doi: 10.1016/j.ygeno.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372:425–432. doi: 10.1038/372425a0. Erratum, Nature 1995;374:479. [DOI] [PubMed] [Google Scholar]
- 41.Farooqi S, O’Rahilly S. Genetics of obesity in humans. Endocr Rev. 2006;27:710–718. doi: 10.1210/er.2006-0040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.