Abstract
Recent work demonstrating the ability of spliceosomes purified after the second catalytic step of splicing to efficiently reverse both steps of the reaction provides answers to several unresolved questions regarding the splicing reaction, and raises many more.
Keywords: ATPases, active site, conformational toggling, reversible splicing, spliced exon reopening
INTRODUCTION
Whereas the evolutionarily related group II self-splicing introns have long been known to be competent to catalyze both the forward and reverse reactions of both splicing steps (Augustin et al. 1990), such reversibility had not been demonstrated for the spliceosome. A recent article by Tseng and Cheng (2008), provides evidence for the reversibility of both catalytic steps of pre-mRNA splicing. By using an epitope-tagged, release-defective mutant form of Prp22, the DEAxD/H box ATPase that facilitates both the second step of splicing and subsequent mRNA release, Tseng and Chen were able to affinity purify near-homogeneous post-catalytic spliceosomes. These purified spliceosome/product complexes, when incubated under mild buffer conditions, can be observed to undergo remarkably efficient reversal of the two steps of splicing. As well as introducing an elegant experimental system that will render feasible important biochemical analysis of the catalytic phase of this complex reaction, these results have far-reaching mechanistic implications for splicing.
MONOVALENT IONS, CONFORMATIONAL TOGGLING, AND THE SIMILARITY OF FLANKING STATES
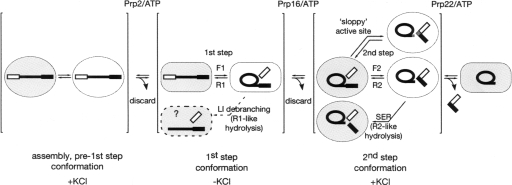
Recent work has suggested that selected spliceosomal interactions are repeatedly disrupted and re-formed during the splicing reaction: for example, the mutually exclusive U2 snRNA stem IIa and IIc structures appear to coincide with alternating “open” and “closed” states of the spliceosome, respectively (Hilliker et al. 2007; Perriman and Ares 2007). The 5′ splice site (5′SS) represents another example of toggling, occupying the same binding site in pre-first-step and post-first-step spliceosomes, and a distinct site for first step catalysis (for review, see Smith et al. 2008). In addition, alleles of the core spliceosomal protein prp8 that suppress mRNA release defects due to prp22 mutations also increase the efficiency of the first catalytic step at the expense of the second (Liu et al. 2007). These data collectively imply that spliceosomal conformations far apart on the reaction coordinate may resemble one another, at least partially. Consistent with disparate states on the splicing pathway sharing conformational similarities, Tseng and Chen observed that F2, the forward reaction of the second step, and R1, the reverse of the first, are both favored by the presence of KCl, while R2 is favored by its absence (Fig. 1). Thus, KCl stabilizes both pre-first-step and post-second-step conformations of the spliceosome relative to the post-first-step, pre-second-step conformation. This observation provides further evidence for the global conformational similarity of the states before the first and after the second catalytic step of splicing. We anticipate that further analysis will highlight the similarities and differences between other spliceosomal states, and clarify the extent to which conformational toggling is a general phenomenon in splicing.
FIGURE 1.
Schematic depiction of the observed and potential reactions occurring during reversible splicing catalysis by compositionally homogeneous spliceosomes, as described by Tseng and Chen (2008). Given the need for catalytic center opening and substrate repositioning between the two catalytic steps (Liu et al. 2007), a complex containing lariat intermediate and free 5′ exon could plausibly be in one of at least four conformations that remain conformationally undefined: post-first step but preopening, open but unrepositioned, open and repositioned, or closed pre-second step. Similarly complex transitions may also be required at other steps shown here as a single transition. KCl stimulates the forward second step and reverse first step: It is unclear, however, precisely which states along the splicing pathway are relatively stabilized or destabilized by KCl.
As noted by Tseng and Chen, the selective stabilization of various spliceosomal conformations via modulation of salt conditions will facilitate the enrichment of defined states for experiments such as snRNA secondary structure mapping; such investigations will add greater temporal resolution to the current view of snRNA–snRNA and snRNA–pre-mRNA interactions. As we have previously noted (Smith et al. 2008), the large number of conformational transitions that appear to be necessary for splicing may render difficult the interpretation of the high-resolution crystal structures we hope will become available in the future. Knowledge of the RNA–RNA interactions particular to each state of the spliceosome will help to alleviate these difficulties and underpin a great deal of future splicing catalysis research.
DIVALENT IONS, EXON REOPENING, AND THE LOOSENING OF ACTIVE SITE SPECIFICITY
Splicing catalysis, which comprises two sequential transesterification reactions, almost certainly makes use of two metal ion catalysis (Steitz and Steitz 1993). As part of their stringent analysis of the requirements for forward and reverse splicing, Tseng and Chen investigated the effects of divalent metal ions on the efficiency of each step. The results indicate that Ca2+ or Mn2+ can substitute for Mg2+ for R1 (first step reversal); Mn2+ can support F2 but with relaxed specificity relative to Mg2+; Mg2+ is required for R2. Mn2+ or high pH, both of which fail to support R2, instead appear to catalyze spliced exon reopening (SER)—a reaction first identified in self-splicing introns (Inoue et al. 1986; Jarrell et al. 1988) and considered to be a reversal of the second step in which water, rather than the 3′ end of the excised intron, acts as the nucleophile.
The distinct catalytic capabilities of divalent metal ions suggest subtle differences in active site requirements for the two steps of splicing, although the effects of different ions could be either direct—resulting from coordination of these ions in the active site itself, or indirect—resulting from more global structural differences due to coordination of these ions at other sites in the spliceosome. An intriguing observation is the reduced fidelity of the Mn2+-catalyzed F2 reaction, as this appears to suggest a “loosening” of the active site when Mn2+ replaces Mg2+. Reduced fidelity is, however, only observed for the forward reaction of the second step, with R2 proceeding as accurately in the presence of low Mn2+ concentrations as in the presence of Mg2+. This disparity is likely to be related to lariat intermediate repositioning between the two steps: for R2, the mRNA is already optimally positioned in the active site, and the nucleophile is the only free 3′OH in the intron; thus, for both the stringent Mg2+ and the sloppy Mn2+ active sites, the same reaction will occur. In the case of F2, the lariat intermediate presumably toggles between its post-first-step and pre-second-step locations: the Mg2+ active site will only facilitate a reaction using the wild-type site, while the Mn2+ site will allow the use of cryptic 3′ splice sites between the branch site and the wild-type 3′SS. An alternative possibility is that the presence of Mn2+ inhibits this post-first-step repositioning; such inhibition could also lead to the observed use of these branch-site-proximal cryptic sites. A fuller understanding of this relaxation of sequence specificity could provide insights into the mechanistic basis of substrate selectivity during splicing under physiological conditions.
We note here that R1, which can also be catalyzed in the presence of Mn2+, is necessarily precise because only one branch and one 5′ exon terminal 3′OH exist to participate in the reaction; the failure of Mn2+ to catalyze positionally promiscuous first-step reversal, therefore, does not suggest that active site loosening by Mn2+ is limited to the second step of splicing.
Tseng and Chen's observation of SER instead of R2 in the presence of high concentrations of Mn2+ is consistent with Mn2+-mediated relaxation of the catalytic center. SER requires activation of water as a nucleophile rather than the 3′OH at the end of the intron (this activation involves the stabilization of a negative charge and is thus equivalent to leaving group stabilization during the true second step), and such activation of an inappropriate group is again indicative of increased tolerance of suboptimal substrates. The observation of R2 and SER as mutually exclusive can also be parsimoniously explained without invoking gross, divalent ion-dependent differences in the catalytic center. The purified spliceosomes are likely competent to carry out both R2 and SER, regardless of divalent ion identity and pH: these reactions are in competition with one another, and in the presence of Mg2+ and moderate pH, the more efficient R2 reaction strongly predominates and therefore excludes detectable SER. Mn2+ and high pH are notable, however, for the efficiency with which they catalyze RNA cleavage. Under conditions shown to favor SER, the 3′ end of the lariat intron is therefore constrained in a region of high local metal ion concentration under conditions favoring hydrolysis: this would be predicted to lead to efficient hydrolytic cleavage in this region of the intron. Such cleavage would result in a truncated lariat intron with a 2′-3′ cyclic phosphate at its 3′ end, consistent with the presence of a band migrating just below the lariat product in reactions favoring SER in Figure 2, D and E, in the study by Tseng and Cheng (2008) in the absence of the aberrant 3′SS use that produces similarly migrating lariat products. The presence of a terminal cyclic phosphate on the lariat intron would render it unable to participate in R2, leaving only the short cleaved 3′ terminal fragment of the exon (which would still possess a 3′OH) or water to act as potential nucleophiles for second step reversal on the mRNA poised in the active site. The homogeneity of the SER product observed by Tseng and Chen in fact suggests that water is the only nucleophile used in this reaction, and this presumably reflects dissociation of the short terminal fragment of the intron following cleavage.
The considerations applied to SER should theoretically also be applicable to reversal of the first step via hydrolysis of the 2′-5′ lariat branch; such reversal would be expected to occur under similar conditions to those favoring SER (i.e., those stimulating RNA hydrolysis). It is with interest that we note the appearance of a specific product migrating just below pre-mRNA under conditions of moderate Mn2+ concentration (see Fig. 2A, lane 9 in Tsen and Cheng 2008) or high pH (see Fig. 1C, lanes 9,10 in Tsen and Cheng 2008) that allow true reversal of the second step, as this product may represent a linear lariat-3′ exon molecule that could arise via a hydrolytic first step in either the forward or the reverse direction.
ATPASES, FIDELITY, SPEED, AND WASTE
The spliceosome undergoes many conformational transitions throughout assembly, catalysis and disassembly. These transitions, although presumably able to occur spontaneously at a slow rate, are facilitated by the action of DExD/H box ATPases (for review, see Smith et al. 2008). Tseng and Chen demonstrated the possibility of undergoing the transition between the two chemical steps of splicing (albeit in reverse) in the absence of Prp16, the ATPase normally responsible for this transition. There are two potential mechanistic interpretations of this Prp16-less reversal. It is possible that Prp16 normally promotes transition into the second step by destabilizing the first step conformation of the spliceosome. Because the reverse splicing reactions observed by Tseng and Chen contain neither Prp16 nor ATP, such reversal would be favored and would therefore represent the truly spontaneous transition from the second to the first step. It is also possible, however, that ATPase binding and/or ATP hydrolysis may act to facilitate spliceosomal transitions by the stabilization of transition states along the splicing pathway. In the event that Prp16 stabilized a transition state between the first and second step conformations, it is possible that its absence for the second-to-first-step transition may be partially complemented by the presence of Prp22, the ATPase normally required for the second catalytic step and mRNA release (Schwer and Gross 1998). The existence of at least some shared binding site elements for Prp16 and Prp22, as would be required for such complementation, is supported by the lack of temporal overlap in the binding of these two ATPases (Brys and Schwer 1996).
The widely accepted kinetic proofreading model holds that the conformational change mediated by each ATP hydrolysis event is in competition with the preceding step on the splicing pathway. ATP hydrolysis results in progression through the pathway if the preceding step has been completed, and otherwise in substrate discard (Burgess and Guthrie 1993; Mayas et al. 2006; Xu and Query 2007). Optimal substrates, which interact most favorably with the catalytic center and therefore undergo each step of the splicing reaction more quickly than suboptimal substrates, are thus selectively driven through the reaction. Kinetic proofreading, if the catalytic rates of the ATPases in question are sufficiently high, allows the extent of spliceosome assembly and catalysis on suboptimal substrates to be minimized, as only optimal substrates will react quickly enough to compete with ATP hydrolysis. As the rate of ATP hydrolysis decreases, the ratio of discard to progression (predominantly on suboptimal substrates, but also on optimal substrates) decreases as substrates are afforded more time to assemble/react. This is consistent with the observation that mammalian DExD/H box ATPases may generally have lower catalytic rates than those from yeast (Xu and Query 2007), commensurate with the relaxation of splicing substrate specificity that allows alternative splicing in higher eukaryotes.
In the extreme case of zero ATPase activity (spontaneous progression), reversible splicing will likely allow equilibrium to be reached prior to spliceosomal transitions. Because spliceosomal complexes containing optimal (and optimally reacted) substrates will be more stable than those containing aberrant substrates/products, the most populated intermediates at equilibrium will be those corresponding to assembly and catalysis on such optimal substrates. The equilibrium scheme will therefore favor the use of intrinsically strong splice sites to an extent determined by the difference in binding energy between the relevant wild-type and cryptic sites. Although, as suggested by Tseng and Chen's work, it is therefore theoretically possible to use reversibility and the resulting equilibrium as a slow proofreading mechanism, this is normally precluded by ATPase-mediated introduction of forward ratcheting and discard into the splicing pathway. In certain situations, however, equilibrium-based maintenance of splicing fidelity may be advantageous. Metazoan genes can be extremely long: a well-known example is the human dystrophin gene, which gives rise to a 14-kb mRNA, is 2.3 Mb long, and has 79 exons that are spliced during the 16 h required for its transcription (Tennyson et al. 1995). In the context of such a long timeframe and energetically costly transcript, it is conceivable that rapid splicing with the concomitant risk of discard is disfavored and that local ATPase concentration/activity may be down-regulated such that splicing events lie closer to equilibrium and fidelity is maintained by reversibility. It is worth noting that up-regulation of ATPase activity could act as an effective and rapidly activatable transcriptional attenuator in this system, as discard between the catalytic steps would cleave the nascent transcript. At the other end of the spectrum lie ribosomal protein and other highly transcribed genes, especially those in rapidly dividing cells: For such genes, the energetic waste associated with substrate discard is likely outweighed by the greatly increased rate of splicing facilitated by ATPases, and thus the more rapid production of functional pre-mRNA.
ACKNOWLEDGMENTS
We thank members of the Konarska laboratory, Charles Query, and Jon Staley for helpful suggestions and comments on the manuscript.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1289808.
REFERENCES
- Augustin, S., Muller, M.W., Schweyen, R.J. Reverse self-splicing of group II intron RNAs in vitro. Nature. 1990;343:383–386. doi: 10.1038/343383a0. [DOI] [PubMed] [Google Scholar]
- Brys, A., Schwer, B. Requirement for SLU7 in yeast pre-mRNA splicing is dictated by the distance between the branchpoint and the 3′ splice site. RNA. 1996;2:707–717. [PMC free article] [PubMed] [Google Scholar]
- Burgess, S.M., Guthrie, C. A mechanism to enhance mRNA splicing fidelity: The RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell. 1993;73:1377–1391. doi: 10.1016/0092-8674(93)90363-u. [DOI] [PubMed] [Google Scholar]
- Hilliker, A.K., Mefford, M.A., Staley, J.P. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes & Dev. 2007;21:821–834. doi: 10.1101/gad.1536107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, T., Sullivan, F.X., Cech, T.R. New reactions of the ribosomal RNA precursor of Tetrahymena and the mechanism of self-splicing. J. Mol. Biol. 1986;189:143–165. doi: 10.1016/0022-2836(86)90387-6. [DOI] [PubMed] [Google Scholar]
- Jarrell, K.A., Peebles, C.L., Dietrich, R.C., Romiti, S.L., Perlman, P.S. Group II intron self-splicing. Alternative reaction conditions yield novel products. J. Biol. Chem. 1988;263:3432–3439. [PubMed] [Google Scholar]
- Liu, L., Query, C.C., Konarska, M.M. Opposing classes of prp8 alleles modulate the transition between the catalytic steps of pre-mRNA splicing. Nat. Struct. Mol. Biol. 2007;14:519–526. doi: 10.1038/nsmb1240. [DOI] [PubMed] [Google Scholar]
- Mayas, R.M., Maita, H., Staley, J.P. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat. Struct. Mol. Biol. 2006;13:482–490. doi: 10.1038/nsmb1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriman, R.J., Ares M., Jr Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes & Dev. 2007;21:811–820. doi: 10.1101/gad.1524307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer, B., Gross, C.H. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 1998;17:2086–2094. doi: 10.1093/emboj/17.7.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, D.J., Query, C.C., Konarska, M.M. “Nought may endure but mutability”: Spliceosome dynamics and the regulation of splicing. Mol. Cell. 2008;30:657–666. doi: 10.1016/j.molcel.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz, T.A., Steitz, J.A. A general two-metal-ion mechanism for catalytic RNA. Proc. Natl. Acad. Sci. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennyson, C.N., Klamut, H.J., Worton, R.G. The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nat. Genet. 1995;9:184–190. doi: 10.1038/ng0295-184. [DOI] [PubMed] [Google Scholar]
- Tseng, C.K., Cheng, S.C. Both catalytic steps of nuclear pre-mRNA splicing are reversible. Science. 2008;320:1782–1784. doi: 10.1126/science.1158993. [DOI] [PubMed] [Google Scholar]
- Xu, Y.Z., Query, C.C. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol. Cell. 2007;28:838–849. doi: 10.1016/j.molcel.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]