Abstract
Here, we report on the discovery of a locus in the human genome, which evolved by gene duplication followed by an internal DNA inversion. This locus exhibits high sequence similarity to the gene for the inducible isoform of NOS protein (NOS2A) and is transcribed into a noncoding RNA containing a region of significant antisense homology with the NOS2A mRNA. We show that this antisense transcript (anti-NOS2A RNA) is expressed in different types of brain tumors, including meningiomas and glioblastomas. More importantly, we demonstrate that the expression profiles of the anti-NOS2A RNA and the NOS2A mRNA exhibit concurrent reciprocal changes in undifferentiated human embryonic stem cells (hESCs) and in hESCs induced to differentiate into neurogenic precursors such as neurospheres. As NOS2A has a role in neurogenesis, our results suggest that the anti-NOS2A RNA is involved in the regulation of neuronal differentiation of hESCs through the modulation of NOS2A gene expression.
Keywords: antisense RNA, DNA inversion, nitric oxide, stem cells, neuronal differentiation
INTRODUCTION
Natural antisense transcripts (NATs) are usually defined as endogenous RNA molecules that are complementary to transcripts of established function. Depending on their origin, NATs can be classified into two major groups: cis-encoded (produced from the same loci as their sense counterparts) and trans-encoded (transcribed from different loci). Recent studies have shown that NATs are far more abundant in eukaryotic systems than previously thought (Riddihough 2005; Ge et al. 2006; Zhang et al. 2007; Tam et al. 2008; Watanabe et al. 2008), and they appear to be particularly prevalent in the nervous system (for review, see Korneev and O'Shea 2005). In light of the proposed important role of NATs in the regulation of gene expression, attention is now focused on specific examples of NATs and their contribution to physiological and pathophysiological processes.
Our previous work on a molluscan model system Lymnaea stagnalis has shown that a transNAT complementary to the nitric oxide synthase (NOS)-encoding mRNA plays an important role in the regulation of the nitric oxide (NO) signaling pathway in the CNS (Korneev et al. 1999; Korneev et al. 2005). We have also demonstrated that this NAT is transcribed from a NOS pseudogene evolved from a duplicated copy of a NOS gene by internal DNA inversion (Korneev and O'Shea 2002).
We have now discovered a comparable locus in the human genome, which also evolved by gene duplication followed by an internal DNA inversion reminiscent of our finding in Lymnaea. This locus has the highest sequence similarity to the gene encoding the inducible isoform of the NOS protein (NOS2A). We named this locus anti-NOS2A, and show that it is transcribed into a noncoding NAT containing a region of significant complementarity to NOS2A mRNA. This human anti-NOS2A RNA is expressed in different types of brain tumors, including meningiomas and glioblastomas. In addition, we demonstrate that the expression profiles of the anti-NOS2A RNA and the NOS2A mRNA exhibit concurrent reciprocal changes in undifferentiated human embryonic stem cells (hESCs) and in hESCs induced to differentiate into neurogenic precursors. These results suggest that the anti-NOS2A RNA plays an important role in the regulation of neuronal differentiation of hESCs through the modulation of NOS2A gene expression.
RESULTS
Human genome contains a locus evolved from a NOS2A gene by partial duplication coupled with internal DNA inversion
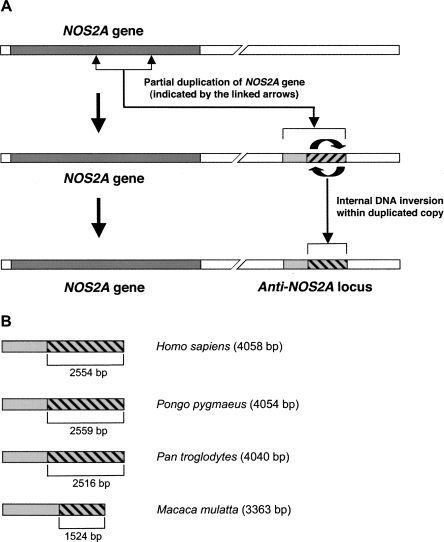
From our molluscan studies we know that NOS-related transNATs can be transcribed from genomic loci evolved by DNA inversions (Korneev and O'Shea 2002). To reveal similarly organized regions in the human genome we first identified all loci exhibiting significant sequence similarity to NOS-encoding mRNAs. The revealed NOS-homologies have been thoroughly examined using the Lasergene software (DNASTAR). Our aim was to identify those loci, in which relatively long regions exhibiting sense homology with NOS-encoding mRNAs coexist with regions showing antisense homology. Remarkably, such an organization, which is indicative of a DNA inversion, was found to be a characteristic feature of one particular locus. This locus is on chromosome 17 at position q23.3 (contig AC053481.13.1.176235), and has high sequence similarity to the gene encoding the inducible isoform of NOS (NOS2A), located on the same chromosome. Hereafter we will refer to this locus as anti-NOS2A, because genomic regions evolved by an internal DNA inversion are a potential source of antisense RNAs (Okano et al. 1988; Tosic et al. 1990; Korneev and O'Shea 2002). No other NOS-homologous loci containing evidence of DNA inversions were found in the human genome. Sequence comparison between the NOS2A gene and the anti-NOS2A locus has enabled us to make the following important observations. First, the anti-NOS2A locus is about 4 kb, which is significantly shorter than the NOS2A gene (∼37 kb). In fact, the homology of the anti-NOS2A locus is restricted to exons 15, 16, 17, 18, and 19 and introns 14, 15, 16, 17, 18, and 19 of the NOS2A gene (Fig. 1A). The average sequence identity between the anti-NOS2A and corresponding regions of the NOS2A gene is about 80%. Thus, we can conclude that the anti-NOS2A locus is the result of a duplication of part of the NOS2A gene. This does not mean, however, that the anti-NOS2A locus necessarily retains the same exon/intron organization as the parent gene. In fact, we showed that this locus is intronless (see the next section). Second, computational analysis has confirmed that the organization of the anti-NOS2A locus has been affected by an internal DNA inversion. The predicted inverted segment is about 2.5 kb, and is homologous to introns 16, 17, 18, and 19 and exons 16, 17, 18, and 19 of the original NOS2A gene (Fig. 1A).
FIGURE 1.
Evolution of anti-NOS2A locus. (A) The chain of evolutionary events suggested by our studies and leading to the creation of the anti-NOS2A locus in the human genome involved partial duplication (indicated by the linked arrows) of an ancestral NOS2A gene coupled to an internal DNA inversion (hatched box) within the duplicated copy (light gray box). Because of such dramatic disruption of the organization, the anti-NOS2A locus cannot produce mature NOS2A mRNA. Instead, it can be transcribed into a NOS2A-related transNAT. (B) Organization of orthologous anti-NOS2A loci in the human (Homo sapiens), orangutan (Pongo pygmaeus), chimpanzee (Pan troglodytes), and rhesus macaque (Macaca mulatta) genomes. Internal DNA inversions are shown by hatched boxes.
Taken together, these observations indicate that the process associated with the evolution of the human anti-NOS2A resembles those associated with the evolution of a previously characterized anti-NOS locus in a mollusc (Korneev and O'Shea 2002). Both involve gene duplication coupled with an internal DNA inversion (Fig. 1A). Such a remarkable evolutionary similarity suggested to us that the human anti-NOS2A locus, if transcribed, could produce functional NOS-related transNATs, as does its molluscan counterpart.
We have also investigated whether similarly organized NOS-homologous loci exist in other mammals. Using a bioinfomatic approach we analyzed the orangutan (Pongo pygmaeus), chimpanzee (Pan troglodytes), and rhesus macaque (Macaca mulatta) genomes and found that the anti-NOS2A locus is conserved in all primates examined (Fig. 1B). The revealed orthologous loci are located in genomic contigs 7902.2 (chromosome 17), 135.50 (chromosome 17), and 1099214711959 (chromosome 16) in orangutan, chimpanzee, and rhesus macaque, respectively. We have, however, failed to identify similar loci in rodents.
Anti-NOS2A locus is transcribed into a NOS2A-related NAT
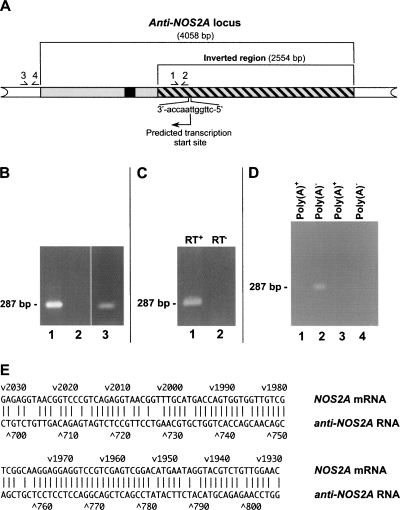
In order to investigate transcriptional status of the anti-NOS2A locus we have analyzed its nucleotide sequence by using Promoter 2.0 software (Knudsen 1999) designed to identify regulatory elements critical for transcription. A region exhibiting the very high likelihood of the presence of a functional promoter has been found within the inverted region and a potential transcription start site has been identified 436 bp upstream of the inversion breakpoint (Fig. 2A). Notably, the promoter was predicted to drive transcription in the opposite orientation with respect to the NOS2A gene, potentially producing an RNA with antisense homology with the NOS2A mRNA. No promoter sequences were identified when the anti-NOS2A locus was analyzed in the opposite orientation.
FIGURE 2.
Anti-NOS2A locus is transcribed into noncoding NOS2A-related NAT. (A) Structural organization of human anti-NOS2A locus. The inverted part of the locus is shown by the hatched box. Primers used in RT-PCR experiments are numbered. The DNA sequence of the anti-NOS2A locus has been analyzed using Promoter 2.0 software (Knudsen 1999). Notably, a region located within the inverted segment has scored 1.082, indicating the very high likelihood of the presence of a functional promoter. The promoter was predicted to drive transcription of the anti-NOS2A locus in the opposite orientation with respect to NOS2A gene. The putative transcription start site and the direction of transcription are shown by an arrow. A DNA region, transcription of which results in the synthesis of RNA fragment antisense to the NOS2A mRNA, is indicated in black. (B) The results of RT-PCR experiments performed on total RNA extracted from human meningioma (lanes 1,2) and glioblastoma (lane 3) using primers 1 and 2 (lanes 1,3) or primers 3 and 4 (lane 2). (C) The results of RT-PCR experiments performed on total RNA from human meningioma using primers 1 and 2. A sequence-specific primer complimentary to a putative RNA containing NOS2A mRNA-homologous region in antisense orientation was used in the RT reaction. Note that there is no product in the experiments in which reverse transcriptase was omitted from the first strand synthesis reaction (lane 2). (D) The results of RT-PCR experiments performed on total RNA from human meningioma using primers 1 and 2 and either poly(A)+ (lanes 1,3) or poly(A)− (lanes 2,4) RNA fractions. Note that there are no products in the experiments in which reverse transcriptase was omitted from the first-strand synthesis reactions (lanes 3,4). (E) Alignment of the antisense region of the anti-NOS2A RNA with its complementary counterpart in the NOS2A mRNA. In this alignment there is >80% complementarity.
To verify the significance of these predictions we employed RT-PCR analysis of RNA extracted from human brain tumors using primers 1 and 2 (Fig. 2A) located downstream of the predicted transcription start site. The results of the experiment shown in Figure 2B fulfill our expectations and demonstrate that the anti-NOS2A locus is indeed transcribed in some types of brain malignancies. Moreover, cloning and sequencing of the PCR product shows that it contains a region exhibiting significant homology with the NOS2A mRNA. In these RT-PCR experiments we used total RNA and a random primer to produce single-stranded cDNA. This does not select for RNAs in a particular orientation, and so the question of the actual orientation of the detected transcript remains unanswered. In order to address this important issue we set up additional RT-PCR experiments in which cDNA was produced using a sequence-specific primer complimentary to putative RNA containing the NOS2A mRNA-homologous region in antisense orientation. A single band of exactly the expected size has been detected (Fig. 2C). This result confirms that the anti-NOS2A locus is transcribed in human brain tumor into a transNAT that contains a region complementary to the conventional NOS2A mRNA. Therefore, hereafter we will refer to this transcript as anti-NOS2A RNA.
Since total RNA contains both polyadenylated [poly(A)+] and nonpolyadenylated [poly(A)−] RNA molecules, it was important to know to which class of RNA the anti-NOS2A RNA belongs. To answer this question we separated total RNA into poly(A)+ and poly(A)− RNA fractions. These fractions were then separately subjected to RT-PCR analysis. The results of the experiment presented in Figure 2D show that the PCR product is detected only when poly(A)− RNA was used as a template in the RT reaction, indicating that the anti-NOS2A RNA belongs to the class of nonpolyadenylated RNAs.
The absence of the poly(A) tail at the 3′ end significantly complicates the task of obtaining the full-length version of the anti-NOS2A RNA using conventional cDNA cloning techniques such as RACE. To solve this problem we employed another RT-PCR-based approach known as “cDNA walking.” Using this strategy we have reconstructed the primary structure of the anti-NOS2A RNA from several overlapping cDNA fragments. Sequence analysis of the anti-NOS2A RNA has resulted in three important findings. First, the anti-NOS2A RNA is a noncoding RNA. Second, there is a perfect uninterrupted match between the anti-NOS2A RNA and the corresponding genomic sequence. This indicates that the anti-NOS2A locus is intronless. Thus, we can conclude that exon/intron organization of the anti-NOS2A locus has been dramatically changed compared to the original NOS2A gene. DNA segments which acted as introns in the NOS2A gene have lost this function in the novel anti-NOS2A locus and become a part of a single “exon.” It is noteworthy that similar transition of introns into exons has been reported by us previously when we analyzed the evolution of NOS-related genes in molluscs. Third, we estimated the size of the anti-NOS2A RNA to be around 1900 nucleotides (nt), since no product was detected in the RT-PCR experiment in which primers 3 and 4 located just outside of the anti-NOS2A locus were used (Fig. 2B). The antisense region of the anti-NOS2A RNA is about 100 nt, and exhibits >80% complementarity to NOS2A mRNA (Fig. 2E).
Anti-NOS2A RNA and NOS2A mRNA exhibit concurrent reciprocal changes in their expression during neuronal differentiation of hESCs
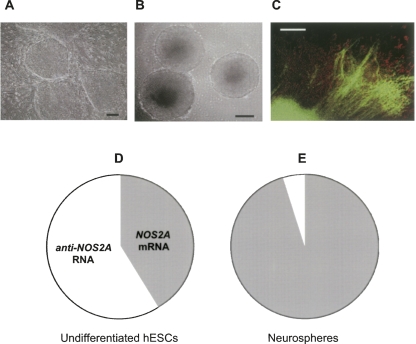
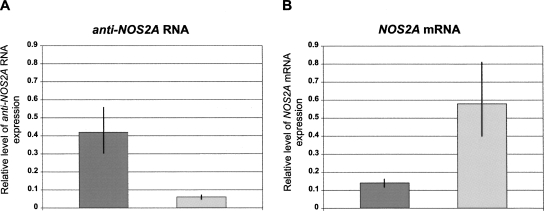
Recent studies indicate that in many types of cancer including certain types of neural malignancies there is a small cell population that exhibits stem cell properties (cancer stem cells) and is responsible for the development and the maintenance of the tumor (Nicolis 2007). This concept recognizes that cancer stem cells share many features with normal stem cells, such as long-term self-renewal and multipotency. In fact, it was shown that when cultured in the presence of mitogenes, cells from primary brain tumors can give rise to neurospheres. Furthermore, there is evidence that molecular mechanisms regulating the balance between self-replication and terminal differentiation are similar in the two systems (Dreesen and Brivanlou 2007). Based on the above, we hypothesized that the anti-NOS2A RNA, which we initially identified in brain tumors, should be also expressed in pluripotent human embryonic stem cells (hESCs). Consequently, we used hESCs as a model system to gain insights into the potential role of the anti-NOS2A RNA in the regulation of NOS2A gene expression during neuronal differentiation. Specifically, by means of real-time RT-PCR we investigated quantitative changes in the expression levels of the anti-NOS2A RNA and NOS2A mRNA in undifferentiated hESCs (Fig. 3A) and in hESCs induced to differentiate into neurogenic precursors such as neurospheres (Fig. 3B). Neurospheres are characterized by a premature neuronal phenotype and contain heterogeneous population of glial cells and neurons (Fig. 3C). Two well-established and characterized cell lines, ESM01 and ESM02, were used for this analysis (Lagarkova et al. 2006). Our quantitative experiments have led to the following important observations. First, we found that the expression level of the anti-NOS2A RNA in undifferentiated ESM01 is almost 1.5 times higher but in neurospheres is ∼20 times lower than the expression level of NOS2A mRNA (Fig. 3D,E). Second, the expression of the anti-NOS2A RNA is ∼7 times higher in the undifferentiated cells than in neurospheres (Fig. 4A). Very similar expression patterns were observed in ESM02 cell line (data not shown). Interestingly, the expression of NOS2A mRNA exhibited the opposite dynamics: namely, it was upregulated in neurospheres compared to the undifferentiated cells (Fig. 4B). From these results we can conclude that neuronal differentiation of hESCs is associated with downregulation of the anti-NOS2A RNA and upregulation of NOS2A mRNA. Thus, there is a clear indication of concurrent reciprocal changes in the expression of the anti-NOS2A RNA and NOS2A mRNA in undifferentiated hESCs and in neurospheres. This suggests an involvement of the anti-NOS2A RNA in neuronal differentiation of hESCs through negative regulation of NOS2A gene expression.
FIGURE 3.
The results of real-time RT-PCR performed on hESCs. (A) Colonies of hESM01 on the feeder layer of mouse embryonic fibroblasts. Scale bar represents 200 μm. (B) Neurospheres derived from hESM01 grown in suspension for 1 wk. Scale bar represents 200 μm. (C) Immunohistochemical analysis of differentiating neurosphere derived from hESM01. Cells were stained with antibodies against human NSE (green) and GFAP (red). Scale bar represents 200 μm. (D, E) Circular diagrams illustrating the ratio between relative levels of NOS2A mRNA (gray sectors) and anti-NOS2A RNA (white sectors) expression in undifferentiated hESM01 (D) and in neurospheres (E) derived from hESM01. The ratios were calculated as 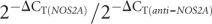 .
.
FIGURE 4.
The expression profiles of the anti-NOS2A RNA and the NOS2A mRNA exhibit concurrent reciprocal changes in undifferentiated hESCs and in hESCs induced to differentiate into neurospheres. (A, B) The relative levels of anti-NOS2A RNA (A) and NOS2A mRNA (B) expression normalized to an endogenous control (GAPDH) and relative to a calibrator in undifferentiated hESM01 (dark gray bars) and in neurospheres derived from hESM01 (light gray bars).
DISCUSSION
In our work on evolution of the NOS gene family in the pond snail Lymnaea stagnalis we found that an ancestral NOS gene was duplicated and that one copy retained its original function, whereas an internal DNA inversion occurred in the other (Korneev and O'Shea 2002). We demonstrated that the mutated copy is transcribed into NOS-related transNAT, which acts as a negative regulator of NOS gene expression (Korneev et al. 1999; Korneev et al. 2005). Here, we report on the discovery in the human genome of the anti-NOS2A locus whose evolution also involved gene duplication followed by an internal DNA inversion. The locus exhibits high sequence identity to the NOS2A gene but cannot produce mature NOS2A mRNA. Because of this, the anti-NOS2A locus can be described formally as an unprocessed NOS2A pseudogene. An additional point emerges from the fact that the orthologous anti-NOS2A locus is present in other primates such as orangutan, chimpanzee, and rhesus macaque, but not in nonprimate mammals examined. This suggests that partial duplication of the ancestral NOS2A gene coupled to inversion mutation occurred at least 25 million years ago.
Importantly, we found that, exactly as its molluscan counterpart, the human anti-NOS2A locus is transcribed into a noncoding NOS-related transNAT. The remarkable similarity in the process by which the NAT-producing loci have evolved in such distantly related organisms suggests strongly that the human anti-NOS2A RNA also plays role in the regulation of NOS gene expression. An important indication that this is so comes from our quantitative experiments in which we studied the expression of both anti-NOS2A RNA and NOS2A mRNA in undifferentiated hESCs and hESCs induced to differentiate into neurogenic precursors such as neurospheres. The rationale for this was that in many systems endogenous NO produced by NOS enzymes is an important component of intracellular pathways controlling neuronal differentiation. In particular, neurogenesis in the mammalian brain is associated with the upregulation of the NOS2A gene expression (Zhu et al. 2003). These findings are in line with the results of our real-time RT-PCR experiments performed on hESCs, which showed a significant increase in the NOS2A gene activity during neuronal differentiation of hESCs. The most exciting outcome of our quantitative studies is, however, the revealed concurrent reciprocal changes in the expression of the anti-NOS2A RNA and NOS2A mRNA in undifferentiated hESCs and in neurospheres. This indicates possible involvement of human anti-NOS2A RNA in the regulation of neuronal differentiation of hESCs through the suppression of NOS2A gene expression.
Another important aspect of the current paper arises from our findings that the anti-NOS2A RNA is expressed in different types of human brain tumor. Experimental work of the past several years has shown that endogenous nitric oxide (NO) produced by NOS2A plays a complex role in tumor biology. Thus, NO production initially induces apoptosis of cells undergoing malignant transformation (Lechner et al. 2005), whereas chronic exposure to NO appears to promote tumor formation (Hussain et al. 2004). These opposing effects probably depend on the local concentration of NOS2A and indicate the existence of molecular machinery delivering fine-tuning of NOS2A gene expression. One potential candidate to fulfill these demands could be a mechanism involving the anti-NOS2A RNA we have discovered.
It is of interest to note that the proposed regulatory role of the anti-NOS2A RNA is also supported, although indirectly, by experiments conducted on myelin-deficient mice. The genome of mutated mice has undergone exactly the same types of rearrangements we discovered in humans, namely, partial duplication of the gene encoding myelin basic protein (MBP) coupled to a DNA inversion occurring in the duplicated copy (Okano et al. 1988, 1991; Tosic et al. 1990). Moreover, the mutated copy was shown to produce a noncoding transNAT that downregulates MBP gene expression leading to a phenotype characteristic for myelin-deficient mice.
Finally, the NOS gene family in mammals consists of three distinct genes encoding three major isoforms of NOS protein: neuronal NOS or NOS1, inducible NOS or NOS2A, and endothelial NOS or NOS3 (for review, see Alderton et al. 2001). The NOS proteins catalyze the production of endogenous NO, an important signaling molecule that is also a reactive and toxic gas. Unlike most other chemical neurotransmitters, it cannot be stored and its production must be tightly controlled by a variety of mechanisms operating at different levels (Bredt 2003). Recently, for example, it was reported that a cisNAT is involved in post-transcriptional regulation of the expression of the NOS3 gene (Robb et al. 2004; Fish et al. 2007). Until now there was no evidence for the existence of NAT-mediated mechanisms controlling the expression of the two other NOS genes. In the light of the data reported here, however, we can suggest that in addition to the NOS3 gene, the expression of the NOS2A gene can also be regulated by a NAT.
MATERIALS AND METHODS
Computational analysis
The human, orangutan, chimpanzee, macaque rhesus, rat, and mouse genomes available on the NCBI server was analyzed by BlastView algorithm provided by the Ensembl project (Hubbard et al. 2007). The organization of the revealed NOS-homologous loci was studied by using the Lasergene software (DNASTAR).
Promoter 2.0 software (Knudsen 1999) was used to predict transcription start sites within the anti-NOS2A locus. On average, the software picks up about 80% of all PolII promoters. Typically, a sequence scoring 0.5–0.8 (marginal prediction) contains about 65% of true promoter sequence. For a sequence scoring 0.8–1.0 (medium likely prediction) this figure is about 80%, and for a region scoring above 1.0 (highly likely prediction) it reaches 95%.
RNA extraction and cDNA synthesis
Total RNA was extracted from human brain tumors and from hESCs by means of Absolutely RNA kit (Stratagene). To remove all traces of genomic DNA the extracted RNA was additionally treated with DNase TURBO according to the manufacture's protocol (Ambion). Some preparations of the total RNA were separated into poly(A)+ and poly(A)− fractions by means of Dynabeads oligo-dT25 (Dynal). Purified RNAs were reverse transcribed using iScript cDNA synthesis kit (Bio-Rad).
Verification of the orientation of RNA transcribed from the anti-NOS2A locus
In these experiments cDNA synthesis was performed using SuperScript II reverse transcriptase (Invitrogen) and a primer (5′-AGGGTAGGAAGCCCAGAG-3′) complimentary to putative RNA containing NOS2A mRNA-homologous region in antisense orientation. After the completion of reaction the primer was removed by means of Chroma Spin-100 columns (Clontech).
Conventional PCR
Synthesized cDNAs were subjected to 35 cycles of PCR using HotStar Taq DNA polymerase (QIAGEN) and primers either 5′-TATATGGACATCAGAAGAAACC-3′ (primer 1) and 5′-TGGAAACTAAGCATATGCTCC-3′ (primer 2) or 5′-GAGACATTCAGAGATGGAG-3′ (primer 3) and 5′-ACAGGTACGAGAAGTTAGG-3′ (primer 4). The identity of PCR products was conformed by sequencing.
“cDNA walking”
The 287 bp RT-PCR product generated by primers 1 and 2 (see Fig. 2) was sequenced, and a second PCR was conducted using a primer targeting the 287 bp cDNA and a primer targeting the anti-NOS2A locus several hundred base pairs downstream of the primer 1. The product of the reaction was sequenced and the whole procedure was then repeated once again. This approach has resulted in the creation of overlapping cDNAs from which the primary structure of the anti-NOS2A RNA was reconstructed.
Real-time PCR
cDNAs produced from RNA preparations extracted from undifferentiated hESCs and from neurospheres were amplified and analyzed on the Mx3000 real-time cycler (Stratagene) using QuantiTect SYBR Green PCR kit (QIAGEN) and the following parameters: denaturation, 94°C, 30 sec; annealing, 52°C, 1 min; extension, 72°C, 30 sec. We used primers 5′-TCCAGACCTCAGGTCATC-3′ and 5′-AGTATAGGCTGAGCTGCC-3′ for the detection of anti-NOS2A RNA; primers 5′-GACATCAACAACAATGTGGAG-3′ and 5′-TTCTGCTGCTTGCTGAGG-3′ for the detection of NOS2A mRNA; primers 5′-AAGGTGAAGGTCGGAGTC-3′ and 5′-GTAAACCATGTAGTTGAGGTC-3′ for the detection of GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA. The identity of all PCR products was confirmed by sequencing. The amount of target transcript (either anti-NOS2A RNA or NOS2A mRNA), normalized to an endogenous reference (GAPDH) and relative to a calibrator was calculated as 2−ΔΔC T (Pfaffl 2001) where ΔΔCT = ΔCT − ΔCT(CAL). ΔCT and ΔCT(CAL) are the differences in threshold cycles for target and reference measured in the samples and in the calibrator, respectively.
Cultivation and differentiation of hESCs
HESCs were cultured in medium consisted of 80% KnockOut DMEM, 20% FBS, 1 mM glutamine, 1% nonessential amino acids, 50 units/mL penicillin, 50 μg/mL streptomycin (all from Invitrogen), 0.1 mM b-mercaptoethanol (Sigma), and 4 μg/mL bFGF (Chemicon) on mitomycin C treated (10 μg/mL, Sigma) mouse embryonic fibroblasts (MEF) using tissue culture dishes precoated with 0.1% gelatin (Merck). MEFs were prepared from day 12.5 p.c. fetuses of F1 (C57BL/6J×CBA/Ca) mice. hESC colonies were replated every 5–6 d by exposure to type IV collagenase (200 U/mL, Invitrogen) followed by mechanical dissociation. Cells between passages 20 and 23 were used for the real-time RT-PCR analysis.
Differentiation of hESCs into neurospheres was performed as described (Gerrard et al. 2005; Itsykson et al. 2005) with some modifications. To induce neuronal differentiation dishes with undifferentiated hESCs grown for 7 d were treated with collagenase IV for 10 min, washed twice with growth medium, and collected. Colonies were transferred to new 35 mm Petri dishes precoated with poly L-lysin and laminin (Sigma) and cultivated in DMEM/F12 (Hyclone) containing N2 and B27 supplements (Invitrogen), 50 ng/mL recombinant human noggin (Chemicon), and 20 ng/mL bFGF (Chemicon). Medium was changed every 2 d. After 2 wk of cultivation rosette-like structures were collected and cultivated in suspension in 12-well plates precoated with 1.5% Agarose (Bio-Rad). After 5 d, cell clumps displayed typical neurosphere morphology. Neurospheres were propagated for 2 wk in DMEM/F12 (Hyclone) containing N2 and B27 supplements (Invitrogen), 20 ng/mL bFGF (Chemicon), and 10 ng/mL EGF (Sigma).
Immunohistochemistry
Cultured cells were fixed in 100% ice-cold methanol for 5 min, washed for 1 h in blocking buffer (PBS, 0.1% Tween-20, 5% FBS, 2% goat serum), and incubated for 1 h at room temperature with primary mouse antihuman neuron-specific enolase (NSE) (1:50; Chemicon) and rabbit antihuman glial fibrillary acidic protein (GFAP) (1:1000; Chemicon) antibodies in PBS, 0.1% Tween 20. Following three washes with PBS, 0.1% Tween 20, cells were incubated with secondary antibodies: Alexa Fluor 488-conjugated goat antimouse IgG and Alexa Fluor 546-conjugated goat antirabbit IgG (diluted 1:800; Molecular Probes).
ACKNOWLEDGMENTS
This work was supported by a Wellcome Trust grant. Funding to pay the Open Access publication charges for this article was provided by Wellcome Trust.
Footnotes
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1084308.
REFERENCES
- Alderton, W.K., Cooper, C.E., Knowles, R.G. Nitric oxide synthase: Structure, function and inhibition. Biochem. J. 2001;357:593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredt, D.S. Nitric oxide signaling specificity—The heart of the problem. J. Cell Sci. 2003;116:9–15. doi: 10.1242/jcs.00183. [DOI] [PubMed] [Google Scholar]
- Dreesen, O., Brivanlou, A.H. Signaling pathways in cancer and embryonic stem cells. Stem Cell Rev. 2007;3:7–17. doi: 10.1007/s12015-007-0004-8. [DOI] [PubMed] [Google Scholar]
- Fish, J.E., Matouk, C.C., Yeboah, E., Bevan, S.C., Khan, M., Patil, K., Ohh, M., Marsden, P.A. Hypoxia-inducible expression of a natural cis-antisense transcript inhibits endothelial nitric-oxide synthase. J. Biol. Chem. 2007;282:15652–15666. doi: 10.1074/jbc.M608318200. [DOI] [PubMed] [Google Scholar]
- Ge, X., Wu, Q., Jung, Y.-C., Chen, J., Wang, S.M. A large quantity of novel human antisense transcripts detected by LongSAGE. Bioinformatics. 2006;22:2475–2479. doi: 10.1093/bioinformatics/btl429. [DOI] [PubMed] [Google Scholar]
- Gerrard, L., Rodgers, L., Cui, W. Differentiation of human embryonic stem cells to neural lineages in adherent culture by blocking bone morphogenetic protein signaling. Stem Cells. 2005;23:1234–1241. doi: 10.1634/stemcells.2005-0110. [DOI] [PubMed] [Google Scholar]
- Hubbard, T.J., Aken, B.L., Beal, K., Ballester, B., Caccamo, M., Chen, Y., Clarke, L., Coates, G., Cunningham, F., Cutts, T., et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain, S.P., Trivers, G.E., Hofseth, L.J., He, P., Shaikh, I., Mechanic, L.E., Doja, S., Jiang, W., Subleski, J., Shorts, L., et al. Nitric oxide, a mediator of inflammation, suppresses tumorigenesis. Cancer Res. 2004;64:6849–6853. doi: 10.1158/0008-5472.CAN-04-2201. [DOI] [PubMed] [Google Scholar]
- Itsykson, P., Ilouz, N., Turetsky, T., Goldstein, R.S., Pera, M.F., Fishbein, I., Segal, M., Reubinoff, B.E. Derivation of neural precursors from human embryonic stem cells in the presence of noggin. Mol. Cell. Neurosci. 2005;30:24–36. doi: 10.1016/j.mcn.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Korneev, S.A., O'Shea, M. Evolution of nitric oxide synthase regulatory genes by DNA Inversion. Mol. Biol. Evol. 2002;19:1228–1233. doi: 10.1093/oxfordjournals.molbev.a004183. [DOI] [PubMed] [Google Scholar]
- Korneev, S.A., O'Shea, M. Natural antisense RNAs in the nervous system. Rev. Neurosci. 2005;16:213–222. doi: 10.1515/revneuro.2005.16.3.213. [DOI] [PubMed] [Google Scholar]
- Korneev, S.A., Park, J.-H., O'Shea, M. Neuronal expression of neural nitric oxide synthase (nNOS) protein is supressed by an antisense RNA transcribed from a NOS pseudogene. J. Neurosci. 1999;19:7711–7720. doi: 10.1523/JNEUROSCI.19-18-07711.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneev, S.A., Straub, V., Kemenes, I., Korneeva, E.I., Ott, S.R., Benjamin, P.R., O'Shea, M. Timed and targeted differential regulation of NOS and antiNOS genes by reward conditioning leading to long-term memory formation. J. Neurosci. 2005;25:1188–1192. doi: 10.1523/JNEUROSCI.4671-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen, S. Promoter 2.0: for the recognition of PolII promoter sequences. Bioinformatics. 1999;15:356–361. doi: 10.1093/bioinformatics/15.5.356. [DOI] [PubMed] [Google Scholar]
- Lagarkova, M.A., Volchkov, P.Y., Lyakisheva, A.V., Philonenko, E.S., Kiselev, S.L. Diverse epigenetic profile of novel human embryonic stem cell lines. Cell Cycle. 2006;5:416–420. doi: 10.4161/cc.5.4.2440. [DOI] [PubMed] [Google Scholar]
- Lechner, M., Lirk, P., Rieder, J. Inducible nitric oxide synthase (iNOS) in tumor biology: The two sides of the same coin. Semin. Cancer Biol. 2005;15:277–289. doi: 10.1016/j.semcancer.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Nicolis, S.K. Cancer stem cells and “stemness” genes in neuro-oncology. Neurobiol. Dis. 2007;25:217–229. doi: 10.1016/j.nbd.2006.08.022. [DOI] [PubMed] [Google Scholar]
- Okano, H., Ikenaka, K., Mikoshiba, K. Recombination within the upstream gene of duplicated myelin basic protein genes of myelin deficient shimld mouse results in the production of antisense RNA. EMBO J. 1988;7:3407–3412. doi: 10.1002/j.1460-2075.1988.tb03214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano, H., Aruga, J., Nakagawa, T., Shiota, C., Mikoshiba, K. Myelin basic protein gene and the function of antisense RNA in its repression in myelin-deficient mutant mouse. J. Neurochem. 1991;56:560–567. doi: 10.1111/j.1471-4159.1991.tb08186.x. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddihough, G. In the forest of RNA dark matter. Science. 2005;309:1507. [Google Scholar]
- Robb, G.B., Carson, A.R., Tai, S.C., Fish, J.E., Singh, S., Yamada, T., Scherer, S.W., Nakabayashi, K., Marsden, P.A. Post-transcriptional regulation of endothelial nitric-oxide synthase by an overlapping antisense mRNA transcript. J. Biol. Chem. 2004;279:37982–37996. doi: 10.1074/jbc.M400271200. [DOI] [PubMed] [Google Scholar]
- Tam, O.H., Aravin, A.A., Stein, P., Girard, A., Murchison, E.P., Cheloufi, S., Hodges, E., Anger, M., Sachidanandam, R., Schultz, R.M., et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosic, M., Roach, A., de Rivaz, J.C., Dolivo, M., Matthieu, J.M. Post-transcriptional events are responsible for low expression of myelin basic protein in myelin deficient mice: Role of natural antisense RNA. EMBO J. 1990;9:401–406. doi: 10.1002/j.1460-2075.1990.tb08124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, T., Totoki, Y., Toyoda, A., Kaneda, M., Kuramochi-Miyagawa, S., Obata, Y., Chiba, H., Kohara, Y., Kono, T., Nakano, T., et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Li, J., Kong, L., Gao, G., Liu, Q.-R., Wei, L. NATsDB: Natural antisense transcripts database. Nucleic Acids Res. 2007;35:D156–D161. doi: 10.1093/nar/gkl782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, D.Y., Liu, S.H., Sun, H.S., Lu, Y.M. Expression of inducible nitric oxide synthase after focal cerebral ischemia stimulates neurogenesis in the adult rodent dentate gyrus. J. Neurosci. 2003;23:223–229. doi: 10.1523/JNEUROSCI.23-01-00223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]