Abstract
Methylation of the exocyclic amino group of guanine is a relatively common modification in rRNA and tRNA. Single methylation (N2-methylguanosine, m2G) is the second most frequently encountered nucleoside analog in Escherichia coli rRNAs. The most prominent case of dual methylation (N2,N2-dimethylguanosine, m2 2G) is found in the majority of eukaryotic tRNAs at base pair m2 2G26:A44. The latter modification eliminates the ability of the N2 function to donate in hydrogen bonds and alters its pairing behavior, notably vis-à-vis C. Perhaps a less obvious consequence of the N2,N2-dimethyl modification is its role in controlling the pairing modes between G and A. We have determined the crystal structure of a 13-mer RNA duplex with central tandem m2 2G:A pairs. In the structure both pairs adopt an imino-hydrogen bonded, pseudo-Watson–Crick conformation. Thus, the sheared conformation frequently seen in tandem G:A pairs is avoided due to a potential steric clash between an N2-methyl group and the major groove edge of A. Additionally, for a series of G:A containing self-complementary RNAs we investigated how methylation affects competitive hairpin versus duplex formation based on UV melting profile analysis.
Keywords: base mismatch; RNA methylation; RNA stability; hydration, X-ray crystallography
INTRODUCTION
Post-translational modifications of rRNA, mRNA, tRNA, and snRNA are ubiquitous and concern both the sugar and nucleobase moieties (Grosjean and Benne 1998; Rozenski et al. 1999). Methylation of bases and the ribose 2′-hydroxyl group are particularly common, but the functional purpose of modifications has remained mysterious in many cases. The rRNAs in Escherichia coli contain five m2G, three m5C, and four 2′-O-methylated residues among the 24 methylated nucleotides (Møller Andersen and Douthwaite 2006). Crystal structures of the E. coli ribosome (Schuwirth et al. 2005) allow detailed insights into the individual environment of the five m2G residues and site-specific RNA methyltransferases involved, and the potential functions of m2G residues in rRNAs have recently been reviewed (Sergiev et al. 2007). The m2G residues map to three clusters of modified nucleotides in the decoding and peptidyltransferase centers and the subunit interface. It is hypothesized that methylation either serves the formation of a hydrophobic contact with protein or RNA or a structural purpose such as the prevention of base triples involving the minor groove (Lesnyak et al. 2007; Sergiev et al. 2007). The m2 2G nucleotide is found in the bend between the dihydro-uridine stem and the anticodon stem in the vast majority of eukaryotic tRNAs (Edqvist et al. 1992; Grosjean and Benne 1998). At that location, m2 2G26 is paired with A44 and flanked by C27:G43 on one side and the m2G10-C25-G45 triple on the other. Both G10:C25 and the adjacent C11:G24 pair serve as identity elements for the dimethylating enzyme targeting G26. It has been pointed out that the presence of m2 2G in cytosolic tRNAs most likely prevents them from folding into atypical structures adopted by mitochondrial tRNAs (mtRNAs) that do not feature the six canonical base pairs in the anticodon stem (Steinberg and Cedergren 1995). This interpretation is supported by the fact that bacterial tRNAs are unable to adopt such alternative folds, thus precluding the need for dimethylation of G and explaining the absence of m2 2G in the tRNAs of bacteria (Edqvist et al. 1995). The presence of two methyl groups—unlike in the case of a single methyl in m2G—virtually eliminates the possibility of pairing with C and, indeed, m2 2G26 pairs exclusively with A, U, or G at position 44. In the alternate conformers, unmethylated G26 always pairs with a C (Steinberg and Cedergren 1995). This notion lends support to the idea that the tRNA-m2 2G26-methyltransferase prevents misfolding by eukaryotic tRNAs into an inactive form and can be considered an RNA chaperone (Rajkowitsch et al. 2007).
Several studies have probed the effects of nucleobase methylations on RNA duplex and hairpin stability. Incorporation of m2G was found to be isoenergetic with G in the duplex context as well as in GNRA (N = any nucleotide and R = purine) tetraloops (Rife et al. 1998). Thus, the m2G analog seemed to be equally stable as either the s-cis (N2-methyl group pointing in the direction of N1) or the s-trans rotamer (N2 methyl group pointing in the direction of N3). On the other hand, it is noteworthy that base methylation affects the equilibrium of the duplex-hairpin conversion with RNA oligonucleotides (Micura et al. 2001). The effects of base methylation on the duplex-hairpin conversion were systematically investigated with the self-complementary sequence r(CGCGAAUUCGCGA), which forms a stable Watson Crick base-paired duplex under a variety of buffer conditions. The sequence is forced to adopt a hairpin conformation if one of the central six nucleotides (N4–N10) is replaced by the corresponding methylated nucleotide, i.e., 1-methylguanosine (m1G), m2 2G, N 6,N 6-dimethyladenosine (m6 2A), or 3-methyluridine (m3U). Conversely, the duplex structure is retained and even stabilized by replacement of a central nucleotide with m2G or N 4-methylcytidine (m4C). By comparison, N 6-methyladenosine (m6A) represented a borderline case. Although generally a duplex-preserving modification, the data indicated that m6A in strand position 5 and at low-strand concentrations is able to bring about duplex-to-hairpin conversion. The role of base methylation on the conformation of the ribosomal helix-45 sequence motif (small subunit), r(GACCm2GGm6 2Am6 2AGGUC), was also assessed (Höbartner et al. 2002). It was demonstrated that tandem m6 2A modification in this oligoribonucleotide prevents duplex formation with complementary strands. Therefore, one can conclude that base methylation does not simply modulate the pairing type, but has the potential to substantially affect RNA structure by formation of different secondary structure motifs.
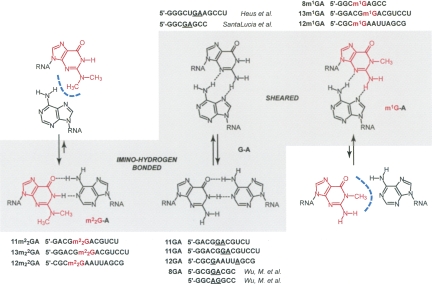
The starting point of the present study is a rational analysis of how methylation at the guanine nucleobase can affect the two most frequently encountered G:A mismatch pairing modes—namely, the sheared G:A and the imino-hydrogen bonded G:A conformations. Figure 1 illustrates how these pairing modes are affected by replacement of G with either m1G or m2 2G. The most obvious consequence for the pairing between G and A of single methylation at the N1 or double methylation at the N2 position is the restriction to a particular conformation. Whereas in the native state the G:A pair can in principle adopt either the sheared or the imino-hydrogen bonded conformations, m1G can only pair with A in the sheared conformation. Conversely, the presence of two methyl groups at N2 limits the relative orientations of the two base moieties to the imino-hydrogen bonded type.
FIGURE 1.
Methylation of guanine influences G:A pairing modes at neutral pH. Whereas in the nonmodified state the G:A pair can, in principle, adopt either the sheared or the imino-hydrogen bonded conformations, m1G can only pair with A in the sheared conformation. Conversely, the presence of two methyl groups at N2 limits the relative orientations of the two base moieties to the imino-hydrogen bonded type. Sequences that have been investigated in the present study are depicted next to their likely pairing mode.
In this context, our aim is to obtain high-resolution structures of RNAs containing methylated G:A base pairs to elucidate their base pairing and stacking interactions at the atomic level. In solution, methylation can affect hairpin/duplex equilibria of self-complementary sequences. However, it is not yet well understood whether this effect is a consequence of methylation itself (leading to increased hydrophobicity, and consequently, altered hydration) or rather the result of a change in the base-pairing mode (resulting in different enthalpic contributions). To shed light on these aspects, we carried out a series of UV-melting profile analyses for self-complementary oligoribonucleotide sequences with G:A mismatch pairs and the corresponding methylated counterparts. Although melting profiles deliver a qualitative rather than a quantitative picture, they nevertheless serve as a useful framework for the main focus of the present work, the crystal structure of a 13-mer RNA with central m2 2G:A pairs that was determined at 1.8 Å resolution. This structure allows detailed insights into the geometry of the m2 2G:A pair and its hydration relative to the native imino-type G:A pair, as well as the differences due to stacking compared with duplexes containing tandem G:A pairs of the sheared type (SantaLucia and Turner 1993; Heus et al. 1997; Jang et al. 2004).
RESULTS AND DISCUSSION
Influence of the m1G and m2 2G modifications paired with A on the thermal stability of RNA and the duplex-hairpin equilibrium
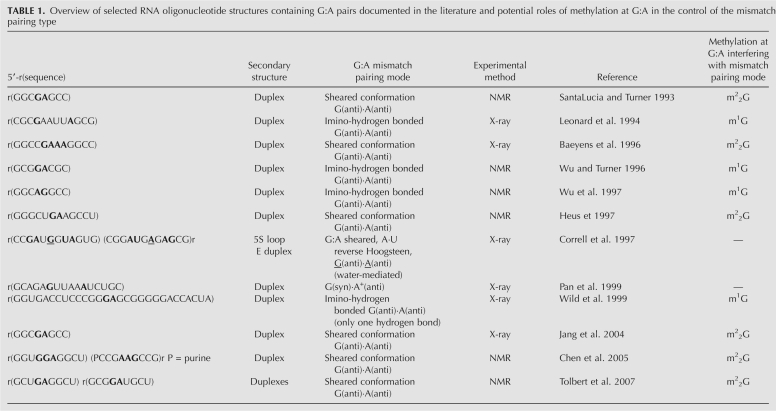
We initiated our investigation by selecting three self-complementary oligoribonucleotide sequences to assess how methylated guanosine residues opposite A (underlined below) affect RNA stability and secondary structure. These are the 11-mer 5′-GACGGACGUCU (11GA), the 12-mer 5′-CGCGAAUUAGCG (12GA), and the 13-mer 5′-GGACGGACGUCCU (13GA) (Fig. 1). All three sequences can, in principle, adopt hairpin and duplex secondary structures (Fig. 2). Both 11GA and 13GA feature 3′-terminal dangling Us and central tandem G:A pairs (duplex) or unpaired G and A within the loop (hairpin). In contrast, 12GA forms a blunt-end duplex or hairpin and single G:A pairs are separated by four central A:U pairs (duplex), or G:A forms the loop-closing base pair stacked onto the stem of the alternative hairpin structure. The corresponding methylated oligonucleotides are referred to as 11m2 2GA, 12m1GA, 12m2 2GA, 13m1GA, and 13m2 2GA (Fig. 1). In both the 11-mer and 13-mer RNAs, the sequence of the central tetramer is 5′-GGAC, and thus, G and A can be expected to pair via imino-hydrogen bonding in the duplex form. Tandem G:A pairs were generally found to exhibit this pairing type when the first G:A was preceded by G (as in 5′-GCGGACGC) or in cases where the positions of tandem G:A pairs were switched (as in 5′-GGCAGGCC) (Wu and Turner 1996). Conversely, tandem G:A pairs in RNAs containing, for example, the tetramers 5′-CGAG (SantaLucia and Turner 1993; Jang et al. 2004) or 5′-UGAA (Heus et al. 1997), displayed the sheared conformation (Fig. 1; Table 1). In the case of the 12GA duplex, the two isolated G:A mismatches adopted the imino-hydrogen bonded conformation as established by X-ray crystallography (Leonard et al. 1994; Li et al. 2007).
FIGURE 2.
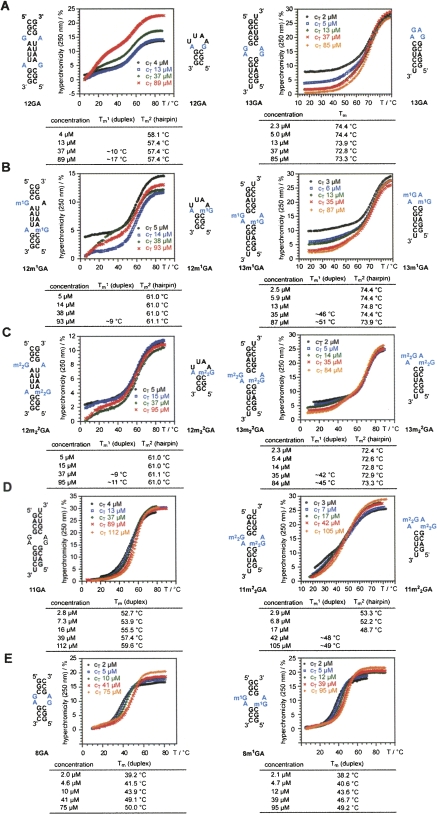
(A–E) UV melting profiles and thermal stabilities of nonmodified and methylated G:A containing oligoribonucleotides (see supporting information for derivative profiles). The secondary structures involved are indicated; for a detailed discussion see the main text. Conditions: 10 mM Na2HPO4, 150 mM NaCl (pH 7.0).
TABLE 1.
Overview of selected RNA oligonucleotide structures containing G:A pairs documented in the literature and potential roles of methylation at G:A in the control of the mismatch pairing type
We carried out concentration-dependent UV-melting experiments with a total of 10 self-complementary RNAs (12GA, 13GA, 12m1GA, 13m1GA, 12m2 2GA, 13m2 2GA, 11GA, 11m2 2GA, 8GA, and 8m1GA) to obtain a qualitative picture of the influence of G/A base-pair methylation on RNA secondary structure and stability (Fig. 2; Supplemental Fig. S1 [derivative profiles]). All RNAs were measured in a concentration range between ∼1 and ∼100 μM. Concentration dependence of Tm values versus concentration independence of Tm values was used as a primary criterion to distinguish between duplex and hairpin formation. In addition, hyperchromicity and the shape of the melting profiles provided further hints as to the (co)existence of hairpin and duplex secondary structures in solution. We are aware that for the quantification of hairpin/duplex ratios, other methods such as gel-shift assays under native conditions (Bernacchi et al. 2005, 2007; Ennifar et al. 2007; Sun et al. 2007) or NMR spectroscopy are required (Micura et al. 2001). Here, the set of RNAs was exclusively investigated by UV absorbance/temperature profiles. Nevertheless, this study provides a useful qualitative picture of how the sequence context in which the GA mismatch is embedded impacts secondary structure and stability of the respective RNAs. For example, 12GA shows a melting transition at 57.4°C that hardly changes with concentration (Fig. 2A, left). This behavior clearly refers to a hairpin transition in solution and is in accordance with the relatively small change in hyperchromicity of about 11%, due to melting of the small number of base pairs in the hairpin stem. At increasing RNA concentrations, a second transition appears for 12GA at around 15°C, indicating competing duplex formation. At first sight, the melting of the duplex appears low when compared with other GA mismatch containing duplexes (Morse and Draper 1995). However, the Watson–Crick paired regions in the 12GA duplex consist of two terminal CGC/GCG tracks and one central AAUU/UUAA track, separated by the G:A mismatches. These Watson–Crick paired duplex regions are at the nucleation limit (three to four nucleotides) (Saenger 1984). Moreover, the bimolecular species is a blunt-end duplex with two terminal G:C pairs, prone to base-pair fraying. This feature, together with a central track that consists of “weak” A:U pairs, may constitute the major reason for the low melting of the duplex form of 12GA.
When G is replaced by m1G, the melting transition for 12m1GA is observed at 61°C (increase of 3.5°C compared with 12GA) and remains concentration independent (Fig. 2B, left). Also, the hyperchromicity is comparable to nonmodified 12GA (∼11%), thus supporting hairpin formation. For higher concentrations (>40 μM) a second transition emerges around 10°C, pointing to duplex formation. Likewise, replacement of G by m2 2G (12m2 2GA) yields a melting behavior comparable to nonmodified 12GA: The N2 doubly methylated guanine increases hairpin stability by the same amount as m1G does (Fig. 2C, left). Concerning the duplex stabilities of 12GA, 12m1GA, and 12m2 2GA, the shapes of the individual transitions at high concentrations allow a rough estimation, suggesting a destabilization of the duplex by methylation.
The 13GA RNA shows a melting behavior that—at first sight—implies a hairpin structure due to concentration independence of the melting process (Fig. 2A, right). A more detailed analysis of the profiles at different concentrations suggests that dominant hairpin formation is encountered only at low concentrations according to the small hyperchromicity of ∼18% (2 μM) compared with ∼28% at the highest concentration measured (85 μM). Moreover, deviation from an ideal sigmoid shape of the profile at 5-μM concentration indicates competition with an alternative structure. The slight “step” around 50°C moves to higher temperatures with increasing RNA concentration and suggests duplex formation with a Tm value in the same range as the hairpin's Tm. This interpretation makes sense when compared with the melting behavior of the corresponding methylated counterparts 13m1GA (Fig. 2B, right) and 13m2 2GA (Fig. 2C, right). For these modified RNAs, the concentration-dependent, “low hyperchromicity” transition is observed at lower temperatures. Thereby, it is significantly separated from the concentration-independent, “high hyperchromicity” transition, allowing more accurate determination of Tm values for the two melting transitions. To summarize, the thermal hairpin stability of 13GA remains rather unaffected by m1G replacement (13m1GA) and is slightly destabilized (1°C) by m2 2G replacement (13m2 2GA). Concerning duplex stabilities within this series, the melting profile analysis suggests significant destabilization, which is more pronounced for replacement of G by m2 2G than for replacement by m1G.
The 13GA sequence differs from 11GA by formation of an additional G:C base pair at the termini of the duplex or respective hairpin. The effects of the shortening are dramatic in that the melting profiles for 11GA show nearly ideal sigmoid shapes together with a clear concentration dependence of the Tm values, characteristic for duplex formation (Fig. 2D, left). Also, the hyperchromicity of about 30% is consistent with the higher number of base pairs that are broken (upon melting of the duplex) and is significantly larger compared with the 18% hyperchromicity observed for the hairpin-forming 13GA RNA (at low concentration) (Fig. 2A, right). Interestingly, replacement of G by m2 2G in 11GA results in an ambivalent melting behavior (reminiscent of that triggered by N6 methylation of adenosine-5 in rCGCGAAUUCGCGA; Micura et al. 2001). At low RNA concentrations, high Tm values are observed with small hyperchromicity. By increasing the concentration of the RNA, Tm values first decrease and then increase again. Such a behavior is characteristic for a mixture of hairpin and duplex structures with duplex Tm values slightly lower than the respective hairpin's Tm values (Micura et al. 2001).
In addition to the series of 11GA, 12GA, 13GA, and the corresponding methylated counterparts, we investigated a short eight-nucleotide, tandem GA sequence motif where hairpin formation is significantly less likely compared with duplex formation because of a very short (and therefore less stable) stem region in the hairpin alternative. Indeed, both 8GA and 8m1GA show nearly ideal sigmoid melting profiles, with a pronounced concentration dependence (Fig. 2E). These RNAs exist as duplexes and their melting profiles are included for comparison with those of longer sequences and support our argumentation above. Both 8GA and 8m1GA show two-state melting behavior (as opposed to most of the RNAs discussed above) and the thermodynamic data can be determined reliably (Supplemental Fig. S2).
Crystal structure of a 13-mer RNA duplex with tandem m2 2G:A pairs
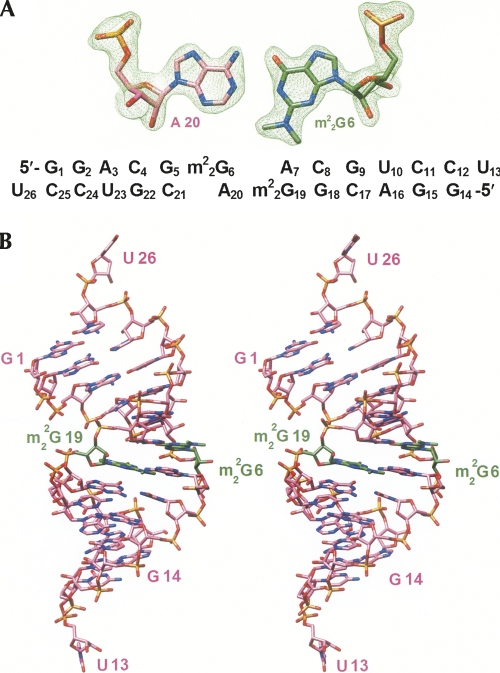
To gain insight into the conformational consequences of tandem m2 2G:A pairs in an RNA duplex and the geometrical differences between G:A and m2 2G:A base pairs, we determined the crystal structure of the 13m2 2GA duplex at 1.8 Å resolution. The structure was determined by the Molecular Replacement technique using a canonical 12-mer duplex with Watson–Crick base pairs as the search model. An example of the quality of the final electron density, as well as the sequence and numbering of residues, are depicted in Figure 3A (residues in strands 1 and 2 of the duplex are numbered G1 to U13 and G14 to U26, respectively). Selected crystal data, data collection, and crystallographic refinement parameters are summarized in Table 2. The A-form duplex with average values of 2.69 Å and 34° for helical rise and twist, respectively, and an inclination of 13.7° between base pairs and helix axis displays an increased diameter (∼2 Å) in the region of the purine:purine pairs. All sugars are in the expected C3′-endo conformation, and the only change at the global level beside the widening in the center is a slight bend into the major groove (Fig. 3B), adjacent to the tandem m2 2G:A pairs. At the G5:C21/m2 2G6:A20 base-pair step, the helix axis is bent by ∼9.5°. Terminal uridines are unstacked and engage in lattice interactions with neighboring duplexes (see Supplemental Fig. S3A,B). The base moiety of U13 is sandwiched between the phosphate group of G2# from a symmetry-related duplex and the 2′-hydroxyl group of C17## from a third duplex (the distances between O1P# and O2′## and the center of the uracil ring are 3.21 Å and 3.08 Å, respectively) (Supplemental Fig. S3A). In addition, the 2′-hydroxyl group of U13 forms a hydrogen bond with the above phosphate oxygen (2.71 Å). At the other end, U26 is tucked against the minor grooves of two neighboring duplexes (Supplemental Fig. S3B). In the case of the first symmetry mate (#), the interactions concern the center of the minor groove, in close vicinity to m2 2G:A pairs. The 2′-hydroxyl group of U26 forms hydrogen bonds with both N3 and O2′ of A7# (2.98 and 3.04 Å, respectively) and, in addition, forms a contact to O2′ of G22## from a second symmetry mate (distance 3.42 Å). The 2′-hydroxyl group of A7# is also involved in a hydrogen bond with O3′ (U26; 2.77 Å). That 3′-terminal hydroxyl group then establishes a further contact with the first symmetry-related duplex (O4′ of C8#; 3.19 Å). In turn, O4′ of U26 lies also relatively close to one of the N2-methyl groups of m2 2G19# (3.42 Å; Supplemental Fig. S3B).
FIGURE 3.
(A) Quality of the final Fourier (2Fo-Fc) sum electron density map and sequence of the 13m2 2GA RNA. (B) Stereo diagram of the overall geometry of the m2 2G-modified duplex across the major and minor grooves; carbon atoms of m2 2G nucleotides are highlighted in green and selected residues are labeled.
TABLE 2.
Selected crystal data and refinement parameters
The presence of tandem m2 2G:A pairs leads to several conformational changes at the local level. It is likely that some of them are not directly related to N2,N2-dimethylation of G, but are mere adjustments of the A-form sugar-phosphate backbone required to accommodate the wider imino-type purine:purine pairs. For example, backbone torsion angles α, β, ε, and ζ of G5 that is located 5′-adjacent to m2 2G:A pairs exhibit significant changes relative to the conformations of corresponding angles in all other residues (see Supplemental material file, output of the program CURVES [Lavery and Sklenar 1989]). The α angle adopts the -sc conformation, but is contracted by ∼10° (−58°), β changes from the ap conformation to +ac (139°), ε (normally ap) becomes ac− (−127°), and ζ opens up from −sc to -ac (−112°). Interestingly, these variations occur in the same region as the kink into the major groove. Thus, it is possible that the angle changes underlie the compression of the major groove, and that bending provides an indication for an increased plasticity of the RNA duplex near tandem G:A pairs. Judging from the so-called local interbase parameters, the duplex is also underwound at that site, as the helical twist between base pairs G5:C21/m2 2G6:A20 is only 14° (the roll of 21° and the shift, Dx, of –1.8 Å there represent the highest values for the respective parameters in the entire duplex). On the other hand, the twist at the adjacent m2 2G6:A20/A7:m2 2G19 step is increased (37°) relative to the average value of 33° for A-form RNA duplexes.
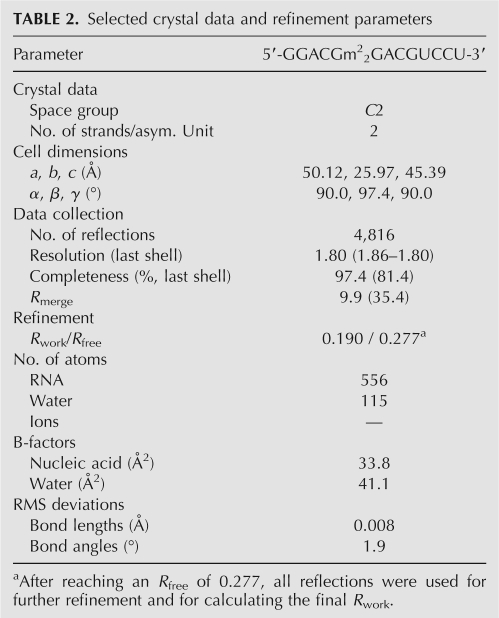
The geometry of m2 2G:A pairs differs significantly from that of G:A base pairs. In order to compare the two, we used the G:A pairs in the 12GA duplex, whose crystal structure had previously been determined at 1.12 Å resolution as a reference (Li et al. 2007). The 12GA duplex is located on a crystallographic twofold rotation axis in space group C2, and therefore the two G:A pairs assume an identical geometry. The two m2 2G:A pairs in the 13m2 2GA duplex, the G:A pair in the 12GA duplex, and a superimposition of the m2 2G6:A20 and the native pair are depicted in Figure 4. A common feature of these purine:purine pairs is the virtual absence of propeller twisting that amounts to about −15° on average for other base pairs in the two duplexes. The lengths of hydrogen bonds between (G)N1-H and N1(A) and (G)O6 and H-N6(A) are practically the same in the G:A pair (2.91 Å and 2.87 Å, respectively) (Fig. 4A). The base-pair opening amounts to −10° (toward the major groove), and no appreciable shearing between the G and A bases was observed. Conversely, the lengths of the above hydrogen bonds in m2 2G:A pairs are different; the (G)N1-H…N1(A) bond is longer than the (G)O6…H-N6(A) bond (3.24 Å vs. 2.75 Å in the m2 2G6:A20 pair and 3.15 Å vs. 2.84 Å in the A7:m2 2G19 pair). The m2 2G6:A20 pair displays an opening of −35° (Fig. 4B,D). The A7:m2 2G19 pair also shows a significant opening of 23° (Fig. 4C). Undoubtedly, the geometric perturbations in the N2,N2-dimethylated G:A pairs help minimize potentially repulsive interactions between methyl groups and atoms from paired or stacked residues. However, the lengthening of hydrogen bonds in m2 2G:A pairs comes at a price and can be expected to reduce RNA thermodynamic stability, consistent with the lower Tm values measured for duplexes with m2 2G:A pairs compared with those harboring G:A pairs.
FIGURE 4.
Geometries and hydration (red spheres) of the G104:A209 (12GA duplex) (Li et al. 2007) (A), m2 2G6:A20 (B), and A7:m2 2G19 (C) base pairs. Hydrogen bonds are dashed lines. (D) Superimposition of the m2 2G6:A20 and G104:A209 pairs.
Water structure around m2 2G:A and G:A pairs and comparison with the geometry of the m2 2G26:A40 pair in tRNAs
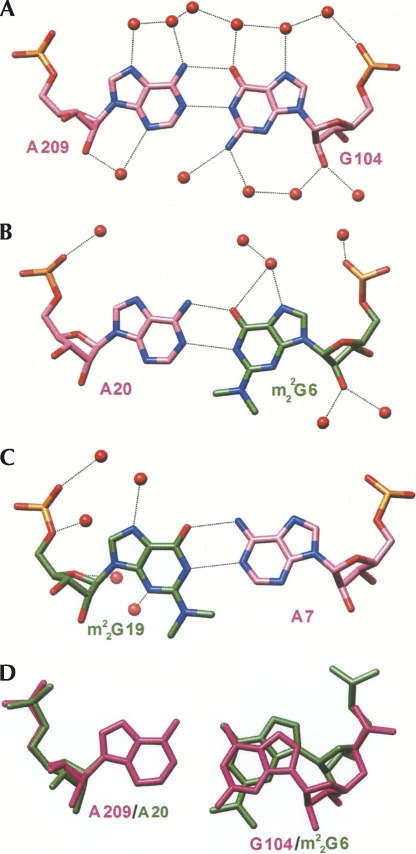
Hydration patterns around the native and N2,N2-dimethylated G:A pairs are depicted in Figure 4. As expected, the minor groove around m2 2G:A pairs is relatively dry. In principle, N3 of A can form a hydrogen bond to a water molecule. However, one of the dangling 3′-terminal uridines of a neighboring duplex resides in the minor groove, in close vicinity to the tandem m2 2G:A pairs (see Supplemental Fig. S3B). Thus, N3 of A7# is engaged in a hydrogen-bonding interaction with O2′ of U26 (see above). Thus, the presence of methyl groups disrupts the water structure in the center of the minor groove. In particular, water bridges with 2′-hydroxyl groups from residues on opposite strands serving as bridgeheads are missing (Egli et al. 1996). The closest contacts involving N2-methyl groups are observed to C2-H of the pairing partner (3.45 Å, m2 2G6…A20; 3.27 Å, m2 2G19…A7) and the O2 keto oxygen atoms of cross-strand cytosines stacked above A7 (3.32 Å, C8…m2 2G19) and A20 (3.81 Å, C21…m2 2G6) (Fig. 5A). By comparison, the major groove exhibits a regular hydration pattern around both the G:A and m2 2G:A pairs (Fig. 4). Some of the observed changes may be due to the different resolutions at which the two structures were determined (1.12 Å vs. 1.8 Å). However, the overall reduced hydration around m2 2G:A pairs is consistent with the detrimental effect of methylation on thermodynamic stability.
FIGURE 5.
Interactions of N2-methyl groups. Stereo diagrams of the tandem m2 2G:A pairs in the 13m2 2GA structure (A) and base pairs m2 2G26:A44 and C27:G43 in tRNAPhe (PDB code 1ehz) (Shi and Moore 2000) (B). Dashed lines are hydrogen bonds (including a possible C-H…π interaction in B) and thin solid lines are van der Waals interactions or putative C-H…O hydrogen bonds (see text for distance information).
The single imino-hydrogen bonded m2 2G26:A44 pair in tRNA (i.e., tRNAPhe [Shi and Moore 2000]) exhibits a geometry that differs significantly from that of the tandem m2 2G:A pairs in the 13m2 2GA duplex. Unlike the tandem pairs that feature minimal propeller twist and buckling, the m2 2G26:A44 pair shows considerable propeller twisting and buckling. In addition, m2 2G26 exhibits a relatively loose stacking interaction with the adjacent C27 residue (Fig. 5B). Instead, the main stacking interaction is between m2 2G26 and m2G10. And, unlike with m2 2G:A pairs in the structure of the 13m2 2GA duplex, the lengths of hydrogen bonds between (m2 2G)N1-H and N1(A) (2.87 Å) and (m2 2G)O6 and H-N6(A) (2.84 Å) are nearly the same. Thus, instead of base-pair shearing and opening as seen with tandem m2 2G:A pairs, propeller twisting and buckling here may serve to relieve a potentially short contact between the N2-methyl group and C2-H of A. No water molecules are found in the vicinity of methyl groups and the shortest contacts are to C2-H of A44 (3.26 Å) and the imidazole ring of m2G10 (3.29 Å to the ring centroid). In the latter case, the N2-methyl group pointing away from A44 sits directly above that ring of m2G10 and is most likely engaged in a stabilizing H…π interaction (Fig. 5B).
CONCLUSIONS
Methylation of nucleobases can affect RNA structure and function in multiple ways, i.e., by controlling secondary and/or tertiary structure and interactions with proteins. We show here that m2 2G has a destabilizing effect on RNA duplexes in cases where the N2,N2-dimethylated G forms an imino-hydrogen bonded pair with A. This destabilization occurs independently of whether m2 2G:A exists in an isolated form embedded in an otherwise canonical Watson–Crick base-paired stem or as part of a tandem in the central section of an RNA duplex. Depending on the sequence context and the length of the RNA, m2 2G can drive the duplex-hairpin equilibrium toward the hairpin. Double methylation of the exocyclic amino group of guanine affects pairing with A in a variety of ways. The crystal structure of an RNA duplex with m2 2G:A pairs determined here reveals subtle changes of the imino-hydrogen-bonded pairing geometry as a result of methylation. The observed differences in the case of tandem m2 2G:A pairs differ somewhat from those seen in the case of the single pair in many eukaryotic tRNAs. Nevertheless, our observations with regard to stability, pairing mode, and pairing geometry of m2 2G:A pairs support the conclusion that this G analog does not just prevent pairing of G26 with C in the tRNA D-stem, but also controls the pairing mode with A. Specifically, m2 2G rules out the common sheared orientation between G and A and limits the pairing mode to the more stable imino-hydrogen bonded form.
MATERIALS AND METHODS
Oligonucleotide synthesis and purification
The modified nucleotide phosphoramidites m1G and m2 2G were synthesized according to published procedures (Höbartner et al. 2003) and were incorporated into oligoribonucleotides by automated RNA solid-phase synthesis. In all cases, the synthesis was at the 1.3 μM scale and the ribose 2′-hydroxyl group of the building blocks was protected by the [(triisopropylsilyl)oxy]methyl (TOM) group (Pitsch et al. 2001). All oligonucleotides were deprotected and cleaved from solid support using the following conditions: (1) CH3NH2 in ethanol (8 M, 700 μL) and CH3NH2 in water (41%, 700 μL) for 6 h; (2) TBAF·3H2O in THF (1 M, 950 μL) for 12 h; and (3) neutralization with triethylammonium acetate buffer in water (1 M, 950 μL). The samples were desalted using a Sephadex G10 column (30 × 1.5 cm), monitored by UV detection at 270 nm and elution with water, followed by evaporation to dryness. For purifications we used ion-exchange chromatography (semipreparative Dionex DNAPac column, 9 × 250 mm at 80°C, flow rate 2 mL/minute, detection at 265 nm) with buffers A and B being 25 mM Tris-HCl, 6 M urea, in water (pH 8.0), and 25 mM Tris-HCl, 0.5 M NaOCl4, 6 M urea, in water (pH 8.0), respectively. Following purification, fractions were desalted once again by loading onto a C18 SepPak cartridge (Waters/Millipore), followed by elution with aqueous 0.1–0.2 M (Et3NH)HCO3 and then water/CH3CN (6:4, v/v). The combined fractions containing the oligonucleotide triethylammonium salts were lyophilized to dryness and analyzed by MALDI-TOF.
UV melting experiments
Absorbance versus temperature profiles were recorded at 250, 260, 265, and 270 nm on a Varian Cary-1 spectrophotometer equipped with a multiple cell holder and a Peltier temperature-control device. Each oligonucleotide was measured at four or five different concentrations ranging from about 1 to 100 μM in buffer solutions of 10 mM Na2HPO4 (pH 7.0) containing 150 mM NaCl. Data were collected after a complete cooling and heating cycle at a rate of 0.7°C/minute. Melting transitions were reversible for all sequences and essentially the same with respect to the four different wavelengths. Tm values were determined by calculating the first derivative (all corresponding curves depicted in the Supporting Information) and represent the mean of at least three individual measurements. Variation of Tm values within such a series was always less than ±0.2 K.
Sample preparation: Oligonucleotides were lyophilized to dryness, dissolved in the corresponding buffer from stock solutions, and subsequently degassed in the quartz cuvette. A layer of silicon oil was placed on the surface of the solution.
Crystallization and diffraction data collection
The concentrations of oligonucleotide stock solutions were adjusted to ∼1 mM in water, and crystallization experiments were performed with the Nucleic Acid Miniscreen (Hampton Research) (Berger et al. 1996) using the hanging-drop vapor diffusion technique. Diffraction-quality crystals could only be grown for the self-complementary 13-mer with central tandem m2 2G:A mismatches. Crystals for diffraction data collection were obtained from 2-μL droplets containing 0.5 mM oligonucleotide, 5% 2-methyl-2,4-pentanediol (MPD), 20 mM sodium cacodylate (pH 7.0), 6 mM spermine-4HCl, and 40 mM sodium chloride, that were equilibrated against a reservoir of 1 mL of 35% MPD at 291 K. Crystals appeared in about a week's time. Crystals were mounted in cryo-loops without further protection and diffraction data were collected at 120 K on the bending magnet beamline of the Southeast Regional Collaborative Access Team (SER-CAT, 22-BM) at the Advanced Photon Source (APS). Data were processed with XGEN (Howard 2000) and selected crystal data and data collection parameters are summarized in Table 2.
Structure determination and refinement
The structure was determined by the Molecular Replacement method with the program MOLREP (CCP4 1994; Vagin and Teplyakov 1997) using a canonical A-RNA dodecamer search model (residues U13 and U26 omitted) that was generated with the program TURBO-FRODO (Cambillau and Roussel 1997). Initial refinement cycles were carried out with the program CNS (Brunger et al. 1998) by performing simulated annealing, followed by a few cycles of gradient minimization and refinement of individual isotropic temperature factors. Water molecules (∼15 in each cycle) were added into regions of superimposed (2Fo−Fc) sum and (Fo−Fc) difference Fourier electron density after the gradient minimization cycles. At this stage, the terminal uridine residues (U13 and U26) could be clearly traced in the electron density map. Further isotropic and TLS refinements (Winn et al. 2001) were performed using the program REFMAC (Murshudov et al. 1997), followed by inspection of the electron density and addition of water molecules. After additional cycles of refinement, the N2,N2-dimethylguanosine residues were added and the dictionary file for REFMAC adapted. The dictionary file was generated with the program PRODRG (version 041117.0531; Schuettelkopf and van Aalten 2004) and the refinement mode was then made anisotropic. After reaching an R free of 0.277, all reflections were included in the final rounds of refinement. Final refinement parameters are listed in Table 2. Illustrations in Figures 3, 4, and 5 were generated with the program UCSF Chimera (Pettersen et al. 2004).
Coordinates and structure factors
Final coordinates and structure factors for the 13m2 2GA duplex have been deposited in the Protein Data Bank (http://www.rcsb.org): PDB ID code 3cjz.
SUPPLEMENTAL DATA
Derivative graphs of UV melting profiles and thermal stabilities of nonmodified and methylated G:A containing oligoribonucleotides, illustrations of packing interactions involving 3′-terminal uridines, CD spectra, and the output file with helical parameters for the 13m2 2GA duplex derived with the program CURVES can be accessed online at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We acknowledge financial support from the US National Institutes of Health (grant no. R01 GM55237 to M.E.), the Austrian Science Fund FWF (no. P17864 to R.M.), and the bm:wf (Gen-AU program; project “Noncoding RNAs” no. P7260-012-011 to R.M.). R.M. thanks Claudia Höbartner (MPI Göttingen) for stimulating discussions. Crystallographic data were collected on the 22-BM beamline of the Southeast Regional Collaborative Access Team (SER-CAT) at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at www.ser-cat.org/members.html. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract no. W-31-109-Eng-38.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1078508.
REFERENCES
- Baeyens, K.J., De Bondt, H.L., Pardi, A., Holbrook, S.R. A curved RNA helix incorporating an internal loop with G.A and A.A non-Watson–Crick base pairing. Proc. Natl. Acad. Sci. 1996;93:12851–12855. doi: 10.1073/pnas.93.23.12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger, I., Kang, C.H., Sinha, N., Wolters, M., Rich, A. A highly efficient 24-condition matrix for the crystallization of nucleic acid fragments. Acta Crystallogr. D Biol. Crystallogr. 1996;52:465–468. doi: 10.1107/s0907444995013564. [DOI] [PubMed] [Google Scholar]
- Bernacchi, S., Ennifar, E., Tóth, K., Walter, P., Langowski, J., Dumas, P. Mechanism of hairpin-duplex conversion for the HIV-1 dimerization initiation site. J. Biol. Chem. 2005;280:40112–40121. doi: 10.1074/jbc.M503230200. [DOI] [PubMed] [Google Scholar]
- Bernacchi, S., Freisz, S., Maechling, C., Spiess, B., Marquet, R., Dumas, P., Ennifar, E. Aminoglycoside binding to the HIV-1 RNA dimerization initiation site: Thermodynamics and effect on the kissing-loop to duplex conversion. Nucleic Acids Res. 2007;35:7128–7139. doi: 10.1093/nar/gkm856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cambillau, C., Roussel, A. Turbo Frodo, Version OpenGL.1. Université Aix-Marseille II; Marseille, France: 1997. [Google Scholar]
- CCP4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Chen, G., Znosko, B.M., Kennedy, S.D., Schroeder, S.J., Krugh, T.R., Turner, D.H. Solution structure of an RNA internal loop with three consecutive sheared GA pairs. Biochemistry. 2005;45:2845–2856. doi: 10.1021/bi048079y. [DOI] [PubMed] [Google Scholar]
- Correll, C.C., Freeborn, B., Moore, P.B., Steitz, T.A. Metals, motifs, and recognition in the crystal structure of a 5S rRNA domain. Cell. 1997;91:705–712. doi: 10.1016/s0092-8674(00)80457-2. [DOI] [PubMed] [Google Scholar]
- Edqvist, J., Grosjean, H., Sträby, K.B. Identity elements for N2-dimethylation of guanosine-26 in yeast tRNAs. Nucleic Acids Res. 1992;20:6575–6581. doi: 10.1093/nar/20.24.6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edqvist, J., Sträby, K.B., Grosjean, H. Enzymatic formation of N2,N2-dimethylguanosine in eukaryotic tRNA: Importance of the tRNA architecture. Biochimie. 1995;77:53–60. doi: 10.1016/0300-9084(96)88104-1. [DOI] [PubMed] [Google Scholar]
- Egli, M., Portmann, S., Usman, N. RNA hydration: A detailed look. Biochemistry. 1996;35:8489–8494. doi: 10.1021/bi9607214. [DOI] [PubMed] [Google Scholar]
- Ennifar, E., Bernacchi, S., Wolff, P., Dumas, P. Influence of C-5 halogenation of uridines on hairpin versus duplex RNA folding. RNA. 2007;13:1445–1452. doi: 10.1261/rna.408507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Benne R., editors. Modification and editing of RNA. ASM Press; Washington, DC: 1998. [Google Scholar]
- Heus, H.A., Wijmenga, S.S., Hoppe, H., Hilbers, C.W. The detailed structure of tandem G.A mismatched base-pair motifs in RNA duplexes is context dependent. J. Mol. Biol. 1997;271:147–158. doi: 10.1006/jmbi.1997.1158. [DOI] [PubMed] [Google Scholar]
- Höbartner, C., Ebert, M.-O., Jaun, B., Micura, R. RNA two-state conformation equilibria and the effect of nucleobase methylation. Angew. Chem. Int. Ed. 2002;41:605–609. [Google Scholar]
- Höbartner, C., Kreutz, C., Flecker, E., Ottenschlaeger, E., Pils, W., Grubmayr, K., Micura, R. The synthesis of 2′-O-[(triisopropylsilyl)oxy]methyl (TOM) phosphoramidites of methylated ribonucleosides (m1G, m2G, m2 2G, m1I, m3U, m4C, m6A, m6 2A) for use in automated RNA solid-phase synthesis. Chemical Monthly. 2003;134:851–873. [Google Scholar]
- Howard, A.J. Data processing in macromolecular crystallography. In: Bourne P.E., Watenpaugh K.D., editors. Crystallographic computing 7: Proceedings from the macromolecular crystallographic computing school. Oxford University Press; Oxford, UK: 2000. pp. 150–165. [Google Scholar]
- Jang, S., Baeyens, K., Jeong, M., SantaLucia J., Jr, Turner, D., Holbrook, S.R. Structure of two RNA octamers containing tandem G.A base pairs. Acta Crystallogr. D Biol. Crystallogr. 2004;60:829–835. doi: 10.1107/S0907444904003804. [DOI] [PubMed] [Google Scholar]
- Lavery, R., Sklenar, H. Defining the structure of irregular nucleic acids: Conventions and principles. J. Biomol. Struct. Dyn. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- Leonard, G.A., McAuley-Hecht, K.E., Ebel, S., Lough, D.M., Brown, T., Hunter, W.N. Crystal and molecular structure of r(CGCGAAUUAGCG): An RNA duplex containing two G(anti).A(anti) base pairs. Structure. 1994;2:483–494. doi: 10.1016/S0969-2126(00)00049-6. [DOI] [PubMed] [Google Scholar]
- Lesnyak, D.V., Osipiuk, J., Skarina, T., Sergiev, P.V., Bogdanov, A.A., Edwards, A., Savchenko, A., Joachimiak, A., Dontsova, O.A. Methyltransferase that modifies guanine 966 of the 16S rRNA. Functional identification and tertiary structure. J. Biol. Chem. 2007;282:5880–5887. doi: 10.1074/jbc.M608214200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, F., Pallan, P.S., Maier, M.A., Rajeev, K.G., Mathieu, S.L., Kreutz, C., Fan, Y., Sanghvi, J., Micura, R., Rozners, E., et al. Crystal structure, stability and in vitro RNAi activity of oligoribonucleotides containing the ribodifluorotoluyl nucleotide: Insights into substrate requirements by the human RISC Ago2 enzyme. Nucleic Acids Res. 2007;35:6424–6438. doi: 10.1093/nar/gkm664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micura, R., Pils, W., Höbartner, C., Grubmayr, K., Ebert, M.-O., Jaun, B. Methylation of the nucleobases in RNA oligonucleotides mediates duplex-hairpin conversion. Nucleic Acids Res. 2001;29:3997–4005. doi: 10.1093/nar/29.19.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller Andersen, N., Douthwaite, S. YebU is a m5C methyltransferase specific for 16 S rRNA nucleotide 1407. J. Mol. Biol. 2006;359:777–786. doi: 10.1016/j.jmb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Morse, S.E., Draper, D.E. Purine-purine mismatches in RNA helices: Evidence for protonated G.A pairs and next-nearest neighbor effects. Nucleic Acids Res. 1995;23:302–306. doi: 10.1093/nar/23.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Pan, B., Mitra, S.N., Sundaralingam, M. Crystal structure of an RNA 16-mer duplex r(GCAGAGUUAAAUCUGC)2 with nonadjacent G(syn).A+(anti) mispairs. Biochemistry. 1999;38:2826–2831. doi: 10.1021/bi982122y. [DOI] [PubMed] [Google Scholar]
- Pettersen, E.F., Goddard, T.D., Huang, C.C., Couch, G.S., Greenblatt, D.M., Meng, E.C., Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pitsch, S., Weiss, P.A., Jenny, L., Stutz, A., Wu, X. Reliable chemical synthesis of oligoribonucleotides (RNA) with 2′-O-[(triisopropylsilyl)oxy]methyl (2′-O-tom) protected phosphoramidites. Helv. Chim. Acta. 2001;84:3773–3795. [Google Scholar]
- Rajkowitsch, L., Chen, D., Stampfl, S., Semrad, K., Waldsich, C., Mayer, O., Jantsch, M.F., Konrat, R., Bläsi, U., Schroeder, R. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- Rife, J.P., Cheng, C.S., Moore, P.B., Strobel, S.A. N2-Methylguanosine is iso-energetic with guanosine in RNA duplexes and GNRA tetraloops. Nucleic Acids Res. 1998;26:3640–3644. doi: 10.1093/nar/26.16.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenski, J., Crain, P.F., McCloskey, J.A. The RNA modification database: 1999 update. Nucleic Acids Res. 1999;27:196–197. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenger, W. Principles of nucleic acid structure. Springer Verlag; New York: 1984. [Google Scholar]
- SantaLucia J., Jr, Turner, D.H. Structure of (rGGCGAGCC)2 in solution from NMR and restrained molecular dynamics. Biochemistry. 1993;32:12612–12623. doi: 10.1021/bi00210a009. [DOI] [PubMed] [Google Scholar]
- Schuettelkopf, A.W., van Aalten, D.M.F. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Schuwirth, B.S., Borovinskaya, M.A., Hau, C.W., Zhang, W., Vila-Sanjurjo, A., Holton, J.M., Doudna Cate, J.H. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Sergiev, P.V., Bogdanov, A.A., Dontsova, O.A. Ribosomal RNA guanine-(N2)-methyltransferases and their targets. Nucleic Acids Res. 2007;35:2295–2301. doi: 10.1093/nar/gkm104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H., Moore, P.B. The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: A classic structure revisited. RNA. 2000;6:1091–1105. doi: 10.1017/s1355838200000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg, S., Cedergren, R. A correlation between N2-dimethylguanosine presence and alternate tRNA conformers. RNA. 1995;1:886–891. [PMC free article] [PubMed] [Google Scholar]
- Sun, X., Li, J.M., Wartell, R.M. Conversion of stable RNA hairpin to a metastable dimer in frozen solution. RNA. 2007;13:2277–2286. doi: 10.1261/rna.433307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert, B.S., Kennedy, S.D., Schroeder, S.J., Krugh, T.R., Turner, D.H. NMR structures of (rGCU GA GGCU)2 and (rGCG GA UGCU)2: Probing the structural features that shape the thermodynamic stability of GA pairs. Biochemistry. 2007;46:1511–1522. doi: 10.1021/bi061350m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin, A., Teplyakov, A. MOLREP: An automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- Wild, K., Weichenrieder, O., Leonard, G.A., Cusack, S. The 2 Å structure of helix 6 of the human signal recognition particle RNA. Structure. 1999;7:1345–1352. doi: 10.1016/s0969-2126(00)80024-6. [DOI] [PubMed] [Google Scholar]
- Winn, M.D., Isupov, M.N., Murshudov, G.N. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D Biol. Crystallogr. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- Wu, M., Turner, D.H. Solution structure of (rGCGGACGC)2 by two-dimensional NMR and the iterative relaxation matrix approach. Biochemistry. 1996;35:9677–9689. doi: 10.1021/bi960133q. [DOI] [PubMed] [Google Scholar]
- Wu, M., SantaLucia J., Jr, Turner, D.H. Solution structure of (rGGCAGGCC)2 by two-dimensional NMR and the iterative relaxation matrix approach. Biochemistry. 1997;36:4449–4460. doi: 10.1021/bi9625915. [DOI] [PubMed] [Google Scholar]