Abstract
Protein synthesis is tightly controlled by assembly of an intricate ribonucleoprotein complex at the m7GTP-cap on eukaryotic mRNAs. Ensuing linear scanning of the 5′ untranslated region (UTR) is believed to transfer the preinitiation complex to the initiation codon. Eukaryotic mRNAs are characterized by significant 5′ UTR heterogeneity, raising the possibility of differential control of translation initiation rate at individual mRNAs. Curiously, many mRNAs with unconventional, highly structured 5′ UTRs encode proteins with central biological roles in growth control, metabolism, or stress response. The 5′ UTRs of such mRNAs may influence protein synthesis rate in multiple ways, but most significantly they have been implicated in mediating alternative means of translation initiation. Cap-independent initiation bypasses strict control over the formation of initiation intermediates at the m7GTP cap. However, the molecular mechanisms that favor alternative means of ribosome recruitment are not understood. Here we provide evidence that eukaryotic initiation factor (eIF) 4G controls cap-independent translation initiation at the c-myc and vascular endothelial growth factor (VEGF) 5′ UTRs in vivo. Cap-independent translation was investigated in tetracycline-inducible cell lines expressing either full-length eIF4G or a C-terminal fragment (Ct) lacking interaction with eIF4E and poly(A) binding protein. Expression of Ct, but not intact eIF4G, potently stimulated cap-independent initiation at the c-myc/VEGF 5′ UTRs. In vitro RNA-binding assays suggest that stimulation of cap-independent translation initiation by Ct is due to direct association with the c-myc/VEGF 5′ UTR, enabling 43S preinitiation complex recruitment. Our work demonstrates that variant translation initiation factors enable unconventional translation initiation at mRNA subsets with distinct structural features.
Keywords: C-myc, VEGF, translation, eIF4G, eIF4E, PABP
INTRODUCTION
Translation control provides a critical level of gene regulation, which has been implicated in the malignant phenotype (Bader and Vogt 2004; Mamane et al. 2004, 2006). It confers the cell's ability to integrate nutrient, stress, and mitogenic signals for global proteome control or for selective translation of specific mRNAs. In general, protein synthesis is modulated at the level of initiation (Gebauer and Hentze 2004). Eukaryotic initiation occurs when the 43S preinitiation complex, comprised of the 40S ribosomal subunit and ternary complex with eIF3, is recruited to the mRNA. Conventionally, this requires 5′ m7GTP-cap interaction with the tripartite eIF4F (Gingras et al. 1999; Hershey and Merrick 2000). eIF4F constituents bind the cap (eIF4E), supply RNA helicase activity (eIF4A), and provide the central scaffold to engage eIF3 and the poly(A) binding protein (eIF4G). Once recruited to the mRNA, ribosomal scanning of the 5′ untranslated region (UTR) ensues until an initiation codon is encountered within a favorable context. Subsequent joining of the large (60S) ribosomal subunit produces an intact 80S particle that is primed for elongation.
A requirement for assembly of the canonical initiation apparatus at the cap tightly controls protein synthesis. However, this restraint is relaxed for certain mRNAs whose 5′ UTRs enable ribosome recruitment in a cap-independent manner. This mechanism is exemplified by uncapped positive-strand RNA virus genomes (Jang et al. 1988; Pelletier and Sonenberg 1988; Tsukiyama-Kohara et al. 1992). The principle of cap-independent translation initiation via an internal ribosomal entry site (IRES), first established with picornaviruses, has since been demonstrated with eukaryotic 5′ UTRs as well (Sachs et al. 1997; Stoneley and Willis 2004; Jackson 2005). There are no a priori structural predictors for IRES capacity, but generally these 5′ UTRs are uncommonly large, are predicted to form complex higher-order structures, and contain numerous upstream AUGs or CUGs.
Intriguingly, irregular 5′ UTRs with a potential for alternative initiation are common with potent regulators of fundamental biological processes, e.g., the c-myc oncogene (Nanbru et al. 1997; Stoneley et al. 1998). Alternative initiation may bypass the restraints of global translation repression and permit prompt selective induction of critical mRNAs with sudden onset of metabolic (Yaman et al. 2003), hypoxic (Stein et al. 1998), or thermal (Hernandez et al. 2004) stress as well as during apoptosis (Stoneley et al. 2000). Despite increasingly compelling evidence for a role of alternative translation initiation in cellular gene expression, less is known about the mechanisms facilitating this event.
In contrast, alternative initiation at viral IRESs has been more thoroughly characterized. Generally, translation of viral RNA occurs in the context of serious irreversible changes to the translation machinery and little regard for host cell survival (Lloyd 2006). For example, enterovirus 2A proteinase (2Apro) cleavage of eIF4GI (Etchison et al. 1982) and -II (Gradi et al. 1998) separates the eIF4E/poly(A) binding protein (PABP)-binding motifs from the eIF3/-4A sites (Fig. 1A; Lamphear et al. 1995), blocking cap-dependent host protein synthesis without affecting viral IRES-mediated translation (Ohlmann et al. 1996). Analogous to eIF4G cleavage by viral proteases, eIF4G degradation and stimulation of m7GTP-cap-independent translation has been observed in situations of cellular stress, e.g., that associated with the onset of apoptosis (Clemens et al. 1998; Bushell et al. 2006).
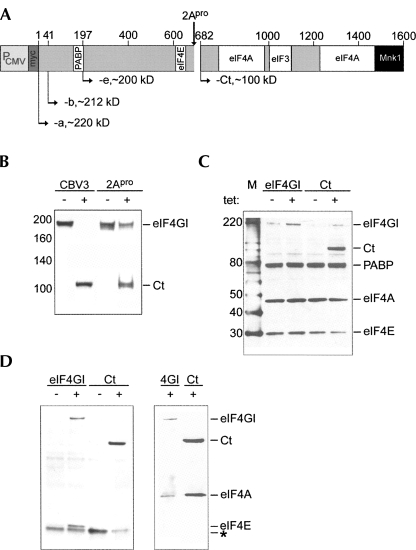
FIGURE 1.
eIF4G cleavage and tet-inducible myc-eIF4G-b and myc-Ct expressing cell lines. (A) Schematic of eIF4GI structure. Binding domains for PABP, eIF4E, -4A, -3, and Mnk1 are indicated. Numbering refers to amino acids in eIF4GI-a (Byrd et al. 2002). (B) eIF4GI immunoblot of lysates from untreated (−), CBV3-infected (CBV3), or 2Apro transfected (2Apro) HeLa cells. (C) eIF4GI-b and Ct expressing cell lines were tet-induced for 16 h and subjected to immunoblot using eIF4GI, -eIF4E-, eIF4A-, and PABP antibodies. (D) Lysates from eIF4GI-b or Ct expressing cell lines were subjected to cap-sepharose pull down (left panel) or immunoprecipitation (IP, right panel) using anti-myc antibody or mouse IgG control (data not shown). Cap-pull downs were probed with eIF4GI- and eIF4E antibodies, while IPs were probed with eIF4GI-, and eIF4A antibodies. The asterisk denotes a nonspecific background band.
We examined the role of eIF4G and variants thereof in the induction of m7GTP-cap-independent translation initiation at the 5′ UTRs of the c-myc oncogene and VEGF. We focused our studies mostly on the c-myc 5′ UTR, because c-myc mRNA remains polysome-bound in enterovirus-infected cells, demonstrating IRES competence (Johannes and Sarnow 1998). C-myc expression is abnormally up-regulated in many cancers (Adhikary and Eilers 2005), its IRES has been directly implicated in aberrant expression in multiple myeloma (Chappell et al. 2000), and c-myc IRES-driven translation persists despite global translation repression in apoptosis (Nevins et al. 2003). Likewise the VEGF 5′ UTR has been shown to harbor IRES activity (Stein et al. 1998), which may operate in hypoxia-induced VEGF expression (Koritzinsky et al. 2006). Employing an inducible in vivo expression system, we found that the c-myc/VEGF 5′ UTRs, but not viral IRESs, are potently stimulated by a C-terminal eIF4G fragment. Trans-activation occurred despite the presence of endogenous eIF4G within an intact translation environment. We demonstrated that induction of cap-independent translation initiation is due to the particular structure of eIF4GI fragments rather than absent interactions with other translation initiation factors. Finally, our experiments suggest that eIF4GI fragments stimulate cap-independent translation initiation by direct interaction with certain 5′ UTRs.
RESULTS
Stable cell lines with inducible expression of eIF4GI proteins
Enterovirus infection selectively favors cap-independent initiation (Lee and Sonenberg 1982; Buckley and Ehrenfeld 1987). Despite a global shut-off of host protein synthesis in enterovirus-infected cells, mRNAs capable of cap-independent protein synthesis continue to be translated (Johannes and Sarnow 1998; Johannes et al. 1999). This scenario, thus, serves as a precedent for studies of the mechanisms inducing alternative translation initiation. Protein synthesis modulation by Enteroviruses has chiefly been attributed to 2Apro-mediated cleavage of eIF4GI and -II (Fig. 1B). Six hours after infection with Coxsackievirus B3 (CBV3) or 12 h after transfection with a 2Apro expression RNA into HeLa cells, eIF4GI is efficiently degraded, giving rise to distinct N- and C-terminal cleavage fragments (Fig. 1B, Ct). Incomplete cleavage observed in transfected cells is likely due to transfection efficiency of <100%. The known effects of 2Apro on cap-independent translation could be due to (1) diminished intact eIF4GI/-II; (2) distinct activity of fragments produced from intact eIF4G; (3) a combination of both; or (4) unrelated events, e.g., cleavage of other proteins by 2Apro (Bovee et al. 1998; Zamora et al. 2002). To distinguish between these possibilities we generated stable HeLa cell lines with tet-inducible expression of myc-tagged eIF4GI-b or the Ct fragment (Fig. 1C). eIF4GI-b is produced by initiation 40 nucleotides (nt) downstream of the AUG for eIF4GI-a and yields more abundant protein than the latter, most likely because the 40 N-terminal amino acids contain 12 prolines (Fig. 1A; Byrd et al. 2002). We specifically investigated the effect of Ct in our approach because its appearance in CBV3-infected cells correlates with stimulation of cap-independent translation at the c-myc 5′ UTR, it contains the eIF3 binding site for 43S complex recruitment, and it can substitute for intact eIF4GI in c-myc 5′ UTR-mediated cap-independent translation in vitro (Hundsdoerfer et al. 2005).
Tet-induction for 16 h resulted in prominent expression of eIF4GI-b and Ct in the respective stable cell lines (Fig. 1C), although expression of the former was consistently less than Ct (Fig. 1C). Levels of other translation factors, e.g., PABP, eIF4A, and eIF4E, were not affected by ectopic expression of myc-eIF4GI-b or -Ct (Fig. 1C). Similarly, the interactions of exogenous eIF4GI and Ct with other translation initiation factors were intact (Fig. 1D). Coimmunoprecipitation using anti-myc-tag antibody confirmed binding of eIF4E to exogenous full-length eIF4GI-b, but not Ct, reflecting the lack of an eIF4E-binding domain in the latter (Fig. 1A). In contrast, eIF4A coimmunoprecipitated with both exogenous eIF4GI and Ct (Fig. 1D), as expected.
Ct is sufficient to stimulate the c-myc/VEGF 5′ UTRs in a cap-independent manner in vivo
To examine the effect of modulating eIF4GI expression or integrity on c-myc 5′ UTR-mediated translation in vivo, we conducted reporter RNA transfections in HeLa cells infected with CBV3, cotransfected with 2Apro expression RNA, or with overexpression of myc-eIF4GI or myc-Ct (Fig. 2A). Translation of an uncapped c-myc 5′ UTR reporter (Fig. 2B) was stimulated significantly with all conditions producing Ct (Fig. 2A). Approximately sixfold induction in CBV3-infected cells exceeded approximately threefold stimulation with 2Apro, correlating with the extent of eIF4GI cleavage (Fig. 1B). Tet-induced expression of exogenous eIF4GI had no effect on cap-independent translation mediated by the c-myc 5′ UTR, but induced Ct expression elevated c-myc 5′ UTR-mediated translation substantially (Fig. 2A). Indeed, stimulation by Ct elevated translation levels of uncapped c-myc 5′ UTR reporters within range of their capped equivalent (Fig. 2A). Since tet induction produced lower levels of full-length eIF4GI-b than Ct, lacking stimulation of cap-independent translation with the former might be a dose effect. Our findings do not support this possibility, because the eIF4GI-e isoform lacks stimulatory activity despite expression levels similar to Ct in transient transfection experiments (Fig. 5A, below). Supplying exogenous Ct in uninfected cells in the presence of intact native eIF4GI produced enhanced stimulation compared to virus- or 2Apro-mediated eIF4G cleavage, approximately eightfold versus approximately six- and approximately threefold, respectively. Reducing intact eIF4G via CBV3 infection or 2Apro expression in induced Ct-expressing cells did not produce incremental stimulation of c-myc 5′ UTR-mediated translation (data not shown), suggesting that Ct expression alone is sufficient for stimulation of cap-independent translation.
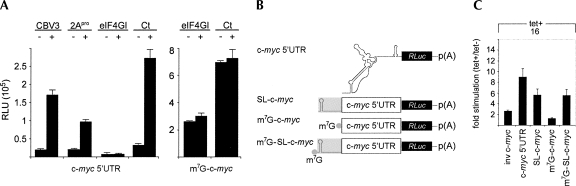
FIGURE 2.
Effect of eIF4G cleavage and inducible eIF4GI-b and Ct expression on c-myc 5′ UTR-mediated translation. (A, left panel) Uncapped c-myc 5′ UTR-driven reporter translation in untreated (−), CBV3-infected (CBV3), 2Apro co-transfected (2Apro) HeLa cells, or stable tet-inducible eIF4GI-b and Ct expressing cell lines. Cells were infected with CBV3 30 min prior to reporter transfection or reporter RNA was cotransfected with 2Apro expression RNA (Dobrikova et al. 2006). Tet induction was initiated 16 h prior to reporter transfection and all cells were lysed 6 h thereafter. (A, right panel) Capped c-myc 5′ UTR reporter translation in tet-inducible eIF4GI-b and Ct expressing cell lines. (B) Structure of c-myc 5′ UTR reporters. All constructs contained the c-myc 5′ UTR. Gray boxes symbolize a stable stem–loop structure followed by a spacer to block scanning (Thoma et al. 2004; Hundsdoerfer et al. 2005). m7G indicates the presence of a cap structure. (C) Fold stimulation of translation upon tet induction of Ct-expressing cells transfected with the indicated reporter RNAs.
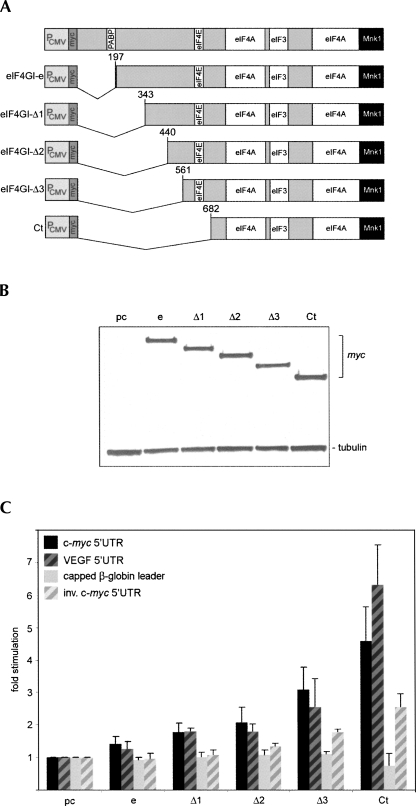
FIGURE 5.
Effect on cap-independent translation of N-terminal eIF4GI deletion variants. (A) Structure of eIF4GI deletion variants. (B) Lysates from C were probed with myc-tag and tubulin antibodies. (C) Twenty-four hours after transfection with pcDNA, eIF4GI-e, eIF4GI-Δ1-3, or Ct expression plasmids, HeLa cells were transfected with uncapped c-myc (black bars) or VEGF (black hatched bars) IRES reporters or capped β-globin (gray bars) or uncapped inverse c-myc IRES (gray hatched bars) reporter constructs and lysed 6 h thereafter.
A critical factor in studies evaluating alternative translation initiation is the overall efficiency of cap-independent protein synthesis. Cap-independent translation at the c-myc 5′ UTR was substantially above background levels in all samples (Fig. 2A). To control for spurious m7GTP-cap-independent initiation, we generated reporters containing the inverted c-myc 5′ UTR. Translation of un-capped mRNA containing the inverse c-myc 5′ UTR barely exceeded background levels (data not shown) and was not significantly stimulated in tet-induced Ct-expressing cells (Fig. 2C). Translation of mRNAs containing the correct c-myc 5′ UTR in Ct-expressing cells exceeded that at the negative control inverse construct 100-fold (data not shown). These observations suggest that stimulation of cap-independent translation by Ct requires the authentic c-myc 5′ UTR and is not due to random initiation at uncapped reporters.
To unambiguously discern cap-dependent from cap-independent initiation, we generated additional reporter constructs. Induction of cap-independent translation initiation is likely to occur when cap-dependent initiation is repressed, for example, with eIF4G cleavage. Therefore, to discourage m7GTP cap-mediated scanning while simultaneously allowing cap-independent translation, we engineered a stable stem–loop (SL) upstream of the c-myc 5′ UTR, following previously established approaches (Fig. 2B; Sherrill et al. 2004; Hundsdoerfer et al. 2005). Capped c-myc reporter translation was unaffected by Ct expression (Fig. 2A), but SL restored induction of cap-independent translation initiation at the c-myc 5′ UTR in the presence of an m7GTP-cap (Fig. 2C). It also diminished the level of stimulation slightly (approximately sixfold versus approximately eightfold), most likely due to unintended effects on the structural arrangement of the c-myc 5′ UTR. Importantly, c-myc 5′ UTR reporters containing SL with or without an m7GTP-cap were stimulated equally at approximately sixfold by Ct (Fig. 2C). Our data show that Ct-mediated stimulation of translation initiation mediated by the c-myc 5′ UTR is operational under conditions of diminished 5′ UTR scanning.
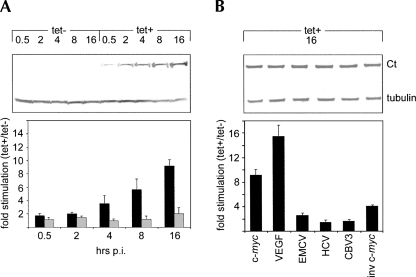
To examine dose dependency and specificity of Ct-mediated stimulation of cap-independent translation, we compared diverse 5′ UTRs of cellular and viral origin in induced cells over time (Fig. 3). Stimulation of the uncapped c-myc 5′ UTR correlated with increasing Ct levels throughout 16 h post-induction (pi) without significant changes in m7GTP-cap-mediated translation at the β-globin leader (Fig. 3A). Comparison of various uncapped reporters containing diverse 5′ UTRs demonstrated that maximum eightfold stimulation of translation at the c-myc 5′ UTR was exceeded only by the VEGF 5′ UTR (∼14-fold). Interestingly, the IRESs of encephalomyocarditis virus (EMCV), hepatitis C virus (HCV), or CBV3 were relatively unaffected (approximately twofold), as were reporters translating under control of the uncapped inverted c-myc 5′ UTR (Fig. 3B).
FIGURE 3.
Dose dependency and specificity of Ct-mediated IRES stimulation. (A) Ct cells were tet-induced from 30 min up to 16 h. At given intervals, uncapped c-myc 5′ UTR (black bars) or m7GTP-capped β-globin leader (gray bars) reporter RNAs were transfected and cells were lysed 6 h thereafter. The lysates were tested by immunoblot with myc-tag or tubulin antibodies (top panel). (B) Ct cells were tet-induced for 16 h and transfected with uncapped reporter RNAs under the control of the indicated sequences: the c-myc- or VEGF- 5′ UTRs; the EMCV-, HCV-, or CBV3 IRESs; the inverted c-myc 5′ UTR. RLuc activity was measured 6 h after transfection and lysates were tested by immunoblot with myc-tag and tubulin antibodies (top panel).
Effect of eIF4G interaction with PABP and eIF4E on cap-independent translation
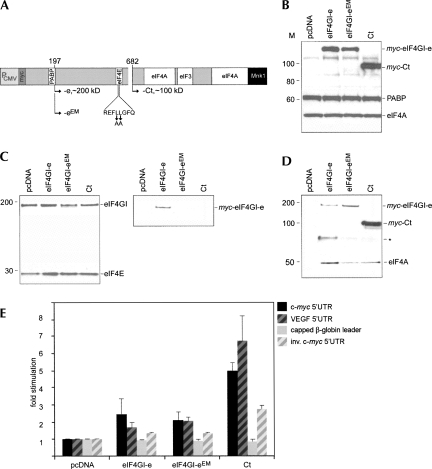
Why does Ct but not full-length eIF4GI stimulate cap-independent translation via the c-myc/VEGF 5′ UTRs? Ct's stimulatory effect could be due to loss of interaction with PABP and eIF4E (Fig. 1A). We tested this possibility by transiently expressing myc-tagged eIF4GI-e (naturally lacking the PABP binding motif) (Byrd et al. 2002), an eIF4I-e variant with mutations in the eIF4E binding motif (eIF4I-eEM) unable to bind to PABP and eIF4E (Mader et al. 1995), and Ct (Fig. 4A). Transient transfections of HeLa cells yielded approximately even levels of all eIF4GI variants (Fig. 4B).
FIGURE 4.
Effect on translation of eIF4GI-e and -4GI-e variants deficient in binding eIF4E. (A) Structure of eIF4GI-e and -4GI-eEM (mutant amino acids in the eIF4E binding domain are indicated by arrows). (B) Hela cells were transfected with pcDNA, myc-eIF4GI-e, -4GI-eEM, or Ct and lysates analyzed by immunoblot for exogenous myc-tagged proteins, PABP, and eIF4A. (C) m7GTP-cap sepharose binding assays of lysates from B. Lysates were incubated with cap-sepharose, and bound proteins were eluted and probed with eIF4E, eIF4GI, or myc-tag antibodies. Endogenous eIF4E/eIF4G are bound by m7GTP-cap sepharose in all samples. Only wild-type exogenous myc-eIF4G-e associates with the cap. (D) Co-IP assays of lysates from B. IP with myc-tag antibody reveals eIF4A binding to exogenous eIF4G variants in all samples. The asterisk indicates a nonspecific background band. (E) HeLa cells were transfected with c-myc (black bars) and VEGF (black hatched bars) IRES reporters or capped β-globin (gray bars) or uncapped inverse c-myc IRES (gray hatched bars) reporter constructs and lysed 6 h thereafter.
We verified endogenous translation initiation factor interactions with exogenous eIF4GI variants using m7-GTP cap-sepharose pull-down (Fig. 4C) or IP assays (Fig. 4D) from transfected cell lysates. As predicted, cap-sepharose pull-down revealed that endogenous eIF4GI and myc-eIF4GI-e interact with cap-eIF4E, while myc-eIF4GI-eEM and myc-Ct failed to do so (Fig. 4C). Moreover, IP with myc-tag antibody confirmed eIF4A interaction with all eIF4G variants (Fig. 4D). Next, we tested the effect of our eIF4GI variants on the translation efficiency of transfected reporter RNAs (Fig. 4E). None of the variants affected the capped β-globin leader reporter, and the inverted c-myc 5′ UTR was only moderately stimulated by Ct (Fig. 4E). Similarly, eIF4GI-e/eEM stimulated translation via the uncapped c-myc/VEGF 5′ UTRs only minimally (approximately twofold), while Ct significantly induced cap-independent translation (approximately sixfold) (Fig. 4E). Transient Ct transfection consistently yielded lower stimulation of the c-myc/VEGF 5′ UTRs than stable inducible expression (approximately five- to eightfold versus approximately eight- to 14-fold, respectively).
Role of the N terminus of eIF4G in cap-independent translation initiation
Since abolishing interaction with PABP or eIF4E itself did not convey stimulatory activity to eIF4GI-e, we evaluated if the 485 amino acids upstream of the 2Apro cleavage site contain inhibitory activity. We generated three staggered N-terminal deletion constructs (Fig. 5A) for transient expression in HeLa cells (Fig. 5B). Stimulation of c-myc/VEGF 5′ UTR-mediated translation increased with successive deletions. However, even eIF4GI-Δ3 (N-terminally extending Ct by 121 amino acids) only produced ∼50% of the stimulation observed with Ct (Fig. 5C). Inverted c-myc 5′ UTR-, and β-globin-leader-driven reporters were minimally affected (Fig. 5C).
Ct preferentially interacts with the c-myc and VEGF 5′ UTRs
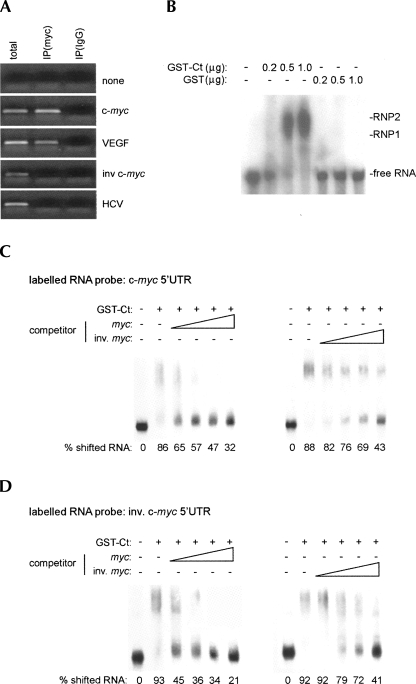
Since eIF4GI is an RNA-binding protein (Goyer et al. 1993) capable of recruiting the 43S preinitiation complex on its own, we speculated that Ct may induce cap-independent translation at the c-myc 5′ UTR through direct association. All regions of eIF4G implicated in RNA binding are contained within Ct (Lomakin et al. 2000). First, to examine binding of Ct to specific RNAs in vivo, tet-induced, myc-Ct-expressing cells were transfected with the indicated reporter RNAs (Fig. 6A). Cell lysates generated from transfected cells were subjected to IP with myc-tag antibody or preimmune serum. The immunoprecipitate was processed to isolate associated RNAs (see Materials and Methods). RT-PCR of RNA coimmunoprecipitated with myc antibody readily amplified a portion of the RLuc ORF from cells transfected with c-myc and VEGF 5′ UTR reporters, but not the inverted c-myc 5′ UTR construct or the HCV IRES (Fig. 6A). We also determined if endogenous c-myc mRNA can bind to inducible Ct. Quantitative RT-PCR of the c-myc transcript and a GAPDH control revealed a sixfold enrichment of c-myc vs. GAPDH template in myc antibody IP (data not shown).
FIGURE 6.
Interaction between Ct and the c-myc 5′ UTR. (A) RT-PCR of RNA co-IPed with myc-tag antibody. RT-PCR from total cellular RNA, myc-tag IP, or isotype controlled nonspecific mouse IgG from lysed tet-induced Ct-expressing cells previously transfected with the indicated reporters. (B) Effect of GST-Ct or GST alone on radioactively labeled c-myc 5′ UTR RNA in EMSA. Twenty nanograms of labeled c-myc 5′ UTR (105 cpm) were incubated with varying amounts of GST-Ct or GST as indicated. (C) Effect of unlabeled competitors on the interaction between GST-Ct and labeled c-myc 5′ UTR RNA and (D) inverse c-myc 5′ UTR in EMSA. Twenty nanograms of labeled RNA were incubated with GST-Ct and increasing amounts of nonradioactive competitor RNA as indicated. Ratios of competitor RNA were 1:1, 1:2.5, 1:5, and 1:10.
To further investigate Ct:RNA interactions, we examined binding of recombinant purified GST-tagged Ct to the c-myc 5′ UTR biochemically by electrophoretic mobility shift assay (Fig. 6B–D, EMSA). Purified recombinant GST-Ct stimulated c-myc 5′ UTR-mediated translation 4.5-fold in HeLa in vitro translation extracts (data not shown). This confirmed the functionality of the purified GST-fusion protein. Then, we tested GST-Ct:c-myc 5′ UTR interactions by EMSA. GST-Ct, but not GST alone, formed ribonucleoprotein (RNP) complexes with the c-myc 5′ UTR, indicating that the GST-tag does not materially alter Ct's RNA-binding capacity (Fig. 6B).
To investigate the basis for Ct's selective binding to certain 5′ UTRs in vivo (Fig. 6A), we conducted competition assays comparing the authentic c-myc 5′ UTR with its inverse counterpart. As expected, RNP complex formation at the c-myc 5′ UTR was abolished by adding increasing amounts of unlabeled c-myc competitor (Fig. 6C, left panel). In contrast, unlabeled inverse c-myc competitor displayed inferior Ct binding, since higher concentrations were needed to compete for binding of the c-myc 5′ UTR to Ct (Fig. 6C, right panel). Conversely, although Ct shifted the inverse c-myc 5′ UTR, indicating the presence of general RNA-binding activity, RNP complex formation was substantially reduced even with the lowest concentration of unlabeled c-myc 5′ UTR competitor tested (Fig. 6D, left panel). Moreover, competition was more pronounced at all competitor RNA concentrations tested, when compared to the authentic c-myc 5′ UTR (Fig. 6, cf. C and D, left panels). Competition for binding to Ct of inverse c-myc 5′ UTR with unlabeled inverse competitor resembled the parallel experiment with the authentic c-myc 5′ UTR (Fig. 6, cf. C and D, right panels). These results suggest that Ct's capacity for interaction with a given 5′ UTR determines its effects on translation.
Ct mutants lacking RNA-binding activity fail to induce cap-independent translation
If Ct exerts stimulatory activity on cap-independent translation via direct association with the c-myc or VEGF 5′ UTRs, abolishing this binding should prevent translation stimulation. Although intrinsic RNA-binding ability of eIF4G has been reported (Goyer et al. 1993), the structural basis for this is not understood. With this in mind, we based our approach on previous speculation about a “classic” RNA recognition motif (RRM; Kenan et al. 1991; Burd and Dreyfuss 1994) in eIF4G. The two putative components of this motif have been altered before to study their influence on eIF4G binding to the EMCV IRES (Lomakin et al. 2000).
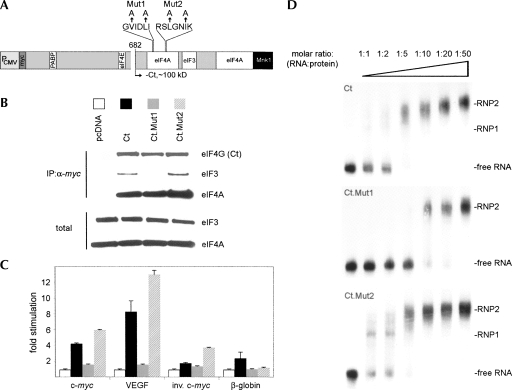
We reengineered these mutations into Ct (Fig. 7A), to investigate their effect on RNA binding and c-myc/VEGF 5′ UTR trans-activation. Since both Mut1 and -2 are situated within the binding site for eIF4A and close to the proposed footprint for eIF3 (Fig. 7A), we tested translation factor interactions with the mutant proteins (Fig. 7B). All Ct variants were expressed at even levels after transfection of cDNA expression plasmids. IP with myc-tag antibody from lysates of transfected cells revealed efficient eIF4A binding to all Ct forms; curiously, Mut1 exhibited significantly less binding to eIF3 than Ct or Mut2 in this assay. It has been observed before that mutations in the proximal eIF4A binding site can affect eIF3 binding as well (Imataka and Sonenberg 1997).
FIGURE 7.
A Ct mutant with reduced RNA binding capacity fails to stimulate cap-independent translation. (A) Structure of eIF4G, Ct, and the location of two separate elements of a putative RNA recognition motif (Lomakin et al. 2000). Altered amino acids in Mut1 and -2 are indicated by arrows. (B) Co-IP of translation factors with myc-Ct or its variants. Immunoblots of eIF4A, eIF3a, and eIF4G from lysates transfected with the indicated expression plasmids. (C) Stimulation of the indicated 5′ UTRs after transfection of RNA reporters into cells expressing the diverse Ct variants (bars are labeled according to B). (D) EMSA of labeled c-myc 5′ UTR with recombinant GST-Ct or its variants as indicated.
Transient expression of both Ct variants had opposing effects on cap-independent translation at the c-myc/VEGF 5′ UTRs. While Mut1 abolished stimulation altogether, Mut2 yielded significantly enhanced stimulation compared to parental Ct (Fig. 7C). There were marginal effects on the inverse c-myc 5′ UTR and capped β-globin reporters with Ct or Mut2. We generated recombinant GST-tagged Mut1 and Mut2 proteins to perform EMSAs. These studies revealed that, generally, stimulation of cap-independent translation via the c-myc 5′ UTR correlated with the ability of recombinant Ct variants to interact with RNA in vitro (Fig. 7D). Considerably higher concentrations (two- to fourfold) of Mut1 versus Ct protein were needed to generate the characteristic RNP complex observed upon interaction of Ct with the c-myc 5′ UTR (Fig. 7D). Notably, Mut2 formed complexes with the c-myc 5′ UTR more readily than either Mut1 or Ct (Fig. 7D). Mut1 displayed altered eIF3 binding activity, while both mutants showed differences in RNA affinity. These results further suggest a correlation between the intrinsic RNA-binding properties of Ct and its ability to stimulate translation initiation.
DISCUSSION
We show that an eIF4GI C-terminal fragment stimulates cap-independent translation initiation at the 5′ UTRs of c-myc and VEGF in vivo. Our data expand previous findings in eIF4GI-depleted HeLa cell extracts in vitro, which demonstrated that c-myc IRES-driven translation can occur independent of full-length eIF4GI and that Ct can functionally replace eIF4GI for translation initiation at the c-myc IRES, but not the m7GTP-cap (Thoma et al. 2004; Hundsdoerfer et al. 2005). Our observations indicate that stimulation of cap-independent translation at the c-myc and VEGF 5′ UTRs is due to direct association with Ct, which occurred despite the presence of endogenous intact eIF4GI. Ct, because it contains all parts of eIF4G implicated in RNA-binding and the recognition motif for eIF3, retains the ability to recruit the 43S preinitiation complex. Translation stimulation at the c-myc 5′ UTR by Ct variants carrying mutations in putative RNA-binding motifs of eIF4G co-varied with the ability of recombinant protein to associate with this RNA element in vitro. This suggests that the level of stimulation of cap-independent translation at individual mRNAs correlates with the relative ability of Ct to associate with their 5′ UTR. Interestingly, Mut1, which was deficient in binding to the c-myc 5′ UTR (and, hence, devoid of stimulatory activity in vivo), also displayed reduced recruitment of eIF3. This suggests that RNA-binding and eIF3-association functions of eIF4G may overlap.
Interestingly, the effect of Ct expression on the c-myc/VEGF 5′ UTRs did not extend to the CBV3 IRES. This confirms earlier findings which show that CBV3 IRES stimulation in infected cells occurs independent of eIF4GI cleavage (Roberts et al. 1998; Dobrikova et al. 2006). This may appear counterintuitive since Ct is a by-product of CBV3 infection. Our observations are discordant with reports of 2Apro-mediated stimulation of viral IRES-mediated translation in vitro (Liebig et al. 1993; Ziegler et al. 1995). However, similar in vitro translation studies yielded contradictory results regarding the magnitude and specificity of the effect of eIF4G cleavage on viral IRES-mediated translation (Liebig et al. 1993; Ziegler et al. 1995; Rifo et al. 2007). We attribute this variance to inconsistent empirical systems, variable structure of reporter constructs, and inherent differences of in vivo vs. in vitro assays. Moreover, we previously reported that correctly configured enteroviral IRES reporters translate as efficiently as capped conventional mRNAs in vivo in the absence of viral alterations of the host cell translation machinery (Dobrikova et al. 2006).
Lastly, CBV3 IRES-mediated translation is responsive to PABP (Bradrick et al. 2007), while translation at the c-myc IRES is not (Thoma et al. 2004), indicating divergent initiation factor involvement. We consistently observed more pronounced effects of Ct on the VEGF 5′ UTR than c-myc, suggesting that even among cellular 5′ UTRs, the response to eIF4GI variants may vary considerably.
What determines Ct's ability to induce cap-independent translation at specific mRNAs? IRESs are determined by their function, not their structure (Wimmer et al. 1993). Therefore, it is impossible to predict a capacity for cap-independent translation initiation a priori. Our findings imply that Ct may exhibit preferential interaction with certain 5′ UTRs in vivo. It will be interesting to decipher whether distinct subclasses of mRNAs can be defined by their capacity for direct interaction with eIF4G variants. If so, cap-independent translation of mRNA subsets may be controlled by the specific conditions regulating eIF4G. For example, in agreement with our findings, increased selective translation of IRES-containing mRNAs by eIF4G overexpression during hypoxia in breast cancer cells has been recently reported (Braunstein et al. 2007).
Interestingly, Ct-mediated stimulation of m7GTP-capped c-myc reporter RNAs was contingent upon discouraging cap-dependent scanning by a stable stem–loop structure inserted upstream of the c-myc 5′ UTR. In agreement with previously published work on alternative translation initiation at eukaryotic mRNAs (Sherrill et al. 2004; Hundsdoerfer et al. 2005), our data show that m7GTP-cap-independent initiation may be operational when conventional cap-dependent translation initiation is hindered.
Since all functional domains of Ct are present in eIF4G, the intact protein could in principle exert similar regulatory properties. For example, it is conceivable that loss of eIF4E interaction frees eIF4G from assembly at the m7GTP-cap, thus increasing the amount of eIF4G available for cap-independent recruitment of ribosomes. However, our studies do not support this hypothesis because eIF4GI-eEM, incapable of both PABP and eIF4E interaction, failed to stimulate cap-independent translation initiation. Alternatively, the N terminus might modulate functions of C-terminal eIF4G through integration of signaling stimuli. For example, binding of p21 activated protein kinase 2 (PAK2) to N-terminal eIF4G produces phosphorylation of Ser-896, which modulates its ability to support cap-dependent translation (Ling et al. 2005). Since successive N-terminal deletions of eIF4GI-e produced incremental c-myc/VEGF 5′ UTR stimulation, we speculate that Ct's unique properties may be due to altered structural arrangements, which may modify its inherent RNA-binding capacity (De Gregorio et al. 1998; Marcotrigiano et al. 2001).
Ct shares ∼28% homology with p97/Dap5. Accordingly, recent reports have shown that p97 stimulates c-myc IRES activity in vitro (Hundsdoerfer et al. 2005), enhances cap-independent translation of pro-survival factors during mitosis (Marash et al. 2008), and modulates alternative translation initiation during endoplasmic reticulum stress (Lewis et al. 2008). Thus, p97 and Ct may exert similar effects on cap-independent translation of c-myc or VEGF mRNA in vivo.
Intuitively, the structural heterogeneity of 5′ UTRs suggests that the prevailing conditions for translation initiation in the cell do not equally apply to the entire transcript pool. Indeed, our studies show how a variant translation initiation factor selectively favors translation of a certain class of messages. While our studies focus on the c-myc/VEGF transcripts, two exemplar mRNAs known for their capacity for cap-independent initiation, this ability may be far more prevalent than currently recognized. Also, it is conceivable that 5′ UTRs that fail common tests for cap-independent initiation competence in vitro, e.g., translation of dicistronic reporters in translation extracts, are stimulated by Ct in vivo. We are currently evaluating these hypotheses. Cap-independent translation may be induced upon eIF4G degradation in tumor cells exposed to acute stress, e.g., ionizing radiation or metabolic crisis, promoting tumor cell survival and treatment resistance. It is also possible that aberrantly generated eIF4G variants may lead to translational deregulation of specific messages in an otherwise intact translation environment.
MATERIALS AND METHODS
Stable cell lines and viruses
pcDNA5/TO/FRT/myc-eIF4GI-b and pcDNA5/TO/FRT/myc-Ct were constructed by subcloning NheI–NotI fragments of myc-tagged eIF4GI-b and Ct from corresponding pcDNA3.1 expression vectors (Dobrikova et al. 2006). A pcDNA5/TO/FRT/luc positive control vector was produced by subcloning an XhoI–NotI Renilla Luciferase (RLuc) fragment from pTNT (Dobrikova et al. 2006). Stable, inducible cell lines were established using the Flp-In T-Rex System (Invitrogen) according to the manufacturer's instructions. Briefly, HeLa R19 cells were transfected with ScaI-digested pFRT/lacZeo, and zeocin-resistant clones were screened by Southern blot for integrants containing a single FRT target site. Selected clones were transfected with BstZ17I-digested pcDNA6/TR, the tetracycline (tet) repressor expression vector. Individual zeocin/blasticidin-resistant clones were tested for tet-inducible RLuc expression after pcDNA5/TO/FRT/Luc transfection. We selected clones exhibiting the lowest basal levels:highest after-induction ratio for Flp-In T-Rex host cell lines. Finally, inducible eIF4GI-b or Ct cell lines were generated by transfecting host cell lines with corresponding vectors followed by blasticidin/hygromycinB selection. Propagation and use of CBV3 and construction of CBV3 2Apro expression vectors are described elsewhere (Dobrikova et al. 2006).
RLuc reporter vectors, in vitro transription, RNA transfection, and RLuc assays
All reporter plasmids were generated by cloning diverse 5′ UTR sequences into the pSVN vector (Invitrogen) described before (Dobrikova et al. 2006), except c-myc/inverse c-myc 5′ UTR reporters, which were cloned into a pUC19 (NEB) backbone. Cloning cassettes consisted of the RLuc open reading frame (ORF) and an encoded 50-mer poly(A) tail; the T7 promoter was included in the forward 5′ UTR primer. The c-myc 5′ UTR was obtained by RT-PCR from HeLa cell total RNA with primers 1 and 2 (Table 1). We used a 5′ UTR segment corresponding to the P2 transcript, which constitutes ∼75%–90% of the c-myc message (Stewart et al. 1984) and exhibits IRES activity (Le Quesne et al. 2001; Cencig et al. 2004; Thoma et al. 2004). The PCR-generated c-myc 5′ UTR fragment, or its inverse generated with primers 3 and 4, was inserted into the pUC19 cassette digested with BamHI/SfuI. To categorically exclude 5′ end-dependent translation at m7GTP-capped or uncapped c-myc 5′ UTR reporters, we followed a strategy described previously (Thoma et al. 2004; Hundsdoerfer et al. 2005). Briefly, a predicted stem–loop structure was introduced upstream of the c-myc 5′ UTR by inserting synthetic oligonucleotides 5–6 into the EcoRI-KpnI sites of the pUC19 cassette. A c-myc 5′ UTR fragment lacking the T7 promoter was amplified with primers 7 and 2 and cloned into KpnI-SfuI sites of the stem–loop-containing reporter construct. To minimize interference of the stem–loop with cap-independent initiation at the c-myc 5′ UTR, a 200-nt segment amplified from the human β-globin ORF (using primers 8, 9) was inserted into a KpnI site to separate the stem–loop from the IRES element as described previously (Thoma et al. 2004; Hundsdoerfer et al. 2005). The VEGF 5′ UTR was PCR-amplified using primers 10 and 11 from BAC RP11 (clone 710L16; Children's Hospital and Research Center, Oakland, CA) and cloned into the pSVN cassette digested with NotI/SfuI. The reporter constructs containing the CBV3 IRES and the β-globin leader (Dobrikova et al. 2006) as well as the EMCV and HCV IRESs (Bradrick et al. 2007) have been described elsewhere.
TABLE 1.
Primers used in this study
Reporter plasmids were linearized and used for in vitro transcription with T7 RNA polymerase (Ambion) to produce RNAs. Capped reporter RNAs were generated by adding m7G(5′)ppp(5′)G RNA cap structure analog (NEB) to in vitro transcription reactions as specified by the manufacturer. RNAs were purified by RNeasy (Quiagen), inspected for quality by agarose gel electrophoresis, and quantified by spectrometric analysis. In vivo Rluc expression assays were performed as described before (Bradrick et al. 2006; Dobrikova et al. 2006). Briefly, HeLa monolayers were transfected with equimolar amounts of reporter RNA using DMRIE-C reagent (Invitrogen). Six hours post-transfection, the cells were lysed with Rluc assay lysis buffer (Promega) and Rluc activity was assayed in a Berthold LB9507 luminometer. At least three independent transfection experiments were carried out, and the data shown represent the average values and standard deviation.
Cloning of eIF4GI mutants and deletion variants, transient exogenous eIF4GI expression, and immunoblot
Myc-tagged eIF4GI-e was generated as described before (Dobrikova et al. 2006). The eIF4E binding site mutation in eIF4GI-eEM was created by substituting Leu-617/Ala and Leu-618/Ala (Mader et al. 1995) by overlapping PCR using primers 12–15. The PCR product was inserted into the vector via PflMI/SbfI restriction sites. eIF4GI deletions, eIF4GI-Δ1-3, were generated by PCR with forward primers 16–18, respectively, and reverse primer 15. The PCR products were inserted into the vector cassette using PflMI and HindIII. Mutations in putative RNA-binding domains of Ct were introduced by overlapping PCR. Ct Mut1 (Val-797/Ala; Leu-800/Ala) and Mut2 (Leu-857/Ala; Ile-860/Ala) were generated using primers 19–20 and 23–24 or primers 21–22 and 23–24, respectively. The PCR products were inserted into the vector using HindIII/NotI. HeLa cells were transfected with the myc-eIF4GI-e, eIF4GI-eEM, eIF4GI-Δ1-3, Ct Mut1/2, or wild-type Ct expression constructs or with pcDNA3.1 vector (1 μg per 60-mm dish) using Lipofectamine 2000 (Invitrogen). Twenty-four hours after DNA transfection, cells were washed with serum-free medium, transfected with reporter RNA, and subjected to RLuc assay and analyzed by immunoblot. Myc-tag antibody (9E10; Sigma), biotinylated anti-mouse IgG (Vector Lab), streptavidin-peroxidase conjugate (Roche), and ECL Western blotting detection reagents (GE Healthcare) were used to detect exogenous eIF4GI.
Immunoprecipitation (IP), m7GTP pull down and translation factor immunoblots
IP was carried out as described previously (Keene et al. 2006). Briefly, cells were scraped in PBS, pelleted by centrifugation, and resuspended in polysome lysis buffer (100 mM KCl, 5 mM MgCl2, 10 mM HEPES at pH 7.4, 0.5% NP40, 2 mM DTT). Lysis was enhanced by rapid freezing in dry ice:ethanol, and cytoplasmic extract was collected by centrifugation. Prior to precipitation, protein G-sepharose beads (GE Healthcare) were washed in NT2 buffer (50 mM Tris at pH 7.4, 150 mM NaCl, 1 mM MgCl2, 0.05% NP40) and coupled to myc-tag antibody or mouse IgG for 2 h at 4°C. Immuno-complexes were precipitated by incubating cell lysate with beads for 2 h at 4°C. The bound proteins were eluted in LDS buffer (Invitrogen), resolved by SDS-PAGE, and analyzed by immunoblot. The bound RNA was extracted with Trizol LS and stored at −80°C. m7GTP pull downs were carried out with m7GTP-sepharose beads (GE Healthcare) using the procedures described for IP. Cell lysates were resolved by SDS gel electrophoresis in precast 4%–12% Bis-Tris NuPAGE gels (Invitrogen) and transferred to PROTRAN nitrocellulose membrane (Whatman). Membranes were incubated with monoclonal antibodies against eIF4A and -4E (#31217 and #1126; Abcam), eIF3a (#2538, Cell Signaling), c-myc and tubulin (#9E10 and #T5168; Sigma), PABP (kindly provided by L. Penalva, University of Texas Health Science Center) or rabbit polyclonal eIF4GI antibody D279 raised against amino acids 1179–1196 of eIF4GI (Dobrikova et al. 2006).
Reverse transcription and quantitative PCR
Total cellular RNA or RNA coimmunoprecipitated with myc or nonspecific mouse IgG from tet-induced Ct-expressing cells was isolated using Trizol (Invitrogen) following the manufacturer's instructions. Reverse transcription was carried out with 500 ng of RNA using AMV-RT (Promega) followed by PCR with primers 25 and 26, to detect Rluc reporter RNA, or quantitative PCR with Platinum Sybr Green qPCR SuperMix-UDG (Invitrogen) using primers 27–28 and 29–30 to detect c-myc and GAPDH mRNA, respectively, in a Roche Light Cycler.
Recombinant proteins and EMSA
Ct or its variants were expressed as glutathione-S-transferase (GST) fusion proteins in BL21 bacteria. For this purpose, Ct was cloned into pGEX-4T1 (Amersham) using XmaI/HinDIII and NotI yielding pGEX-Ct. Escherichia coli BL21 cells transformed with pGEX-Ct or the vector backbone were induced with IPTG (0.1 mM) and cultured for 3 h. Cells were lyzed according to the manufacturer's instructions. Proteins were purified using a GSTrapFF column (Amersham) and dialyzed against hypotonic buffer (10 mM HEPES at pH 7.5, 10 mM KoAC, 0.5 mM MgOAc, 1 mM DTT, 5% glycerol). Recombinant proteins were used in EMSA as follows. Essentially, EMSAs were performed as described before (Gamarnik and Andino 1997). [α32P]-UTP-labeled RNA probes were generated from c-myc or inverse c-myc 5′ UTR templates by in vitro transcription using T7 polymerase (Ambion). Unlabeled competitors were generated by in vitro transcription in the absence of [α32P-UTP]. The binding reactions were carried out in binding buffer (40 mM KCl, 5 mM HEPES at pH 7.5, 2 mM MgCl2, 4% glycerol, 2 mM DTT) with 20 ng [32P]-labeled RNA in 20 μL final volume. The mix was incubated for 10 min at room temperature and analyzed by electrophoresis in 4% native polyacrylamide gels. Gels were prerun for 30 min at 4°C at 100 mV, then 15 μL of samples were loaded and electrophoresis was allowed to proceed for 2 h 20 min at constant voltage. Recombinant Gst-Ct, Gst alone, or RNA competitors were included in the preincubation reaction as described in each case. The gels were dried and visualized by autoradiography. The proportion of free RNA in each lane was determined by PhosphorImager quantification to calculate the percentage of shifted RNA in RNPs.
ACKNOWLEDGMENTS
We thank Luis O. Penalva for kindly providing the PABP antibody. This work was supported by DOD predoctoral fellowship BC050992 (C.K.), a grant from the Susan G. Komen Foundation (M.G.), and Public Health Service grant CA124756 (M.G.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1171808.
REFERENCES
- Adhikary, S., Eilers, M. Transcriptional regulation and transformation by Myc proteins. Nat. Rev. Mol. Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- Bader, A.G., Vogt, P.K. An essential role for protein synthesis in oncogenic cellular transformation. Oncogene. 2004;23:3145–3150. doi: 10.1038/sj.onc.1207550. [DOI] [PubMed] [Google Scholar]
- Bovee, M.L., Marissen, W.E., Zamora, M., Lloyd, R.E. The predominant elF4G-specific cleavage activity in poliovirus-infected HeLa cells is distinct from 2A protease. Virology. 1998;245:229–240. doi: 10.1006/viro.1998.9171. [DOI] [PubMed] [Google Scholar]
- Bradrick, S.S., Walters, R.W., Gromeier, M. The hepatitis C virus 3′-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucleic Acids Res. 2006;34:1293–1303. doi: 10.1093/nar/gkl019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradrick, S.S., Dobrikova, E., Kaiser, C., Shveygert, M., Gromeier, M. Poly(A)-binding protein is differentially required for translation mediated by viral internal ribosomal entry sites. RNA. 2007;13:1582–1593. doi: 10.1261/rna.556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein, S., Karpisheva, K., Pola, C., Goldberg, J., Hochman, T., Yee, H., Cangiarella, J., Arju, R., Formenti, S.C., Schneider, R.J. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol. Cell. 2007;28:501–512. doi: 10.1016/j.molcel.2007.10.019. [DOI] [PubMed] [Google Scholar]
- Buckley, B., Ehrenfeld, E. The cap-binding protein complex in uninfected and poliovirus-infected HeLa cells. J. Biol. Chem. 1987;262:13599–13606. [PubMed] [Google Scholar]
- Burd, C.G., Dreyfuss, G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Bushell, M., Stoneley, M., Kong, Y.W., Hamilton, T.L., Spriggs, K.A., Dobbyn, H.C., Qin, X., Sarnow, P., Willis, A.E. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Mol. Cell. 2006;23:401–412. doi: 10.1016/j.molcel.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Byrd, M.P., Zamora, M., Lloyd, R.E. Generation of multiple isoforms of eukaryotic translation initiation factor 4GI by use of alternate translation initiation codons. Mol. Cell. Biol. 2002;22:4499–4511. doi: 10.1128/MCB.22.13.4499-4511.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencig, S., Nanbru, C., Le, S.Y., Gueydan, C., Huez, G., Kruys, V. Mapping and characterization of the minimal internal ribosome entry segment in the human c-myc mRNA 5′ untranslated region. Oncogene. 2004;23:267–277. doi: 10.1038/sj.onc.1207017. [DOI] [PubMed] [Google Scholar]
- Chappell, S.A., LeQuesne, J.P., Paulin, F.E., deSchoolmeester, M.L., Stoneley, M., Soutar, R.L., Ralston, S.H., Helfrich, M.H., Willis, A.E. A mutation in the c-myc-IRES leads to enhanced internal ribosome entry in multiple myeloma: A novel mechanism of oncogene de-regulation. Oncogene. 2000;19:4437–4440. doi: 10.1038/sj.onc.1203791. [DOI] [PubMed] [Google Scholar]
- Clemens, M.J., Bushell, M., Morley, S.J. Degradation of eukaryotic polypeptide chain initiation factor (eIF) 4G in response to induction of apoptosis in human lymphoma cell lines. Oncogene. 1998;17:2921–2931. doi: 10.1038/sj.onc.1202227. [DOI] [PubMed] [Google Scholar]
- De Gregorio, E., Preiss, T., Hentze, M.W. Translational activation of uncapped mRNAs by the central part of human eIF4G is 5′ end-dependent. RNA. 1998;4:828–836. doi: 10.1017/s1355838298980372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrikova, E.Y., Grisham, R.N., Kaiser, C., Lin, J., Gromeier, M. Competitive translation efficiency at the picornavirus type 1 internal ribosome entry site facilitated by viral cis and trans factors. J. Virol. 2006;80:3310–3321. doi: 10.1128/JVI.80.7.3310-3321.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchison, D., Milburn, S.C., Edery, I., Sonenberg, N., Hershey, J.W. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 1982;257:14806–14810. [PubMed] [Google Scholar]
- Gamarnik, A.V., Andino, R. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- Gebauer, F., Hentze, M.W. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras, A.C., Raught, B., Sonenberg, N. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- Goyer, C., Altmann, M., Lee, H.S., Blanc, A., Deshmukh, M., Woolford J.L., Jr, Trachsel, H., Sonenberg, N. TIF4631 and TIF4632: Two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol. Cell. Biol. 1993;13:4860–4874. doi: 10.1128/mcb.13.8.4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradi, A., Svitkin, Y.V., Imataka, H., Sonenberg, N. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. 1998;95:11089–11094. doi: 10.1073/pnas.95.19.11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez, G., Vazquez-Pianzola, P., Sierra, J.M., Rivera-Pomar, R. Internal ribosome entry site drives cap-independent translation of reaper and heat shock protein 70 mRNAs in Drosophila embryos. RNA. 2004;10:1783–1797. doi: 10.1261/rna.7154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey, J.W., Merrick, W.C. Pathway and mechanism of initiation of protein synthesis. In: Sonenberg N., et al., editors. Translational control of gene expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2000. pp. 637–654. [Google Scholar]
- Hundsdoerfer, P., Thoma, C., Hentze, M.W. Eukaryotic translation initiation factor 4GI and p97 promote cellular internal ribosome entry sequence-driven translation. Proc. Natl. Acad. Sci. 2005;102:13421–13426. doi: 10.1073/pnas.0506536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imataka, H., Sonenberg, N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, R.J. Alternative mechanisms of initiating translation of mammalian mRNAs. Biochem. Soc. Trans. 2005;33:1231–1241. doi: 10.1042/BST0331231. [DOI] [PubMed] [Google Scholar]
- Jang, S.K., Krausslich, H.G., Nicklin, M.J., Duke, G.M., Palmenberg, A.C., Wimmer, E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes, G., Sarnow, P. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA. 1998;4:1500–1513. doi: 10.1017/s1355838298981080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes, G., Carter, M.S., Eisen, M.B., Brown, P.O., Sarnow, P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl. Acad. Sci. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene, J.D., Komisarow, J.M., Friedersdorf, M.B. RIP-Chip: The isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protocols. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- Kenan, D.J., Query, C.C., Keene, J.D. RNA recognition: Towards identifying determinants of specificity. Trends Biochem. Sci. 1991;16:214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- Koritzinsky, M., Magagnin, M.G., van den Beucken, T., Seigneuric, R., Savelkouls, K., Dostie, J., Pyronnet, S., Kaufman, R.J., Weppler, S.A., Voncken, J.W., et al. Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. EMBO J. 2006;25:1114–1125. doi: 10.1038/sj.emboj.7600998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamphear, B.J., Kirchweger, R., Skern, T., Rhoads, R.E. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 1995;270:21975–21983. doi: 10.1074/jbc.270.37.21975. [DOI] [PubMed] [Google Scholar]
- Le Quesne, J.P., Stoneley, M., Fraser, G.A., Willis, A.E. Derivation of a structural model for the c-myc IRES. J. Mol. Biol. 2001;310:111–126. doi: 10.1006/jmbi.2001.4745. [DOI] [PubMed] [Google Scholar]
- Lee, K.A., Sonenberg, N. Inactivation of cap-binding proteins accompanies the shut-off of host protein synthesis by poliovirus. Proc. Natl. Acad. Sci. 1982;79:3447–3451. doi: 10.1073/pnas.79.11.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, S.M., Cerquozzi, S., Graber, T.E., Ungureanu, N.H., Andrews, M., Holcik, M. The eIF4G homolog DAP5/p97 supports the translation of select mRNAs during endoplasmic reticulum stress. Nucleic Acids Res. 2008;36:168–178. doi: 10.1093/nar/gkm1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebig, H.D., Ziegler, E., Yan, R., Hartmuth, K., Klump, H., Kowalski, H., Blaas, D., Sommergruber, W., Frasel, L., Lamphear, B. Purification of two picornaviral 2A proteinases: Interaction with eIF-4 γ and influence on in vitro translation. Biochemistry. 1993;32:7581–7588. doi: 10.1021/bi00080a033. [DOI] [PubMed] [Google Scholar]
- Ling, J., Morley, S.J., Traugh, J.A. Inhibition of cap-dependent translation via phosphorylation of eIF4G by protein kinase Pak2. EMBO J. 2005;24:4094–4105. doi: 10.1038/sj.emboj.7600868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, R.E. Translational control by viral proteinases. Virus Res. 2006;119:76–88. doi: 10.1016/j.virusres.2005.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin, I.B., Hellen, C.U., Pestova, T.V. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 2000;20:6019–6029. doi: 10.1128/mcb.20.16.6019-6029.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader, S., Lee, H., Pause, A., Sonenberg, N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 γ and the translational repressors 4E-binding proteins. Mol. Cell. Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamane, Y., Petroulakis, E., Rong, L., Yoshida, K., Ler, L.W., Sonenberg, N. eIF4E–from translation to transformation. Oncogene. 2004;23:3172–3179. doi: 10.1038/sj.onc.1207549. [DOI] [PubMed] [Google Scholar]
- Mamane, Y., Petroulakis, E., LeBacquer, O., Sonenberg, N. mTOR, translation initiation and cancer. Oncogene. 2006;25:6416–6422. doi: 10.1038/sj.onc.1209888. [DOI] [PubMed] [Google Scholar]
- Marash, L., Liberman, N., Henis-Korenblit, S., Sivan, G., Reem, E., Elroy-Stein, O., Kimchi, A. DAP5 promotes cap-independent translation of Bcl-2 and CDK1 to facilitate cell survival during mitosis. Mol. Cell. 2008;30:447–459. doi: 10.1016/j.molcel.2008.03.018. [DOI] [PubMed] [Google Scholar]
- Marcotrigiano, J., Lomakin, I.B., Sonenberg, N., Pestova, T.V., Hellen, C.U., Burley, S.K. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol. Cell. 2001;7:193–203. doi: 10.1016/s1097-2765(01)00167-8. [DOI] [PubMed] [Google Scholar]
- Nanbru, C., Lafon, I., Audigier, S., Gensac, M.C., Vagner, S., Huez, G., Prats, A.C. Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J. Biol. Chem. 1997;272:32061–32066. doi: 10.1074/jbc.272.51.32061. [DOI] [PubMed] [Google Scholar]
- Nevins, T.A., Harder, Z.M., Korneluk, R.G., Holcik, M. Distinct regulation of internal ribosome entry site-mediated translation following cellular stress is mediated by apoptotic fragments of eIF4G translation initiation factor family members eIF4GI and p97/DAP5/NAT1. J. Biol. Chem. 2003;278:3572–3579. doi: 10.1074/jbc.M206781200. [DOI] [PubMed] [Google Scholar]
- Ohlmann, T., Rau, M., Pain, V.M., Morley, S.J. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 1996;15:1371–1382. [PMC free article] [PubMed] [Google Scholar]
- Pelletier, J., Sonenberg, N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- Rifo, R.S., Ricci, E.P., Décimo, D., Moncorgé, O., Ohlmann, T. Back to basics: The untreated rabbit reticulocyte lysate as a competitive system to recapitulate cap/poly(A) synergy and the selective advantage of IRES-driven translation. Nucleic Acids Res. 2007;35:e121. doi: 10.1093/nar/gkm682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, L.O., Seamons, R.A., Belsham, G.J. Recognition of picornavirus internal ribosomal entry sites within cells; influence of cellular and viral proteins. RNA. 1998;4:520–529. doi: 10.1017/s1355838298971989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs, A.B., Sarnow, P., Hentze, M.W. Starting at the beginning, middle, and end: Translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- Sherrill, K.W., Byrd, M.P., Van Eden, M.E., Lloyd, R.E. BCL-2 translation is mediated via internal ribosome entry during cell stress. J. Biol. Chem. 2004;279:29066–29074. doi: 10.1074/jbc.M402727200. [DOI] [PubMed] [Google Scholar]
- Stein, I., Itin, A., Einat, P., Skaliter, R., Grossman, Z., Keshet, E. Translation of vascular endothelial growth factor mRNA by internal ribosome entry: Implications for translation under hypoxia. Mol. Cell. Biol. 1998;18:3112–3119. doi: 10.1128/mcb.18.6.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, T.A., Pattengale, P.K., Leder, P. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell. 1984;38:627–637. doi: 10.1016/0092-8674(84)90257-5. [DOI] [PubMed] [Google Scholar]
- Stoneley, M., Willis, A.E. Cellular internal ribosome entry segments: Structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- Stoneley, M., Paulin, F.E., Le Quesne, J.P., Chappell, S.A., Willis, A.E. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- Stoneley, M., Chappell, S.A., Jopling, C.L., Dickens, M., MacFarlane, M., Willis, A.E. c-myc protein synthesis is initiated from the internal ribosome entry segment during apoptosis. Mol. Cell. Biol. 2000;20:1162–1169. doi: 10.1128/mcb.20.4.1162-1169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma, C., Bergamini, G., Galy, B., Hundsdoerfer, P., Hentze, M.W. Enhancement of IRES-mediated translation of the c-myc and BiP mRNAs by the poly(A) tail is independent of intact eIF4G and PABP. Mol. Cell. 2004;15:925–935. doi: 10.1016/j.molcel.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Tsukiyama-Kohara, K., Iizuka, N., Kohara, M., Nomoto, A. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer, E., Hellen, C.U., Cao, X. Genetics of poliovirus. Annu. Rev. Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- Yaman, I., Fernandez, J., Liu, H., Caprara, M., Komar, A.A., Koromilas, A.E., Zhou, L., Snider, M.D., Scheuner, D., Kaufman, R.J., et al. The zipper model of translational control: A small upstream ORF is the switch that controls structural remodeling of an mRNA leader. Cell. 2003;113:519–531. doi: 10.1016/s0092-8674(03)00345-3. [DOI] [PubMed] [Google Scholar]
- Zamora, M., Marissen, W.E., Lloyd, R.E. Multiple eIF4GI-specific protease activities present in uninfected and poliovirus-infected cells. J. Virol. 2002;76:165–177. doi: 10.1128/JVI.76.1.165-177.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler, E., Borman, A.M., Kirchweger, R., Skern, T., Kean, K.M. Foot-and-mouth disease virus Lb proteinase can stimulate rhinovirus and enterovirus IRES-driven translation and cleave several proteins of cellular and viral origin. J. Virol. 1995;69:3465–3474. doi: 10.1128/jvi.69.6.3465-3474.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]