Abstract
Leaderless mRNAs are translated in the absence of upstream signals that normally contribute to ribosome binding and translation efficiency. In order to identify ribosomal components that interact with leaderless mRNA, a fragment of leaderless cI mRNA from bacteriophage λ, with a 4-thiouridine (4S-U) substituted at the +2 position of the AUG start codon, was used to form cross-links to Escherichia coli ribosomes during binary (mRNA+ribosome) and ternary (mRNA+ribosome+initiator tRNA) complex formation. Ribosome binding assays (i.e., toeprints) demonstrated tRNA-dependent binding of leaderless mRNA to ribosomes; however, cross-links between the start codon and 30S subunit rRNA and r-proteins formed independent of initiator tRNA. Toeprints revealed that a leaderless mRNA's 5′-AUG is required for stable binding. Furthermore, the addition of a 5′-terminal AUG triplet to a random RNA fragment can make it both competent and competitive for ribosome binding, suggesting that a leaderless mRNA's start codon is a major feature for ribosome interaction. Cross-linking assays indicate that a subset of 30S subunit r-proteins, located at either end of the mRNA tunnel, contribute to tRNA-independent contacts and/or interactions with a leaderless mRNA's start codon. The interaction of leaderless mRNA with ribosomes may reveal features of mRNA binding and AUG recognition that are distinct from known signals but are important for translation initiation of all mRNAs.
Keywords: translation initiation, ribosome recognition, leaderless mRNA
INTRODUCTION
Translation initiation involves the assembly of ribosomes on mRNA and is the rate-limiting step in bacterial protein synthesis. The conventional pathway that leads to the formation of translation initiation complexes involves the selection of an mRNA's translation initiation region (TIR) and initiator tRNA by the 30S ribosomal subunit, with the aid of three initiation factors (IF1, IF2, and IF3) (for review, see Laursen et al. 2005). Previous reports revealed that 30S subunits are able to bind to Shine–Dalgarno (SD)-leadered mRNA in the absence of initiator tRNA (Ringquist et al. 1993; Benelli et al. 2003), and the stabilization of these binary complexes is presumably enabled by the complementary SD–anti-Shine–Dalgarno (ASD) base-pairing interaction between mRNA and the 30S subunit's 16S rRNA (Shine and Dalgarno 1974). In the presence of initiator tRNA, the SD-ASD pairing contributes to stabilization of the mRNA–ribosome–initiator tRNA ternary complex and to P-site placement of the start codon, which then proceeds into translation initiation. Photocross-linking (Rinke-Appel et al. 1991; La Teana et al. 1995; Sergiev et al. 1997), biochemical, immuno- and cryo-EM (Shatsky et al. 1991; Frank et al. 1995), and X-ray crystallographic studies (Ogle et al. 2001; Yusupova et al. 2001; Berk et al. 2006; Selmer et al. 2006) have established a high resolution model for the path of SD-leadered mRNA through the bacterial ribosome (Laursen et al. 2005).
An increasing number of non-SD-led genes (Chang et al. 2006), including genes encoding leaderless mRNA completely lacking a 5′ untranslated region (UTR) (Janssen 1993; Wu and Janssen 1996, 1997; Tedin et al. 1997; Grill et al. 2000, 2001; Moll et al. 2002; and references therein), continue to be identified. Leaderless mRNAs have been reported to follow a novel pathway of translation initiation involving 70S ribosomes (Balakin et al. 1992; O'Donnell and Janssen 2002; Moll et al. 2004; Udagawa et al. 2004) or 30S subunits free of IF3, which is reported to discriminate against a 5′-terminal start codon (Moll et al. 1998; Tedin et al. 1999; Grill et al. 2000, 2001). Toeprint assays have demonstrated tRNA-dependent ribosome binding to leaderless mRNA, and an AUG start codon is required for efficient ribosome binding and translation in Escherichia coli (Van Etten and Janssen 1998; O'Donnell and Janssen 2001). The mRNA features or signals present in non-SD-led genes that are important for ribosome association and P-site placement of the start codon are not well understood.
To identify specific ribosomal contacts that might contribute to the binding and possible recognition of a leaderless mRNA's start codon, we used a fragment of bacteriophage λ's cI leaderless mRNA (Ptashne et al. 1976) with a 4-thiouridine (4S-U; Favre 1990) substitution at the +2 position of the AUG start codon. Photoactivation of the 4S-U-containing mRNA in complex with E. coli ribosomes is predicted to form cross-links to rRNA and r-proteins within the cross-linking radius of 4S-U (i.e., ∼4 Å). Although 70S ribosomes bound more leaderless mRNA than 30S subunits bound, all identified cross-links occurred at sites within the 30S subunit. A cross-link was localized near the 3′ end of 16S rRNA and several cross-linked 30S subunit r-proteins were identified by mass spectrometry. While ribosome binding for toeprint and electrophoretic gel mobility shift assays required initiator tRNA for stability, 4S-U cross-links to 30S subunit rRNA and r-proteins did not require initiator tRNA. Furthermore, the addition of a 5′ AUG triplet to a random RNA fragment made it competent for ribosome binding, suggesting that the presence of a 5′ AUG might be sufficient for the interactions that lead to translation of leaderless mRNA. Our results suggest that a leaderless mRNA's 5′-AUG is involved in tRNA-independent interactions with the ribosome that contributes to initial mRNA binding.
RESULTS
The model 4S-U RNA contains the 5′-terminal 20 nucleotides (nt) of the λ cI leaderless mRNA with a photoactivatable 4S-U at the +2 position of the AUG start codon. 4S-U RNA bound ribosomes and competed with leaderless mRNA for ribosome binding, suggesting that ribosomes recognized it as a leaderless mRNA. Photoactivation of 4S-U RNA bound to ribosomes formed cross-links to closely associated rRNA and a subset of r-proteins. While initiator tRNA was required for stable association of 4S-U RNA with ribosomes, the initial interaction(s) between 4S-U RNA and ribosomes was tRNA independent. Studies with several competitor RNAs (listed in Table 1) indicated that a leaderless mRNA's 5′-terminal AUG was required and possibly sufficient for initial contact of the mRNA with ribosomes.
TABLE 1.
Competitor RNAs
4S-U RNA as a model leaderless mRNA
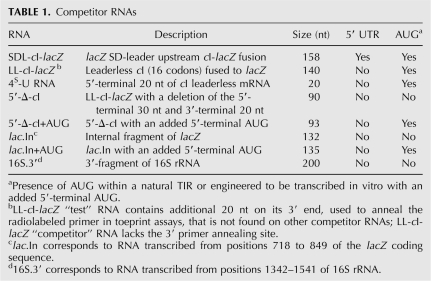
Gel electrophoretic mobility shift assays confirmed that stable ribosome binding to 32P-labeled 4S-U RNA required the presence of tRNAMet f . A greater amount of 4S-U RNA bound to 70S ribosomes (Fig. 1A, lane 4) than to 30S subunits (Fig. 1A, lane 6), in agreement with Figure 1B (cf. lanes 3 and 7) and previous toeprint assays where leaderless mRNA bound E. coli 70S ribosomes more strongly than 30S subunits (Balakin et al. 1992; O'Donnell and Janssen 2002). In addition, toeprint assays revealed that excess unlabeled 4S-U RNA competed for LL-cI-lacZ binding to E. coli 70S ribosomes (Fig. 1B, cf. lanes 3 and 4) and 30S subunits (Fig. 1B, cf. lanes 7 and 8), whereas a competitor RNA lacking start site information (lac.In) had comparatively modest effects on binding (Fig. 1B, lanes 5,9). These results suggest that the 4S-U RNA 20-mer functions as a leaderless mRNA in tRNA-dependent ternary complex formation with 70S ribosomes and 30S subunits.
FIGURE 1.
Gel mobility shift and toeprint assays demonstrate 4S-U RNA binds to 30S subunits and 70S ribosomes. (A) Gel mobility shift assays. 32P-labeled 4S-U RNA was incubated with 70S ribosomes or 30S subunits in the presence or absence of tRNAMet f . (Lane 1) ethidium bromide stain to show 4S-U RNA shifts to rRNA (ribosome) position (→). (B) Toeprint assays. Pre-bound 4S-U RNA or lac.In competitor RNAs (40×) was tested for their ability to block 30S subunit or 70S ribosome binding to LL-cI-lacZ mRNA (1×); arrow indicates toeprint signals at +16. Reaction components are indicated for each lane (A,B).
4S-U RNA cross-links to rRNA and r-proteins from 30S subunits
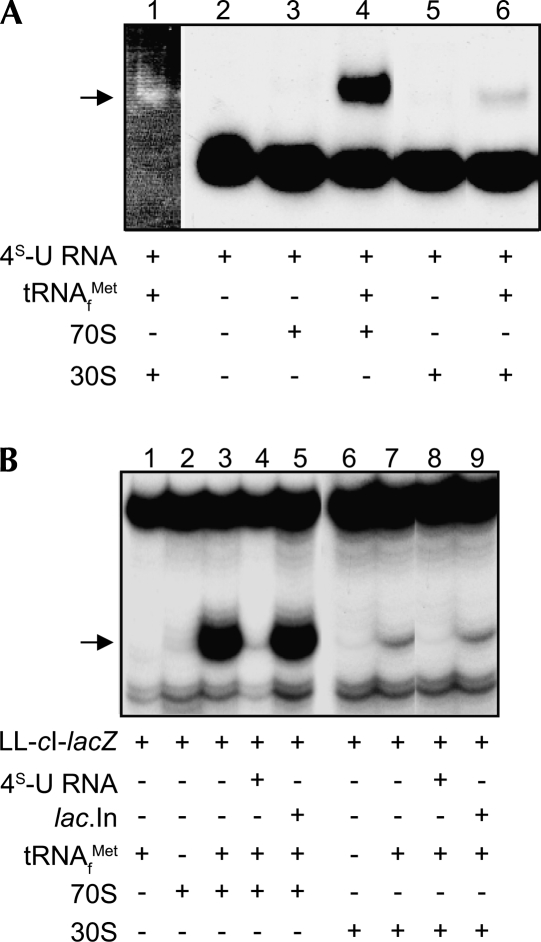
Cross-links between 4S-U RNA and rRNA in ternary complexes containing 32P-labeled 4S-U RNA+ribosomes+tRNAMet f were detected only after UV-activation (Fig. 2A, cf. lane 3 with 2 and lane 6 with 5). The migratory positions of 16S and 23S rRNAs were determined by ethidium bromide staining of gel lanes containing 30S subunits or 70S ribosomes (Fig. 2A, lanes 4,7, respectively) and revealed that 4S-U RNA cross-links were associated with the 16S rRNA.
FIGURE 2.
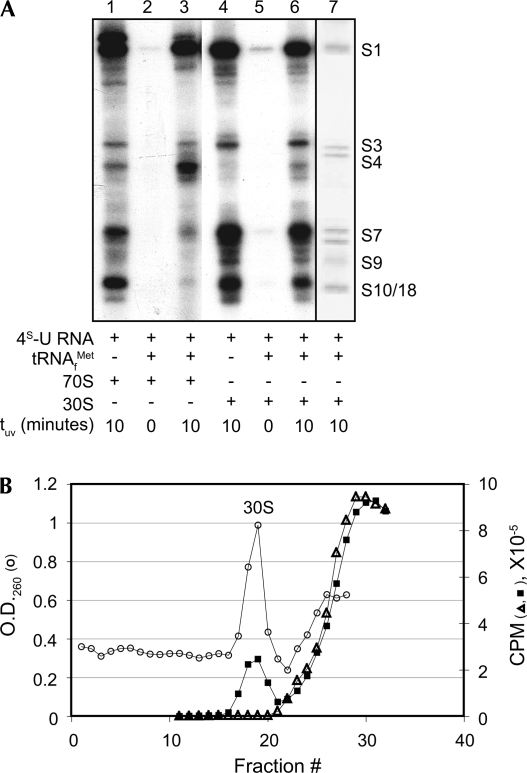
4S-U RNA forms cross-links to rRNA and r-proteins. Reaction complexes were photoactivated and analyzed for cross-links to rRNA (A) and r-proteins (B). (A, lanes 4,7) ethidium bromide stains of 30S subunit and 70S ribosome rRNAs; arrow indicates cross-links to 30S subunit rRNA. (B, lanes 1,2) Coomassie protein stains of molecular weight size markers (size in kDa on left) and 70S ribosomes, respectively. (Lanes 3–6) autoradiographic exposures showing UV-dependence of 4S-U RNA cross-links to 30S subunits and 70S ribosomes. (Lane 6) an autoradiographic exposure of Coomassie stained lane 2. Reaction components and UV photoactivation times are indicated for each lane in A and B. (C) Sucrose gradient profile of 32P-labeled 4S-U RNA cross-linked to 70S ribosomes, after splitting into 30S and 50S subunits. (○), O.D.260 trace of ribosomal subunits; (■), radioactivity trace of 32P-labeled 4S-U RNA.
Cross-links between 4S-U RNA and r-proteins in ternary complexes that contain 32P-labeled 4S-U RNA+ribosomes+tRNAMet f were detected by SDS-PAGE only after UV-activation. Coomassie staining visualized total proteins (Fig. 2B, lane 2), and autoradiography revealed proteins cross-linked to 32P-labeled 4S-U RNA (Fig. 2B, lanes 4,6). Comparison of the cross-linked protein profile from 30S complexes (Fig. 2B, lane 4) to 70S complexes (Fig. 2B, lane 6) suggested that all detected contact proteins were in the 30S subunit, albeit the abundance of certain protein cross-links from 70S complexes was reduced. Cross-linked r-protein bands were not visualized if samples were digested with proteinase K prior to electrophoresis (data not shown).
Splitting UV-irradiated 70S ternary complexes into 30S and 50S subunits (Stade et al. 1989) revealed that the radiolabeled 4S-U RNA cosedimented with 30S subunits, but not with 50S subunits, confirming that cross-links formed exclusively to the small subunit of 70S ribosomes (Fig. 2C). Although 30S subunits bound less leaderless mRNA than did 70S ribosomes, our observations that all cross-links occurred to components of the 30S subunit (here and below), combined with the reduced complexity of the 30S subunit relative to 70S ribosomes, prompted us to concentrate our analysis on cross-links to 30S subunits.
4S-U RNA cross-links to 16S rRNA near position 1530
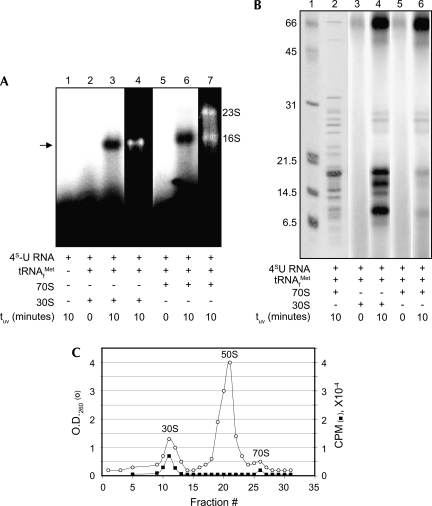
After purification through successive sucrose (Fig. 3A) and SDS-glycerol density gradients (Fig. 3B), 16S rRNA was analyzed with RNase H to localize the 16S rRNA position(s) cross-linked to 4S-U RNA. DNA oligonucleotides were annealed to specific sites within 16S rRNA. RNase H digestion of RNA:DNA hybrids released a radioactive product corresponding in size to the 3′ fragment of the digested 16S rRNA (Fig. 3C, lanes 3–6; Fig. 3D, lanes 2–6) except when the central base of the hybrid site was position 1530 (Fig. 3D, lane 7). Digestion of hybrids containing a complementary DNA 10-mer, with its central base at position 1530, yielded a single radioactive band (Fig. 3C, lane 7) close in size to the uncut 16S rRNA (Fig. 3C, lane 2). The close proximity of position 1530 to the 16S rRNA 3′ end (i.e., position 1542) prevented identification of the cross-linked nucleotide(s) by primer extension analysis. These data indicate that 4S-U RNA cross-links to a nucleotide(s) near the 3′ end of the 16S rRNA close to position 1530, similar to the position reported to be cross-linked by the start codon of a leadered mRNA (Rinke-Appel et al. 1994; La Teana et al. 1995).
FIGURE 3.
RNase H analysis localizes cross-links within 16S rRNA. (A) Sucrose gradient profile of 32P-labeled 4S-U RNA cross-linked to 30S subunits. (○), O.D.260 trace of ribosomal subunits; (■), radioactivity trace of 32P-labeled 4S-U RNA. (B) Pooled fractions containing 32P-labeled 4S-U RNA+tRNAMet f+30S ternary complexes from A were centrifuged in SDS-glycero1 gradients to separate 16S rRNA from r-proteins. (C,D) Polyacrylamide gel electrophoresis of gradient-purified cross-linked 16S rRNA after RNase H analysis. Each lane represents a single RNase H cleavage reaction. The diagrams below the autoradiograms identify the central base of the annealed oligonucleotide (vertical arrows) together with the lane and size (in nucleotides) of the 4S-U RNA-containing cleavage product (horizontal arrows). The dotted vertical lines indicate the 1530 position at which the 32P-labeled cross-link no longer migrates with the predicted 3′ cleavage fragment. (C) RNase H digestion of cross-linked complexes with oligos annealed at 16S rRNA positions: (lane 2) no oligo; (lane 3) 550; (lane 4) 705; (lane 5) 815; (lane 6) 925; (lane 7) 1530. (D) RNase H digestion of cross-linked complexes with oligos annealed at 16S rRNA positions: (lane 2) 1492; (lane 3) 1500; (lane 4) 1510; (lane 5) 1520; (lane 6) 1525; (lane 7) 1530. (Lane 1) DNA markers with size (in nucleotides) indicated. Cross-linked 4S-U RNA cause fragments to migrate ∼20 nt larger than predicted.
4S-U RNA cross-links to several 30S subunit r-proteins
Gradient-purified complexes of 4S-U RNA cross-linked to r-proteins (as above; see Fig. 3A,B) were annealed to a biotinylated complementary DNA oligonucleotide, followed by affinity purification with streptavidin-coated magnetic beads and SDS-PAGE analysis. The affinity-purified r-protein profile visualized by Coomassie staining (Fig. 4A, lane 7) was similar to the autoradiographic profile (Fig. 4A, lane 6). Coomassie stained bands were excised, digested with trypsin, and analyzed by mass spectrometry.
FIGURE 4.
Cross-links of 4S-U RNA to r-proteins in the presence and absence of initiator tRNA. (A, lanes 1–6) cross-links of radiolabeled 4S-U RNA to r-proteins, with (lanes 1,3,4, 6) or without (lanes 2,5) exposure to UV light. (Lane 7) gradient-purified, affinity selected Coomassie stained r-proteins identified by mass spectrometry and indicated on the right as S1, S3, S4, S7, S9, and S10/18. Reaction components are indicated for each lane. (B) Sucrose gradient profiles of 32P-labeled 4S-U RNA + 30S subunit binary complexes in the absence (△) or presence (■) of photactivation. (○), O.D.260 trace of 30S subunits; (△, ■), radioactivity trace of 32P-labeled 4S-U RNA.
S1, S3, S4, S7, and S9 were identified as cross-linked r-proteins through MALDI peptide mass fingerprinting. S7 was identified as a doublet, as reported previously (La Teana et al. 1995). S1, S7, and S10 were identified through LC-MS/MS peptide sequence tags. S18 was sometimes identified in gel slices containing S10. For all mass spectral analyses, the identified proteins were found to be the most probable match to the experimental data, with MALDI data yielding multiple peptide matches and LC-MS/MS sequence tags yielding significant fragment ion assignments (Supplemental Table S1; Figs. S1–S3).
4S-U RNA cross-links to ribosomes independent of initiator tRNA
4S-U RNA and 30S subunits from photoactivated binary complexes sedimented together in sucrose gradients (Fig. 4B), suggesting a tRNA-independent association of leaderless mRNA with 30S subunits. The cross-link profile to 16S rRNA, as described above (Fig. 3), was the same in the presence or absence of tRNAMet f (data not shown); UV-dependent cross-links to 30S subunit r-proteins were similar in the presence or absence of of tRNAMet f (Fig. 4A, lanes 4,6), indicating that the 5′ AUG's contact with 30S subunit rRNA and r-proteins were not dependent on initiator tRNA. Cross-links to 70S ribosomal r-proteins were similar to 30S subunit r-proteins in the absence of initiator tRNA (Fig. 4A, cf. lanes 1 and 4); however, the 70S cross-link profile was influenced significantly by the presence of initiator tRNA (Fig. 4A, lane 3), presumably reflecting a tRNA-dependent change in start codon position. The tRNA-dependent increase in S4 cross-links, as seen in Figure 4A (lanes 3,6), was not consistently observed (e.g., Fig. 2B, lane 6). As reported by Berk et al. (2006), no r-proteins would be found within the 4S-U cross-linking radius (∼4 Å) of a start codon in the P-site, suggesting that P-site stabilization of codon–anticodon pairing in a 70S ternary complex reduces contact with a subset of r-proteins. These results indicate that initiator tRNA, which is required for stable ribosome binding in gel shift and toeprint assays (Fig. 1A,B), is not required for the interaction and cross-link formation between 4S-U RNA and rRNA or r-proteins.
A leaderless mRNA's 5′-terminal AUG facilitates initial ribosomal recognition
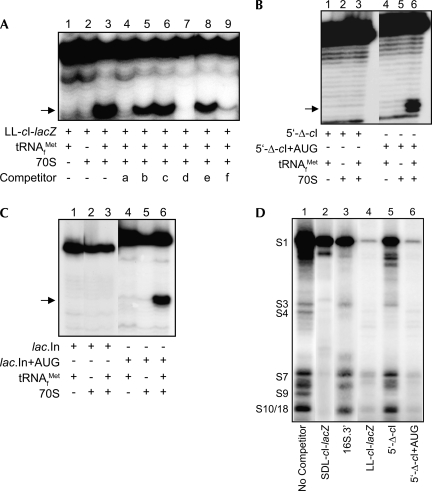
Several RNAs (Table 1) were tested in toeprint assays for their ability to compete with LL-cI-lacZ mRNA for binding to 70S ribosomes. Briefly, ribosomes were bound with excess competitor RNA and then assayed for their ability to form ternary complexes with LL cI-lacZ test mRNA (annealed with a radiolabeled primer). The LL-cI-lacZ RNA competitor (lacking the 3′ primer annealing site) and the 4S-U RNA competitor contained authentic start sites from a leaderless mRNA, while the SDL-cI-lacZ RNA competitor contained the start site from a canonical SD-leadered mRNA. The 4S-U RNA and LL-cI-lacZ competitors significantly blocked binding of the LL-cI-lacZ test mRNA to 70S ribosomes (Fig. 5A, cf. lanes 3 and 4,7). The SDL-cI-lacZ competitor caused only a marginal reduction in 70S ribosome binding (Fig. 5A, cf. lanes 3 and 5), presumably reflecting the inability of canonical SD-leadered mRNA to form ternary complexes with 70S ribosomes. The 5′-Δ-cI competitor RNA, which is identical to the LL-cI-lacZ competitor (Fig. 5A, lane 7) but lacked the 5′-terminal 10 codons with start site information, was unable to block 70S ribosome binding to LL-cI-lacZ mRNA (Fig. 5A, cf. lanes 3 and 8); a similar result was observed with a 3′ fragment of 16S rRNA (i.e., 16S.3′ RNA), representing a noncoding competitor RNA (Fig. 5A, cf. lanes 3 and 6). However, addition of an AUG triplet to the 5′ terminus of 5′-Δ-cI (i.e., 5′-Δ-cI+AUG) resulted in a competitor RNA that nearly eliminated LL-cI-lacZ mRNA binding to 70S ribosomes (Fig. 5A, cf. lanes 3 and 9). These results indicate that addition of a 5′-terminal AUG to the noncompetitive 5′-Δ-cI RNA converts it to an RNA that competes with a bona fide leaderless mRNA for 70S ribosome binding.
FIGURE 5.
Competitor RNAs affect ribosome binding and 4S-U cross-links to r-proteins. (A,B,C) Toeprint assays. Arrow indicates toeprint signal at +16. Reaction components are indicated. (A) Prebound competitor RNAs (40×) were tested for their ability to block 70S ribosome and 30S subunit binding to LL-cI-lacZ mRNA (1×). Competitors: a, 4S-U RNA; b, SDL-cI-lacZ; c, 16S.3′; d, LL-cI-lacZ; e, 5′-Δ-cI; f, 5′-Δ-cI+AUG. (B,C) Competitor RNAs: 5′-Δ-cI versus 5′-Δ-cI+AUG (B) and lac.In versus lac.In+AUG (C) were tested for their ability to bind 70S ribosomes. (D) 32P-labeled 4S-U RNA was cross-linked to 30S subunits in the presence of the indicated prebound competitors (20×). Ribosomal proteins identified by mass spectrometry are indicated.
To determine whether the addition of a 5′-terminal AUG to a noncompetitive RNA can make it competent to bind ribosomes, the 70S ribosome binding to 5′-Δ-cI and lac.In competitor RNAs, with (Fig. 5 B,C, lanes 4–6) or without (Fig. 5B,C, lanes 1–3) an added 5′-terminal AUG, were compared. Both RNAs were able to form ternary complexes with 70S ribosomes only if they contained the added 5′-terminal AUG (Fig. 5B,C, cf. lanes 3 and 6). Taken together, these results suggest that a 5′-terminal AUG is sufficient for an RNA molecule to form a ternary complex with tRNA and ribosomes and are consistent with a previous report demonstrating the importance of a 5′-AUG for ribosome binding and translation of leaderless mRNA (Van Etten and Janssen 1998; O'Donnell and Janssen 2001).
RNAs used to compete ribosome binding in toeprint reactions (Fig. 5A; Table 1) were also used to compete cross-links between 4S-U RNA and 30S subunit r-proteins (a representative gel from multiple independent assays is shown in Fig. 5D). Prebinding subunits with SDL-cI-lacZ or LL-cI-lacZ competitors, which contain authentic translation start sites, substantially reduced cross-link formation between the radiolabeled 4S-U RNA and all contacting r-proteins (Fig. 5D, cf. lanes 1 and 2,4, respectively). Prebinding subunits with the 16S.3′ or 5′-Δ-cI competitor RNAs that lack translation start site information had comparatively minor effects on cross-links to several of the r-proteins (Fig. 5D, lanes 3,5, respectively). The reduced ability of the 16S.3′ and 5′-Δ-cI RNAs to protect the r-proteins from 4S-U RNA cross-links suggests that the protection provided by the SDL- and LL-cI-lacZ RNAs results from specific ribosome recognition of their translational start sites. Addition of a 5′-terminal AUG to the 5′-Δ-cI (i.e., 5′-Δ-cI+AUG) RNA reduced all cross-links (Fig. 5D, lane 6), similar to the competition seen with prebound LL-cI-lacZ RNA (Fig. 5D, lane 4), suggesting that this RNA now contains the necessary information for specific ribosome recognition as a leaderless mRNA; the toeprint assays in Figure 5B (lane 6) also suggest that this RNA is recognized as a leaderless mRNA by the ribosome. These results suggest that ribosomes bind leaderless mRNA through recognition of their 5′-AUG and that the ribosome is able to distinguish mRNAs by the presence or absence of a 5′-terminal AUG.
DISCUSSION
This study provides novel evidence that interactions other than codon–anticodon pairing contribute to the initial ribosomal interaction with a leaderless mRNA's 5′-AUG. Our results suggest that, in the absence of a SD-leader sequence, a 5′-terminal AUG provides a major feature for ribosome recognition and binding to a leaderless mRNA. Although gel shift and toeprint assays have demonstrated the importance of initiator tRNA in ternary complex stabilization, cross-linking experiments revealed that the initial contact and interaction between the ribosome and a leaderless mRNA's 5′-AUG occurs through the 30S subunit and is independent of initiator tRNA. Together with earlier reports that codon–anticodon pairing is insufficient for expression (Van Etten and Janssen 1998) and that non-AUG start codons (i.e., GUG, UUG, CUG) do not support efficient ribosome binding and expression (O'Donnell and Janssen 2001), our results provide additional support that a 5′ AUG is specifically required for efficient expression of leaderless mRNA in E. coli.
Toeprint assays revealed that the addition of a 5′-terminal AUG to RNA fragments that were otherwise incapable of ribosome binding resulted in RNAs (i.e., lac.IN+AUG and 5′-Δ-cI+AUG) that were both competitive for ribosome binding and competent for tRNA-dependent ternary complex formation. Also, fusion of the lac.In+AUG sequence to a lacZ reporter gene provides for β-galactosidase production (data not shown), thereby indicating that addition of the 5′ AUG results in formation of translationally active complexes in vivo.
Position of cross-links on the 30S subunit crystal structure
The rRNA cross-link detected in this study was localized near the 16S rRNA position 1530 and agrees closely with cross-links reported for the start codon of a SD-leadered mRNA (Rinke-Appel et al. 1994; La Teana et al. 1995). While it is not clear how this rRNA position would contact the start codon of a leaderless or a SD-leadered mRNA, the consistency of this interaction with both leadered and leaderless mRNAs implies an involvement of this position in the initial interaction of the 30S subunit with a start codon. Alternatively, the 3′ tail of 16S rRNA is reported to fold back into the 30S P-site (Wimberly et al. 2000; Yusupova et al. 2001), thereby making it possible that the start codon contacts the 1530 region as the mRNA replaces the 3′ tail in the mRNA binding tunnel.
The P- and A-site mRNA codons and tRNA anticodon loops do not contact any ribosomal proteins when in a ternary initiation complex (Schluenzen et al. 2000; Berk et al. 2006), suggesting that the tRNA-independent cross-links detected here might result from the “trapping” of transient mRNA-ribosome interactions that occur independent of codon–anticodon pairing. As revealed by the crystal structure of ribosomes complexed with mRNA and P-site tRNA (Yusupova et al. 2001; Berk et al. 2006), the 4S-U RNA's 5′-AUG formed cross-links to r-proteins near the ribosome's mRNA tunnel, with the S7/S18 contacts occurring near the upstream (E-site proximal) end of the tunnel and the S3/S4/S10 contacts occurring near the downstream (A-site proximal) end of the tunnel. The most abundant cross-links occurred to S1 and S7, suggesting that these proteins have the closest or most stable contacts to 4S-U RNA's start codon (thereby increasing the likelihood of a successful cross-link). Previous reports that S1 is dispensable for translation of leaderless mRNAs (Tedin et al. 1997; Moll et al. 2002) do not rule out the possibility that S1, when present, binds the 5′ end of leaderless mRNAs and contributes to their association with ribosomes.
Implications for ribosome recognition and binding of leaderless mRNA
The data presented in this study suggest that ribosomes interact with leaderless mRNA through recognition of a 5′-AUG start codon. The short cross-linking radius of 4S-U (∼4 Å) indicates that the rRNA and r-protein contacts reported in this study are in close association with a leaderless mRNA's 5′ AUG. Observations that the same subset of r-proteins are cross-linked on 30S subunits and 70S ribosomes, combined with a report that similar contacts to rRNA and r-proteins occur with a leadered mRNA's start codon (La Teana et al. 1995), argue that these interactions are not random associations of mRNA with ribosomes. In addition, the protection of specific r-proteins from 4S-U RNA cross-links by mRNA fragments containing authentic translational start sites, as well as our observations of ribosome binding and translation of random RNA fragments after addition of a 5′ AUG, suggests that the ribosome recognizes and interacts specifically with the 5′-terminal start codon of a leaderless mRNA. Furthermore, the observed reduction in 4S-U RNA cross-links when 30S subunits were prebound with leaderless or SD-leadered mRNA implies that interaction with a leaderless mRNA requires specific ribosome contacts that are not available when the mRNA tunnel is occupied.
Our data suggest that a subset of 30S r-proteins positioned at or near the mRNA binding tunnel provides some of the initial contacts with a 5′-terminal AUG start codon. Whether contact with these proteins results from a nonspecific affinity for single-stranded RNA or reflects r-protein involvement in 5′-AUG recognition remains to be determined. S1's high affinity for mRNA 5′-termini (Sukhodolets et al. 2006) and its high reactivity for RNA with a 5′-AUG observed in this study suggests a role for S1 in the initial binding of leaderless mRNA and possibly positioning the mRNA near, or within, the binding tunnel. Once at the mRNA tunnel, and without the SD-ASD interaction to help position the mRNA, contacts between the leaderless mRNA and several r-proteins that surround the tunnel might function to increase the local concentration of leaderless mRNA at the tunnel and favor the adjustment of the 5′-AUG into a stable codon–anticodon pairing at the P-site. The high abundance of cross-links to S7 and its position near the E/P-site boundary (Yusupova et al. 2001) implies that S7 might contribute to placement of the 5′-AUG near the P-site until codon–anticodon pairing stabilizes the complex. In support of this, S7 was also reported to form strong cross-links to the +2 position of an AUG start codon from a SD-leadered mRNA when the start codon was positioned near the ribosome decoding site (La Teana et al. 1995). Furthermore, hydrogen bonding between G926 of 16S rRNA and the phosphate 5′ of the P-site's +1 nucleotide (Berk et al. 2006) also might contribute to stabilization of a leaderless mRNA's start codon in the P-site.
The expression of leaderless mRNAs in Bacteria, Archaea, and Eukarya suggests that the ability to bind and translate these mRNAs represents a common, possibly conserved, function of all ribosomes (Janssen 1993; Moll et al. 2002; Andreev et al. 2006; Brenneis et al. 2007). Evidence that 5′-AUG recognition is a conserved function of perhaps all ribosomes suggests that this capability may have persisted since early evolution of the translational apparatus. Additional sequence upstream of the mRNA's coding sequence may have evolved in order to provide additional mRNA–ribosome interaction for regulated, precise, and efficient translation as the number and sequences of mRNAs became increasingly complex. The interactions that contribute to recognition and placement of a leaderless mRNA's start codon in the ribosomal P-site might also play a role in ribosome positioning of a leadered mRNA's start codon; for example, following the SD-ASD interaction, contacts with S7 and/or S18 near the P-site may contribute to start codon recognition and may be a determinant of translational efficiency for mRNAs with cognate or noncognate start codons.
MATERIALS AND METHODS
Reagents
A 4S-U RNA 20-mer (Dharmacon), corresponding to the 5′ end of λ cI leaderless mRNA with 4S-U at the +2 position (5′-A4S-UGAGCACAAAAAAGAAACC-3′), was phosphorylated prior to use with T4 polynucleotide kinase (NEB) and ATP or radiolabeled [γ-32P]ATP (6000 Ci/mmol; 150 mCi/mL; Perkin Elmer). DNA oligonucleotides were purchased commercially. E. coli MRE600 tRNAMet f , proteinase K, and RNase A (Sigma-Aldrich), T7 RNA polymerase (NEB), Pfu DNA polymerase (Stratagene), RNase H (TAKARA), AMV reverse transcriptase (Life Sciences), streptavidin-coated magnetic beads (Dynal), and RNase-free DNase I (Ambion) were used as recommended.
Ribosome isolation
Isolation of E. coli MRE600 70S ribosomes and 30S ribosomal subunits was done as previously described (Martin-Farmer and Janssen 1999). 30S subunits were purified over two successive sucrose gradients to ensure purity and absence of tRNA.
Gel shift assays
Binding reaction mixtures (15 μL) containing 32P-labeled 4S-U RNA (33 nM), 70S ribosomes or 30S subunits, tRNAMet f (at a 1:20:40 ratio), and 1XSB (60 mM NH4Cl, 10 mM Tris acetate at pH 7.4, 10 mM Mg acetate, 6 mM β-mercaptoethanol) were incubated at 37°C for 30 min and transferred to ice. The reaction mixtures were then subjected to electrophoresis in 1.5% agarose gels at 4°C, and the migratory position of 32P-labeled 4S-U RNA was visualized by autoradiography. Ethidium bromide staining of rRNA confirmed that the shifted position of 32P-labeled 4S-U RNA corresponded to bound ribosomes.
Primer extension inhibition (toeprint) experiments
RNAs used in toeprint experiments were synthesized in vitro with T7 RNA polymerase and gel purified as described (Brock et al. 2007); in vitro synthesized transcripts contain an additional G, present at the 5′ terminus, as the initiating nucleotide of T7 RNA polymerase. Toeprint experiments measuring the relative amount of 70S ribosomes or 30S subunits bound to a cI leaderless “test” mRNA (Table 1, LL-cI-lacZ; O'Donnell and Janssen 2002), in the presence or absence of various “competitor” RNAs (Table 1), were conducted as described by Brock et al. (2007). Briefly, competitor RNAs (1.2 μM = 40×) were prebound to 70S ribosomes or 30S subunits in the presence of tRNAMet f and 1XSB at 37°C for 25 min and transferred to ice. In reactions lacking competitor, H2O compensated the volume. The cI leaderless test mRNA (30 nM = 1×) with a 32P-labeled primer annealed to the 3′ end was then added, establishing a test mRNA:ribosome:tRNA ratio of 1:6:12, and reaction mixtures (15 μL) were incubated at 37°C for 15 min. The reaction mixtures were placed on ice, and 1.5 μl of reverse transcriptase (1 U/μL) was added to each mixture; this was followed by incubation at 37°C for 10 min. Reaction mixtures were finally ethanol precipitated and subjected to electrophoresis in 6% polyacrylamide–7 M urea gels. Toeprint assays with other test RNAs were performed under similar conditions.
Formation of 4S-U mRNA–ribosome cross-links
All cross-linking experiments were performed in the dark as described (Yu 2000). Binding reaction mixtures (55 μL) containing 32P-labeled 4S-U RNA (0.1 μM), 70S ribosomes or 30S subunits, and the presence or absence of tRNAMet f (1:0.5:1 ratio), as well as 1XSB, were incubated at 37°C for 25 min and transferred to ice. The binding reaction mixtures were spotted onto a Petri dish suspended in ice and exposed to UV for 10 min with a hand-held lamp (Mineralight lamp, model UVGL-25; 366 nm wavelength setting) placed 4 cm from the samples. Cross-linked 16S rRNA and r-proteins were then purified by successive sucrose (10%–30%) and SDS-glycerol (10%–30%) gradient centrifugations and analyzed (see Results). The total cross-links to rRNA and r-protein were determined to be 15% (±3%) of the input 4S-U RNA. Additionally, increasing the 4S-U RNA:ribosome ratio from 2:1 to 10:1 did not change the cross-link profile to r-proteins, suggesting that contact occurred to a specific nonrandom subset of r-proteins.
Analysis of cross-link site(s) in 16S rRNA
The region of the 16S rRNA sequence containing a 4S-U RNA cross-link was localized by annealing 10-15-mer DNA oligonucleotides along the 16S rRNA and cleaving at RNA–DNA hybrid sites with RNase H as described (Brimacombe et al. 1990; Rinke-Appel et al. 1994). Briefly, gradient-purified 16S rRNA (1 pmol) was heat-denatured at 95°C for 10 min and placed on ice. A 10-15-mer DNA oligonucleotide (1 pmol) complementary to a specific sequence of the 16S rRNA sequence was annealed at 70°C for 2 min followed by 55°C for 30 min. Conditions were then adjusted to 37°C and 10 mM Mg+2 for RNase H digestion (1 U for 1 h). RNase H cleavage products were separated on 6% polyacrylamide gels and visualized by autoradiography. The proximity of the 4S-U RNA cross-link to the 16S rRNA 3′ end was determined as described (Rinke-Appel et al. 1994) and involved a series of complementary DNA oligonucleotides and “walking” along 16S rRNA toward the 3′ end until RNase H digestion released the 4S-U RNA cross-link with the 5′ cleavage product.
Purification and analysis of cross-linked r-proteins
In order to identify cross-linked r-proteins, large scale binding reactions (0.4 mL) were prepared containing nonradiolabeled 4S-U RNA (0.1 μM), 70S ribosomes or 30S subunits, tRNAMet f (1:0.5:1 ratio), and 1XSB. Gradient-purified r-proteins were affinity selected by annealing the cross-linked 4S-U RNA to a complementary biotinylated (5′ end) DNA oligonucleotide, followed by binding the annealed complex to streptavidin-coated magnetic beads. Bound proteins were eluted from the beads by boiling in 0.1% SDS/10 mM EDTA for 5 min, and precipitated with cold 5% TCA. The r-proteins were then separated by SDS-PAGE, visualized by Coomassie staining, and identified by mass spectrometry.
Mass spectrometry
Protein bands were excised from gels and digested overnight with trypsin (sequencing grade, Promega) after reduction and alkylation (Suh and Limbach 2004). Tryptic peptide samples were analyzed using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) or capillary liquid chromatography/tandem mass spectrometry.
All MALDI-TOF MS experiments were performed on a Bruker Reflex IV MALDI-TOF from Bruker Daltonics having a 3-m effective flight path, a two-stage gridless ion reflector, pulsed ion extraction, and a nitrogen laser (λ = 337 nm). All MALDI spectra were acquired in positive polarity and in reflectron mode. The instrument parameters were: IS1, 20 kV; IS2, 16.30 kV; lens voltage 9.4 kV; and reflector voltage 23 kV. A dried droplet sample spotting approach was used. The α-cyano-4-hydroxycinnamic acid (CHCA) matrix was prepared by making a saturated solution of CHCA in equal volumes of 0.1% TFA and acetonitrile. For MALDI analysis, equal volumes of MALDI matrix and sample were combined and 1 μL was spotted on the MALDI target plate. Vendor-supplied Flex control and X-mass software were used for data acquisition and processing, respectively. Typically, 200 laser shots were co-added per spectrum. The instrument was calibrated internally with angiotensin, neurotensin, and bradykinin, which bracketed the m/z range of interest. Mascot was used to search the MALDI data against the SwissProt database.
Capillary liquid chromatography/tandem mass spectrometry experiments were performed using a Thermo Surveyor MS pump coupled with a Micro AS autosampler connected in-line to a Thermo LTQ-FT (Thermo Scientific) mass spectrometer equipped with an ESI source. Reversed phase chromatography was performed on a PicoFrit C18, 0.075 mm × 100 mm column (New Objective). Mobile phase buffers consisted of A: 0.1% formic acid–5% acetonitrile, B: 0.1% formic acid–95% acetonitrile. A linear gradient, consisting of a 5-min hold at 1% B, was then ramped to 60% B at 1% B/min, followed by a ramp to 80% B in 5 min, then returned to 1% B in 20 min. The mass spectrometer was used in data dependent mode with a survey scan in the FT section of the instrument followed by 6 MS/MS scans in the ion-trap. Minimum signal intensity for triggering MS/MS was 1000 counts and isolation width was 2.00 Da. Source voltage was 2.00 kV and capillary temperature 175°C. The parent mass and sequence information obtained for tryptic peptides was used to search the UniProt database using the SEQUEST search engine.
Cross-links to r-proteins in the presence of competitor RNAs
Experiments measuring the relative amounts of cross-linked r-proteins from 30S subunits in the presence or absence of various competitor RNAs (Table 1) were performed by prebinding competitor RNAs (2 μM = 20×) to 30S subunits (0.05 μM = 0.5×) in the presence of tRNAMet f and 1XSB at 37°C for 25 min, and transferring to ice. In reactions lacking competitor, H2O compensated the volume. 32P-labeled 4S-U-test RNA (0.1 μM = 1×) was then added, establishing a test mRNA:ribosome:tRNA ratio of 1:0.5:1, and reaction mixtures (20 μL) were incubated at 37°C for 15 min and transferred to ice. Reactions were irradiated as described above and analyzed by SDS-PAGE.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Karthik Krishnan and Moojin Suh for intellectual and technical contributions to the development of this work and Anton Vila-Sanjurjo for comments on the manuscript. This work was supported by grants GM065120 (G.R.J), RR019900 (P.A.L), and GM58843 (P.A.L) from the National Institutes of Health.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1089208.
REFERENCES
- Andreev, D.E., Terenin, I.M., Dunaevsky, Y.E., Dmitriev, S.E., Shatsky, I.N. A leaderless mRNA can bind to mammalian 80S ribosomes and direct polypeptide synthesis in the absence of translation initiation factors. Mol. Cell. Biol. 2006;26:3164–3169. doi: 10.1128/MCB.26.8.3164-3169.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakin, A.G., Skripkin, E.A., Shatsky, I.N., Bogdanov, A.A. Unusual ribosome binding properties of mRNA encoding bacteriophage λ repressor. Nucleic Acids Res. 1992;20:563–571. doi: 10.1093/nar/20.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benelli, D., Maone, E., Londei, P. Two different mechanisms for ribosome/mRNA interaction in archaeal translation initiation. Mol. Microbiol. 2003;50:635–643. doi: 10.1046/j.1365-2958.2003.03721.x. [DOI] [PubMed] [Google Scholar]
- Berk, V., Zhang, W., Pai, R.D., Cate, J.H.D. Structural basis for mRNA and tRNA positioning on the ribosome. Proc. Natl. Acad. Sci. 2006;103:15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenneis, M., Hering, O., Lange, C., Soppa, J. Experimental characterization of cis-acting elements important for translation and transcription of halophilic Archaea. PLoS Genet. 2007;3:2450–2467. doi: 10.1371/journal.pgen.0030229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimacombe, R., Greuer, B., Gulle, H., Kosack, M., Mitchell, P., Osswald, M., Stade, K., Stiege, W. New techniques for the analysis of intra-RNA and RNA protein cross-linking data from ribosomes. In: Spedding G., editor. Ribosomes and protein synthesis—A practical approach. Oxford University Press; Oxford: 1990. pp. 131–159. [Google Scholar]
- Brock, J.B., Paz, R.L., Cottle, P., Janssen, G.R. Naturally occurring adenines within mRNA coding sequences affect ribosome binding and expression in Escherichia coli . J. Bacteriol. 2007;189:501–510. doi: 10.1128/JB.01356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, B., Halgamuge, S., Tang, S. Analysis of SD sequences in completed microbial genomes: Non-SD-led genes are as common as SD-led genes. Gene. 2006;373:90–99. doi: 10.1016/j.gene.2006.01.033. [DOI] [PubMed] [Google Scholar]
- Favre, A. Photochemistry and the nucleic acids. In: Morrison H., editor. Bioorganic photochemistry. John Wiley & Sons; New York: 1990. pp. 379–425. [Google Scholar]
- Frank, J., Zhu, J., Penczek, P., Li, Y., Srivastava, S., Verschoor, A., Radermacher, M., Grassucci, R.K., Lata, R.K., Agrawal, R.K. A model of protein synthesis based on cryo-electron microscopy of the E. coli ribosome. Nature. 1995;376:441–444. doi: 10.1038/376441a0. [DOI] [PubMed] [Google Scholar]
- Grill, S., Gualerzi, C.O., Londei, P., Blasi, U. Selective stimulation of translation of leaderless mRNA by initiation factor 2: Evolutionary implications for translation. EMBO J. 2000;19:4101–4110. doi: 10.1093/emboj/19.15.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill, S., Moll, I., Hasenohrl, D., Gualerzi, C.O., Blasi, U. Modulation of ribosomal recruitment to 5′-terminal start codons by translation initiation factors IF2 and IF3. FEBS Lett. 2001;495:167–171. doi: 10.1016/s0014-5793(01)02378-x. [DOI] [PubMed] [Google Scholar]
- Janssen, G.R. Eubacterial, archaeabacterial, and eukaryotic genes that encode leaderless mRNA. In: Baltz R., et al., editors. Industrial microorganisms: Basic and applied molecular genetics. ASM Press; Washington, DC: 1993. pp. 59–67. [Google Scholar]
- La Teana, A., Gualerzi, C.O., Brimacombe, R. From stand-by to decoding site. Adjustment of the mRNA on the 30S ribosomal subunit under the influence of initiation factors. RNA. 1995;1:772–782. [PMC free article] [PubMed] [Google Scholar]
- Laursen, B.S., Sorensen, H.P., Mortensen, K.K., Sperling-Petersen, H.U. Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 2005;69:101–123. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Farmer, J.A., Janssen, G.R. A downstream CA repeat sequence increases translation from leadered and unleadered mRNA in Escherichia coli . Mol. Microbiol. 1999;31:1025–1038. doi: 10.1046/j.1365-2958.1999.01228.x. [DOI] [PubMed] [Google Scholar]
- Moll, I., Resch, A., Blasi, U. Discrimination of 5′-terminal start codons by translation initiation factor 3 is mediated by ribosomal protein S1. FEBS Lett. 1998;436:213–217. doi: 10.1016/s0014-5793(98)01131-4. [DOI] [PubMed] [Google Scholar]
- Moll, I., Grill, S., Gualerzi, C.O., Blasi, U. Leaderless mRNAs in bacteria: Surprises in ribosomal recruitment and translational control. Mol. Microbiol. 2002;43:239–246. doi: 10.1046/j.1365-2958.2002.02739.x. [DOI] [PubMed] [Google Scholar]
- Moll, I., Hirokawa, G., Kiel, M.C., Kaji, A., Blasi, U. Translation initiation with 70S ribosomes: An alternative pathway for leaderless mRNAs. Nucleic Acids Res. 2004;32:3354–3363. doi: 10.1093/nar/gkh663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell, S.M., Janssen, G.R. The initiation codon effects ribosome binding and translational efficiency in Escherichia coli of cI mRNA with or without the 5′ untranslated leader. J. Bacteriol. 2001;183:1277–1283. doi: 10.1128/JB.183.4.1277-1283.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell, S.M., Janssen, G.R. Leadeless mRNAs bind 70S ribosomes more strongly than 30S ribosomal subunits in Escherichia coli . J. Bacteriol. 2002;184:6730–6733. doi: 10.1128/JB.184.23.6730-6733.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle, J.M., Brodersen, D.E., Clemons, W.M., Tarry M.J., Jr, Carter, A.P., Ramakrishnan, V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- Ptashne, M., Backman, M.K., Humayum, M.Z., Jeffery, A., Maurer, R., Meyer, B., Sauer, R.T. Autoregulation and function of a repressor in bacteriophage lambda. Science. 1976;194:156–161. doi: 10.1126/science.959843. [DOI] [PubMed] [Google Scholar]
- Ringquist, S., MacDonald, M., Gibson, T., Gold, L. Nature of the ribosomal mRNA track: Analysis of ribosome-binding sites containing different sequences and secondary structures. Biochemistry. 1993;32:10254–10262. doi: 10.1021/bi00089a048. [DOI] [PubMed] [Google Scholar]
- Rinke-Appel, J., Junke, N., Stade, K., Brimacombe, R. The path of mRNA through the Escherichia coli ribosome; site-directed crosslinking of mRNA analogues carrying a photo-reactive label at various points 3′ to the coding site. EMBO J. 1991;10:2195–2202. doi: 10.1002/j.1460-2075.1991.tb07755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke-Appel, J., Junke, N., Brimacombe, R., Lavrik, I., Dokudovskaya, S., Dontsova, O., Bogdanov, A.A. Contacts between 16S ribosomal RNA and mRNA, within the spacer region separating the AUG initiator codon and the Shine–Dalgarno sequence; a site-directed cross-linking study. Nucleic Acids Res. 1994;22:3018–3025. doi: 10.1093/nar/22.15.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluenzen, F., Tocilj, A., Zarivach, R., Harms, J., Gluehmann, M., Janell, D., Bashan, A., Bartels, H., Agmon, I., Franceschi, F., et al. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Selmer, M., Dunham, C.M., Murphy, F.V., Weixlbaumer, A., Petry, S., Kelley, A.C., Weir, R.J., Ramakrishnan, W.V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Sergiev, P.V., Lavrik, I.N., Wlasoff, V.A., Dokudovskaya, S.S., Dontsova, O.A., Bogdanov, A.A., Brimacombe, R. The path of mRNA through the bacterial ribosome: A site-directed crosslinking study using new photoreactive derivatives of guanosine and uridine. RNA. 1997;3:464–475. [PMC free article] [PubMed] [Google Scholar]
- Shatsky, I.N., Bakin, A.V., Bogdanov, A.A., Vasiliev, V.D. How does the mRNA pass through the ribosome? Biochimie. 1991;73:937–945. doi: 10.1016/0300-9084(91)90135-n. [DOI] [PubMed] [Google Scholar]
- Shine, J., Dalgarno, L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: Complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade, K., Rinke-Appel, J., Brimacombe, R. Site-directed cross-linking of mRNA analogues to the Escherichia coli ribosome; identification of 30S ribosomal components that can be cross-linked to the mRNA at various points 5′ with respect to the decoding site. Nucleic Acids Res. 1989;17:9889–9908. doi: 10.1093/nar/17.23.9889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh, M.J., Limbach, P.A. Investigation of methods suitable for the matrix-assisted laser desorption/ionization mass spectrometric analysis of proteins from ribonucleoprotein complexes. Eur. J. Mass Spectrom. 2004;10:89–99. doi: 10.1255/ejms.626. [DOI] [PubMed] [Google Scholar]
- Sukhodolets, M.V., Garges, S., Adhya, S. Ribosomal protein S1 promotes transcriptional cycling. RNA. 2006;12:1505–1513. doi: 10.1261/rna.2321606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedin, K., Resch, A., Blasi, U. Requirements for ribosomal protein S1 for translation initiation of mRNAs with and without 5′-leader sequence. Mol. Microbiol. 1997;25:189–199. doi: 10.1046/j.1365-2958.1997.4421810.x. [DOI] [PubMed] [Google Scholar]
- Tedin, K., Moll, I., Grill, S., Resch, A., Gualerzi, C.O., Blasi, U. Translation initiation factor 3 antagonizes authentic atart codon selection on leaderless mRNAs. Mol. Microbiol. 1999;31:67–77. doi: 10.1046/j.1365-2958.1999.01147.x. [DOI] [PubMed] [Google Scholar]
- Udagawa, T., Shimizu, Y., Ueda, T. Evidence for the translation initiation of leaderless mRNAs by intact 70S ribosomes without its dissociation into subunits in eubacteria. J. Biol. Chem. 2004;279:8539–8546. doi: 10.1074/jbc.M308784200. [DOI] [PubMed] [Google Scholar]
- Van Etten, W., Janssen, G.R. An AUG initiation codon, not codon–anticodon complementarity, is required for the translation of unleadered mRNA in Escherichia coli . Mol. Microbiol. 1998;27:987–1001. doi: 10.1046/j.1365-2958.1998.00744.x. [DOI] [PubMed] [Google Scholar]
- Wimberly, B.T., Broderson, D.E., Clemons W.M., Jr, Morgan-Warren, R.J., Carter, A.P., Vonrhein, C., Hartsch, T., Ramakrishnan, V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- Wu, C.J., Janssen, G.R. Translation of vph mRNA in Streptomyces lividans and Escherichia coli after removal of the 5′-untranslated leader. Mol. Microbiol. 1996;22:339–355. doi: 10.1046/j.1365-2958.1996.00119.x. [DOI] [PubMed] [Google Scholar]
- Wu, C.J., Janssen, G.R. Expression of a streptomycete leaderless mRNA encoding chloramphenicol acetyltransferase in Escherichia coli . J. Bacteriol. 1997;179:6824–6830. doi: 10.1128/jb.179.21.6824-6830.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Y. Site-specific 4-thiouridine incorporation into RNA molecules. Methods Enzymol. 2000;318:71–87. doi: 10.1016/s0076-6879(00)18045-0. [DOI] [PubMed] [Google Scholar]
- Yusupova, G.Z., Yusupov, M.M., Cate, J.H.D., Noller, H.F. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]