Abstract
Ribonucleoprotein complexes (RNP) remodeling by DEAD-box proteins is required at all stages of cellular RNA metabolism. These proteins are composed of a core helicase domain lacking sequence specificity; flanking protein sequences or accessory proteins target and affect the core's activity. Here we examined the interaction of eukaryotic initiation factor 4AI (eIF4AI), the founding member of the DEAD-box family, with two accessory factors, eIF4B and eIF4H. We find that eIF4AI forms a stable complex with RNA in the presence of AMPPNP and that eIF4B or eIF4H can add to this complex, also dependent on AMPPNP. For both accessory factors, the minimal stable complex with eIF4AI appears to have 1:1 protein stoichiometry. However, because eIF4B and eIF4H share a common binding site on eIF4AI, their interactions are mutually exclusive. The eIF4AI:eIF4B and eIF4AI:eIF4H complexes have the same RNase resistant footprint as does eIF4AI alone (9–10 nucleotides [nt]). In contrast, in a selective RNA binding experiment, eIF4AI in complex with either eIF4B or eIF4H preferentially bound RNAs much longer than those bound by eIF4AI alone (30–33 versus 17 nt, respectively). The differences between the RNase resistant footprints and the preferred RNA binding site sizes are discussed, and a model is proposed in which eIF4B and eIF4H contribute to RNA affinity of the complex through weak interactions not detectable in structural assays. Our findings mirror and expand on recent biochemical and structural data regarding the interaction of eIF4AI's close relative eIF4AIII with its accessory protein MLN51.
Keywords: eIF4AI, eIF4B, eIF4H, eIF4AIII, DEAD-box, translation initiation
INTRODUCTION
DExH/D-box proteins, a group of RNA-stimulated ATPases belonging to helicase superfamily 2 (SF2) (Singleton et al. 2007), play numerous essential roles in cellular RNA metabolism (Tanner and Linder 2001). Within the larger protein group, the DExH, DEAH, and DEAD subfamilies are set apart by distinct signatures of seven diagnostic SF2 motifs within a shared helicase core. The core consists of two similar RecA-like structural units connected by a flexible linker (Jankowsky and Fairman 2007). These structural units can adopt either an open conformation with relatively low affinity for ATP and RNA or a more closed conformation that brings together the conserved helicase motifs to facilitate substrate binding (Theissen et al. 2008).
DExH/D-box proteins are thought to remodel RNAs and ribonucleoprotein (RNP) complexes through modulation of RNA:RNA or RNA:protein interactions (Staley and Guthrie 1998; Schwer 2001; Tanner and Linder 2001; Jankowsky and Bowers 2006). Such structural rearrangements can result from translocation of the protein along a single RNA strand (as has been demonstrated for the DExH-box proteins HCV NS3 and NPH-II; Kawaoka et al. 2004; Beran et al. 2006; Jankowsky and Fairman 2007) or by kinking of the phosphodiester backbone (as observed in crystal structures of the DEAD-box proteins eIF4AIII and VASA; Andersen et al. 2006; Bono et al. 2006; Sengoku et al. 2006) resulting in local structural perturbations (Yang et al. 2007). Because the helicase core lacks sequence specificity, most DExH/D-box proteins contain extra sequences flanking the core and/or act in concert with one or more accessory factors; these extra cis- or trans-acting sequences are thought to target the helicase core to specific RNAs and/or alter its RNP remodeling activity (Kossen et al. 2002; Pang et al. 2002; Silverman et al. 2003).
Currently one of the best-understood examples of how accessory factors modulate DEAD-box protein activity is provided by eukaryotic translation initiation factor 4AIII (eIF4AIII; henceforth, 4AIII). Implicated in ribosome biogenesis (Kressler et al. 1997) and translational regulation (Weinstein et al. 1997; Li et al. 1999), 4AIII is perhaps best known as the RNA binding anchor of the exon junction complex (EJC) (Shibuya et al. 2004), a group of proteins deposited on mRNAs as a consequence of pre-mRNA splicing. The minimal EJC core is a heterotetramer consisting of 4AIII and its binding partners MLN51 and the Magoh:Y14 heterodimer (Tange et al. 2005). On its own, 4AIII displays minimal RNA binding, ATPase, and helicase activities, but all three are enhanced by MLN51 (Ballut et al. 2005; Noble and Song 2007). In the presence of the nonhydrolyzable ATP analog AMPPNP, 4AIII and MLN51 form a stable RNA binding complex detectable by both electrophoretic mobility shift assay (EMSA) and RNA pull down. In the presence of ATP, however, stable complex formation requires Magoh:Y14, which potently inhibit 4AIII's ATPase activity (Ballut et al. 2005). Recent crystal structures of the tetrameric EJC core in complex with RNA and AMPPNP show that MLN51's SELOR domain (speckle localizer and RNA binding module) promotes RNA binding of 4AIII through interactions with both 4AIII's N- and C-terminal domains as well as the bound RNA. In contrast, the Magoh:Y14 heterodimer binds on the opposite side of 4AIII from the RNA binding site, via interactions primarily with 4AIII's C-terminal domain and the interdomain linker (Andersen et al. 2006; Bono et al. 2006).
A close relative of 4AIII is eIF4AI (65% identical; henceforth, 4AI), the founding member of the DEAD-box protein family (Rogers et al. 2002). 4AI's proposed physiological role, and that of its highly homologous sibling eIF4AII (91% identical; Nielsen and Trachsel 1988), is to unwind RNA structures in the 5′-UTR of mRNAs to facilitate 40S ribosomal subunit loading and subsequent AUG start codon identification. In vivo, 4AI constitutes one subunit of eIF4F, which also includes the 7-methyl-G cap binding protein eIF4E and the scaffolding protein eIF4G (4G) (Kapp and Lorsch 2004). Within the context of eIF4F, 4AI has an increased capacity to unwind RNA duplexes in vitro (Rogers et al. 2001). NMR analysis has indicated that the main site of interaction between 4AI and the middle domain of 4G occurs on the surface of 4AI's C-terminal RecA-like domain. Through additional interactions with 4AI's N-terminal domain, 4G has been proposed to act as a soft clamp to stabilize 4AI's closed, RNA binding conformation (Oberer et al. 2005).
Two other initiation factors, eIF4B (4B) and eIF4H (4H), have also been shown to increase 4AI's in vitro helicase activity (Rogers et al. 2001). However, how and where 4B and 4H interact with 4AI is not understood, nor is it clear how these interactions promote helicase activity and translation initiation. In this article, we investigate interactions between 4AI and its accessory proteins 4B and 4H. We find that, like 4AIII and VASA (Ballut et al. 2005; Sengoku et al. 2006), 4AI can form a stable complex with RNA in the presence of AMPPNP. Both 4B and 4H can add to this complex, but because they share a common binding site on 4AI, their interactions are mutually exclusive. Using a competitive binding assay, we find that the minimal preferred binding site sizes of 4AI and its accessory factor complexes are surprisingly large. Nonetheless, the RNase-resistant footprint sizes of 4AI-containing complexes are virtually indistinguishable from a complex containing 4AIII and MLN51. Possible explanations for the apparent site size disparities are discussed, along with parallels between 4AI and 4AIII with regard to how their respective accessory factors serve to modulate various DEAD-box protein activities.
RESULTS
eIF4AI stably associates with RNA in the presence of AMPPNP
To determine whether 4AI could form a stable complex with RNA in the presence of AMPPNP, akin to the complexes previously observed for 4AIII (Ballut et al. 2005; Andersen et al. 2006; Bono et al. 2006) and VASA (Sengoku et al. 2006), we performed pull-down experiments using a biotinylated 32-nucleotide (nt) RNA (Fig. 1A). Whereas no 4AI precipitated in either the absence of nucleotide or the presence of ATP (Fig. 1A, lanes 1,2), a significant fraction did remain associated with immobilized RNA in the presence of AMPPNP (Fig. 1A, lane 3). Control experiments lacking RNA confirmed that this AMPPNP-dependent precipitation occurred via the RNA and not through direct binding of 4AI to the streptavidin beads (data not shown).
FIGURE 1.
Nucleotide dependence of eIF4AI containing-complexes. (A) SDS-PAGE of proteins bound and unbound to streptavidin beads from reaction mixes containing biotinylated 32-nt RNA (1 μM), 4AI (1 μM), indicated nucleotides (1.5 mM), and BSA (indicated by *; 0.1 mg/mL). (MW) Molecular weight markers with sizes indicated on left. (B) EMSA of reactions containing 32P-labeled 32-nt RNA (30 nM) plus 4AI (250 or 750 nM), full-length 4B (250 or 750 nM), and/or nucleotides (1 mM) as indicated. (C) Same as B, except using 4AI and/or 4H. (D, top) Schematic diagram of full-length 4B domain structure, ΔRRM-4B (amino acids 178–611) and C-term-4B (amino acids 332–611). (Bottom) EMSA as in B and C of reactions containing 4AI, full-length 4B, ΔRRM-4B, and/or C-term-4B as indicated. All reactions contained AMPPNP. (*) 4AI:C-term-4B species. (E) SDS-PAGE of proteins bound and unbound to Ni-coated beads in the presence of 32-nt RNA (1 μM) and/or AMPPNP (1.5 mM) as indicated. (Lanes 1–3) His6-4AI plus untagged ΔRRM-4B; (lanes 4–6) His6-4H plus untagged 4AI. All proteins were 1 μM. (MW) Molecular weight markers with sizes indicated on left.
Stable binding of 4AI to a 32-nt RNA could also be observed in EMSA (Fig. 1B,C). At high protein:RNA ratios (20-fold excess), the gel shift appeared as a doublet. The slower migrating species likely represents a complex containing more than one bound 4AI per RNA, as it was not observed at lower protein:RNA ratios (data not shown). As was observed in the pull-down assay, shifted complexes were only seen when AMPPNP was present.
eIF4B and eIF4H form stable complexes with eIF4AI, RNA, and AMPPNP
We next tested the abilities of 4B and 4H to form stable complexes with 4AI and/or RNA. In the EMSA format, addition of either factor to reactions containing 4AI, RNA, and AMPPNP resulted in supershifted complexes (Fig. 1B,C). When compared to 4AI alone, a much greater fraction of the RNA was shifted in the 4AI:4B and 4AI:4H complexes, consistent with higher RNA affinities. As for the complex of 4AI alone with RNA, these slower migrating 4AI:4B and 4AI:4H species required the noncleavable nucleotide analog—they were not observed either in the absence of nucleotide or in the presence of ATP.
Consistent with previous reports (Methot et al. 1994; Naranda et al. 1994; Richter-Cook et al. 1998), the EMSA also showed that both 4B and 4H have some capacity to interact with RNA on their own (resulting in a diffuse high mobility shift near the free RNA in Fig. 1B, lanes 10,12,13; Fig. 1C, lanes 8–12). In contrast to the 4AI-dependent supershifts, however, this weak binding activity was nucleotide independent. In the case of 4B, its inherent RNA binding activity proved problematic for pull-down assays. To circumvent this, we turned to N-terminally truncated versions of 4B. A previous truncation analysis had shown that just the C-terminal 358 amino acids of 4B could enhance the RNA binding and helicase activity of 4AI, albeit to a diminished degree (Methot et al. 1994). Consistent with this, we found that the 280 C-terminal amino acids of 4B (C-term-4B) were capable of supershifting the 4AI:RNA complex, but much less robustly than full-length 4B (Fig. 1D, lane 7, species denoted by *). This result nonetheless indicates that the minimal 4AI-interaction region on 4B is within the 280 C-terminal amino acids. In contrast to the more severely truncated form of 4B, we found that a version lacking just the N-terminal RRM domain (ΔRRM-4B=amino acids 178–611) yielded very similar RNA supershifts in the presence of 4AI and AMPPNP (Fig. 1D; data not shown) to those formed by the full-length protein. Because ΔRRM-4B proved easier to purify in higher yields and seemed less prone to self-aggregation than the full-length protein (data not shown), we used it in all subsequent experiments unless otherwise noted.
We next wanted to determine whether stable interaction between 4AI and 4B or 4H required RNA and/or nucleotide. To do so, we used a pull-down assay employing His6-tagged proteins (Fig. 1E). His6-tagged 4AI and untagged ΔRRM-4B (Fig. 1E, lanes 1–3) or untagged 4AI and His6-tagged 4H (Fig. 1E, lanes 4–6) were incubated with a 32-mer RNA, AMPPNP, or both and bound to Ni-coated beads. In both cases, the untagged protein was precipitated only in the presence of both RNA and AMPPNP. We conclude that stable association between 4AI and its accessory proteins 4B and 4H occurs only in the context of a four-way complex containing the two polypeptides, RNA and AMPPNP.
eIF4B and eIF4H stimulate the helicase activity of eIF4AI but not eIF4AIII
Having shown that 4B and 4H directly associate with 4AI in the context of an RNA:AMPPNP complex, we next wanted to examine the specificity of these interactions. A previous report had suggested that 4B enhancement of helicase activity is not limited to 4AI, but can extend to 4AI's close relative 4AIII (Li et al. 1999). In our hands, however, at protein concentrations capable of promoting complete strand separation by 4AI, neither ΔRRM-4B nor 4H had any effect on the helicase activity of 4AIII (Fig. 2A). Indeed, 4AIII alone (even at concentrations up to 5 μM; data not shown) exhibited no detectable strand separation activity in our assays.3 However, the failure of ΔRRM-4B or 4H to enhance 4AIII's helicase activity is not due to the latter's complete inactivity, because addition of 4AIII's known binding partner MLN51 did lead to increased strand separation as has been previously reported (Noble and Song 2007). Unexpectedly, this same concentration of MLN51 similarly enhanced 4AI's helicase activity, suggesting that the mechanism by which MLN51 modulates helicase activity is not dependent on specific interaction with a DEAD-box protein binding partner (see Discussion). In contrast, our data indicate that 4B and 4H are highly specific for 4AI over 4AIII.
FIGURE 2.
Mapping the eIF4B and eIF4H binding sites on eIF4AI. (A) Helicase assays containing 32P-labeled duplex RNA±indicated DEAD-box proteins (4AI or 4AIII) and accessory factors (ΔRRM-4B, 4H, or MLN51). After 0, 4, 10, and 15 min incubation at 35°C, samples were quenched with SDS and electrophoresed in a nondenaturing polyacrylamide gel to separate duplex and single-stranded RNAs (indicated at right). (Lane 1) duplex alone incubated at 35°C for 15 min; (lane 2) duplex incubated at 95°C prior to electrophoresis to completely separate the strands. (Δ) Duplex degradation product. (B, top) schematic diagram of 4A3133 chimera. Numbers indicate quadrant boundaries. (Bottom) Same as Figure 1A, except pull-down reactions contained 4AI, 4AIII, the 4A3133 chimera, and/or ΔRRM-4B as indicated. All reactions contained BSA (*) and AMPPNP (1.5 mM). (C) Same as Figure 1, B and C, except that all reactions contained AMPPNP (1.5 mM) and the indicated proteins at 1 μM. (D) Same as C, except reactions contained 4AI and ΔRRM-4B and/or 4H at indicated concentrations and RNA was in excess (200 μM).
eIF4B and eIF4H interact with the same region of eIF4AI
Consistent with the specificity of ΔRRM-4B and 4H for 4AI in the helicase assay, we could detect no stable association of either protein with 4AIII (Fig. 2B, lane 2; Fig. 2C, lane 7) under conditions where 4AI:ΔRRM-4B and 4AI:4H complexes were readily observable (Fig. 2B, lane 1; Fig. 2C, lane 6). We took advantage of this affinity difference to map the 4B and 4H binding sites on 4AI, using a set of N-terminally His6-tagged 4AI/4AIII chimeras previously employed to map the MLN51 interaction site on 4AIII (Ballut et al. 2005). In these constructs, the DEAD-box protein is divided into four roughly equal quadrants (dubbed regions 1–4 from the N terminus), and chimeras were constructed by swapping regions of one protein for the other. Thus, for example, 4A3133 contains regions 1, 3, and 4 from 4AIII and region 2 from 4AI (Fig. 2B).
Preliminary experiments using Ni-NTA resin revealed that ΔRRM-4B was able to weakly associate with His6-4A1131 and His6-4A1133 in the presence of RNA and AMPPNP, but not with His6-4A1331, His6-4A3313, or His6-4A1311 (data not shown). This implicated 4AI quadrant 2 as the 4B binding site. This result was verified using an RNA pull-down assay containing the 4A3133 chimera, ΔRRM-4B, and AMPPNP (Fig. 2B, lane 3). Quadrant 2 also proved to be the binding site for 4H as determined by EMSA using the 4A3133 chimera (Fig. 2C, lane 8).
Having shown that 4B and 4H both target the same region of 4AI, we next wondered whether their interactions with 4AI were mutually exclusive or could occur simultaneously. To address this, we used EMSA in the presence of excess RNA (>100-fold over protein) to ensure that only one protein complex could form per RNA molecule. Under these conditions, when ΔRRM-4B was titrated into reactions containing constant concentrations of 4AI and 4H, no new bands of either intermediate or decreased mobility relative to the 4AI:4H and 4AI:ΔRRM-4B complexes were observed (Fig. 2D). This indicates that the interactions between 4AI, 4B, and 4H are mutually exclusive. We conclude that 4B and 4H share a common binding site in the N-terminal half of 4AI.
eIF4AI, eIF4AI:4B, and eIF4AI:4H complexes yield similarly sized RNase resistant footprints
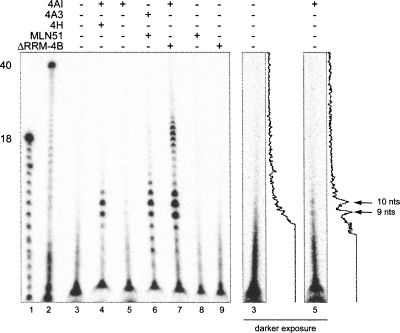
Recent X-ray crystal structures of the EJC core show that 4AIII contacts a total of six nucleotides, with the SELOR domain of MLN51 also touching the 5′-most nucleotide within the 4AIII binding site (Andersen et al. 2006; Bono et al. 2006). Consistent with this, RNase footprinting analysis of 4AIII in complex with amino acids 137–283 of MLN51 (the SELOR domain) yielded a stable RNA binding site size of 7 nt (Ballut et al. 2005). To determine the stable RNA binding site sizes of the 4AI, 4AI:ΔRRM-4B, and 4AI:4H complexes, we subjected samples containing various combinations of proteins plus AMPPNP and an unlabeled 40-nt RNA to micrococcal nuclease (MNase) digestion. After inactivation of MNase with EGTA, RNAs were labeled with γ32P-ATP and polynucleotide kinase and analyzed by denaturing PAGE (Fig. 3).
FIGURE 3.
Micrococcal nuclease (MNase) footprinting of eIF4AI and eIF4AIII complexes. Denaturing PAGE of RNAs recovered after MNase digestion of reactions containing poly(U)40, AMPPNP and indicated proteins (lanes 3–9). (Lanes 1,2) poly(U)18 and poly(U)40 hydrolysis ladders. (Right panels) Darker exposure of lanes 3 and 5 with densitometry traces.
In our hands, a control reaction containing 4AIII in association with amino acids 1–400 of MLN51 yielded a 9–10-nt footprint (Fig. 3, lane 6). This slightly larger footprint than was previously reported (Ballut et al. 2005) could either be due to the larger fragment of MLN51 utilized here or a difference in the nucleases employed. In any event, this size footprint was very similar to the footprints observed for the 4AI-containing complexes. Indeed, all samples containing 4AI yielded observable footprints, albeit to different degrees. Consistent with the stronger affinity of the 4AI:4B and 4AI:4H complexes for RNA than 4AI alone (see Fig. 1), the 4AI:ΔRRM-4B and 4AI:4H footprints were substantially stronger than was the footprint for 4AI alone (Fig. 3, lanes 4,5,7). Nonetheless, both the 4AI alone and 4AI:4H samples yielded almost identical footprints, centered around 9–10 nt. This same pattern was also prominent in the 4AI:4B lane, which additionally exhibited a second and larger footprint centered around 18–20 nt. Since the protein concentrations were roughly equivalent to that of the RNA in this experiment, a likely explanation for this larger footprint was the presence of two adjacent 4AI:4B complexes that protected a larger region of the RNA from MNase digestion. Consistent with this idea, 4B has been shown to self-associate via its DRYG domain (Methot et al. 1996b). Control reactions containing only 4B or MLN51 yielded no detectable footprints (Fig. 3, lanes 8–9).
Taken together, our footprinting data indicate that whereas 4B and 4H clearly contribute to the affinity of 4AI for RNA, they do not extend the region of RNA protected from nuclease digestion beyond that protected by 4AI alone. Further, the footprints of 4AI-containing complexes are nearly identical to the footprint of the 4AIII:MLN51 complex.
eIF4AI:4B and eIF4AI:4H prefer longer RNAs than eIF4AI in a selective binding assay
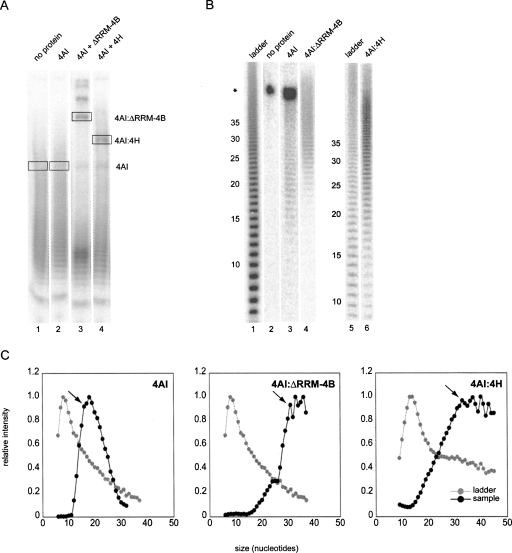
Although neither ΔRRM-4B nor 4H extended the RNase resistant footprint of 4AI, both proteins exhibit some RNA binding affinity of their own (see Fig. 1B,C; Methot et al. 1994; Naranda et al. 1994; Richter-Cook et al. 1998), and the 4AI:4B and 4AI:4H complexes appear to bind more tightly to RNA than 4AI alone (Fig. 1B,C; Methot et al. 1994). In an attempt to assess whether the inherent RNA binding affinity of 4B and 4H contributes in a functional way, we employed a selective RNA binding assay to determine the preferred RNA binding site sizes for 4AI and the 4AI:4B and 4AI:4H complexes. To do so, we first generated a hydrolysis ladder from poly(U) RNA such that lengths between 6 and 40 nt were well populated (Fig. 4B, lanes 1,5). We then performed gel shift assays as before, but using this hydrolysis ladder (Fig. 4A). In the presence of 4AI, 4AI+ΔRRM-4B, or 4AI+4H, slower migrating species were observed identical to those previously observed with the 32-nt RNA (Fig. 1B,C; data not shown). To determine which size RNAs were preferentially bound in each complex, we cut out individual bands from the native gel and ran the extracted RNAs on a denaturing gel (Fig. 4B). To control for possible preferential loss of small RNA fragments during ethanol precipitation, the input hydrolysis ladder was precipitated in parallel to experimental samples prior to denaturing electrophoresis. We then plotted the relative intensity of each band in the denaturing gel (normalized to the most intense band analyzed in that lane) as a function of RNA length along with an analogous representation of the hydrolysis ladder (Fig. 4C).
FIGURE 4.
Preferred RNA binding site sizes of eIF4AI complexes. (A) Representative EMSA of reactions containing 32P-labeled poly(U) hydrolysis ladder, AMPPNP (1.5 mM), and indicated proteins (1 μM). (Boxes) Gel slices excised for subsequent denaturing analysis. (B) Representative denaturing PAGE of RNAs recovered from slices such as those indicated in A, alongside the hydrolysis ladder used in that experiment (lanes 1,5). (*) Nonshifted bands comigrating with 4AI complexes. (C) Plots of relative band intensities versus RNA fragment length for hydrolysis ladders (gray circles) and indicated complexes in panel B (black circles). Minimum preferred RNA length is indicated by arrow.
In all experiments (Fig. 4B; data not shown), the band intensity distributions of the mobility-shifted species differed markedly from those of the hydrolysis ladders. Whereas both hydrolysis ladders shown (Fig. 4B, lanes 1,5) were predominated by oligonucleotides in the 6–12-nt range, fragments of this size were barely detectable in samples derived from regions of the native gel containing RNA:protein complexes (Fig. 4B, lanes 3,4,6). For example, RNAs from the 4AI gel shift band peaked in abundance around 18 nt, with the intensity of species shorter than 16 nt decreasing sharply such that oligos less than 12 nt were hardly detectable (Fig. 4C). In contrast, extraction of a gel slice at the same position as the 4AI shift but from a sample containing no input protein (Fig. 4A, lane 1) yielded none of the smaller RNAs obtained from the 4A gel shift band, but only a tightly grouped multiplet of ∼45–50 nt (Fig. 4B, lane 2, species denoted by *). This multiplet, which represented unbound RNAs comigrating with the 4AI:RNA complex, was also apparent in the 4AI gel shift sample (Fig. 4B, lane 3). We therefore conclude that the appearance in the denaturing gel of any RNA fragment <45 nt derived from the 4AI gel shift sample was due to specific binding of that RNA by 4AI and its subsequent retardation in the native gel.
For each protein complex, we estimated its preferred RNA binding site size as the oligonucleotide length below which binding sharply decreased (arrows in Fig. 4C). For each complex, data from multiple experiments were combined to generate an average preferred binding site size (Table 1). This size was ∼17 nt for 4AI alone. The site sizes for the fastest migrating 4AI:ΔRRM-4B and 4AI:4H complexes were considerably larger (∼30 and ∼33 nt, respectively). Attempts to analyze the more slowly migrating 4AI:ΔRRM-4B and 4AI:4H species in the native gel (presumably containing multiple protein complexes per bound RNA molecule; Fig. 4A, lanes 3,4, shifts above boxed species) hinted at even larger site sizes (data not shown). However, we did not pursue these further. Nonetheless, our data clearly indicate that the preferred RNA binding site sizes of 4AI and the 4AI:ΔRRM-4B and 4AI:4H complexes are significantly larger than their RNase-resistant footprints. Furthermore, the preferred binding site sizes of the 4AI:ΔRRM-4B and 4AI:4H complexes are substantially larger than that of 4AI alone.
TABLE 1.
RNase protected site sizes and preferred RNA binding site sizes of eIF4AI containing complexes
The minimal eIF4AI:4B:RNA complex contains one molecule of each protein
One possible explanation for the discrepancies between the RNA footprint and preferred binding site sizes of complexes containing 4AI is that the stoichiometry of these complexes is something other than 1:1. To address the number of 4B molecules in the minimal 4AI:4B complex, we used EMSA under conditions of excess RNA (100-fold over protein) so that only one protein complex could form per RNA molecule. Under these conditions, when full-length 4B was titrated into reactions containing constant concentrations of 4AI and ΔRRM-4B, no new bands of different mobility relative to the 4AI:4B and 4AI:ΔRRM-4B complexes were observed (Fig. 5A). We conclude that there is only one 4B molecule in the minimal 4AI:4B complex.
FIGURE 5.
The stoichiometry of eIF4AI:eIF4B complexes. (A) EMSA as in Figure 1B,C, except all reactions contained AMPPNP (1.5 mM), 4AI, ΔRRM-4B, and/or 4B at indicated concentrations, and RNA was in excess (200 μM). (B) Same as Figure 1E, except all reactions contained AMPPNP (1.5 mM), the indicated proteins (1 μM), and RNA was in excess (10 μM).
To address the number of 4AI molecules in the 4AI:4B complex, we used a protein pull-down assay containing His6-tagged-4AI, untagged ΔRRM-4B, a 32- or 12-nt RNA, and AMPPNP, plus untagged 4AI in equimolar amounts relative to the His6-tagged-4AI. Under conditions of excess RNA (5- to 10-fold over protein), ΔRRM-4B was precipitated, but not the untagged 4AI (Fig. 5B, lanes 4,5). This suggests that there is only one 4AI molecule stably associated with each 4AI:4B complex.
DISCUSSION
We find here that, in the presence of a nonhydrolyzable ATP analog, 4AI can form stable complexes with RNA and its accessory proteins 4B and 4H. Further, both accessory factors interact with the second quandrant of 4AI in a mutually exclusive manner. This suggests that in vivo, 4B and 4H compete with one another for interaction with and regulation of 4AI. Although 4B and 4H appear to be ubiquitously expressed (Nomura et al. 1994; Richter et al. 1999), tissue-specific variations in their mRNA levels suggest that individual tissues contain unique ratios of the two proteins. For example, 4H transcript levels are twice those of 4B in the brain, whereas 4B transcripts are circa four times those of 4H in skeletal muscle (Richter et al. 1999). 4B and 4H could facilitate translation of different transcripts, or they could represent different control points for signal transduction pathways affecting translation initiation. With regard to the latter, 4B is a known phosphorylation target of the mTOR/PI3K and MAPK signal transduction pathways (Gingras et al. 2001). In response to hormones and nutrient sufficiency, increased 4B phosphorylation promotes its association with the translation preinitiation complex, where it is thought to enhance translation initiation by stimulating the activity of 4AI (Holz et al. 2005). In vivo, 4H may also be phosphorylated (Rush et al. 2005; Tao et al. 2005; Molina et al. 2007), but the kinases involved have yet to be identified, and nothing is yet known about the effects of phosphorylation on 4H activity. In the future it will certainly be of interest to determine how the phosphorylation states of 4B and 4H affect their interaction with and functional modulation of 4AI. It will also be of interest to investigate whether they interact similarly with eIF4AII to modulate its function.
Other than both containing a well-defined RNA recognition motif (RRM) (Milburn et al. 1990; Richter-Cook et al. 1998), 4B and 4H bear very little sequence similarity to one another. 4B's RRM has been proposed to interact with 18S rRNA and thereby promote association of the small ribosomal subunit with cap-bound translation initiation factors (Methot et al. 1996a). Consistent with this idea, our truncation analysis indicated that this RRM is unnecessary for stable interaction of 4B with 4AI. Rather, the minimal 4AI-binding region in 4B is located within its C-terminal 280 amino acids, a region that bears no apparent sequence similarity to 4H (Fig. 1D). Thus, although 4B and 4H both interact with the same surface of 4AI (Fig. 2), they likely do so using different structural motifs. This trend can be extended to MLN51, which binds the same region on 4AIII as 4B and 4H bind to 4AI, yet lacks any known sequence or structural homolog (Degot et al. 2002).
Initially, pull-down experiments similar to those performed here indicated that the SELOR domain of MLN51 interacts stably with the second quadrant of 4AIII in the context of an RNA- and AMPPNP-dependent complex (Ballut et al. 2005). Subsequent crystal structures revealed additional contacts between the SELOR domain and 4AIII's C-terminal RecA-like domain (Andersen et al. 2006; Bono et al. 2006). Thus, by interacting with both RecA-like domains of 4AIII, MLN51 likely increases 4AIII's affinity for RNA by stabilizing the closed RNA binding conformation. As noted in the Introduction, a similar mechanism has been proposed for how the middle domain of 4G promotes 4AI's helicase activity (Oberer et al. 2005). Interactions with both RecA-like domains of 4AI may also be key to how 4H modulates 4AI's RNA binding and helicase activities. Whereas our data reveal a site of stable interaction between 4H and the second half of 4AI's N-terminal RecA-like domain, NMR analysis indicates that 4H's C-terminal peptide additionally contacts 4AI's C-terminal RecA-like domain (A. Marintchev and G. Wagner, pers. comm.). Thus, an emerging theme for how accessory proteins can increase the RNA binding affinity and/or helicase activity of their DEAD-box partner is by interacting with both RecA-like domains and stabilizing the closed conformation. In the future, it will be of interest to determine whether 4B makes additional contacts with 4AI's C-terminal RecA-like domain and acts in a manner analogous to that proposed for 4H and 4G.
Although our data indicate that 4B and 4H exhibit a high degree of specificity for 4AI, 4AI's helicase activity could also be enhanced by MLN51, albeit to a lesser degree than MLN51's effect on 4AIII (Fig. 2). MLN51 might enhance 4AI's helicase activity via the same mechanism as 4B and 4H involving specific protein:protein contacts; however, a previous study failed to detect any stable interaction between 4AI and the MLN51 SELOR domain (Ballut et al. 2005). Although it is possible that the longer version of MLN51 employed here does interact directly with 4AI, the effect of MLN51 on 4AI's helicase activity could also reflect an entirely different mechanism. For example, MLN51 could bind and sequester the ssRNA that has been separated by the DEAD-box protein. Consistent with this idea, we observed that the first 400 amino acids of MLN51 had much more significant ssRNA binding activity on their own than did either 4B or 4H (data not shown). Indeed, this high intrinsic RNA binding activity prevented us from testing whether this longer version of MLN51 forms any stable RNA-dependent complexes with 4AI.
Another analogy that can be drawn between MLN51 and 4B/4H is in how they contribute to RNA binding by their respective DEAD-box protein without making extensive additional stable contacts with the RNA. In the case of MLN51, X-ray crystal structures of the EJC core showed that MLN51's SELOR domain contacts only 1 of the 6 nt interacting with 4AIII (Andersen et al. 2006; Bono et al. 2006). The Magoh:Y14 heterodimer binds on the surface of 4AIII opposite from the RNA binding site and thus makes no visible RNA:protein contacts. Consistent with what can be seen in the structures, previous RNase A footprinting analysis of 4AIII complexes had revealed a 7-nt footprint in the presence of the MLN51 SELOR domain, which extended to 8–9 nt with addition of the Magoh:Y14 heterodimer (Ballut et al. 2005). The extra 1–3 nt of footprint compared to the crystal structures are likely explained by steric constraints imposed by the bound proteins on RNase accessibility.
Using micrococcal nuclease (MNase) we here observed a 9–10-nt footprint for the 4AIII:MLN51 complex (Fig. 3). This same size footprint was observed for 4AI alone and 4AI in complex with either ΔRRM-4B or 4H. A larger 18–20-nt footprint was also observed in the 4AI:4B reactions. Because this larger footprint is exactly double the 9–10-nt footprint, the most likely explanation is that it represents the presence of two 4AI:4B complexes immediately juxtaposed on the RNA, possibly through self-dimerization of 4B (see below). In any event, the footprinting data indicate that, like MLN51's interaction with 4AIII, neither 4H nor 4B significantly extends the size of the nuclease-resistant RNA binding site beyond that inherent to 4AI. Thus the only protein:RNA contacts stable enough to resist RNase degradation in the 4AI:4B and 4AI:4H complexes likely occur through 4AI. If, like MLN51, 4B and/or 4H do contribute additional stable contacts to the RNA, these contacts are likely limited to the nucleotides already in contact with 4AI.
In contrast to the footprinting results, our preferential binding (affinity) experiments using a population of differently sized oligonucleotides revealed a preference for longer RNAs for all complexes (Fig. 4; Table 1). Whereas 4AI alone preferentially bound oligos ≥17 nt, the 4AI:4B and 4AI:4H complexes preferred even longer RNAs (≥30 nt and ≥33 nt, respectively). Although it might seem surprising that RNAs much longer than the physical footprint were preferentially selected in a binding assay, nucleic acid binding proteins can exhibit increased apparent affinity for longer RNAs simply due to statistical factors (Kelly et al. 1976). That is, additional nucleotides flanking the protein's footprint can contribute to apparent binding by increasing the number of registers in which the protein can interact with the nucleic acid lattice. In our hydrolysis ladder experiment, shorter RNAs (e.g., 9–10 nt) may have lost out to competition from slightly longer RNAs that allowed for multiple modes of binding, thereby overestimating the length of nucleotides actually accommodated by the protein.
Regardless of whether the preferential binding assay overestimates the true site size of the protein, the 4AI:4B and 4AI:4H complexes preferentially selected significantly longer RNAs (≥30–33 nt) than did 4AI alone (≥17 nt) despite yielding the same sized RNase resistant footprint (9–10 nt) as 4AI alone. One possible explanation for this difference is that the larger complexes represent protein multimers rather than 1:1 complexes. Indeed, 4B has previously been shown to self-associate via its DRYG domain (Methot et al. 1996b). Consistent with a tendency to self-aggregate, we observed that upon gel filtration, a significant fraction of either 4B alone or 4B in complex with 4AI eluted in the void volume, with the remainder spread across the entire elution profile (data not shown). This prevented us from using gel filtration to obtain a reliable molecular weight estimate of the 4AI:4B and 4AI:ΔRRM-4B complexes. Nonetheless, the pull-down and EMSA data shown in Figure 5 indicate that the minimal 4AI:4B complex contains a single molecule of each protein. Although we were unable to perform a similar experiment for 4H, molecular weight estimation by gel filtration and sucrose gradients of 4AI:4H in complex with a 12-mer RNA (data not shown) gave a molecular weight consistent with a 1:1 ratio of 4AI:4H. While this does not rule out the possibility of higher order 4AI:4H complexes, it does confirm that 4H and 4AI also minimally form a 1:1 complex.
In the preferential binding assay, the RNA site sizes of ≥30 and ≥33 nt were determined from the highest mobility species (Fig. 4A), which most likely represent 1:1 4AI:4B and 4AI:4H complexes, respectively. If these high mobility complexes do contain only a single molecule of each protein, then an alternate explanation for why 4AI:4B and 4AI:4H have a much larger preferred binding site size than 4AI alone is that, in addition to the primary footprint imparted by 4AI, the accessory factor makes additional, transient interactions with the flanking nucleotides on longer RNAs. These additional weak interactions would not be protected in a footprinting experiment, but could still contribute significantly to the overall binding energy of a protein complex for longer RNAs. Consistent with this idea, both 4B and 4H have sequence elements indicative of RNA binding and demonstrable, though weak, affinity for RNA (Fig. 1; Methot et al. 1994; Naranda et al. 1994; Richter-Cook et al. 1998). Thus, in addition to stabilizing 4AI's closed conformation as proposed above, weak interactions between 4B or 4H and flanking nucleotides could further increase the affinity of these complexes for RNA. In the future, it will be of interest to determine if such interactions exist, to what extent they contribute to RNA binding by the complex, and whether this is also a general feature of DEAD-box protein accessory factors.
MATERIALS AND METHODS
Plasmids
Plasmids containing human eIF4AI in pET3 (Pause and Sonenberg 1992), human His6-eIF4AI in pET15 (Oberer et al. 2005), human His6-eIF4H in pET15 (Doepker et al. 2004), and the His6-4AI-4AIII chimeras in pET28 (Ballut et al. 2005) were generous gifts from Nahum Sonenberg (McGill University), Gerhard Wagner (Harvard Medical School), Jim Smiley (University of Alberta), and Hervé Le Hir (Centre de Génétique Moléculaire, CNRS), respectively. Human eIF4AIII (Shibuya et al. 2004), human eIF4B (a generous gift from Nahum Sonenberg; Pause and Sonenberg 1992), and its truncations ΔRRM-4B (amino acids 178–611) and C-term-4B (amino acids 332–611) were subcloned into a modified pET28 vector (Novagen; T.Ø. Tange, unpubl.) containing TEV protease cleavable Protein A and His6 tags at the N- and C-terminal ends of the ORF, respectively. MLN51 (amino acids 1–400, amplified from a HeLa cell library; T.Ø. Tange, unpubl.) was subcloned into a modified pET28 vector (E. Hallacli and T.Ø. Tange, unpubl.) containing TEV protease cleavable GST and His6 tags at the N- and C-terminal ends of the ORF, respectively. Double tagging was necessary to remove partially degraded or partially translated proteins, and the removal of the C-terminal His6 tag was necessary to prevent nonspecific RNA binding by 4AI and 4AIII (data not shown).
Protein expression and purification
All plasmids were transformed into BL21-CodonPlus (DE3) Escherichia coli (Stratagene). Bacterial cultures were grown to OD600 ∼0.6 at 37°C. His6-4AI, 4AIII, His6-4AI-4AIII chimeras, 4B, ΔRRM-4B, C-term-4B, and His6-4H were induced with 0.1 mM IPTG for 1 to 3 h at 37°C, 4AI was induced with 1 mM IPTG for 2–3 h at 37°C, and MLN51 was induced with 0.5 mM IPTG overnight at 22°C. After pelleting, cells were sonicated in their respective purification buffers (described below) with protease inhibitors (Roche), except cells expressing 4AI, which were sonicated in purification buffer containing 200 mM KCl. Untagged 4AI was purified based on a previously described protocol (Lorsch and Herschlag 1998), except only two columns were employed: Blue Sepharose (GE Healthcare), followed by a MonoQ (GE Healthcare). His6-4AI, His6-4AI-4AIII chimeras, and His6-4H were purified on Ni-NTA agarose resin (Qiagen) following the manufacturer's instructions, except using a binding and washing buffer containing 50 mM HEPES (pH 7.5), 400 mM NaCl, 20 mM imidazole, and 3 mM β-mercaptoethanol. Bound proteins were eluted with the same buffer using a 20–400 mM imidazole gradient. The double-tagged proteins 4AIII, 4B, ΔRRM-4B, and C-term-4B were first purified using Ni-NTA agarose resin as above, then on IgG-beads (GE Healthcare) following the manufacturer's protocol, except using a binding and washing buffer containing 50 mM HEPES (pH 7.5), 300 mM NaCl, 0.5 mM EDTA, 10% glycerol, and 3 mM β-mercaptoethanol, followed by His6-TEV Protease (Invitrogen) cleavage following the manufacturer's instructions. The His6-TEV protease was removed by incubation with Ni-NTA agarose resin (Qiagen). Purification of double-tagged MLN51 was hindered by dimerization of degraded or partially translated protein through the GST tag. MLN51 was therefore purified using Ni-NTA agarose as above, except the chaotropic salt potassium thiocyanate (KSCN) was added to 1.5 M prior to binding to the Ni-NTA resin and was present during subsequent washes. Bound protein was eluted with 200 mM imidazole. MLN51 was further purified on a Glutathione Sepharose 4B resin (GE Healthcare) following the manufacturer's instructions and then cleaved by His6-TEV protease as above. The protease was removed by running the cleavage reaction on a MonoQ column (GE Healthcare) in 50 mM Tris (pH 8), 20 mM NaCl, 0.5 mM EDTA, and 3 mM DTT. MLN51 eluted at 275 mM. All proteins were stored in 20 mM Tris (pH 7.5), 10% glycerol, 100 mM KCl, 0.1 mM EDTA, and 2 mM DTT, except 4AI and MLN51, which were stored in the same buffer but without EDTA (both buffers referred to a Storage Buffer: SB).
Pull downs
RNA pull-down reactions typically contained 1 μM proteins, 1 μM 32 nt RNA (5′-GGACUACUACUACUACUAAUGCACCGUAAAGC-3′; Integrated DNA Technologies) with a 5′ biotin attached via an 18-atom internal PEG linker, 1.5 mM Mg2+•NTP, and 0.1 mg/mL BSA in Binding and Washing Buffer 1 (BWB1: 20 mM Tris at pH 7.5, 100 mM KCl, 10% glycerol, 2 mM DTT, 0.1% NP-40). Following 30 min incubation at room temperature, reactions were mixed with prewashed magnetic streptavidin beads (Dynabeads M-280 Streptavidin, Invitrogen) and rotated for 30 min at 4°C. The beads were then washed three times with BWB1, eluted by boiling in SDS-loading buffer, and unbound- and eluted-samples analyzed by SDS-PAGE.
Protein pull-down reactions were similar, except the RNA did not contain the 5′ biotin nor the PEG linker, and Binding and Washing Buffer 2 was used (BWB2: 20 mM Tris at pH 7.5, 100 mM KCl, 10% glycerol, 2 mM DTT, 0.1% Tween-20, and 20 mM imidazole). The beads were eluted with BWB2 containing 250 mM imidazole. In cases where a 12-nt RNA was used, the sequence was as in the helicase assay (see below).
Electrophoretic mobility shift assays
EMSA reactions (Fig. 1B,C,D) were performed initially in low pH, low salt buffer (Buffer B; 25 mM MES at pH 6, 15 mM KOAc, 2.5 mM MgCl2), because earlier studies found 4AI to bind RNA more tightly under these conditions (Lorsch and Herschlag 1998). However, when AMPPNP is present, this buffer is no longer required for efficient RNA binding (data not shown). Storage buffer without glycerol was used in all subsequent EMSA experiments (Figs. 2C,D, 4A, 5A). Reactions containing the indicated proteins, 0.1 mg/mL BSA, 32P-end-labeled RNA, and nucleotide were incubated for 20 min at room temperature. Glycerol was added to 8% prior to loading samples onto a 7.5%, 0.5× TBE nondenaturing polyacrylamide gel. The sequence of the 32-nt RNA used was as in the pull-down assays (see above).
Helicase assay
The duplex was composed of 32-nt RNA (see above) and 12-nt RNA (5′-GCUUUACGGUGC-3′; both from Integrated DNA Technologies). 32P-5′-end-labeled 12-nt RNA and unlabeled 32-nt RNA were mixed in a 1.75:1 molar ratio, and hybridized duplexes prepared as previously described (Peck and Herschlag 2003). Helicase assays were performed in Buffer B with 0.1–0.2 nM duplex, 1 mM Mg2+•ATP, 0.5 mg/mL BSA, and 1 μM of the indicated proteins. Reactions were incubated at 35°C, and at the indicated times, aliquots were added to an equal volume of loading buffer (1% SDS, 50 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue, 20% glycerol). Single stranded and duplex RNAs were resolved on 15% nondenaturing polyacrylamide gels.
RNase footprinting
Ten-microliter reactions containing 2 μM 40-mer poly(U) (Integrated DNA Technologies), 1 mM AMPPNP, and 0.5 mg/mL BSA in SB (minus glycerol) and the following proteins where indicated: 2 μM 4AI, 4H, 4B or 1 μM 4AIII and MLN51, were incubated for 20 min at room temperature. MNase was added followed by incubation for 20 min at 25°C. MNase was stopped by addition of EGTA to a final concentration of 10 mM. Remaining RNAs were 5′-end labeled with polynucleotide kinase and γ32P-ATP (Perkin Elmer), extracted, precipitated, and resolved on a 22.5% denaturing polyacrylamide gel. Densitometry traces were obtained using ImageJ (Abramoff et al. 2004).
Preferred RNA binding site size
Poly(U) RNA (GE Healthcare) was incubated at 85°C for 18 min in 50 mM sodium carbonate (pH 9.2) to generate a hydrolysis ladder. Following neutralization with 130 mM Tris (final concentration; pH 7.5), the ladder was 5′-end labeled with polynucleotide kinase and γ32P-ATP (PerkinElmer) and spun through a Centri-spin 10 column (Princeton Separations). Following EMSA with the hydrolysis ladder, shifted bands were excised from the native gel and extracted overnight in gel extraction buffer containing 0.3 M sodium acetate (pH 5.5), 1 mM EDTA, and 10% phenol (pH 4.3). Extracted RNAs were then ethanol precipitated in the presence of 10 mM MgCl2 and 50 μg yeast tRNA, resuspended in formamide loading buffer, and separated on an 18% denaturing polyacrylamide gel. Phosphorimages were analyzed using ImageQuant software (Molecular Dynamics/GE Healthcare).
ACKNOWLEDGMENTS
We thank E. Jankowsky, J. Lorsch, and G. Wagner for helpful discussions and communicating results prior to publication. We are grateful to H. Le Hir, J. Smiley, N. Sonenberg, and G. Wagner for their generous gifts of plasmids. We are thankful to Debbie Bakes for purifying the His6-eIF4H used throughout this study and for conducting preliminary studies with this protein. We are grateful to Thomas Ø. Tange for the constructs used in eIF4B and MLN51 subcloning and to Erinc Hallacli for subcloning MLN51. We thank Beth Stroupe for her advice and discussions throughout the work described here and Niko Grigorieff for generously allowing us to conduct experiments in his laboratory. Lastly, we are thankful to all the members of the Moore laboratory for their continuous support. This work was supported in part by NIH R01-GM35007 (M.J.M.) and NIH Training Grant GM07596 (A.C.B). M.J.M. is an HHMI Investigator.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1049608.
The differences in 4AIII's basal helicase activity and its ability to be stimulated by 4B could be the result of two mutations, P210S and R370Q, that were inadvertently introduced into the 4AIII cDNA by PCR cloning in the Li et al. (1999) paper. The version of 4AIII used here does not contain these mutations.
REFERENCES
- Abramoff, M.D., Magelhaes, P.J., Ram, S.J. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Andersen, C.B., Ballut, L., Johansen, J.S., Chamieh, H., Nielsen, K.H., Oliveira, C.L., Pedersen, J.S., Seraphin, B., Le Hir, H., Andersen, G.R. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- Ballut, L., Marchadier, B., Baguet, A., Tomasetto, C., Seraphin, B., Le Hir, H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat. Struct. Mol. Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- Beran, R.K., Bruno, M.M., Bowers, H.A., Jankowsky, E., Pyle, A.M. Robust translocation along a molecular monorail: The NS3 helicase from hepatitis C virus traverses unusually large disruptions in its track. J. Mol. Biol. 2006;358:974–982. doi: 10.1016/j.jmb.2006.02.078. [DOI] [PubMed] [Google Scholar]
- Bono, F., Ebert, J., Lorentzen, E., Conti, E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Degot, S., Regnier, C.H., Wendling, C., Chenard, M.P., Rio, M.C., Tomasetto, C. Metastatic Lymph Node 51, a novel nucleo-cytoplasmic protein overexpressed in breast cancer. Oncogene. 2002;21:4422–4434. doi: 10.1038/sj.onc.1205611. [DOI] [PubMed] [Google Scholar]
- Doepker, R.C., Hsu, W.L., Saffran, H.A., Smiley, J.R. Herpes simplex virus virion host shutoff protein is stimulated by translation initiation factors eIF4B and eIF4H. J. Virol. 2004;78:4684–4699. doi: 10.1128/JVI.78.9.4684-4699.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras, A.C., Raught, B., Sonenberg, N. Regulation of translation initiation by FRAP/mTOR. Genes & Dev. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- Holz, M.K., Ballif, B.A., Gygi, S.P., Blenis, J. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell. 2005;123:569–580. doi: 10.1016/j.cell.2005.10.024. [DOI] [PubMed] [Google Scholar]
- Jankowsky, E., Bowers, H. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res. 2006;34:4181–4188. doi: 10.1093/nar/gkl410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky, E., Fairman, M.E. RNA helicases—One fold for many functions. Curr. Opin. Struct. Biol. 2007;17:316–324. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Kapp, L.D., Lorsch, J.R. The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- Kawaoka, J., Jankowsky, E., Pyle, A.M. Backbone tracking by the SF2 helicase NPH-II. Nat. Struct. Mol. Biol. 2004;11:526–530. doi: 10.1038/nsmb771. [DOI] [PubMed] [Google Scholar]
- Kelly, R.C., Jensen, D.E., von Hippel, P.H. DNA “melting” proteins. IV. Fluorescence measurements of binding parameters for bacteriophage T4 gene 32-protein to mono-, oligo-, and polynucleotides. J. Biol. Chem. 1976;251:7240–7250. [PubMed] [Google Scholar]
- Kossen, K., Karginov, F.V., Uhlenbeck, O.C. The carboxy-terminal domain of the DExDH protein YxiN is sufficient to confer specificity for 23S rRNA. J. Mol. Biol. 2002;324:625–636. doi: 10.1016/s0022-2836(02)01140-3. [DOI] [PubMed] [Google Scholar]
- Kressler, D., de la Cruz, J., Rojo, M., Linder, P. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae . Mol. Cell. Biol. 1997;17:7283–7294. doi: 10.1128/mcb.17.12.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q., Imataka, H., Morino, S., Rogers G.W., Jr, Richter-Cook, N.J., Merrick, W.C., Sonenberg, N. Eukaryotic translation initiation factor 4AIII (eIF4AIII) is functionally distinct from eIF4AI and eIF4AII. Mol. Cell. Biol. 1999;19:7336–7346. doi: 10.1128/mcb.19.11.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch, J.R., Herschlag, D. The DEAD box protein eIF4A. 1. A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry. 1998;37:2180–2193. doi: 10.1021/bi972430g. [DOI] [PubMed] [Google Scholar]
- Methot, N., Pause, A., Hershey, J.W., Sonenberg, N. The translation initiation factor eIF-4B contains an RNA-binding region that is distinct and independent from its ribonucleoprotein consensus sequence. Mol. Cell. Biol. 1994;14:2307–2316. doi: 10.1128/mcb.14.4.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methot, N., Pickett, G., Keene, J.D., Sonenberg, N. In vitro RNA selection identifies RNA ligands that specifically bind to eukaryotic translation initiation factor 4B: The role of the RNA recognition motif. RNA. 1996a;2:38–50. [PMC free article] [PubMed] [Google Scholar]
- Methot, N., Song, M.S., Sonenberg, N. A region rich in aspartic acid, arginine, tyrosine, and glycine (DRYG) mediates eukaryotic initiation factor 4B (eIF4B) self-association and interaction with eIF3. Mol. Cell. Biol. 1996b;16:5328–5334. doi: 10.1128/mcb.16.10.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milburn, S.C., Hershey, J.W., Davies, M.V., Kelleher, K., Kaufman, R.J. Cloning and expression of eukaryotic initiation factor 4B cDNA: Sequence determination identifies a common RNA recognition motif. EMBO J. 1990;9:2783–2790. doi: 10.1002/j.1460-2075.1990.tb07466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina, H., Horn, D.M., Tang, N., Mathivanan, S., Pandey, A. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. 2007;104:2199–2204. doi: 10.1073/pnas.0611217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranda, T., Strong, W.B., Menaya, J., Fabbri, B.J., Hershey, J.W. Two structural domains of initiation factor eIF-4B are involved in binding to RNA. J. Biol. Chem. 1994;269:14465–14472. [PubMed] [Google Scholar]
- Nielsen, P.J., Trachsel, H. The mouse protein synthesis initiation factor 4A gene family includes two related functional genes which are differentially expressed. EMBO J. 1988;7:2097–2105. doi: 10.1002/j.1460-2075.1988.tb03049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble, C.G., Song, H. MLN51 stimulates the RNA-helicase activity of eIF4AIII. PLoS ONE. 2007;2:e303. doi: 10.1371/journal.pone.0000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, N., Miyajima, N., Sazuka, T., Tanaka, A., Kawarabayasi, Y., Sato, S., Nagase, T., Seki, N., Ishikawa, K., Tabata, S. Prediction of the coding sequences of unidentified human genes. I. The coding sequences of 40 new genes (KIAA0001-KIAA0040) deduced by analysis of randomly sampled cDNA clones from human immature myeloid cell line KG-1. DNA Res. 1994;1:27–35. doi: 10.1093/dnares/1.1.27. [DOI] [PubMed] [Google Scholar]
- Oberer, M., Marintchev, A., Wagner, G. Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes & Dev. 2005;19:2212–2223. doi: 10.1101/gad.1335305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang, P.S., Jankowsky, E., Planet, P.J., Pyle, A.M. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J. 2002;21:1168–1176. doi: 10.1093/emboj/21.5.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause, A., Sonenberg, N. Mutational analysis of a DEAD box RNA helicase: The mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck, M.L., Herschlag, D. Adenosine 5′-O-(3-thio)triphosphate (ATPγS) is a substrate for the nucleotide hydrolysis and RNA unwinding activities of eukaryotic translation initiation factor eIF4A. RNA. 2003;9:1180–1187. doi: 10.1261/rna.2103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, N.J., Rogers G.W., Jr, Hensold, J.O., Merrick, W.C. Further biochemical and kinetic characterization of human eukaryotic initiation factor 4H. J. Biol. Chem. 1999;274:35415–35424. doi: 10.1074/jbc.274.50.35415. [DOI] [PubMed] [Google Scholar]
- Richter-Cook, N.J., Dever, T.E., Hensold, J.O., Merrick, W.C. Purification and characterization of a new eukaryotic protein translation factor. Eukaryotic initiation factor 4H. J. Biol. Chem. 1998;273:7579–7587. doi: 10.1074/jbc.273.13.7579. [DOI] [PubMed] [Google Scholar]
- Rogers G.W., Jr, Richter, N.J., Lima, W.F., Merrick, W.C. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J. Biol. Chem. 2001;276:30914–30922. doi: 10.1074/jbc.M100157200. [DOI] [PubMed] [Google Scholar]
- Rogers G.W., Jr, Komar, A.A., Merrick, W.C. eIF4A: The godfather of the DEAD box helicases. Prog. Nucleic Acid Res. Mol. Biol. 2002;72:307–331. doi: 10.1016/s0079-6603(02)72073-4. [DOI] [PubMed] [Google Scholar]
- Rush, J., Moritz, A., Lee, K.A., Guo, A., Goss, V.L., Spek, E.J., Zhang, H., Zha, X.M., Polakiewicz, R.D., Comb, M.J. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- Schwer, B. A new twist on RNA helicases: DExH/D box proteins as RNPases. Nat. Struct. Biol. 2001;8:113–116. doi: 10.1038/84091. [DOI] [PubMed] [Google Scholar]
- Sengoku, T., Nureki, O., Nakamura, A., Kobayashi, S., Yokoyama, S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- Shibuya, T., Tange, T.O., Sonenberg, N., Moore, M.J. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat. Struct. Mol. Biol. 2004;11:346–351. doi: 10.1038/nsmb750. [DOI] [PubMed] [Google Scholar]
- Silverman, E., Edwalds-Gilbert, G., Lin, R.J. DExD/H-box proteins and their partners: Helping RNA helicases unwind. Gene. 2003;312:1–16. doi: 10.1016/s0378-1119(03)00626-7. [DOI] [PubMed] [Google Scholar]
- Singleton, M.R., Dillingham, M.S., Wigley, D.B. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- Staley, J.P., Guthrie, C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Tange, T.O., Shibuya, T., Jurica, M.S., Moore, M.J. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA. 2005;11:1869–1883. doi: 10.1261/rna.2155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner, N.K., Linder, P. DExD/H box RNA helicases: From generic motors to specific dissociation functions. Mol. Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- Tao, W.A., Wollscheid, B., O'Brien, R., Eng, J.K., Li, X.J., Bodenmiller, B., Watts, J.D., Hood, L., Aebersold, R. Quantitative phosphoproteome analysis using a dendrimer conjugation chemistry and tandem mass spectrometry. Nat. Methods. 2005;2:591–598. doi: 10.1038/nmeth776. [DOI] [PubMed] [Google Scholar]
- Theissen, B., Karow, A.R., Kohler, J., Gubaev, A., Klostermeier, D. Cooperative binding of ATP and RNA induces a closed conformation in a DEAD box RNA helicase. Proc. Natl. Acad. Sci. 2008;105:548–553. doi: 10.1073/pnas.0705488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein, D.C., Honore, E., Hemmati-Brivanlou, A. Epidermal induction and inhibition of neural fate by translation initiation factor 4AIII. Development. 1997;124:4235–4242. doi: 10.1242/dev.124.21.4235. [DOI] [PubMed] [Google Scholar]
- Yang, Q., Del Campo, M., Lambowitz, A.M., Jankowsky, E. DEAD-box proteins unwind duplexes by local strand separation. Mol. Cell. 2007;28:253–263. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]