Abstract
The human CD45 gene encodes a protein–tyrosine phosphatase that exhibits differential isoform expression in resting and activated T cells due to alternative splicing of three variable exons. Previously, we have used biochemical methods to identify two regulatory proteins, hnRNP L and PSF, which contribute to the activation-induced skipping of CD45 via the ESS1 regulatory element in variable exon 4. Here we report the identification of a third CD45 regulatory factor, hnRNP L-like (hnRNP LL), via a cell-based screen for clonal variants that exhibit an activation-like phenotype of CD45 splicing even under resting conditions. Microarray analysis of two splicing-altered clones revealed increased expression of hnRNP LL relative to wild-type cells. We further demonstrate that both the expression of hnRNP LL protein and its binding to ESS1 are up-regulated in wild-type cells upon activation. Forced overexpression of hnRNP LL in wild-type cells results in an increase in exon repression, while knock-down of hnRNP LL eliminates activation-induced exon skipping. Interestingly, analysis of the binding of hnRNP L and hnRNP LL to mutants of ESS1 reveals that these proteins have overlapping, but distinct binding requirements. Together, these data establish that hnRNP LL plays a critical and unique role in the signal-induced regulation of CD45 and demonstrate the utility of cell-based screens for the identification of novel splicing regulatory factors.
Keywords: CD45, alternative splicing, hnRNP LL, hnRNP L, screening
INTRODUCTION
Alternative splicing is the process by which exons of a given gene are differentially included or excluded from the final message, such that a single gene can be processed into multiple discrete mRNAs (Black 2003; Matlin et al. 2005). Originally thought to affect only a subset of transcripts, it is now known that 60%–80% of genes undergo some form of alternative splicing, often in a manner dictated by cell type or environmental conditions (Modrek et al. 2001; Black 2003; Johnson et al. 2003; Matlin et al. 2005). Each of these variant mRNAs typically encodes a unique protein of distinct function, thus changes in the splicing pattern of a gene can have a profound effect on protein expression and cellular activity (Modrek and Lee 2002). Current estimates, in fact, suggest that at least 15% of disease-causing mutations in humans alter cellular physiology as a consequence of altering pre-mRNA splicing patterns (Matlin et al. 2005).
Alternative splicing is typically controlled by the activity of splicing regulatory proteins that bind to cis-acting auxilary sequences located within or flanking a variable exon (Black 2003; Matlin et al. 2005). Splicing enhancer sequences, and their cognate binding proteins, promote exon inclusion via recruitment of the splicing machinery or “spliceosome,” while splicing silencers disrupt assembly of the splicing machinery and result in exon skipping. Mutations in such enhancer or silencer sequences, as well as mutations in the trans-acting factors that bind these sequences, often lead to significant defects in splicing patterns and alterations in protein expression (Faustino and Cooper 2003). Therefore, identification of the cis and trans-regulatory elements that control the splicing of a given gene is essential for interpreting how that gene will be expressed in particular cell types or under differing environmental conditions.
While alternative splicing is known to occur in all tissues, it is especially prevalent in the nervous and immune systems, where functional diversity is of particular importance (Modrek et al. 2001). With respect to the immune system, a number of functionally relevant genes have been shown to undergo alternative splicing in distinct lymphocyte populations, including several for which isoform expression specifically is regulated in response to antigen stimulation of T cells (Lynch 2004; Ip et al. 2007). A particularly well-documented example of activation-induced splicing in T cells is that of CD45, which encodes a hematopoietic-specific transmembrane protein tyrosine phosphatase. Three variable exons of the CD45 gene (exons 4–6) are inducibly skipped upon T cell activation, leading to decreased phosphatase activity and maintenance of T cell homeostasis (Hermiston et al. 2002; Lynch 2004). The physiologic importance of this change is evidenced by the fact that naturally occurring polymorphisms within the CD45 gene that disrupt signal-induced alternative splicing correlate with susceptibility to a wide range of autoimmune diseases and viral infection in humans (Jacobsen et al. 2000; Tchilian et al. 2001; Tackenberg et al. 2003; Dawes et al. 2006).
Of the three variable exons in CD45, exon 4 is the most robustly regulated and is the exon for which physiologic consequences of regulation have been most clearly demonstrated (Jacobsen et al. 2000; Lynch and Weiss 2000, 2001; Rothrock et al. 2003). In previous work, we identified an exonic splicing silencer (ESS1) within CD45 variable exon 4, which mediates both partial exon repression in resting cells and increased exon skipping upon cellular stimulation (Rothrock et al. 2003). Using a variety of biochemical assays, we recently showed that the partial exon repression in resting cells is conferred via the binding of hnRNP L to ESS1, while cellular activation results in the additional recruitment of PSF to this regulatory sequence, and functional cooperation between hnRNP L and PSF, to yield hyper-repression of exon 4 (Rothrock et al. 2005; Melton et al. 2007). However, in these previous studies we could not rule out the involvement of additional proteins in the regulation of CD45 splicing.
As a complement to our biochemical analysis of CD45 regulatory proteins, we have recently developed a splicing reporter system to identify potential CD45 regulatory factors in vivo (Levinson et al. 2006). Here we report the use of our novel screening approach to identify clonal cell lines that display an “activation-like” phenotype of CD45 splicing (i.e., efficient exon 4 repression), even under resting growth conditions. Strikingly, of the lines we have characterized further, the two with the strongest repression of CD45 exon 4 have increased expression of the hnRNP L homolog hnRNP L-like (hnRNP LL or LL). Further studies reveal that both the expression of hnRNP LL protein and its binding to ESS1 are up-regulated in wild-type (WT) cells upon activation; while the expression and binding of hnRNP L itself remains unchanged under these conditions. Forced overexpression of hnRNP LL results in an increase in exon repression, while knock-down of hnRNP LL eliminates activation-induced exon skipping. Remarkably, analysis of the binding of hnRNP L versus LL to mutants of ESS1 reveals that these proteins have overlapping, but distinct binding requirements. In particular, a mutation in ESS1 that primarily disrupts activation-induced repression abolishes binding of hnRNP LL, while having no effect on the association of hnRNP L. Together, these data demonstrate that hnRNP LL plays a critical and unique role in the signal-induced regulation of CD45. Moreover, as the function of hnRNP LL is not simply redundant to the activity of its more abundant counterpart hnRNP L, these data also suggest important functional distinctions between these two highly related proteins.
RESULTS
A cell-based screen identifies clones with altered splicing phenotype
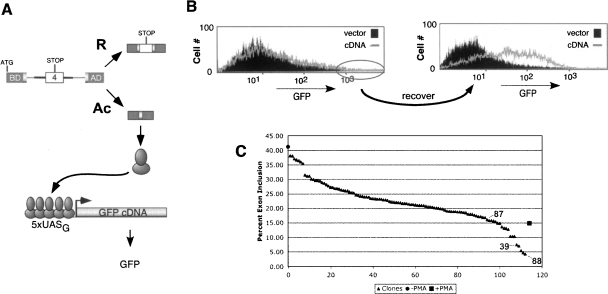
The identification of proteins that regulate splicing in mammals has historically relied primarily on biochemical techniques. While such methods have been successful for isolating proteins that interact directly with RNA, complementary approaches such as genetic screens would be of value in identifying transacting factors not amenable to biochemical detection. However, a significant limitation to the use of cell-based screens in the study of splicing regulation is that many instances of alternative splicing, including that of CD45, involve changes in isoform expression of <10-fold, such that signal-to-noise discrimination can be difficult (Black 2003). Our lab recently developed a dual-reporter system in which transcriptional synergy is used to amplify the detection of changes in alternative splicing (Levinson et al. 2006). In this system, CD45 variable exon 4 is inserted between exons encoding the transcriptional activator Gal4-VP16, such that full-length Gal4-VP16 protein is expressed only when exon 4 is skipped (Fig. 1A). This transcription factor then drives expression of green fluorescent protein (GFP) from a promoter known to respond synergistically to Gal4-VP16 (5xUASG) to yield a proportional, but significantly enhanced change in GFP expression relative to the fold change in splicing (Levinson et al. 2006). For this reporter system we have used the JSL1 cell line that we have previously shown to recapitulate activation-induced skipping of CD45 exon 4 when induced with the phorbolester PMA (Lynch and Weiss 2000). Through screening of JSL1 clones transfected with the dual-reporter plasmids we identified reporter line 3.14, in which the typical three- to fivefold increase in exon 4 skipping induced upon PMA treatment yields a 30- to 50-fold increase in GFP expression (Levinson et al. 2006). This 3.14 reporter cell line has been the basis of our further studies to identify variant clones that display an activation-like pattern of exon 4 splicing (i.e., high GFP) even under resting conditions.
FIGURE 1.
Dual reporter assay of CD45 splicing identified clones with “activated” splicing phenotype. (A) Scheme of dual-reporter system. (B) Flow cytometry analysis of GFP expression in mock-infected resting 3.14 cells (filled), initial cDNA infected cell population (line, left), or resting cell population after recovery from the initial sort (line, right). Circle on left indicates cells that were gated and collected in the initial sort. (C) Graphic representation of exon 4 inclusion in reporter minigene in 106 GFP+ clones (▲), compared with resting (●) or stimulated (■) 3.14 cells. Exon 4 inclusion in all cases was quantitated by low-cycle RT–PCR. Triangles corresponding to data from the three clones chosen for further study are indicated.
A library containing cDNAs from activated cells was engineered in a retroviral expression construct and used to infect resting 3.14 reporter cells. Three days after infection, cells were sorted by flow cytometry for GFP expression. The cells with the highest GFP expression (top 3%–5%) were collected, expanded under resting conditions, and sorted a second time into 96-well plates for the isolation of individual clones (Fig. 1B; see also Materials and Methods). The retroviral infection prior to sorting was intended to enrich for cells in which expression of a novel cDNA caused an activation-like phenotype of CD45 splicing. However, the design of our screen also permitted the isolation of cells in which splicing was altered due to insertional mutagenesis or through natural drift or variation in the 3.14 cell population. A further possible source of GFP-high cells in this screen was from direct alteration of GFP expression or stability. From an initial infection of 70 million cells, 106 GFP-positive clones were obtained. To differentiate between true splicing-altered clones versus those that simply displayed an increase in global transcription or GFP stability, RT–PCR was performed to directly assay the splicing of the exon 4 containing reporter minigene. Strikingly, we observe that 14 clones exhibit equivalent or stronger levels of exon repression than 3.14 cells stimulated with PMA, while an additional 24 clones have at least a twofold increase in exon repression over unstimulated cells (Fig. 1C). While all of these 38 clones are potentially of interest, we initially performed a detailed analysis on three clones that represent a range of exon 4 repression (Fig. 1C, clones 39,87,88).
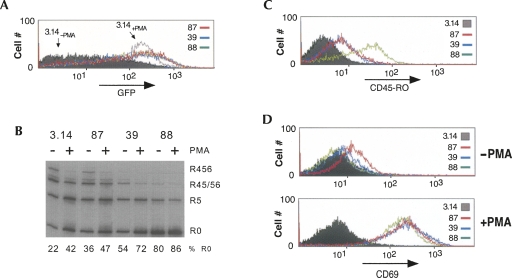
Consistent with the isolation of these clones in the GFP expression screen and with the splicing of the minigene, clones 39, 87, and 88 express GFP to a level similar to stimulated 3.14 cells (Fig. 2A, +PMA). To confirm that the altered splicing of the minigene in these clones was not due to a cis-mutation in the minigene construct or some unanticipated minigene-specific regulatory event, we also analyzed the splicing of the endogenous CD45 gene. As shown in Figure 2B, PMA stimulation of 3.14 cells induces skipping of the three variable exons of CD45, resulting in an accumulation of the smallest CD45 isoform (R0) and a corresponding decrease in the larger isoforms (Fig. 2B, R456, R45/56). Our previous studies have shown that exons 5 and 6 contain the same core regulatory motif as exon 4 (ARS repeats found within the ESS1 sequence, see below) and are likely regulated by overlapping mechanisms, such that we predicted that these exons would be regulated similarly in the clones. Consistent with this prediction and with the splicing results observed with the minigene reporter, the expression of R0 in clone 87 in the absence of PMA is between that of resting and stimulated 3.14 cells, while splicing of CD45 in clones 39 and 88 is even more skewed toward exon skipping (R0) than is observed in 3.14 cells upon stimulation (Fig. 2B). A similar increase in R0 expression in the clones relative to 3.14 cells can also be observed at the protein level using flow cytometry with an antibody that specifically recognizes the R0 isoform (Fig. 2C). Intriguingly, while the clones all have higher levels of basal variable exon exclusion than resting cells, at least for clones 39 and 87, these levels further increase in response to PMA activation (Fig. 2B, cf. +PMA and −PMA for a given clone). This suggests that the individual clones may have alterations in only one of multiple PMA-induced pathways that serve to increase exon repression. Alternatively, the clones may exhibit only a partial activation of a signaling pathway, which is further activated with PMA stimulation.
FIGURE 2.
Individual clones show activated pattern of CD45 splicing without up-regulation of general activation markers. (A) Flow cytometry analysis of GFP expression in isolated resting clones compared with that in resting (−PMA) or stimulated (+PMA) 3.14 cells. (B) RT–PCR analysis of splicing of the endogenous CD45 gene in 3.14 cells and clones. Quantitation of R0 isoform relative to total CD45 RNA is indicated. This and all other quantification in this study are derived from at least two to three independent experiments with deviations of <l15% of average value. (C) Flow cytometry analysis of CD45R0 expression in isolated clones compared with that in 3.14 cells, all under resting conditions. (D) Flow cytometry analysis of CD69 expression in clones versus 3.14 cells grown under resting or stimulated conditions.
CD45 alternative splicing is dependent on a signaling pathway that is initially triggered by the activation of PKC and Ras (Lynch and Weiss 2000); thus, it is possible that our screen will uncover proteins that function at an apical step in this signaling pathway. To test the extent of basal signaling in the clones, we monitored up-regulation of CD69, an event that occurs downstream of Ras activation in normal T cells. Neither clone 39 nor 88 show any significant alteration of CD69 expression relative to 3.14 cells, suggesting that the alteration in these clones is at least downstream of Ras (Fig. 2D). However, we do note some up-regulation of CD69 in clone 87 under resting conditions, as will be discussed below. As a positive control, addition of PMA to all of the cell lines tested results in an equivalent increase in CD69 on the cell surface, demonstrating that the signaling pathways necessary for PMA-induced up-regulating CD69 expression are intact in each of the variant clones (Fig. 2D, +PMA).
HnRNP LL is up-regulated in two activation-phenotype clones and upon T cell stimulation
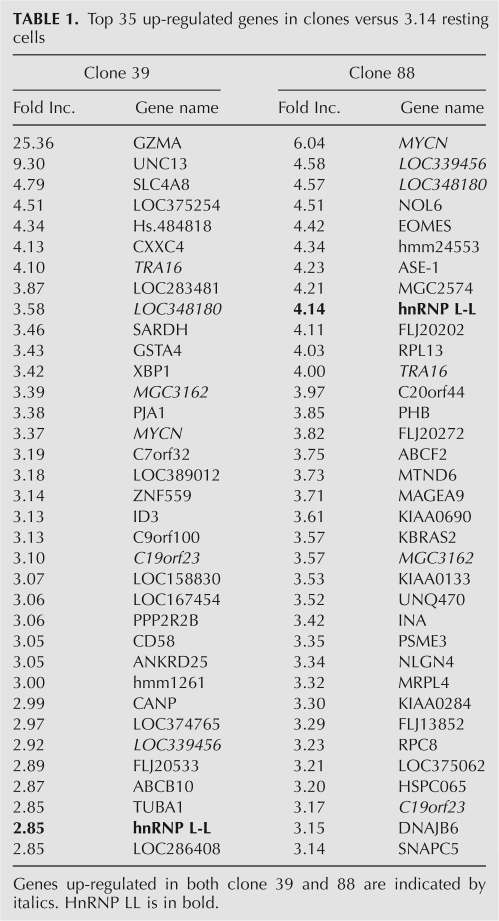
Having identified clonal populations that have altered CD45 splicing, the next goal was to identify the causal functional change in these cells. Despite numerous attempts, none of the retroviral-expressed cDNAs that were recovered from clones 39, 87, and 88 represented full-length open reading frames, and none of these cDNAs were able to induce a change in CD45 splicing when reintroduced into 3.14 or JSL1 cells (data not shown). As discussed above, we recognized that our screening approach was such that we might expect clonal variation or other genetic perturbation apart from introduced cDNAs to be the source of the altered splicing phenotype. Therefore, to identify potential causal genetic changes in a more unbiased manner, we analyzed mRNA expression levels from clones 39, 87, and 88 relative to resting and activated 3.14 cells by standard microarray (see Materials and Methods). The microarray predicted ∼100 genes to be up- or down-regulated by >2.5-fold between each of the clones and resting 3.14 cells (see Supplementary Data for full list), and we find several genes that are up-regulated in both clone 39 and 88 (Table 1, italics; see, also, Discussion). However, the gene that most caught our attention is hnRNP L-like (hnRNP LL), which was among the top 10 most up-regulated genes in our strongest clone (88) and among the top 35 most up-regulated genes in clone 39 (Table 1). HnRNP LL is a close homolog of hnRNP L, which we have already shown to play an essential role in CD45 splicing regulation (Rothrock et al. 2005; Tong et al. 2005; Melton et al. 2007). Moreover, previous mass spectrometry analysis of proteins affinity purified using the core silencer element of exon 4 (ESS1) had suggested hnRNP LL to be a putative ESS1 binding protein, although it was detected at a level much less than hnRNP L itself (C. Rothrock, A. Melton, and K.W. Lynch, unpubl.).
TABLE 1.
Top 35 up-regulated genes in clones versus 3.14 resting cells
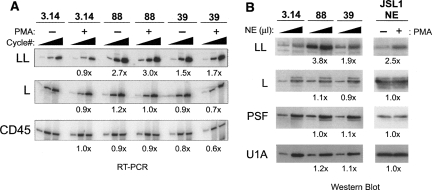
RT–PCR and Western blot analysis confirmed that hnRNP LL is indeed strongly up-regulated at both the message (∼3×) and protein (∼4×) level in clone 88 relative to the parental 3.14 or WT JSL1 cells. Up-regulation of hnRNP LL in clone 39 is more modest (1.5- to twofold) but is still detectable, especially given the slight overall decrease in mRNA levels in this clone (Fig. 3A,B, see CD45 and hnRNP L RNA controls). Clone 87, in contrast, showed no difference in hnRNP LL mRNA or protein relative to 3.14, consistent with our lack of detecting hnRNP LL in the microarray analysis of this clone (data not shown). The potential implications of this difference between clone 87 and 39/88 will be discussed below, but for the purposes of the present study we have not pursued clone 87 further.
FIGURE 3.
HnRNP LL is up-regulated in clones 39 and 88 as well as upon PMA stimulation of WT cells. (A) RT–PCR analysis of hnRNP LL mRNA at 20, 24, and 28 cycles, hnRNP L mRNA at 12, 16, and 20 cycles, or 16, 20, and 24 cycles for CD45 mRNA. Fold increase of signal relative to 3.14 resting cells shown is the average difference quantitated for at least the two lowest cycle points. Constitutive exons 8–10 from the endogenous CD45 gene are used as a control. (B) Western blot analysis of hnRNP LL, L, and PSF in nuclear extracts (NE) from 3.14 cells and clones grown under resting conditions, as well as from parental JSL1 cells grown under resting (−PMA) or activated (+PMA) conditions. All extracts were normalized for total protein level prior to loading and loaded at 5 or 15 μg of total protein per lane. Expression of LL, L, PSF, or U1A (loading control) were quantitated by densitometry at both loading points and averaged to give number shown.
The microarray analysis also did not reveal hnRNP LL to be up-regulated upon stimulation of WT 3.14 cells, and we detect no increase in hnRNP LL message by RT–PCR analysis of resting and activated 3.14 cells (Fig. 3A). However, importantly, we do find that stimulation results in a two- to threefold increase in hnRNP LL protein expression in nuclear extract derived from both JSL1 and 3.14 cells, whereas expression of the other characterized ESS1-regulatory proteins, hnRNP L and PSF, are unchanged (Fig. 3B). Since we have previously demonstrated that these nuclear extracts faithfully recapitulate signal-induced regulation of CD45 (Melton et al. 2007), the increase in hnRNP LL expression in these extracts is consistent with a functional role of this protein in CD45 exon repression. The comparison between steady hnRNP LL mRNA levels and increased nuclear expression is also suggestive of differential localization of hnRNP LL upon stimulation. However, we cannot detect any significant expression of hnRNP LL in cytoplasmic extracts under either resting or stimulated conditions (data not shown). Therefore, we assume that the expression of hnRNP LL is likely regulated at a post-transcriptional level upon T cell activation (see Discussion).
HnRNP LL binds specifically to the ESS1 RNA and causes CD45 exon 4 skipping
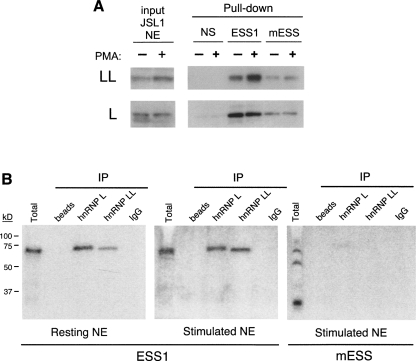
To confirm that hnRNP LL does indeed bind specifically to the ESS1 regulatory element of CD45 exon 4, as suggested by previous mass-spectrometry results, biotinylated RNAs were coupled to beads and used to purify bound proteins from nuclear extracts prepared from resting or stimulated JSL1 cells (Fig. 4A, −PMA,+PMA). For these experiments the RNAs used were the WT ESS1 RNA, a mutant version that is deficient in both basal and activation-induced repressive activity (mESS) (see below, and Rothrock et al. 2003; Melton et al. 2007), or an unrelated silencer sequence (NS) (see Melton et al. 2007). Analysis of the resulting proteins by Western blot demonstrates that hnRNP LL has a similar profile of specificity for these RNAs, as we have previously shown for hnRNP L, as both proteins associate much more strongly with the WT ESS1 RNA than with either control (Fig. 4A; Rothrock et al. 2005). Notably, the binding of hnRNP LL to ESS1 is further enhanced in the stimulated extract as compared with the resting extract, consistent with its corresponding increase in expression (Fig. 4A). In contrast, the binding of hnRNP L to ESS1 remains the same between resting and stimulated extracts, as we have demonstrated previously (Melton et al. 2007).
FIGURE 4.
Binding of hnRNP LL to ESS1 increases relative to hnRNP L upon cellular stimulation. (A) Western blot of hnRNP L and LL following affinity purification (“pull-down”) with biotinylated ESS1, mutant (mESS) or nonspecific (NS) 60-mer RNAs from JSL1 nuclear extract (NE) prepared from resting (−PMA) or activated (+PMA) cells. (B) UV cross-linking experiments done with uniformly labeled ESS1 or mESS RNA and nuclear extract from resting or stimulated cells. As indicated, parallel cross-linked samples were subject to immunoprecipitation with antibodies to hnRNP L, LL, or an IgG control prior to running of the SDS-PAGE.
As further confirmation of the association and specificity of hnRNP LL with ESS1 RNA, we have utilized a UV cross-linking assay. Radiolabeled ESS1 RNA was incubated under standard splicing conditions with nuclear extract derived from resting or stimulated JSL1 cells. Following treatment with UV light and subsequent digestion of the RNA, proteins that were bound to the RNA remain covalently cross-linked to 32P-labeled nucleotides, such that they can be visualized following resolution on an SDS–polyacrylamide gel. Consistent with our previous studies, we observe a single prominent cross-linked species that migrates at ∼70 kD in both resting and stimulated extracts (Fig. 4B, total lanes). Although we have previously shown that antibodies specific to hnRNP L precipitate this cross-linked protein (Rothrock et al. 2005; Tong et al. 2005), such experiments cannot detect whether there are additional cross-linked proteins that do not react with a given antibody. In particular, since hnRNP LL migrates only marginally smaller than hnRNP L, these studies do not allow us to exclude the possibility that hnRNP LL also is a component of the 70-kD band observed in total extract. Therefore, we repeated the immunoprecipitation of the cross-linked reaction with antibodies to hnRNP L or LL (Fig. 4B, IP lanes). As anticipated, antibodies specific for hnRNP L precipitate a similar amount of RNA cross-linked protein from resting or stimulated extract. Interestingly, antibodies specific for hnRNP LL also precipitate cross-linked protein, but in this case the amount of RNA-bound protein is significantly increased in the stimulated extract. Importantly, neither hnRNP L nor LL associate strongly with the mESS control RNA, as indicated by the decrease or absence of both immunoprecipitated proteins as well as loss of the 70 kD species in total extract when the mESS RNA is incubated with either resting or stimulated extract (Fig. 4B, mESS; data not shown).
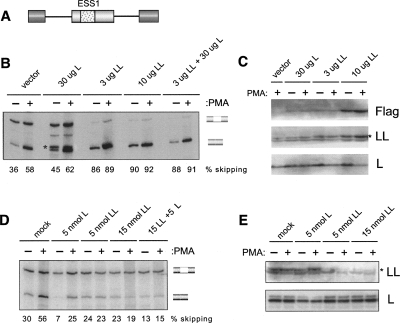
The expression pattern of hnRNP LL, as well as its binding specificity for ESS1, strongly implicate this protein in the signal-induced repression of CD45 exon 4 and suggest that its up-regulation in clones 88 and 39 contribute to the observed increased repression of the CD45 variable exons in these cells. To more directly test this predicted function of hnRNP LL, we first transiently overexpressed a Flag-tagged version of hnRNP LL in the JSL1 cells together with an exon 4 minigene (Fig. 5A). Remarkably, even modest overexpression of hnRNP LL is sufficient to result in a large increase in exon 4 skipping in resting cells (∼40% to ∼85% exon skipping, Fig. 5B,C). In contrast, transfection of a construct encoding hnRNP L causes only a marginal change in splicing, and does not synergize with hnRNP LL. This lack of a significant effect of cotransfected hnRNP L is not inconsistent with its observed role in CD45 splicing in other assays (Fig. 5D; Rothrock et al. 2005; Melton et al. 2007) as the recombinant hnRNP L is expressed about 10-fold less efficiently than hnRNP LL (Fig. 5C), and the endogenous level of hnRNP L is greater than LL, making true overexpression of hnRNP L technically infeasible. Furthermore, the amount of hnRNP L expression construct we have to transfect in order to see any detectable transgene expression is sufficiently high that we observe reduced cellular viability, further complicating the interpretation of hnRNP L overexpression. Therefore, we cannot draw any conclusions from the attempted overexpression of hnRNP L other than to contrast it to the specific result we obtain with hnRNP LL. We also note that overexpression of hnRNP LL does not notably alter the endogenous expression of hnRNP L in these experiments or in clones 39 and 88 (Figs. 3B, 5C). This is distinct from the cross-regulation of hnRNP L and LL observed by others in Hela cells (Hung et al. 2008), but strengthens the conclusion that the effect of hnRNP LL expression on CD45 is direct and not a consequence of perturbing the expression of hnRNP L.
FIGURE 5.
HnRNP LL functionally contributes to repression of CD45 exon 4. (A) Schematic of the WT ESS1 exon 4 minigene described previously (Melton et al. 2007) used in Figures 5 and 6. (B) RT–PCR of WT minigene expression following transient cotransfection in JSL1 cells with constructs expressing Flag-hnRNP L, Flag-hnRNP LL, or Flag vector alone. Percent exon 4 skipping is calculated from two to three independent experiments. Standard error in each case is <15% of exon-skipped value. (*) Cryptic product observed variably with reduced cell viability. (C) Total protein extracts from transfections in B blotted for Flag, hnRNP LL, or hnRNP L. (*) The band corresponding to hnRNP LL, lower band is a nonspecific signal observed on occasional blots. (D) RT–PCR of WT minigene expression following transient cotransfection with morpholino oligomers (MO) that block translation of hnRNP L or LL. Percent exon 4 skipping is calculated as in B. (E) Total protein extracts from transfections in D blotted for hnRNP L or LL. (*) The band corresponding to hnRNP LL.
While the data in Figure 5, B and C, confirm that overexpression of hnRNP LL can force a change in CD45 splicing, these data do not demonstrate that hnRNP LL is an important part of the normal response of CD45 splicing to cellular activation. To test this critical aspect of hnRNP LL function we used morpholino oligomers to knock down protein expression in the JSL1 cells. As we have shown previously (Melton et al. 2007), a ∼50% decrease in hnRNP L expression in JSL1 cells results in a marked overall decrease in exon 4 skipping in both resting and stimulated cells, but does not reduce the fold increase in exon repression induced upon activation (Fig. 5D,E). In striking contrast, depletion of hnRNP LL has only a modest effect on exon 4 skipping in resting cells, but completely abolishes any increase in exon skipping normally induced by PMA treatment (Fig. 5D,E). Moreover, when combined, depletion of hnRNP LL together with L reduces exon 4 skipping to a minor background level regardless of activation state. The observation that depletion of hnRNP LL primarily influences exon 4 expression in activated cells is consistent with the already low level of hnRNP LL expression in resting cells, and provides strong evidence that the increased expression and recruitment of hnRNP LL to the ESS1 sequence is an essential component of the mechanism of activation-induced alternative splicing of CD45.
HnRNP LL binding and functional specificity is distinct from that of hnRNP L
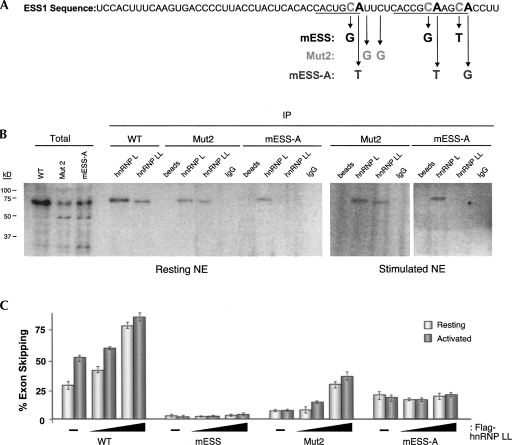
As described above, we have previously implicated both hnRNP L and PSF in the repression of CD45 exon 4. Given our new identification of hnRNP LL as an additional CD45 regulatory protein, we next wanted to begin to investigate how hnRNP LL binding and function relates to that which we have formerly demonstrated for hnRNP L and PSF. The ESS1 regulatory element contains a core sequence (ARS motif) that is common among all three CD45 variable exons (Rothrock et al. 2003). In our earlier studies we have investigated the effects of several mutations within and surrounding this ARS core in the ESS1 RNA. As described above (Fig. 4), mutation of the C residues in the core ARS motif (Fig. 6A, mESS) essentially abolishes all exon 4 skipping in both resting and stimulated cells (Rothrock et al. 2003, 2005; Melton et al. 2007). In contrast, mutation of the pyrimidine residues between the ARS repeats (Fig. 6A, Mut2) or mutation of the conserved As in the ARS motif (Fig. 6A, mESS-A) have a more profound disruption of silencing in activated cells than in resting cells (Melton et al. 2007, and see below). We have further shown that the binding of hnRNP L is disrupted by the mESS mutation, but not by Mut2; while PSF association is abolished by the Mut2 mutation, but not by mESS (Melton et al. 2007); however, neither protein showed sensitivity to the mESS-A mutation (data not shown; see below).
FIGURE 6.
Specificity of hnRNP LL binding to ESS1 is consistent with a unique role in signal-induced exon repression. (A) Sequence of WT ESS1 with location and identity of mutations indicated. Underlined residues correspond to the ARS core motif repeats. (B) UV cross-linking and immunoprecipitation as done in Figure 4 for WT and mutant ESS1 RNAs. (C) RT–PCR analysis of minigenes with WT or indicated mutant ESS1 sequence inserted into ESS1 location of construct shown in Figure 5A. Minigenes were cotransfected with 1 or 3 μg of Flag-hnRNP LL, and quantitation of RT–PCR was as done for Figure 5. Overall exon skipping was somewhat lower in these experiments than in Figure 5 due to inherent variation in transient transfections, but data shown in each figure are from parallel experiments and are thus internally controlled.
To survey the effect of these three mutations on the binding of hnRNP LL, we repeated the UV cross-linking assays. Similar to hnRNP L, we find that the binding of hnRNP LL is markedly reduced by the mESS mutation (Fig. 4), but is largely unaffected by the Mut2 sequence variation (Fig. 6B). Surprisingly, however, while hnRNP L retains full binding activity on an RNA probe containing the mESS-A mutation, hnRNP LL shows no ability to bind this RNA in either resting or stimulated extracts (Fig. 6B, mESS-A). As a further confirmation of the binding specificity of hnRNP LL, we returned to the overexpression experiments using lower input DNA than in Figure 5 so as to not saturate the splicing profiles. Even at these lower amounts of hnRNP LL, we observe a clear increase in exon 4 skipping using the WT ESS1 minigene (Fig. 6C, WT). We also observe a dose-dependent increase in skipping of an exon containing the Mut2 version of ESS1 upon cotransfection of hnRNP LL (Fig. 6C, Mut2), although the overall level of exon skipping is decreased in this construct, consistent with the effect of this mutation on some aspect of CD45 regulation (Melton et al. 2007). In contrast, coexpression of hnRNP LL has no effect on the splicing of either the mESS or mESS-A minigenes (Fig. 6C, mESS,mESS-A). This result is particularly striking for the mESS-A construct in which exon skipping in resting cells is only marginally affected compared with wild type, but no increase in skipping is observed upon either cellular stimulation or cotransfection with hnRNP LL.
Parallel experiments with hnRNP L or PSF were uninformative, as we are unable to achieve overexpression for either of these proteins (Fig. 5; Melton et al. 2007) and knock down experiments such as in Figure 5D similarly are uninterpretable, as the basal skipping in the mutants is already so low that we cannot easily detect an effect even with hnRNP LL. However, the data in Figure 6, B and C, clearly demonstrate that both the binding and function of hnRNP LL are specifically disrupted upon mutation of the C or A residues in ESS1, while the Mut2 mutation disrupts function in a manner independent of this protein. In combination with our previously published work, this distinct specificity of hnRNP LL strongly suggests that this protein plays a critical and nonredundant role with hnRNP L and PSF in the regulation of CD45 splicing.
DISCUSSION
Cell-based screens are only now emerging as a useful tool for identifying splicing regulatory factors (Newman et al. 2006; Kuroyanagi et al. 2007; Ohno et al. 2008). One of the obstacles to the more widespread utilization of screens for splicing regulators is that the typically small expression differences between competing isoforms hinders the robustness of such assays and the feasibility of high-throughput approaches. Here we describe the use of our unique dual-reporter system to identify bonafide mutant clones expressing an activation phenotype of CD45 splicing from a screen of ∼70 million initial cells. Importantly, characterization of two of these mutant clones allowed the identification of a novel regulator of CD45 splicing, hnRNP LL, further emphasizing the utility of such screens.
Analysis of clones 39 and 88 initially suggested that hnRNP LL is an important factor in CD45 splicing regulation, since increased expression of this protein correlated with an increase in CD45 variable exon skipping. Subsequent investigation demonstrated that overexpression of hnRNP LL in WT cells indeed induces skipping of exon 4, while knock down of hnRNP LL causes an increase in exon inclusion primarily in activated cells. This effect of hnRNP LL on CD45 splicing appears direct, as hnRNP LL binds specifically to the ESS1 regulatory sequence, and modulation of the expression of hnRNP LL does not alter expression of hnRNP L in these T cells. Moreover, the fact that hnRNP LL protein expression increases upon stimulation of the JSL1 or 3.14 cells strongly suggests that hnRNP LL is an important component of the signal-induced increase in CD45 exon skipping. Interestingly, the increase in hnRNP LL protein upon stimulation in JSL1 cells does not correlate with an apparent increase in overall message level or change in subcellular localization, suggesting that hnRNP LL is regulated at the post-transcriptional level following T cell activation. The nature of this regulation will be an interesting topic of future study.
One question that arises from this data is how the activity of hnRNP LL relates to our previous characterization of hnRNP L and PSF as general and signal-induced repressors, respectively, of CD45 exon 4 inclusion. Strikingly, our studies presented here combined with our previous studies (Melton et al. 2007) demonstrate that the binding and function of hnRNP L, LL, and PSF are differentially dependent on specific nucleotides in the ESS1 splicing regulatory element. Mutation of the C residues in the ARS repeats (mESS) disrupts both basal and activation-induced silencing and abolishes binding of both hnRNP L and LL; however, only binding of hnRNP LL is disrupted upon mutation of the A residues (mESS-A), which predominantly diminishes silencing under activated conditions. This demonstrates that hnRNP L and LL have overlapping, but distinct binding specificity and is consistent with hnRNP L contributing the majority of the repressive activity under resting conditions, while binding of hnRNP LL is only significant for activation-induced silencing. Interestingly, mutation of the U residues between the ARS motifs (Mut2) does not diminish binding of hnRNP LL, even though this mutation also disrupts activation-induced silencing and abolishes binding of PSF (Melton et al. 2007). Therefore, the binding of hnRNP LL and PSF are separable from one another, even though both are required for maximal activation-induced exon silencing. Whether increased expression of hnRNP LL and increased recruitment of PSF are controlled by the same or distinct signaling pathways and how these proteins interact with each other and with hnRNP L on the RNA is the subject of an ongoing investigation.
The differential specificity and activity observed for hnRNP LL versus hnRNP L suggests that instead of hnRNP LL playing a minor redundant role to hnRNP L, these proteins are true paralogs with related but diverged functions. HnRNP LL and L have 58% overall identity and a similar size. The RNA-binding domains (RRMs) in these proteins are particularly conserved, consistent with the overlapping binding specificity that we observe (Figs. 4, 6); however, there are some divergent residues in these domains that may account for the differential sensitivity of hnRNP L and LL to the mESS-A mutation. Furthermore, the glycine and proline-rich domains in hnRNP L that may be important for function and/or fine tuning of binding specificity are less pronounced in hnRNP LL.
Interestingly, hnRNP LL was initially cloned from bone-marrow stem cells (Shur et al. 2004) and is most abundant in lymphoid cells and testis (http://symatlas.gnf.org/SymAtlas/). HnRNP L is also preferentially expressed in lymphoid cells, but has a higher ubiquitous expression than hnRNP LL across all cell types (http://symatlas.gnf.org/SymAtlas/). In addition, while knock down of hnRNP L in Hela cells alters the splicing of several genes, no aberrant splicing events were detected upon parallel knock down of hnRNP LL (Hung et al. 2008). Therefore, the primary function of hnRNP LL may be to regulate tissue-specific splicing events in a subset of cell types, including T cell-specific splicing events such as that of CD45. Such a relationship between highly similar splicing regulatory proteins is reminiscent of what has been observed for the ubiquitous hnRNP protein PTB and its neural-specific paralog nPTB, which have been shown to have redundant activity on some substrates (Spellman et al. 2007) but distinct effects on neural differentiation and brain-specific splicing events (Boutz et al. 2007; Makeyev et al. 2007). We also cannot rule out that hnRNP LL has cellular functions beyond that of splicing. HnRNP L itself has been shown to influence mRNA polyadenylation, export, translation, and stability (Liu and Mertz 1995; Hahm et al. 1998; Shih and Claffey 1999; Hiu et al. 2003; Guang et al. 2005; Hung et al. 2008), and many of these additional activities may also be shared by hnRNP LL. However, to date, all of our evidence points to hnRNP L and LL directly affecting the splicing of CD45.
In conclusion, the studies we describe here, together with our previous work, demonstrate that hnRNP LL, hnRNP L, and PSF are essential regulators of CD45 splicing. However, this does not preclude the involvement of other factors, either directly on the CD45 RNA or in the upstream events that lead to increased expression of hnRNP LL and/or recruitment of PSF upon activation. In particular, the fact that clone 87 does not have increased expression of hnRNP LL, but does show up-regulation of CD69 (Fig. 2D), suggests that this clone may have an increase in tonic signaling due to a change in apical signaling proteins. Moreover, we note that both clone 88 and 39 show significant up-regulation of several genes in addition to hnRNP LL (Table 1, italics), which may further contribute to CD45 splicing regulation. While preliminary studies of some of these additional genes (e.g., n-Myc) has not revealed a functional role in CD45 splicing (A. Tong and K.W. Lynch, unpubl.), continued analysis of these clones, as well as the other mutant clones that have been identified in this screen, is likely to reveal additional insight into the signaling pathways and regulatory mechanisms that control CD45 isoform expression in response to T cell activation.
MATERIALS AND METHODS
Cell culture and screening
Generation of the 3.14 reporter cell line was described previously (Levinson et al. 2006) from the JSL1 parent cell line (Lynch and Weiss 2000). Cells were plated at a density of 0.3 million/mL and grown in the presence or absence of 20 ng/mL PMA for 48–72 h before harvesting cells for analysis by flow cytometry or RT–PCR. Flow cytometry analysis was done on a FACSCalibur (BD Biosciences) as described previously (Sheives and Lynch 2002), using either endogenous GFP fluorescence or staining with PE-conjugated anti-CD45RO or anti-CD69 antibody (BD Biosciences). Cell sorting was done on a FACStar collecting the brightest 3% on the GFP channel as a pool for the primary sort. Following recovery in 10% FCS in RPMI for 1–2 wk, the cells were sorted again, again collecting the top 5% GFP-expressing clones into 96-well plates to isolate single colonies. These single colonies were expanded and analyzed as individual clonal lines. Transfection of JSL1 cells and vectors used in the transfections were as described previously (Melton et al. 2007).
RT–PCR analysis
Total RNA was harvested from cells using RNABee (Teltest) and analyzed by low-cycle RT–PCR with 32P-end labeled primers as described previously (Rothrock et al. 2003). Primers for RT–PCR analysis of the 3.14 reporter minigene were specific to the Gal4-globin exon 1 (5′-CGCGGATCCGCAGCCGTTACTGCCCTGTGGGGCAAGG) or globin exon 2-VP16 (5′-GCTCTAGAGGCTGCGGAACCCGGACCC) junctions of the minigene, and do not cross-react with any endogenous sequences (Levinson et al. 2006). Primers used to amplify endogenous CD45 were E3 (5′-GGCAAGCCCAACACCTTCCCCCACTG) or E8 (5′-GCTACTCTCCATCTAAGCCAACATGTG) and E9/10 (5′-CTGAAACTTTTCAACCCCTGGTGGCAC). Primers to analyze total mRNA expression for hnRNP L and hnRNP LL are as follows: hnRNP L-E8 (5′-CCCACCTCAACAACAACTTCATGTTTGGGC); hnRNP L-E12R (5′-CGCTCTTGGATTCCCACTCCAGCAGTCC); LL-short-F (5′-GAGCTGGAGGATGGTACCAGCAGC); LL-short-R (5′-GCAAAGCTTCAATGTATAGGGATTGGAACC). The WT and mutant minigenes and primers for analysis are as described previously (Rothrock et al. 2003; Melton et al. 2007). RT–PCR products were suspended in formamide buffer and analyzed on 5% denaturing polyacrylamide gels. Quantitation was performed by densitometry using a Typhoon PhosphorImager (Amersham Biosciences).
Microarray analysis
Total RNA was harvested from clones 39, 87, and 88 grown under resting conditions, as well as 3.14 cells grown under standard resting or stimulated conditions. cDNA was synthesized from total RNA, purified, and hybridized to an Illumina Human-WG6 BeadChip 48K array at the UTSW Microarray Core facility. The arrays were then washed, stained, and scanned according to the manufacturer's protocol. Genes that differed by more than 2.5-fold in intensity between any one sample and the 3.14 resting reference sample were recorded for subsequent analysis.
RNA-binding assays
Direct RNA affinity purifications were performed as described in Melton et al. (2007). Briefly, biotinylated 60 nt RNAs corresponding to the ESS, mESS, or a nonspecific RNA (Dharmacon) were incubated with JSL1 nuclear extract under standard splicing conditions. RNA–protein complexes were then isolated by incubation with streptavidin-agarose beads (Pierce), washed in buffer containing 100 mM KCl., and eluted in SDS-PAGE loading buffer. UV Cross-linking experiments were done as described in Rothrock et al. (2005) by incubating 32P-labeled RNA probes with nuclear extract derived from resting or activated JSL1 cells (Melton et al. 2007) under standard splicing conditions. Reactions were then cross-linked using UV light (254 nm), digested with RNase T1 and RNase A, and resolved by SDS-PAGE. Immunoprecipitation after cross-linking and RNase digestion was done as described previously (Lynch and Maniatis 1996) with antibodies described below.
Morpholino knock down
Antisense knock down of proteins was performed by electroporation of 20 million JSL1 cells with the indicated amount of morpholino oligonucleotide: hnRNP L=CGCCCGCCGCCGCCATCTTCACCAT or hnRNP LL=AGGAAGAGGAGGAGGACATGGCGGC (Gene-Tools, Inc.). Cells were allowed to recover overnight in RPMI+10% fetal calf serum before stimulation with PMA and then harvested for RNA and protein 48 h after treatment with PMA (20 ng/mL).
Antibodies
Antibodies used for the various applications were as follows: anti-hnRNP L (4D11, Abcam), anti-hnRNP LL (Aviva), anti-PSF (6D7, Abnova), U1A (kind gift of Iain Mattaj), anti-Flag (Cell Signaling). The lack of cross-reactivity of antibodies for hnRNP L and LL is suggested by the data shown and confirmed by use of recombinant protein (data not shown). For Western blotting, antibody dilutions and probing conditions were done according to company recommendations for each antibody.
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank the Dallas Cell Analysis Facility and the UTSW Microarray Core Facility for excellent assistance with cell sorting and array analysis, respectively. We also thank Alan Tong and Noah Levinson for technical assistance. This work was supported by R01 GM067719 to K.W.L., an NRSA fellowship to J.D.T. (F32 GM083620), and the Welch Foundation (I-1634).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1212008.
NOTE ADDED IN PROOF
After acceptance of this article, Oberdoerffer et al. (2008) published a study showing that both hnRNP and PSF promote cell surface expression of the CD45 RO protein isoform in activated T cells, consistent with our previous and current data demonstrating a direct effect of these proteins on CD45 splicing (hnRNP L was not able to be knocked down in their study, and thus not directly tested). In addition, they propose that the hnRNP LL protein increases upon T cell stimulation, which our data here demonstrate.
REFERENCES
- Black, D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Boutz, P.L., Stoilov, P., Li, Q., Lin, C.H., Chawla, G., Ostrow, K., Shiue, L., Ares M., Jr, Black, D.L. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes & Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes, R., Hennig, B., Irving, W., Petrova, S., Boxall, S., Ward, V., Wallace, D., Macallan, D.C., Thursz, M., Hill, A., et al. Altered CD45 expression in C77G carriers influences immune function and outcome of hepatitis C infection. J. Med. Genet. 2006;43:678–684. doi: 10.1136/jmg.2005.040485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustino, N.A., Cooper, T.A. Pre-mRNA splicing and human disease. Genes & Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- Guang, S., Felthauser, A.M., Mertz, J.E. Binding of hnRNP L to the pre-mRNA processing enhancer of the herpes simplex virus thymidine kinase gene enhances both polyadenylation and nucleocytoplasmic export of intronless mRNAs. Mol. Cell. Biol. 2005;25:6303–6313. doi: 10.1128/MCB.25.15.6303-6313.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm, B., Kim, Y.K., Kim, J.H., Kim, T.Y., Jang, S.K. Heterogeneous nuclear ribonucleoprotein L interacts with the 3′ border of the internal ribosomal entry site of hepatitis C virus. J. Virol. 1998;72:8782–8788. doi: 10.1128/jvi.72.11.8782-8788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermiston, M.L., Xu, Z., Majeti, R., Weiss, A. Reciprocal regulation of lymphocyte activation by tyrosine kinases and phosphatases. J. Clin. Invest. 2002;109:9–14. doi: 10.1172/JCI14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui, J., Reither, G., Bindereif, A. Novel functional role of CA repeats and hnRNP L in RNA stability. RNA. 2003;9:931–936. doi: 10.1261/rna.5660803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung, L.H., Heiner, M., Hui, J., Schreiner, S., Benes, V., Bindereif, A. Diverse roles of hnRNP L in mammalian mRNA processing: A combined microarray and RNAi analysis. RNA. 2008;14:284–296. doi: 10.1261/rna.725208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip, J.Y., Tong, A., Pan, Q., Topp, J.D., Blencowe, B.J., Lynch, K.W. Global analysis of alternative splicing during T-cell activation. RNA. 2007;13:563–572. doi: 10.1261/rna.457207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, M., Schweer, D., Ziegler, A., Gaber, R., Schock, S., Schwinzer, R., Wonigeit, K., Lindert, R.B., Kantarci, O., Hemmer, B., et al. A point mutation in PTPRC is associated with the development of multiple sclerosis. Nat. Genet. 2000;26:495–499. doi: 10.1038/82659. [DOI] [PubMed] [Google Scholar]
- Johnson, J.M., Castle, J., Garrett-Engele, P., Kan, Z., Loerch, P.M., Armour, C.D., Santos, R., Schadt, E.E., Stoughton, R., Shoemaker, D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Kuroyanagi, H., Ohno, G., Mitani, S., Hagiwara, M. The Fox-1 family and SUP-12 coordinately regulate tissue-specific alternative splicing in vivo. Mol. Cell. Biol. 2007;27:8612–8621. doi: 10.1128/MCB.01508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson, N., Hinman, R., Patil, A., Stephenson, C.R., Werner, S., Woo, G.H., Xiao, J., Wipf, P., Lynch, K.W. Use of transcriptional synergy to augment sensitivity of a splicing reporter assay. RNA. 2006;12:925–930. doi: 10.1261/rna.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., Mertz, J.E. HnRNP L binds a cis-acting RNA sequence element that enables intron-independent gene expression. Genes & Dev. 1995;9:1766–1780. doi: 10.1101/gad.9.14.1766. [DOI] [PubMed] [Google Scholar]
- Lynch, K.W. Consequences of regulated pre-mRNA splicing in the immune system. Nat. Rev. Immunol. 2004;4:931–940. doi: 10.1038/nri1497. [DOI] [PubMed] [Google Scholar]
- Lynch, K.W., Maniatis, T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes & Dev. 1996;10:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- Lynch, K.W., Weiss, A. A model system for the activation-induced alternative-splicing of CD45 implicates protein kinase C and Ras. Mol. Cell. Biol. 2000;20:70–80. doi: 10.1128/mcb.20.1.70-80.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, K.W., Weiss, A. A CD45 polymorphism associated with Muliple Sclerosis disrupts an exonic splicng silencer. J. Biol. Chem. 2001;276:24341–24347. doi: 10.1074/jbc.M102175200. [DOI] [PubMed] [Google Scholar]
- Makeyev, E.V., Zhang, J., Carrasco, M.A., Maniatis, T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin, A.J., Clark, F., Smith, C.W. Understanding alternative splicing: Towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Melton, A.A., Jackson, J., Wang, J., Lynch, K.W. Combinatorial control of signal-induced exon repression by hnRNP L and PSF. Mol. Cell. Biol. 2007;27:6972–6984. doi: 10.1128/MCB.00419-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek, B., Lee, C. A genomic view of alternative splicing. Nat. Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- Modrek, B., Resch, A., Grasso, C., Lee, C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, E.A., Muh, S.J., Hovhannisyan, R.H., Warzecha, C.C., Jones, R.B., McKeehan, W.L., Carstens, R.P. Identification of RNA-binding proteins that regulate FGFR2 splicing through the use of sensitive and specific dual color fluorescence minigene assays. RNA. 2006;12:1129–1141. doi: 10.1261/rna.34906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdoerffer, S., Moita, L.F., Neems, D., Freitas, R.P., Hacohen, N., Rao, A. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science. 2008;321:686–691. doi: 10.1126/science.1157610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, G., Hagiwara, M., Kuroyanagi, H. STAR family RNA-binding protein ASD-2 regulates developmental switching of mutually exclusive alternative splicing in vivo. Genes & Dev. 2008;22:360–374. doi: 10.1101/gad.1620608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothrock, C., Cannon, B., Hahm, B., Lynch, K.W. A conserved signal-responsive sequence mediates activation-induced alternative splicing of CD45. Mol. Cell. 2003;12:1317–1324. doi: 10.1016/s1097-2765(03)00434-9. [DOI] [PubMed] [Google Scholar]
- Rothrock, C.R., House, A.E., Lynch, K.W. HnRNP L represses exon splicing via a regulated exonic splicing silencer. EMBO J. 2005;24:2792–2802. doi: 10.1038/sj.emboj.7600745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheives, P., Lynch, K.W. Identification of cells deficient in signaling-induced alternative splicing by use of somatic cell genetics. RNA. 2002;8:1473–1481. [PMC free article] [PubMed] [Google Scholar]
- Shih, S.C., Claffey, K.P. Regulation of human vascular endothelial growth factor mRNA stability in hypoxia by heterogeneous nuclear ribonucleoprotein L. J. Biol. Chem. 1999;274:1359–1365. doi: 10.1074/jbc.274.3.1359. [DOI] [PubMed] [Google Scholar]
- Shur, I., Ben-Avraham, D., Benayahu, D. Alternatively spliced isoforms of a novel stromal RNA regulating factor. Gene. 2004;334:113–121. doi: 10.1016/j.gene.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Spellman, R., Llorian, M., Smith, C.W. Crossregulation and functional redundancy between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol. Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tackenberg, B., Nitschke, M., Willcox, N., Ziegler, A., Nessler, S., Schumm, F., Oertel, W.H., Hemmer, B., Sommer, N. CD45 isoform expression in autoimmune myasthenia gravis. Autoimmunity. 2003;36:117–121. doi: 10.1080/0891693031000084369. [DOI] [PubMed] [Google Scholar]
- Tchilian, E.Z., Wallace, D.L., Dawes, R., Imami, N., Burton, C., Gotch, F., Beverley, P.C. A point mutation in CD45 may be associated with an increased risk of HIV-1 infection. AIDS. 2001;15:1892–1894. doi: 10.1097/00002030-200109280-00024. [DOI] [PubMed] [Google Scholar]
- Tong, A., Nguyen, J., Lynch, K.W. Differential expression of CD45 isoforms is controlled by the combined activity of basal and inducible splicing-regulatory elements in each of the variable exons. J. Biol. Chem. 2005;280:38297–38304. doi: 10.1074/jbc.M508123200. [DOI] [PubMed] [Google Scholar]