Abstract
Adenosine deaminases that act on RNA (ADARs) are editing enzymes that convert adenosine to inosine in double-stranded RNA (dsRNA). ADARs sometimes target codons so that a single mRNA yields multiple protein isoforms. However, ADARs most often target noncoding regions of mRNAs, such as untranslated regions (UTRs). To understand the function of extensive double-stranded 3′ UTR structures, and the inosines within them, we monitored the fate of reporter and endogenous mRNAs that include structured 3′ UTRs in wild-type Caenorhabditis elegans and in strains with mutations in the ADAR genes. In general, we saw little effect of editing on stability or translatability of mRNA, although in one case an ADR-1 dependent effect was observed. Importantly, whereas previous studies indicate that inosine-containing RNAs are retained in the nucleus, we show that both C. elegans and Homo sapiens mRNAs with edited, structured 3′ UTRs are present on translating ribosomes.
Keywords: ADAR, RNA editing, double-stranded RNA, post-transcriptional regulation, translation
INTRODUCTION
Adenosine deaminases that act on RNA, or ADARs, are present in all animals, where they bind dsRNA and catalyze the hydrolytic deamination of adenosine (A) to inosine (I) (Bass 2002; Valente and Nishikura 2005; Jepson and Reenan 2007). The conversion of A to I alters a double-stranded structure by replacing stable AU base pairs with IU mismatches (Bass and Weintraub 1988; Wagner et al. 1989). Further, as inosine is read as guanosine by the translational machinery, ADARs can alter the protein coding information of an mRNA.
The first RNAs found to be “edited” by ADARs were identified serendipitously and contained inosines in codons (Sommer et al. 1991; Kohler et al. 1994; Burns et al. 1997). Subsequently, a systematic approach to identify ADAR substrates was performed with RNA isolated from human brain and Caenorhabditis elegans. Surprisingly, this study identified mRNAs that exclusively contained inosines in noncoding regions, such as untranslated regions (UTRs) and introns (Morse and Bass 1999; Morse et al. 2002). More recent bioinformatics studies gave similar results and predicted that >5% of human mRNAs contain ADAR editing sites in their noncoding regions (Athanasiadis et al. 2004; Blow et al. 2004; Kim et al. 2004; Levanon et al. 2004). Despite the vast number of mRNAs with edited noncoding regions, the function of these editing events is unknown.
The number of A to I editing events that occur within a dsRNA is determined by its structure (Bass 2002). ADARs act selectively at codon editing sites, where the structure is usually disrupted by mismatches, bulges, and loops. In contrast, RNA that is completely, or largely, double stranded, such as that within 3′ UTR structures, is deaminated nonselectively, at the extreme showing 50%–60% A to I conversion. Several studies suggest that such extensively edited RNAs are retained in the nucleus. For example, the early and late transcripts of polyoma virus are transcribed in opposing directions, and at certain times of infection overlap to create regions of complementarity. The resulting dsRNA is targeted by ADARs, and the extensively edited RNA is retained in the nucleus (Kumar and Carmichael 1997). Similarly, a mouse mRNA with inosines in its 3′ UTR (CTN-RNA) was observed in the nucleus, sequestered in subnuclear paraspeckle structures (Prasanth et al. 2005). In this case, a stress-induced cleavage event is proposed to remove the inosine-containing region of the mRNA to allow its export to the cytoplasm.
A current paradox in the field centers on the numerous mRNAs with extensively edited structures in their UTRs. Many of these mRNAs encode essential proteins, and if they are retained in the nucleus, how are they translated? Are the inosine-containing regions removed from these mRNAs to allow export to the cytoplasm? To address these questions, we studied the fate of C. elegans mRNAs with edited structures in their 3′ UTRs. In contrast to mammals, ADARs are not essential in C. elegans, and this allowed us to directly compare the fate of mRNAs in wild-type and ADAR mutant strains. We did not observe cleavage of mRNAs with structured 3′ UTRs, and editing did not significantly alter mRNA stability or translatability. Importantly, in contrast to previous studies that suggest edited RNA is retained in the nucleus, we find that both C. elegans and Homo sapiens mRNAs with edited, structured 3′ UTRs are present on translating ribosomes.
RESULTS
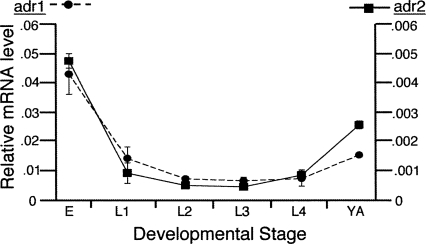
C. elegans expresses two ADAR genes, adr-1 and adr-2, and we began our studies of the fates of edited mRNAs by monitoring ADAR mRNA levels during the C. elegans lifecycle (Fig. 1). RNA was extracted from specific developmental stages of wild-type worms, and ADAR mRNA levels were determined using quantitative real-time PCR (qRT-PCR). For both adr-1 and adr-2 mRNA, the highest expression was observed in embryos (Fig. 1). In addition, for both ADARs, mRNA levels decreased dramatically after embryogenesis, remained relatively constant during larval stages, and increased again at the onset of adulthood. Consistent with the mRNA expression, a polyclonal antibody of ADR-1 showed the highest protein levels in embryos (data not shown). Although a 10-fold difference in mRNA levels of adr-1 compared with adr-2 suggests that adr-1 is more abundant, the lack of an ADR-2 antibody prevented confirmation of a difference at the protein level. Since ADARs were expressed at the highest levels in embryo and adult worms, these stages were used for subsequent analyses.
FIGURE 1.
mRNA levels of C. elegans ADARs. Points represent average mRNA level in RNA isolated from two independent developmental time-course experiments using wild-type worms. Relative expression of adr-1 (circle, dashed line) and adr-2 (square, solid line) was quantified by qRT-PCR (error bars=SEM) with normalization to the average of three genes, gpd-3, ama-1, and fasn-1, that were previously determined to be expressed at relatively constant levels throughout development (Jiang et al. 2001). Y-axes are 10-fold different and indicate the relative level of adr-1 (left) and adr-2 (right) mRNA. X-axis denotes worm stages as embryo (E), larval (L1–L4), and young adult (YA).
Analyses of reporter mRNAs containing edited, structured 3′ UTRs
To understand the effect of edited, structured 3′ UTRs on the fate of mRNA, we first employed an in vivo reporter assay. The sequence encoding red fluorescent protein (RFP) was cloned upstream of five C. elegans 3′ UTRs known to be edited (Morse and Bass 1999; Morse et al. 2002). These 3′ UTRs were chosen to represent a range of double-stranded lengths and a number of editing sites, including the shortest (unc-64, 85 base pairs [bp]) and longest (C35E7.6, 660 bp) 3′ UTR structures, and likewise, one containing very few (pop-1, seven sites) and many (C35E7.6, 188 sites) editing sites (Supplemental Fig. 1).
As a control, the sequence encoding green fluorescent protein (GFP) was cloned upstream of the 3′ UTR of the unc-54 gene. The 3′ UTR of unc-54 is not edited by ADARs and does not form a highly base-paired structure like that observed in ADAR substrates. As C. elegans ADARs were previously suggested to function in the nervous system (Tonkin et al. 2002), both reporters were cloned downstream from a neuron-specific promoter, rab-3, and co-expressed. Some of the transgenic animals were created by co-injection of the reporters into wild-type worms to give rise to extrachromosomal arrays, while others were created by integration of both reporters into the genome; although the expression level differs between the integrated constructs and those expressed extrachromosomally, identical results were obtained for the different transgenes.
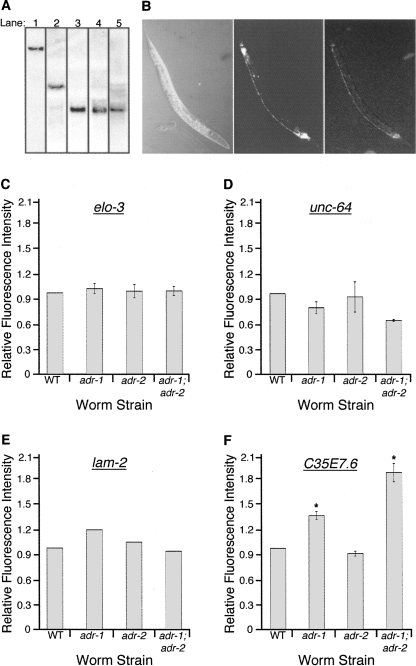
Northern analyses of total RNA isolated from wild-type strains, using probes to the rfp protein coding sequence, revealed that the reporter mRNAs migrated at their predicted size (RFP open-reading frame [ORF] fused to 3′ UTR) (Fig. 2A). The detection of only one mRNA species in each transgenic animal indicates that, at least under normal growth conditions, the reporter mRNA does not undergo a post-transcriptional cleavage to remove the 3′ UTR as was reported for mouse CTN-RNA (Prasanth et al. 2005). Furthermore, amplification and sequencing of the 3′ UTR regions of unc-64, lam-2, C35E7.6, and pop-1 fused to rfp indicate that the reporter 3′ UTR fusions are edited at the same sites as the endogenous genes (data not shown).
FIGURE 2.
In vivo reporter assay for mRNAs with edited 3′ UTRs. (A) Northern blots were performed on total RNA from wild-type strains expressing both rab3∷gfp∷unc-54 (3′ UTR) and the following: (Lane 1) rab3∷rfp∷C35E7.6 (3′ UTR), (lane 2) rab3∷rfp∷unc-64 (3′ UTR), (lane 3) rab3∷rfp∷elo-3 (3′ UTR), (lane 4) rab3∷rfp∷pop-1 (3′ UTR), (lane 5) rab3∷rfp∷lam-2 (3′ UTR). Blots were hybridized with a probe for the rfp coding sequence. (B) Nomarski, GFP, and RFP images (left to right) of the wild-type strain expressing the transgenic rab3∷gfp∷unc-54 (3′ UTR) and rab3∷rfp∷C35E7.6 (3′ UTR) reporters. (C–F) For various strains, the height of the bar represents the fluorescence intensity of reporter proteins synthesized from mRNA with structured 3′ UTRs relative to that derived from mRNA with the control 3′ UTR, normalized to a wild-type (WT) value of 1. Error bars show SEM for multiple trials, where each trial (n) included 13 young adult worms. Analyses were performed on worms expressing the following transgenes: (C) rab3∷rfp∷elo-3 (3′ UTR) and rab3∷gfp∷unc-54 (3′ UTR), n = 3; (D) rab3∷rfp∷unc-64 (3′ UTR) and rab3∷gfp∷unc-54 (3′ UTR), n = 2; (E) rab3∷gfp∷lam-2 (3′ UTR) and rab3∷rfp∷unc-54 (3′ UTR), n = 1; (F) rab3∷rfp∷C35E7.6 (3′ UTR) and rab3∷gfp∷unc-54 (3′ UTR), n = 3. (*) Significant differences of P = 0.001. Both sets of adr alleles were used for analyzing reporters of elo-3, unc-64, and C35E7.6, and were found to give similar results; thus, only a single set of alleles was used in the lam-2 analysis. In C, D, and F, data shown for adr-1 were performed with the tm668 allele, and for adr-2 with the ok735 allele; in E, adr-1 and adr-2 indicate the gv6 and gv42 alleles, respectively.
Reporter proteins fused to each of the five ADAR substrate 3′ UTRs were clearly expressed in wild-type animals (Fig. 2B). In hopes of revealing effects of editing, we monitored reporter expression in wild-type as well as adr mutant animals. Wild-type strains expressing both the reporter with the control, unstructured 3′ UTR, and the reporter with a highly base-paired 3′ UTR, were crossed with the adr-1;adr-2 mutant strain, and the homozygous single and double mutants containing the reporter constructs were isolated from the F1 progeny. Using confocal microscopy, fluorescence intensity was monitored to compare expression of the reporters in the various strains. Fluorescence from the reporter with the structured 3′ UTR was divided by that from the reporter with the unstructured 3′ UTR to determine a ratio, and these values were normalized to wild type. For three of the four reporters, we did not observe significant differences in protein expression between wild-type and ADAR mutant strains (Fig. 2C–E), indicating that editing in these 3′ UTRs did not affect translatability. This was consistent with Western blot analysis of endogenous POP-1, which showed similar levels of protein expression in all strains (data not shown). However, for the reporter ORF fused to the 3′ UTR of C35E7.6, we observed a reproducible increase of 1.4-fold in the adr-1 mutant and 1.9-fold in the adr-1;adr-2 mutant (Fig. 2F), with no significant change in the mRNA levels of the reporter (data not shown). Similarly, an ADR-1 dependent increase in protein expression was obtained when the C35E7.6 (3′ UTR) reporter transgene was introduced into animals containing different adr mutant alleles (Supplemental Fig. 2). Previous studies show that ADR-2 is the active deaminase in C. elegans, and worms deficient for adr-2 completely lack all editing (Tonkin et al. 2002). While ADR-1 can modulate editing, it has no deaminase activity on its own. Thus, since the reporter fused to the C35E7.6 (3′ UTR) was expressed at wild-type levels in the adr-2 mutant, differences observed in adr-1 and adr-1;adr-2 strains cannot be due to alterations in editing. In sum, while some ADR-1 dependent differences were observed, our data indicate that mRNAs with highly structured 3′ UTRs are translated similarly in wild-type animals and those lacking all ADAR editing.
mRNAs with structured 3′ UTRs are loaded onto polysomes
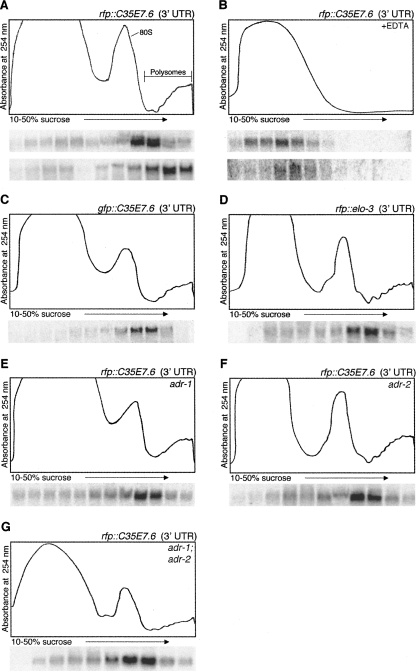
Our analyses of mRNAs with reporter ORFs fused to various 3′ UTRs showed that these mRNAs were translated in wild-type and ADAR mutant animals. Although Northern analyses indicated the reporter mRNA was full-length, we considered the possibility that there were low levels of mRNAs lacking the structured 3′ UTR, and that these truncated mRNAs gave rise to the observed translation. To verify that the mRNAs with structured 3′ UTRs were translated, we analyzed whether the transgenic mRNA was associated with polyribosomes (polysomes). To focus on mRNAs that were actively undergoing translation, cytoplasmic worm extracts were isolated in the presence of the translation inhibiting drug, cycloheximide, which blocks elongation and thus allows for isolation of ribosome–mRNA complexes. To provide further evidence of the polysome association of the mRNAs, extracts were also prepared in the absence of cycloheximide but in the presence of EDTA. The addition of EDTA chelates Mg2+, resulting in ribosomal subunit dissociation and release of mRNA. After sucrose density ultracentrifugation of the extracts, the absorbance of the gradient fractions at 254 nm was compared with the migration of the mRNAs as monitored by Northern blotting. The full-length rfp∷C35E7.6 (3′ UTR) mRNA and the control gfp∷unc-54 (3′ UTR) mRNA were detected predominantly with polysomes (Fig. 3A). Further, in the presence of EDTA, both transgenic mRNAs no longer sediment near the bottom of the gradient, consistent with release of the mRNAs from polysomes (Fig. 3B). Together, these data indicate that C. elegans transgenic mRNAs with structured 3′ UTRs are translated.
FIGURE 3.
Polysome association of C. elegans transgenic mRNAs containing structured 3′ UTRs. Cytoplasmic extracts from young adult worms were isolated in the presence of the translation inhibiting drug, cycloheximide, (A, C–G) or EDTA (B) and separated by sucrose density sedimentation. An absorbance trace (254 nm) across the gradient appears above Northern blots of total RNA isolated from each fraction. The particular structured 3′ UTR being monitored is indicated by the transgene name in the upper right corner of each UV trace. In A and B, mRNA for the structured 3′ UTR, rab3∷rfp∷C35E7.6 (3′ UTR), and control 3′ UTR, rab3∷gfp∷unc-54 (3′ UTR), were both monitored; the upper Northern blot was hybridized with a probe for the rfp coding sequence (structured 3′ UTR), stripped, and re-hybridized with a probe for the gfp coding sequence (lower blot; control 3′ UTR). Blots for the transgenic mRNAs with structured 3′ UTRs monitored in C–G were hybridized with a probe for the gfp coding sequence (C) or the rfp coding sequence (D–G).
Although the coding regions of both gfp and rfp mRNAs are similar in length, we noted that the rfp∷C35E7.6 (3′ UTR) mRNA consistently migrated less deeply in the sucrose gradient, indicating it was associated with lighter polysomes (Fig. 3A). To determine whether this was an effect of the structured 3′ UTR or the coding region, a transgenic animal expressing the 3′ UTR of C35E7.6 fused to gfp was analyzed (Fig. 3C). Interestingly, similar to the rfp∷C35E7.6 (3′ UTR), the gfp∷C35E7.6 (3′ UTR) migrated with the lighter polysomes, indicating that the structured 3′ UTR affected polysome loading of the mRNA. We further determined that this effect of the 3′ UTR did not depend on the length of the double-stranded structure (Fig. 3D) or the presence of either ADAR protein (Fig. 3E–G).
Analysis of endogenous C. elegans mRNAs with edited 3′ UTRs
To validate our observations made with the transgenic reporters, we extended our analyses to endogenous mRNAs. The mRNA levels of C35E7.6, syntaxin (unc-64), laminin-γ (lam-2), elo-3, and pop-1 in embryos and young adults were determined by qRT-PCR for wild-type animals and those lacking the adr genes. We did not observe significant differences in mRNA levels of edited and unedited endogenous mRNAs (Fig. 4A,B). In embryos, the unc-64 mRNA levels were determined to be 30% lower in the adr-1 and adr-1;adr-2 mutant strains compared with wild-type (Fig. 4A). However, this decrease in mRNA expression was not detected in the other adr mutant alleles (data not shown), suggesting the decrease was not a direct effect of the adr mutations.
FIGURE 4.
Levels of C. elegans mRNAs containing structured 3′ UTRs in animals deficient for ADARs. (A) Relative mRNA expression in embryos was quantified by qRT-PCR in the following strains: wild-type (black), adr-1(tm668) (light gray), adr-2(ok735) (white), and adr-1(tm668);adr-2(ok735) (dark gray). Graph illustrates the average fold-change of each mRNA relative to gpd-3 mRNA and normalized to the wild-type level (n = 4, error bars=SEM). (B) Relative mRNA expression as in A for the young adult stage.
Endogenous C. elegans mRNAs with edited 3′ UTRs are present on polysomes
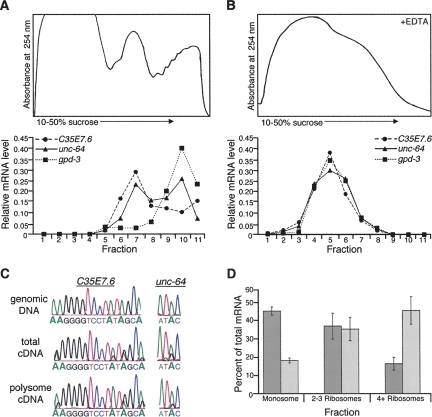
The presence of the transgenic mRNAs with structured 3′ UTRs on polysomes suggested that, in contrast to the model of nuclear retention suggested for edited RNAs (Zhang and Carmichael 2001), edited 3′ UTRs in C. elegans reach the cytoplasm. To provide further evidence of the presence of mRNAs with edited 3′ UTRs on polysomes, we extended our analysis to endogenous ADAR substrate mRNAs. As described above, cytoplasmic extracts were prepared either in the presence of cycloheximide or EDTA and then subjected to ultracentrifugation. Due to the lower abundance of the endogenous mRNAs compared with the transgenic mRNAs, the mRNA present in each fraction was determined using qRT-PCR, a more sensitive technique than Northern analysis. Similar to results obtained with the reporter mRNA, ∼40% of the endogenous C35E7.6 mRNA and 50% of the endogenous unc-64 mRNA were detected in the polysome portion of the gradient in the presence of cycloheximide, but not in the presence of EDTA (Fig. 5A,B).
FIGURE 5.
Polysome association of endogenous C. elegans mRNAs containing edited 3′ UTRs. Cytoplasmic extracts from wild-type young adult worms were isolated in the presence of cycloheximide (A) or EDTA (B) and separated by sucrose density sedimentation. The absorbance trace at 254 nm was monitored for each gradient (top panel) and mRNA levels quantified for each fraction (lower graphs). mRNA levels were determined using qRT-PCR for C35E7.6, unc-64, and gpd-3 and plotted as the fraction of mRNA present. (C) C. elegans genomic DNA, cDNA from total RNA, and cDNA from the polysome portion of the gradient were amplified and sequenced to detect editing events in the 3′ UTRs of C35E7.6 and unc-64. Data for fraction 9 are shown, but editing was detected in fractions 7–11 (data not shown). (D) The percents of total unc-64a (dark gray) and unc-64b (light gray) mRNA, quantified by qRT-PCR for two independent polysome profiles, were plotted against the number of ribosomes in each fraction (error bars=SEM).
To directly test whether edited mRNA was present on polysomes, the RNA from the polysome-containing fractions of the gradient was subjected to reverse transcription and PCR to prepare cDNA. As reverse transcriptase reads inosine as guanosine, editing sites within an mRNA appear as adenosine to guanosine changes in cDNA. Representative chromatograms for both C35E7.6 and unc-64 sequences from total cDNA, and cDNA amplified from the polysome portion of the gradient, illustrate the presence of multiple editing events (Fig. 5C, see G peaks within A peaks). Further, for C35E7.6, the PCR product included a portion of the coding region contiguous with the 3′ UTR, confirming that the polysome-associated edited 3′ UTR is part of the intact mRNA. Together, these data indicate that C. elegans mRNAs containing inosines in their 3′ UTRs are loaded on polysomes. This observation provides the first evidence that mRNAs with edited 3′ UTRs are in the cytoplasm.
Again, as observed with the reporter mRNAs, a greater fraction of the mRNAs with structured 3′ UTRs associated with lighter polysome fractions compared with control mRNAs with unstructured 3′ UTRs. The number of ribosomes associated with an mRNA, and thus its mobility in a polysome profile, can be affected by many things, including the length of the open-reading frame. To more carefully control for such differences, we monitored two splice-isoforms of unc-64 mRNA that differ in the inclusion/exclusion of an edited structure within the 3′ UTR (Fig. 5D). The unc-64a mRNA contains the edited, structured 3′ UTR analyzed earlier, while the unc-64b mRNA excludes this structure due to an alternative splicing event involving the last coding exon. Again, the majority of the splice-isoform containing the structured 3′ UTR (Fig. 5D, unc-64a, dark gray) was associated with fewer ribosomes than that lacking the structure (Fig. 5D, unc-64b, light gray).
Mammalian mRNAs with edited 3′ UTRs are also on polysomes
Our data indicate that C. elegans mRNAs with inosines in their 3′ UTRs are exported to the cytoplasm and translated. Since previous studies demonstrated that, in mammals, inosine-containing RNAs are retained in the nucleus (Kumar and Carmichael 1997; Zhang and Carmichael 2001; Prasanth et al. 2005), we wanted to determine whether mammalian mRNAs with edited 3′ UTRs also exist in the cytoplasm. We chose to examine the fates of two human mRNAs, NDUFC2 (NADH:ubiquinone oxidoreductase subunit B14.5B) and PSMB2 (HsC7-1), which were previously shown to contain multiple editing sites within the inverted Alu elements that comprise their structured 3′ UTRs (Supplemental Fig. 1, Morse et al. 2002). Cytoplasmic extracts were prepared from HeLa cells since this human cell line was previously demonstrated to retain inosine-containing RNA in the nucleus (Zhang and Carmichael 2001). As before, extracts were prepared in the presence of either cycloheximide or EDTA. After sucrose density gradient ultracentrifugation, the mRNA level in each fraction of the gradient was determined using qRT-PCR. At least two-thirds of both NDUFC2 and PSMB2 mRNA co-migrated with the polysomes (Fig. 6A). When ribosomes were disrupted with EDTA, the mRNAs no longer sedimented near the bottom of the gradient, consistent with release from polysomes (Fig. 6B). Furthermore, amplification and sequencing of the cDNA from the polysome fractions of the gradient confirmed that the 3′ UTRs of ribosome-associated NDUFC2 and PSMB2 mRNAs were edited (Fig. 6C). Additionally, as the amplified cDNA included both the coding region of the genes and the 3′ UTRs, the edited NDUFC2 and PSMB2 mRNAs present in polysomes are full-length mRNAs (data not shown). Thus, similar to what we observed for C. elegans, H. sapiens mRNAs with edited 3′ UTRs are loaded onto polysomes.
FIGURE 6.
Mammalian mRNAs with edited 3′ UTRs are present on polysomes. Cytoplasmic extracts from HeLa cells were isolated in the presence of cycloheximide (A) or EDTA (B) and separated by sucrose density sedimentation. mRNA levels were determined by qRT-PCR for NDUFC2 and PSMB2 and GAPDH and plotted as described in Fig. 5. (C) HeLa genomic DNA, cDNA from total RNA, and cDNA from the polysome portion of the gradient were amplified and sequenced to detect the editing events present in the 3′ UTRs of NDUFC2 and PSMB2. Data for fraction 9 are shown, but editing was detected in fractions 7–11 (data not shown).
DISCUSSION
In this study, we analyzed the fate of mRNAs that contain extensive double-stranded structures within their 3′ UTRs. We found that editing within structured 3′ UTRs does not affect mRNA stability or translatability. Further, we find that both C. elegans and H. sapiens mRNAs with edited, structured 3′ UTRs are present on translating ribosomes. Our studies provide the first evidence for a cytoplasmic localization of highly edited mRNAs, and thus contrast with previous reports suggesting that such mRNAs are retained in the nucleus (Zhang and Carmichael 2001; Prasanth et al. 2005).
Do inosine-containing mRNAs have different fates?
A possible explanation for the difference between our results and those of previous studies is that inosine within an RNA has different effects depending on the RNA. In this scenario, the cellular machinery would somehow distinguish between the endogenous, 3′ UTR-edited mRNAs we observed to be translated, and RNAs previously observed to be retained in the nucleus. The latter include the early transcripts of polyoma virus (Kumar and Carmichael 1997), synthetic constructs injected into Xenopus laevis oocytes (Zhang and Carmichael 2001), and the mammalian CTN-RNA (Prasanth et al. 2005). It is easy to imagine that the endogenous mRNAs we analyzed might be distinguished from a viral or synthetic RNA. However, it is harder to envision why the C. elegans and H. sapiens mRNAs we observed on polysomes would be subject to a different fate than the mouse CTN-RNA observed to be retained in the nucleus. All of these endogenous mRNAs have highly structured 3′ UTRs that are edited. Since it is clear that mRNAs with only a few inosines in their coding regions are exported to the cytoplasm and translated (e.g., Sommer et al. 1991; Lomeli et al. 1994; Morse and Bass 1997), it has been proposed that mRNAs retained in the nucleus are more extensively edited (Zhang and Carmichael 2001). CTN-RNA has ∼20% of the adenosines in its 3′ UTR structure edited (Prasanth et al. 2005). We analyzed 3′ UTR structures with a wide range of adenosine deamination (4.3%–40.3%), including those of C. elegans C35E7.6 and unc-64 mRNA, which show ≥20% of their adenosines edited, and that of PSMB2 mRNA, which exhibits 21% editing in human brain and 13% editing in HeLa cells (Figs. 5C, 6C; Morse and Bass 1999; Morse et al. 2002).
In the previous studies, inosine-containing RNAs were retained in the nucleus, but the retention was not proven to be dependent on the presence of inosine. For example, the analyses did not include comparisons to ADAR mutant cells. Thus, it remains possible that nuclear retention is mediated by a feature of RNA other than the presence of inosines. The extensive double-stranded structure in the 3′ UTR of the nuclear-retained CTN-RNA is an obvious feature, but since the mRNAs we studied had similar structured 3′ UTRs, this feature cannot solely be responsible for nuclear retention. Further, the structures in the mammalian mRNAs we studied, like CTN-RNA, were formed by pairing of Alu elements.
Nuclear-retention of inosine-containing RNA is thought to be mediated by the protein p54nrb. In vitro, p54nrb cross-links to dsRNA that has been deaminated by an ADAR, and further, preferentially binds in vitro transcripts synthesized in the presence of ITP instead of GTP (Zhang and Carmichael 2001). In addition, transfection of T7-tagged p54nrb into NIH-3T3 cells, followed by immunoprecipitation with a T7 antibody, brings down CTN-RNA (Prasanth et al. 2005). However, exactly what p54nrb recognizes in an inosine-containing RNA is unclear, and again, the nuclear retention of specific RNAs has not been directly proven to be mediated by p54nrb, for example, by demonstrating a lack of retention in cells deficient for p54nrb. The current understanding of the role of p54nrb is further complicated by the fact that it is reported to bind both RNA and DNA (Yang et al. 1993) and functions in a variety of cellular processes, including transcription, nuclear RNA processing, and DNA relaxation (Shav-Tal and Zipori 2002). Clearly, further studies are necessary to conclusively determine whether p54nrb has a role in nuclear retention of inosine-containing RNAs.
Finally, it is also possible that nuclear retention only occurs during certain conditions, and that these conditions did not exist in our studies. Many possibilities can be imagined, including nuclear sequestration in response to developmental stage, tissue, or stress. In fact, stress induces the export of CTN-RNA to the cytoplasm, and according to the authors' model, this relocalization is triggered by a cleavage event that removes the inosine-containing region of the 3′ UTR (Prasanth et al. 2005). However, as yet, the cleavage site within the 3′ UTR has not been mapped, so we cannot be certain that the cytoplasmic CTN-RNA is free of inosines. Of course, in this model it would be the lack of stress that promotes nuclear retention, and we are still left with the mystery of why the mRNAs we studied had a different fate from those of previous studies.
Does C. elegans ADR-1 play a role in the cytoplasm?
Studies of adr mutant strains indicate that ADR-2 is an active deaminase that is responsible for editing in C. elegans, and that ADR-1 has no deaminase activity on its own (Tonkin et al. 2002). Consistent with the latter, sequence comparisons show that ADR-1 lacks certain amino acids that are important for catalysis in other ADARs. However, we observed that, compared with wild-type animals, animals containing a deletion in the adr-1 gene showed an increase in protein expression from a reporter ORF fused to the C35E7.6 (3′ UTR) (Fig. 2F). In these experiments the presence of ADR-1 in the wild-type strain, which shows robust editing, or the adr-2 strain, which lacks all editing, was sufficient to reduce translation of the C35E7.6 (3′ UTR) reporter (Fig. 2F, WT and adr-2, respectively). Thus, our studies indicate that ADR-1 has roles that are completely independent of editing. ADR-1 possesses two double-stranded RNA binding motifs (dsRBMs), and possibly its roles relate to its ability to bind dsRNA. There are many examples of dsRNA binding proteins (dsRBPs) that lack catalytic domains and mediate their functions by dsRNA binding (Chang and Ramos 2005).
To gain insights into potential functions of ADR-1, we examined its subcellular localization (Supplemental Fig. 3). While ∼60% of ADR-1 was in the nuclear fraction, a large portion (∼35%) was in the ribosome-containing pellet of the cytoplasm. The presence of C. elegans ADR-1 in both the cytoplasm and nucleus is reminiscent of the localization of human ADAR-1 to both compartments (Patterson and Samuel 1995; Eckmann et al. 2001; Desterro et al. 2003), and indicates that future studies aimed at deciphering the biologic role of ADR-1 should consider nuclear as well as cytoplasmic functions. Furthermore, the dsRBPs NF90 and Staufen-2 transport structured RNAs across the nuclear membrane and into the cytoplasm (Brownawell and Macara 2002; Gwizdek et al. 2004; Macchi et al. 2004), and in future studies it will be important to examine whether C. elegans ADR-1 has a similar role in nucleo-cytoplasmic transport of dsRNA. Finally, the co-sedimentation of ADR-1 with the ribosome-containing portion of the gradient is intriguing given that we observed that animals lacking ADR-1 showed increased expression of reporters containing the C35E7.6 (3′ UTR). In future studies it will be interesting to determine if ADR-1 itself binds structured 3′ UTRs to modulate translation.
What is the function of double-stranded structures in 3′ UTRs and editing within them?
We have accomplished the goal of our study, and shed light on a long-time paradox in the field, by demonstrating that C. elegans and H. sapiens mRNAs with edited, structured 3′ UTRs are exported to the cytoplasm and translated. However, we are still left with the question as to the role of the structured 3′ UTRs and the editing sites within them. Could the altered mobility of mRNAs with structured 3′ UTRs in polysome profiles be a clue in regard to this question?
In C. elegans, whether monitoring transgenic or endogenous mRNAs, we observed that a greater fraction of mRNAs with structured 3′ UTRs were associated with lighter polysome fractions compared with mRNAs with unstructured 3′ UTRs (Figs. 3, 5). This association was very reproducible and independent of the ORF or the length of the 3′ UTR structure (Fig. 3C,D). The position of an mRNA in a polysome profile reflects the number of ribosomes it is associated with and, thus, how efficiently it is being translated. In this light, the majority of the C. elegans unc-64a isoform, which contains a double-stranded structure in its 3′ UTR, is poorly translated compared with the unc-64b splice form that excludes the 3′ UTR structure (Fig. 5D).
Our favorite model is one in which the double-stranded region of a 3′ UTR serves as a binding site for dsRBPs that regulate translation. There is precedence for dsRBPs functioning in this regard. For example, the dsRBP NF90 increases VEGF translation by binding to a structure within its 3′ UTR to promote polysome loading (Vumbaca et al. 2008). In addition, dsRBPs sometimes function in localization to allow translation in particular subcellular locations. For example, in both the mammalian nervous system and the developing Drosophila embryo, the dsRBP Staufen binds to the 3′ UTR of certain mRNAs to promote their localization (Roegiers and Jan 2000). Of particular relevance to our studies, in the sensory neurons of Aplysia, Staufen is involved in localizing syntaxin mRNA via its 3′ UTR (Liu et al. 2006).
Of course, if the structured 3′ UTRs are involved in translational control, it is likely that the process will be regulated, and again there is precedence. For example, the NF-90-mediated translational up-regulation of VEGF occurs specifically in response to hypoxic conditions (Vumbaca et al. 2008). Aplysia Staufen promotes movement of syntaxin mRNA from one axonal region to another upon stimulation by serotonin (Liu et al. 2006). While we have observed a state of low translational efficiency only for C. elegans mRNAs with structured 3′ UTRs, in our analysis of mRNAs in HeLa cells, we found that NDUFC2 and PSMB2 mRNAs, which contain structured 3′ UTRs, were on heavier polysomes, indicative of high translational efficiency (Fig. 6A). It will be interesting to examine these mRNAs in other cells to determine if translational efficiency is always enhanced for human mRNAs with structured 3′ UTRs, or if the polysome occupancy observed in the tumor-derived HeLa cells reflects some unique aspect of these cells.
Editing within 3′ UTRs is unlikely to be important for regulating translation under normal growth conditions, as we did not observe differences in translation or polysome association between wild-type animals and those that lacked all ADAR editing activity (Figs. 2C–F, 3E–G). However, it remains possible that editing plays a role in responding to certain conditions, for example, to alter binding of a dsRBP to a structured 3′ UTR. Identifying conditions in which polysome occupancy changes will be critical in understanding the biological function of structured 3′ UTRs and the inosines within them.
While this manuscript was in review, another study reporting that mRNAs with inverted Alu elements are retained in the mammalian nucleus was published (Chen et al. 2008). A decrease in reporter protein expression when the 3′ UTR contained a pair of inverted Alu elements was observed and attributed to nuclear retention of the mRNA. The inverted Alu elements are edited by ADARs, and thus the authors conclude that editing correlates with nuclear retention (Chen et al. 2008). However, again, the cellular localization of the reporter was not examined in the absence of editing. Thus, to date, our study remains the only one to compare expression of edited RNAs between wild-type and ADAR mutant animals. Our C. elegans reporter assay did not detect an increase in protein expression in the absence of editing (Fig. 2C–F), and thus, in our estimation, the function of editing in 3′ UTRs remains unclear.
Interestingly, while the recent study showed that a greater fraction of a reporter mRNA with a structured 3′ UTR was retained in the nucleus compared with that lacking a structured 3′ UTR, 35%–40% of the reporter mRNA with the edited, structured 3′ UTR was in the cytoplasm (Chen et al. 2008). Thus, these results are actually consistent with our observation of endogenous mRNAs with edited 3′ UTRs in the mammalian cytoplasm (Fig. 6). However, in contrast to our conclusion that structured 3′ UTRs affect polysome loading, the authors note that differences were not observed between polysome profiles for a reporter with a structured 3′ UTR compared with a reporter with an unstructured 3′ UTR (Chen et al. 2008). Possibly, a more in-depth analysis of polysome loading for the mammalian reporters will reveal effects similar to those we observe. Regardless, while nuclear retention is proposed to cause the gene silencing effect of the Alu elements, our studies suggest that effects on polysome loading may also contribute to the decreased protein expression.
MATERIALS AND METHODS
Worm strains and culture
The following strains were maintained under standard laboratory conditions (Brenner 1974): Bristol strain N2, BB2 adr-1(gv6), BB3 adr-2(gv42), BB4 adr-1(gv6); adr-2(gv42), BB19 adr-1(tm668), BB20 adr-2(ok735), BB21 adr-1(tm668); adr-2(ok735), BB66 uuEx13[rab3p∷rfp∷lam-2 (3′ UTR), rab3p∷gfp∷unc-54 (3′ UTR)], BB67 uuEx14[rab3p∷rfp∷elo-3 (3′ UTR), rab3p∷gfp∷unc-54 (3′ UTR)], BB68 uuEx15[rab3p∷rfp∷unc-64 (3′ UTR), rab3p∷gfp∷unc-54 (3′ UTR)], BB69 adr-1(tm668); uuEx15, BB70 adr-2(ok735); uuEx15, BB71 adr-1(tm668); adr-2(ok735); uuEx15, BB72 uuEx16[rab3p∷rfp∷C35E7.6 (3′ UTR), rab3p∷gfp∷unc-54 (3′ UTR)], BB73 adr-1(gv6); uuEx16, BB74 adr-2(gv42); uuEx16, BB75 adr-1(gv6); adr-2(gv42); uuEx16, BB76 xtIs4[rab3p∷rfp∷C35E7.6 (3′ UTR), rab3p∷gfp∷unc-54 (3′ UTR), unc-119 genomic rescue], BB77 adr-1(tm668); xtIs4, BB78 adr-2(ok735); xtIs4, BB79 adr-1(tm668); adr-2(ok735); xtIs4, BB80 unc-119 (ed3); xtIs12[rab3p∷rfp∷elo-3 (3′ UTR), rab3p∷gfp∷unc-54 (3′ UTR), unc-119 genomic rescue], BB81 adr-1(tm668); unc-119 (ed3); xtIs12, BB82 adr-2(ok735); unc-119 (ed3); xtIs12, BB83 adr-1(tm668); adr-2(ok735); unc-119 (ed3); xtIs12, BB85 unc-119 (ed3); xtIs8[rab3p∷gfp∷C35E7.6 3′ UTR, rab3p∷rfp∷unc-54 3′ UTR, unc-119 genomic rescue], BB84 uuEx17[rab3p∷gfp∷lam-2 (3′ UTR), rab3p∷rfp∷unc-54 (3′ UTR)], BB86 adr-1(gv6); uuEx17, BB87 adr-2(gv42); uuEx17, BB88 adr-1(gv6); adr-2(gv42); uuEx17. Genotypes of each worm were determined using single-worm PCR.
Embryos were isolated from a well-fed liquid culture of gravid worms using standard hypochlorite treatment (Emmons et al. 1979). Developmentally staged cultures were obtained by hatching eggs overnight in M9 buffer, transferring synchronized L1 worms to NGM plates seeded with Escherichia coli OP50, and incubating at 20°C for 6–8 h (L1), 18–20 h (L2), 28–30 h (L3), 40–42 h (L4), 50–54 h (young adults), and 60 h (gravid adults).
Transgenics
The rab3 promoter (∼1200 bp) was amplified from C. elegans genomic DNA and inserted upstream of the sequence encoding either green fluorescent protein or red fluorescent protein in the pPD95.77 vector (J. Habig and B. Bass, University of Utah). C. elegans 3′ UTRs were amplified from cosmids (Caenorhabditis Genetics Center) immediately downstream of the stop codon through the predicted poly-A site and then inserted immediately downstream of either fluorescent protein.
Integrated reporter strains were made by biolistic transformation (Wilm et al. 1999) of a plasmid containing the promoter-fluorescent protein-3′ UTR sequences cloned on either side of an unc-119 genomic sequence, to allow rescue of unc-119(ed3) (W. Davis and E. Jorgensen, University of Utah).
RNA isolation, Northern analysis, and qRT-PCR
RNA was isolated from worm pellets or liquid fractions using Trizol (Invitrogen). RNA for qRT-PCR was further purified by treating with TURBO DNase (Ambion) followed by RNeasy chromatography (Qiagen).
Northern analyses used standard protocols for 1% formaldehyde agarose gel electrophoresis and nylon membrane blotting. The rfp and gfp Northern probes were synthesized using in vitro transcription (Strip-EZ T7 kit, Ambion) of a PCR template (rfp nt. 1–687, gfp nt. 557–701).
To synthesize cDNA for qRT-PCR, 2 μg of DNase-treated total RNA was reverse transcribed with SuperScript II reverse transcriptase (Invitrogen) and gene-specific primers to the coding region of the mRNA. Samples were treated with RNaseH (New England Biolabs) for 30 min and either used directly (polysome gradients) or diluted 2.5-fold with ddH20. Five microliters of cDNA was analyzed per qRT-PCR in a Lightcycler 2.0 instrument using the Lightcycler FastStart DNA MasterPLUS Syber Green I Kit (Roche). qRT-PCR primers spanned at least one exon boundary and produced products of ∼150 bp. Quality of the qRT-PCR products was assessed using melting curve analysis and gel electrophoresis.
Polysome extract preparation and sucrose gradient ultracentrifugation
Cytoplasmic extracts were prepared from a synchronous young adult worm liquid culture. Worms were washed three times with CB buffer (20 mM HEPES pH 7.5, 5 mM MgCl2, 10 mM KCl, 1 mM EGTA, 10% glycerol, 0.2 mg/mL heparin, 80 U/mL of RNasin, Roche complete protease inhibitor tablet) in the presence of 500 μg/mL cycloheximide (CBC) or without cycloheximide. Worms were resuspended in three volumes of CBC or CB buffer and dropped into liquid nitrogen. The frozen worms were then crushed with a mortar and pestle on a dry ice/ethanol bath. After thawing on ice, the extract was spun at maximum in an Eppendorf microfuge for 10 min at 4°C. The CBC supernatant was immediately loaded onto an 8-mL 10%–50% sucrose gradient and centrifuged in an SW41Ti rotor (Beckman) at 41,000 rpm for 3 h at 4°C. EDTA (final concentration 0.15 M) was added to the extract made in the absence of cycloheximide and incubated for 30 min at 4°C, then loaded onto the gradients and centrifuged as described above. Gradients were scanned at 254 nm and fractionated in 1-mL increments with an ISCO gradient collector.
Cytoplasmic HeLa cell extracts were prepared similar to as described above for the worm extracts. Six to eight 10-mm dishes of HeLa cells grown in Dulbecco's Modified Eagle's Medium supplemented with 10% fetal bovine serum were incubated in the presence of 100 μg/mL cycloheximide for 15 min. Cells were scraped from the plate in PBS, pH 7.5 and centrifuged at 500g for 5 min. The cell pellet was resuspended in 1 mL of CBC buffer+0.2% NP-40 and vortexed for 30 min. Cells treated with EDTA were prepared similarly, except without treatment with cycloheximide. The extracts were cleared of debris by spinning for 5 min at 700g in an Eppendorf microfuge at 4°C. Extracts were immediately loaded onto sucrose gradients, centrifuged, and fractionated as described above.
Cell imaging and quantification
Confocal immunofluorescence images were acquired using either Fluoview software on a FV300 IX81 Olympus microscope or Lasersharp 2000 software on a MRC1024 Biorad microscope. Using the Volocity 3-D quantitation program (Perkin-Elmer), the levels of RFP and GFP were measured throughout the worm.
Antibody production and cell fractionation
A fusion of the C-terminal 346 amino acids of ADR-1 to glutathione S-transferase was purified from E. coli cells and injected into rabbits for antibody generation (Covance).
Embryo cell fractionations were performed as described (Chen et al. 2000). Equivalent amounts of each fraction were subjected to SDS-PAGE, transferred to PVDF (Millipore), and immunoblotted for rabbit acetyl histone-H4 (Millipore), cytoplasmic mouse Hsp90 (Stressgen), or ADR-1 using the SuperSignal West Femto ECL detection system (Pierce).
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Colin Thacker of the University of Utah Worm Core for providing integrated worm strains and Chris Rodesch of the University of Utah Cell Imaging Facility for providing assistance with fluorescence microscopy and data processing. This work was supported by funds to B.L.B. from the National Institutes of Health (GM044073), a postdoctoral fellowship to H.A.H from the Helen Hay Whitney Foundation (F-392); B.L.B. is a Howard Hughes Medical Institute Investigator.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1165008.
REFERENCES
- Athanasiadis, A., Rich, A., Maas, S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, B.L. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, B.L., Weintraub, H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988;55:1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- Blow, M., Futreal, P.A., Wooster, R., Stratton, M.R. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S. The genetics of Caenorhabditis elegans . Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownawell, A.M., Macara, I.G. Exportin-5, a novel karyopherin, mediates nuclear export of double-stranded RNA binding proteins. J. Cell Biol. 2002;156:53–64. doi: 10.1083/jcb.200110082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns, C.M., Chu, H., Rueter, S.M., Hutchinson, L.K., Canton, H., Sanders-Bush, E., Emeson, R.B. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- Chang, K.Y., Ramos, A. The double-stranded RNA-binding motif, a versatile macromolecular docking platform. FEBS J. 2005;272:2109–2117. doi: 10.1111/j.1742-4658.2005.04652.x. [DOI] [PubMed] [Google Scholar]
- Chen, F., Hersh, B.M., Conradt, B., Zhou, Z., Riemer, D., Gruenbaum, Y., Horvitz, H.R. Translocation of C. elegans CED-4 to nuclear membranes during programmed cell death. Science. 2000;287:1485–1489. doi: 10.1126/science.287.5457.1485. [DOI] [PubMed] [Google Scholar]
- Chen, L.L., Decerbo, J.N., Carmichael, G.G. Alu element-mediated gene silencing. EMBO J. 2008;12:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterro, J.M., Keegan, L.P., Lafarga, M., Berciano, M.T., O'Connell, M., Carmo-Fonseca, M. Dynamic association of RNA-editing enzymes with the nucleolus. J. Cell Sci. 2003;116:1805–1818. doi: 10.1242/jcs.00371. [DOI] [PubMed] [Google Scholar]
- Eckmann, C.R., Neunteufl, A., Pfaffstetter, L., Jantsch, M.F. The human but not the Xenopus RNA-editing enzyme ADAR1 has an atypical nuclear localization signal and displays the characteristics of a shuttling protein. Mol. Biol. Cell. 2001;12:1911–1924. doi: 10.1091/mbc.12.7.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmons, S.W., Klass, M.R., Hirsh, D. Analysis of the constancy of DNA sequences during development and evolution of the nematode Caenorhabditis elegans . Proc. Natl. Acad. Sci. 1979;76:1333–1337. doi: 10.1073/pnas.76.3.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwizdek, C., Ossareh-Nazari, B., Brownawell, A.M., Evers, S., Macara, I.G., Dargemont, C. Minihelix-containing RNAs mediate exportin-5-dependent nuclear export of the double-stranded RNA-binding protein ILF3. J. Biol. Chem. 2004;279:884–891. doi: 10.1074/jbc.M306808200. [DOI] [PubMed] [Google Scholar]
- Jepson, J.E., Reenan, R.A. RNA editing in regulating gene expression in the brain. Biochim. Biophys. Acta. 2007 doi: 10.1016/j.bbagrm.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Jiang, M., Ryu, J., Kiraly, M., Duke, K., Reinke, V., Kim, S.K. Genome-wide analysis of developmental and sex-regulated gene expression profiles in Caenorhabditis elegans . Proc. Natl. Acad. Sci. 2001;98:218–223. doi: 10.1073/pnas.011520898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D.D., Kim, T.T., Walsh, T., Kobayashi, Y., Matise, T.C., Buyske, S., Gabriel, A. Widespread RNA editing of embedded Alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler, M., Kornau, H.C., Seeburg, P.H. The organization of the gene for the functionally dominant α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor subunit GluR-B. J. Biol. Chem. 1994;269:17367–17370. [PubMed] [Google Scholar]
- Kumar, M., Carmichael, G.G. Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proc. Natl. Acad. Sci. 1997;94:3542–3547. doi: 10.1073/pnas.94.8.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon, E.Y., Eisenberg, E., Yelin, R., Nemzer, S., Hallegger, M., Shemesh, R., Fligelman, Z.Y., Shoshan, A., Pollock, S.R., Sztybel, D., et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- Liu, J., Hu, J.Y., Wu, F., Schwartz, J.H., Schacher, S. Two mRNA-binding proteins regulate the distribution of syntaxin mRNA in Aplysia sensory neurons. J. Neurosci. 2006;26:5204–5214. doi: 10.1523/JNEUROSCI.4917-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomeli, H., Mosbacher, J., Melcher, T., Hoger, T., Geiger, J.R., Kuner, T., Monyer, H., Higuchi, M., Bach, A., Seeburg, P.H. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- Macchi, P., Brownawell, A.M., Grunewald, B., DesGroseillers, L., Macara, I.G., Kiebler, M.A. The brain-specific double-stranded RNA-binding protein Staufen2: Nucleolar accumulation and isoform-specific exportin-5-dependent export. J. Biol. Chem. 2004;279:31440–31444. doi: 10.1074/jbc.C400226200. [DOI] [PubMed] [Google Scholar]
- Morse, D.P., Bass, B.L. Detection of inosine in messenger RNA by inosine-specific cleavage. Biochemistry. 1997;36:8429–8434. doi: 10.1021/bi9709607. [DOI] [PubMed] [Google Scholar]
- Morse, D.P., Bass, B.L. Long RNA hairpins that contain inosine are present in Caenorhabditis elegans poly(A)+ RNA. Proc. Natl. Acad. Sci. 1999;96:6048–6053. doi: 10.1073/pnas.96.11.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse, D.P., Aruscavage, P.J., Bass, B.L. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc. Natl. Acad. Sci. 2002;99:7906–7911. doi: 10.1073/pnas.112704299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, J.B., Samuel, C.E. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: Evidence for two forms of the deaminase. Mol. Cell. Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth, K.V., Prasanth, S.G., Xuan, Z., Hearn, S., Freier, S.M., Bennett, C.F., Zhang, M.Q., Spector, D.L. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Roegiers, F., Jan, Y.N. Staufen: A common component of mRNA transport in oocytes and neurons? Trends Cell Biol. 2000;10:220–224. doi: 10.1016/s0962-8924(00)01767-0. [DOI] [PubMed] [Google Scholar]
- Shav-Tal, Y., Zipori, D. PSF and p54nrb/NonO—Multi-functional nuclear proteins. FEBS Lett. 2002;531:109–114. doi: 10.1016/s0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- Sommer, B., Kohler, M., Sprengel, R., Seeburg, P.H. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- Tonkin, L.A., Saccomanno, L., Morse, D.P., Brodigan, T., Krause, M., Bass, B.L. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans . EMBO J. 2002;21:6025–6035. doi: 10.1093/emboj/cdf607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente, L., Nishikura, K. ADAR gene family and A-to-I RNA editing: Diverse roles in posttranscriptional gene regulation. Prog. Nucleic Acid Res. Mol. Biol. 2005;79:299–338. doi: 10.1016/S0079-6603(04)79006-6. [DOI] [PubMed] [Google Scholar]
- Vumbaca, F., Phoenix, K.N., Rodriguez-Pinto, D., Han, D.K., Claffey, K.P. Double-stranded RNA-binding protein regulates vascular endothelial growth factor mRNA stability, translation, and breast cancer angiogenesis. Mol. Cell. Biol. 2008;28:772–783. doi: 10.1128/MCB.02078-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner, R.W., Smith, J.E., Cooperman, B.S., Nishikura, K. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc. Natl. Acad. Sci. 1989;86:2647–2651. doi: 10.1073/pnas.86.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm, T., Demel, P., Koop, H.U., Schnabel, H., Schnabel, R. Ballistic transformation of Caenorhabditis elegans . Gene. 1999;229:31–35. doi: 10.1016/s0378-1119(99)00043-8. [DOI] [PubMed] [Google Scholar]
- Yang, Y.S., Hanke, J.H., Carayannopoulos, L., Craft, C.M., Capra, J.D., Tucker, P.W. NonO, a non-POU-domain-containing, octamer-binding protein, is the mammalian homolog of Drosophila nonAdiss. Mol. Cell. Biol. 1993;13:5593–5603. doi: 10.1128/mcb.13.9.5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Carmichael, G.G. The fate of dsRNA in the nucleus: A p54nrb-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]