Abstract
The poly(A) tail is a crucial determinant in the control of both mRNA translation and decay. Poly(A) tail length dictates the triggering of the degradation of the message body in the major 5′ to 3′ and 3′ to 5′ mRNA decay pathways of eukaryotes. In the 5′ to 3′ pathway oligoadenylated but not polyadenylated mRNAs are selectively decapped in vivo, allowing their subsequent degradation by 5′ to 3′ exonucleolysis. The conserved Lsm1p-7p-Pat1p complex is required for normal rates of decapping in vivo, and the purified complex exhibits strong binding preference for oligoadenylated RNAs over polyadenylated or unadenylated RNAs in vitro. In the present study, we show that two lsm1 mutants produce mutant complexes that fail to exhibit such higher affinity for oligoadenylated RNA in vitro. Interestingly, these mutant complexes are normal with regard to their integrity and retain the characteristic RNA binding properties of the wild-type complex, namely, binding near the 3′-end of the RNA, having higher affinity for unadenylated RNAs that carry U-tracts near the 3′-end over those that do not and exhibiting similar affinities for unadenylated and polyadenylated RNAs. Yet, these lsm1 mutants exhibit a strong mRNA decay defect in vivo. These results underscore the importance of Lsm1p-7p-Pat1p complex–mRNA interaction for mRNA decay in vivo and imply that the oligo(A) tail mediated enhancement of such interaction is crucial in that process.
Keywords: Lsm1p-7p-Pat1p complex, mRNA decay, Sm-like, decapping, oligo(A) tail
INTRODUCTION
The poly(A) tail length is an important determinant of mRNA function. Presence of long poly(A) tails at the 3′-end facilitates the translation of the mRNA by recruiting the poly(A) binding protein, which interacts with the eIF4G subunit of the cap binding complex (Tarun and Sachs 1996; Wells et al. 1998; Mangus et al. 2003; Amrani et al. 2008). On the other hand, such long 3′-poly(A) tails typically render the mRNAs resistant to degradation in eukaryotes wherein mRNAs carrying a short 3′-oligo(A) tail are susceptible to decay. Thus, in both of the major eukaryotic mRNA decay pathways, namely, the 3′ to 5′ and 5′ to 3′ pathways, the initial step is poly(A) shortening or deadenylation, which results in the production of oligoadenylated mRNA from polyadenylated mRNA (Tucker et al. 2001; Coller and Parker 2004; Meyer et al. 2004; Parker and Song 2004; Yamashita et al. 2005).
In the 5′ to 3′ pathway, poly(A) shortening of the mRNA triggers the removal of cap structure at the 5′-end of the oligoadenylated mRNA, which then permits the 5′ to 3′ exonucleolytic degradation of the mRNA body by the Xrn1p exonuclease (Decker and Parker 1993; Hsu and Stevens 1993; Muhlrad et al. 1994; Beelman et al. 1996; Dunckley and Parker 1999). Decapping is a very precisely controlled step in this pathway, and several factors are known to affect rates of decapping in vivo (Coller and Parker 2004). The translation initiation machinery including the poly(A) binding protein is antagonistic to decapping (Caponigro and Parker 1995; Coller et al. 1998; Schwartz and Parker 1999; Wilusz et al. 2001; Ramirez et al. 2002; Khanna and Kiledjian 2004). On the other hand, several other factors like Pat1p, Dhh1p, Lsm1p-7p complex, and Edc proteins function as activators of decapping and hence are required for normal rates of decapping in vivo (Boeck et al. 1998; Bonnerot et al. 2000; Bouveret et al. 2000; Tharun et al. 2000; Coller et al. 2001; Dunckley et al. 2001; Fischer and Weis 2002; Kshirsagar and Parker 2004). These activators of decapping along with the decapping enzyme, 5′ to 3′ exonuclease Xrn1p, mRNAs targeted for decay, and mRNA decay intermediates of the 5′ to 3′ pathway are localized in discrete cytoplasmic sites called P-bodies, which are conserved in all eukaryotes (Ingelfinger et al. 2002; Sheth and Parker 2003; Teixeira et al. 2005).
The Lsm1p-7p complex made of seven Sm-like proteins, Lsm1p through Lsm7p, is highly conserved in all eukaryotes (Salgado-Garrido et al. 1999; Bouveret et al. 2000; Tharun et al. 2000). Lsm1p is the key subunit that distinguishes this complex from the related U6 snRNP associated nuclear complex, Lsm2p-8p, which is made of the seven Sm-like proteins Lsm2p through Lsm8p (Achsel et al. 1999; Mayes et al. 1999; Bouveret et al. 2000). The Lsm1p-7p complex physically interacts with other decay factors Pat1p, Dhh1p, and Xrn1p (Bonnerot et al. 2000; Bouveret et al. 2000; Tharun et al. 2000; Coller et al. 2001). Importantly, it preferentially associates in vivo with a pool of oligoadenylated mRNPs that is bound to the decapping enzyme but not the translation factors (Tharun et al. 2000; Tharun and Parker 2001). Thus an mRNP transition involving removal of translation factors and recruitment of decay factors seems to be important to trigger decapping (Tharun et al. 2000; Tharun and Parker 2001). Decapping may also involve additional events, like transport of the mRNP to P-bodies. However, the precise nature and order of the events constituting decapping is not clear.
Recently we showed that the purified Lsm1p-7p-Pat1p complex has a strong binding preference for oligoadenylated RNAs over unadenylated and polyadenylated RNAs in vitro (Chowdhury et al. 2007). This is consistent with its in vivo role as decapping activator in the 5′ to 3′ pathway wherein oligoadenylated mRNAs are selectively decapped. In the present study we have analyzed two lsm1 mutants (lsm1-9 and lsm1-14) (Tharun et al. 2005) that produce mutant complexes that fail to show binding preference for oligoadenylated RNAs. These mutant complexes are normal with regard to their integrity and are similar to the wild-type complex in several aspects, like binding near the 3′-end of the RNA, having higher affinity for unadenylated RNAs carrying U-tracts near their 3′-ends over those that do not, and exhibiting similar affinities for unadenylated and polyadenylated RNAs. Nevertheless, mRNA decay is severely impaired in these mutant cells, implying that the ability of the Lsm1p-7p-pat1p complex to bind preferentially to oligoadenylated RNAs is crucial for mRNA decapping in vivo.
RESULTS
Purified mutant complexes contain all the component proteins
The lsm1-9 and lsm1-14 mutants bear lesions in the predicted RNA contacting surfaces of Lsm1p and are inhibited in mRNA decay in vivo (Tharun et al. 2005). The decay defect is not due to the decreased accumulation of the mutant Lsm1p proteins in these cells (Tharun et al. 2005). In both mutants, Lsm2p, Lsm7p, and (mutant) Lsm1p localize to the P-bodies just as in wild-type cells (Tharun et al. 2005). This suggested that the Lsm1p-7p complex integrity is not impaired in these mutants because such integrity is critical for localization to P-bodies of the Lsm1p through Lsm7p proteins (Ingelfinger et al. 2002). To confirm this, we purified the Lsm1p-containing complex from each of these mutants following the strategy we used in the past (Chowdhury et al. 2007). Briefly, we used yeast strains expressing FLAG-lsm1 (wild type or mutant) and LSM5-6xHis as the only copies of these genes from their native promoters and purified the complex by sequential affinity chromatography exploiting the two epitope tags. SDS–polyacrylamide gel electrophoretic (SDS-PAGE) analysis revealed that the band pattern of the mutant complexes is very similar to that of the wild-type complex (Fig. 1). Mass spectrometry analysis of the complexes isolated from lsm1-9 and lsm1-14 cells revealed, based on tryptic peptides with unambiguous sequence match, that each of them contain all seven Lsm proteins and Pat1p (5, 4, 3, 2, 1, 3, 2, and 3 peptides for Lsm1p through Lsm7p and Pat 1p, respectively, in the case of the complex isolated from lsm1-9 cells and 4, 8, 4, 3, 2, 4, 2, and 13 peptides for Lsm1p through Lsm7p and Pat1p proteins in the case of complex purified from lsm1-14 cells) (see Supplemental Table 1). Measurements of MFA2pG mRNA half-life revealed that epitope tagging did not affect the severity of the mRNA decay defect of the lsm1-9 and lsm1-14 alleles (Tharun et al. 2005; data not shown).
FIGURE 1.
Lsm1p-7p-Pat1p complexes isolated from lsm1-9 and lsm1-14 mutants show a band pattern similar to the wild-type complex in SDS-PAGE. Lsm1p-7p-Pat1p complexes purified from LSM1, lsm1-9, and lsm1-14 cells (lanes 1,2,3, respectively) as described in Materials and Methods were subjected to SDS-PAGE separation followed by visualization of the bands by silver staining. Positions of the molecular weight markers are shown on the left. Identity of the protein bands is indicated on the right wherein asterisks mark the higher mobility species of Pat1p (Chowdhury et al. 2007).
Purified mutant complexes are able to bind RNA
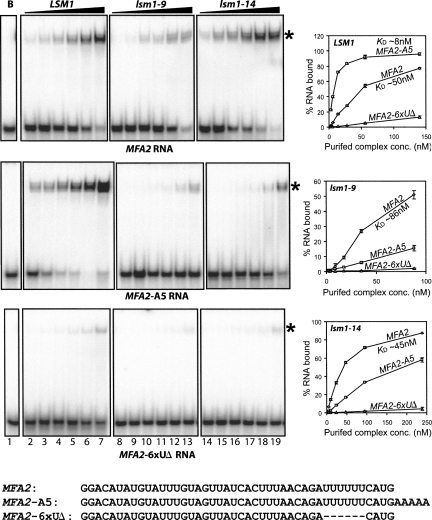
The fact that the mutant complexes purified from the lsm1-9 and lsm1-14 cells are not significantly affected in their integrity raised the possibility that these complexes may be able to bind RNA. Therefore, we studied the RNA binding characteristics of these purified mutant complexes. First, the binding affinity of these complexes for radiolabeled MFA2 RNA (made by in vitro transcription using α-32P-UTP) (see Materials and Methods) was studied using gel mobility shift assays. The MFA2 RNA contains the 3′-most 42 nucleotides (nt) of MFA2 3′ UTR preceded by two G's at the 5′-end (see Fig. 2; Chowdhury et al. 2007). RNA binding reactions were carried out using a constant amount of the substrate RNA and increasing concentrations of the purified complex such that the molar concentration of the RNA is several-fold lower than the lowest molar concentration of the purified complex used in the experiment. As seen in Figure 2, the MFA2 RNA is bound quite well by the mutant complexes. While this RNA was bound by the wild-type complex and Lsm1-14p containing complex with comparable affinities (apparent K D of ∼50 nM and ∼45 nM, respectively), the affinity was moderately decreased in the case of Lsm1-9p containing complex (apparent K D of ∼86 nM). These results and additional studies with PGK1 RNA (see below) (Fig. 3) together reveal that, although the RNA binding ability of the mutant complexes is decreased, it is not fully abolished.
FIGURE 2.
Mutant Lsm1p-7p-Pat1p complexes purified from lsm1-9 and lsm1-14 cells are unable to recognize oligoadenylated status of RNAs. Gel mobility shift assays were carried out as described in Materials and Methods using MFA2, MFA2-A5, or MFA2-6xUΔ (phosphorimages in the first, second, and third panels, respectively, from top) RNA with BSA (lane 1) or increasing concentrations of the complex purified from LSM1 (lanes 2–7), lsm1-9 (lanes 8–13), or lsm1-14 (lanes 14–19) cells. RNA binding was quantitated using a PhosphorImager, and plots of the fraction of RNA bound versus the concentration of the purified complex are shown on the right. Sequences of the RNAs used are shown in the bottom. Asterisks indicate the position of the gel shifted RNA in the gel pictures.
FIGURE 3.
Inability of the mutant complexes to recognize the 3′-oligo(A) tail is not RNA specific. Gel mobility shift assays were carried out as described in Materials and Methods using PGK1 (lanes 1–7) or PGK1-A5 (lanes 8–14) RNA with BSA (lanes 1,14) or increasing concentrations of the complex purified from LSM1, lsm1-9, or lsm1-14 cells (phosphorimages in top, middle, and bottom panels, respectively). Plots of the fraction of RNA bound versus the concentration of the purified complex are shown on the right. Sequences of the RNAs used are shown in the bottom.
The mutant complexes do not exhibit enhanced affinity toward oligoadenylated RNAs over unadenylated RNAs
A unique RNA binding property of the wild-type Lsm1p-7p-Pat1p complex is its strong binding preference for oligoadenylated RNA over unadenylated and polyadenylated RNA (Chowdhury et al. 2007). Therefore, in order to test if the mutant complexes purified from lsm1-9 and lsm1-14 cells possess this property, we compared their affinities for unadenylated and oligoadenylated RNA substrates using gel shift assays. As shown in Figure 2 and consistent with our earlier studies (Chowdhury et al. 2007), the wild-type complex showed several-fold higher affinity (apparent KD of ∼8 nM) for the MFA2-A5 RNA (MFA2 RNA with a 3′-A5 tail; see Fig. 2; Chowdhury et al. 2007) than for MFA2 RNA. However, the complex purified from neither lsm1-9 nor lsm1-14 cells exhibited such higher affinity for MFA2-A5 RNA over the MFA2 RNA. Thus, comparison of relative affinities for the MFA2 and MFA2-A5 RNAs reveals an important difference between the wild-type and mutant complexes; i.e., addition of 3′-oligo(A) tail stimulates RNA binding by the wild-type but not mutant complexes. Such impairment of the mutant complexes is not specific to MFA2 RNA, because similar results were also obtained when the affinities of these complexes for PGK1 (3′-most 42 nt of PGK1 3′UTR preceded by two G's; see Fig. 3; Chowdhury et al. 2007) and PGK1-A5 (PGK1 RNA with a 3′-A5 tail; see Fig. 3) RNAs were compared. As shown in Figure 3 and as expected, the wild-type complex had a much higher affinity for PGK1-A5 RNA than for PGK1 RNA (apparent K D of ∼5 nM and ∼54 nM, respectively). However, the complex purified from lsm1-14 cells bound to both PGK1 and PGK1-A5 RNAs with comparable affinities (apparent K D of 74 nM and 90 nM, respectively). The complex purified from lsm1-9 mutant also exhibited similar behavior. Additional studies also revealed that the complex purified from lsm1-14 cells has comparable affinities for the unadenylated and 3′-pentaadenylated versions of RPL41A RNA (3′-most 42 nt of RPL41A 3′UTR preceded by two G's) (see Supplemental Fig. 1; Chowdhury et al. 2007). Thus, although the mutant complexes are able to bind RNA, their ability to preferentially bind to oligoadenylated RNA (over unadenylated RNA) is impaired, suggesting that they are different from the wild-type complex in the manner in which they bind the RNA. The lack of enhanced affinity of the mutant complexes for the oligoadenylated RNAs was reproducibly observed in multiple experiments.
Our results show that the mutant complexes have a significantly lower affinity for the MFA2-A5 RNA compared to MFA2 RNA (Fig. 2). In order to test if this is due to a specific inhibition by the 3′-oligo(A) tail of the RNA binding by the mutant complexes, we compared the affinities of the mutant complexes for MFA2 and MFA2-CAGAC RNAs. The MFA2-CAGAC RNA is nothing but MFA2 RNA with the pentanucleotide CAGAC added to its 3′-end. We observed that just like the MFA2-A5 RNA, the MFA2-CAGAC RNA was also bound with lower affinity than the MFA2 RNA by the mutant complexes (Fig. 4). Thus, addition of not only a 5-nt-long oligo(A) tail but also a 5-nt-long tail of non-oligo(A) sequence at the 3′-end of MFA2 RNA inhibits the binding of that RNA by the mutant complexes. This suggests that the weaker binding of the MFA2-A5 RNA than the MFA2 RNA by the mutant complexes does not involve a specific recognition of the oligo(A) tail by the mutant complexes. Consistent with this, the addition of 3′-A5 tail does not cause a strong inhibition of binding by the mutant complexes in the case of PGK1 and RPL41A RNAs (Fig. 3; Supplemental Fig. 1). Importantly, the 3′-oligo(A) tail-mediated stimulation of RNA binding by the wild-type complex is specific to oligo(A) and replacement of the oligo(A) tail with non-oligo(A) sequence like CAGAC (Fig. 4) or other type of homo-oligomeric tails (Chowdhury et al. 2007) fails to cause such stimulation.
FIGURE 4.
Binding of MFA2 RNA by the mutant complexes is also impaired by addition of a pentanucleotide non-oligo(A) tail at the 3′-end. Gel mobility shift assays were carried out as described in Materials and Methods using MFA2 or MFA2-CAGAC RNAs (indicated below the phosphorimages) with BSA or increasing concentrations of the complex purified from LSM1, lsm1-9, or lsm1-14 cells (indicated above the phosphorimages). Fraction of RNA bound in the different reactions (quantitated using a PhosphorImager) is shown as a bar diagram in the lower panel. The numbers of the lanes in the upper panel that correspond to each pair of bars in the bar diagram are shown below the bars. In each set of reactions (LSM1, lsm1-9, or lsm1-14), the fraction of RNA bound is normalized to the value obtained with the MFA2 RNA at the highest concentration of the purified complex used.
Binding of unadenylated RNA by the mutant complexes is facilitated by the presence of U-tract near the 3′-end
We showed recently that in the case of RNAs lacking an A-tail, the Lsm1p-7p-Pat1p complex has a binding preference for those that carry an uninterrupted stretch of ≥6 U residues near their 3′-ends (Chowdhury et al. 2007). Therefore we studied the effect of lsm1-9 and lsm1-14 mutations on this property of the Lsm1p-7p-Pat1p complex by determining the relative affinities for the MFA2 and MFA2-6xUΔ RNAs of the complexes purified from the LSM1, lsm1-9, and lsm1-14 cells. The MFA2-6xUΔ RNA is an altered version of the MFA2 RNA in which the stretch of six U residues near the 3′-end has been deleted, and as a result this RNA binds several-fold weaker than the MFA2 RNA to the wild-type Lsm1p-7p-Pat1p complex (Chowdhury et al. 2007). As seen in Figure 2, the complexes purified from both mutants (lsm1-9 and lsm1-14) exhibited several-fold lower affinity for the MFA2-6xUΔ RNA than the MFA2 RNA just like the complex purified from the wild-type cells. Strengths of the secondary structures predicted by the Mfold program (Zuker 2003) for these two RNAs were similar and low (−2.5 kcal/mol and −2.3 kcal/mol, respectively), suggesting that secondary structural differences are less likely to be the cause of the binding difference. Thus, similar to the wild-type complex, the mutant complexes also exhibit a binding preference for RNAs that carry a U-tract near the 3′-end compared to those that do not. This was further supported by additional studies, which revealed that similar to the wild-type complex (Chowdhury et al. 2007), the mutant complexes also bound the RPP2B and TEF1 RNAs (which are poor binding substrates for the wild-type complex because they lack an uninterrupted stretch of ≥6 U's near the 3′-end) (Chowdhury et al. 2007) with much lower affinity than the MFA2 RNA (Supplemental Fig. 2; data not shown).
Mutant complexes bind near the 3′-end of the RNA
The purified wild-type Lsm1p-7p-Pat1p complex binds near the 3′-end of the RNA, consistent with its ability to sense changes at the 3′-end, e.g., presence or absence of oligo(A) tail (Chowdhury et al. 2007). Therefore, we asked if the mutant complexes purified from lsm1-9 and lsm1-14 cells [which are unable to recognize the 3′-oligo(A) tail] also bind near the 3′-end of the RNA. We carried out oligonucleotide inhibition experiments with the mutant complex purified from lsm1-14 cells using the MFA2(u) RNA as described earlier (Chowdhury et al. 2007). The MFA2(u) RNA is same as MFA2 RNA except for the presence of three additional U's at the 3′-end (Chowdhury et al. 2007). As shown in Figure 5A (upper panel), the binding of the mutant complex to the MFA2(u) RNA could be inhibited by annealing to that RNA an oligonucleotide complementary to its 3′-region. Oligonucleotides complementary to other regions of this RNA did not significantly inhibit the binding, although they were able to anneal to the RNA as efficiently as the oligo spanning the 3′-region as revealed by sensitivity to RNase-H of the oligo annealed RNAs (Fig. 5A, lower panel). Similar results were also obtained with the complex purified from lsm1-9 cells (data not shown). These results suggest that the mutant complexes bind near the 3′-end of the RNA similar to the wild-type complex.
FIGURE 5.
Like the wild-type complex, the mutant complexes also bind near the 3′-end of the RNA and exhibit similar affinities for unadenylated and polyadenylated RNAs. (A) Mutant complex purified from lsm1-14 cells binds near the 3′-end of the RNA. Radiolabeled MFA2(u) RNA was annealed to DNA oligonucleotides oST214, oST215, oST216, a nonspecific DNA oligonucleotide, or no oligonucleotide (indicated above the lanes in upper panel) and then the annealed RNA (at a final concentration of 0.333 nM) was used for gel shift assays (top panel) as described in Materials and Methods with the complex isolated from lsm1-14 cells (at a final concentration of 143 nM) or subjected to RNase-H treatment followed by separation on denaturing gels and autoradiography (middle panel). Regions of MFA2(u) RNA spanned by the oligonucleotides oST214, oST215, and oST216 are shown schematically in the bottom panel. Positions of the gel shifted RNA and the 50-mer marker are indicated on the right in the top and middle panels, respectively. (B) Mutant complexes (isolated from lsm1-9 and lsm1-14 cells) bind to unadenylated and polyadenylated RNA with similar affinities like the wild-type complex. Gel mobility shift assays were carried out using MFA2(u) or MFA2(u)A55 RNA (indicated below the lanes in the upper panel) with BSA (lanes marked “B”) or increasing concentrations (14 nM, 56 nM, and 140 nM) of the complex purified from LSM1, (lanes 1,2,3) lsm1-9 (lanes 4,5,6), or lsm1-14 (lanes 7,8,9) cells. Asterisks mark the positions of minor bands of unbound RNA possibly representing alternate secondary structures. Fraction of RNA bound in the different reactions (quantitated using a PhosphorImager) is shown as a bar diagram in the lower panel. The numbers of the lanes in the upper panel that correspond to each pair of bars in the bar diagram are shown below the bars. In each set of reactions (LSM1, lsm1-9, or lsm1-14), the fraction of RNA bound is normalized to the value obtained with the MFA2(u) RNA at the highest concentration of the purified complex used.
Mutant complexes exhibit comparable affinities for unadenylated and polyadenylated RNAs similar to wild-type complex
The wild-type Lsm1p-7p-Pat1p complex binds unadenylated and polyadenylated RNAs with equal affinities but binds oligoadenylated RNA with higher affinity (Chowdhury et al. 2007). Since addition of a 3′-pentaadenylate tail to the RNA failed to enhance its binding by the mutant complexes (Figs. 2, 3), we asked if presence of a longer A-tail would have an effect. So, we compared the affinity of the wild-type and mutant complexes for MFA2(u) RNA and the MFA2(u)A55 RNA (MFA2(u) RNA carrying an A55 tail). The wild-type complex bound to both of these RNAs with similar affinities consistent with our earlier results (Chowdhury et al. 2007), and, importantly, the mutant complexes behaved in the same way as well (Fig. 5B). Similar studies showed that addition of an A-tail of intermediate length (20 A residues) also failed to enhance the RNA binding by the mutant complexes (data not shown). These results suggest that the inability of the mutant complex to recognize the pentaadenylate tail is not because its length is insufficient.
Unadenylated mRNA is not a good substrate for Lsm1p-mediated decay in vivo
The results presented above show that, unlike the wild-type Lsm1p-7p-Pat1p complex, the mutant complexes (made in lsm1-9 and lsm1-14 cells) do not exhibit a binding preference for oligoadenylated RNA over unadenylated RNA. However, the mutant complexes are not affected in their integrity and retain important RNA binding properties of the wild-type complex, like binding near the 3′-end and recognition of U-tracts near the 3′-end. Given the fact that the lsm1-9 and lsm1-14 cells are impaired in mRNA decay in vivo, these observations suggest that the presence of 3′-oligo(A) tail on the mRNA is likely to be very important for Lsm1p-mediated decay of the mRNA in vivo. In order to test this idea, we studied the decay of an unadenylated mRNA whose 3′-end was generated via ribozyme cleavage in vivo. We used the construct expressing (from GAL promoter) the mRNA carrying Protein A and GFP coding regions separated by the hammerhead ribozyme sequence (see Fig. 6; Meaux and Van Hoof 2006). Upon ribozyme cleavage in vivo, a 5′-fragment of this mRNA coding for Protein A and lacking poly(A) tail is generated (Meaux and Van Hoof 2006). We determined the half-life of this unadenylated mRNA in lsm1Δ, wild-type, ski2Δ and ski2Δ lsm1Δ cells. As seen in Figure 6, this mRNA decays with a short half-life of 3 min in wild-type cells, and its stability is not significantly affected in lsm1Δ cells. Since oligoadenylated mRNAs are known to be significantly stabilized in lsm1Δ cells (Boeck et al. 1998; Bouveret et al. 2000; Tharun et al. 2000; Muhlrad and Parker 2005), this observation implies that the presence of 3′-oligo(A) tail is important for Lsm1p-mediated decay of mRNA in vivo. Consistent with this, the unadenylated Protein A mRNA decays primarily via the 3′ to 5′ decay pathway as revealed by its clear stabilization (half-life of 8 min) in the ski2Δ cells wherein the 3′ to 5′ pathway of decay is blocked. Earlier studies also showed that the 3′ to 5′ pathway is the major mode of decay of this unadenylated mRNA (Meaux and Van Hoof 2006). Our studies presented here and earlier (Chowdhury et al. 2007) show that although the purified wild-type Lsm1p-7p-Pat1p complex has a higher affinity toward oligoadenylated RNA (than unadenylated RNA), its affinity for unadenylated RNA is also quite significant. This suggests that unadenylated mRNA is unlikely to be completely immune to Lsm1p-mediated decay in vivo and that such decay is likely to be at least a minor mode of decay for the unadenylated mRNA. In order to test this, we asked if Lsm1p becomes important for the decay of the unadenylated Protein A mRNA when the 3′ to 5′ decay is blocked. As shown in Figure 6, in a ski2Δ lsm1Δ strain this mRNA decays with a much longer half-life (20 min) compared to ski2Δ strain. Thus, absence of Lsm1p affects the stability of the unadenylated Protein A mRNA only under the conditions in which 3′ to 5′ decay is already blocked (ski2Δ background) but not under normal conditions (wild-type background). This suggests that the decay of unadenylated mRNA is much less efficient via the Lsm1p-mediated 5′ to 3′ pathway. Since it is known that the Lsm1p-mediated 5′ to 3′ decay is a major mode of decay for oligoadenylated mRNAs in yeast (Boeck et al. 1998; Bouveret et al. 2000; Tharun et al. 2000; Muhlrad and Parker 2005), these observations support the notion that the presence of 3′-oligo(A) tail is crucial for the normal rates of Lsm1p-mediated 5′ to 3′ decay of mRNAs in vivo.
FIGURE 6.
LSM1 is necessary for the decay of unadenylated mRNA in vivo when 3′ to 5′ decay is blocked. A schematic diagram of the reporter gene construct used for this experiment (Meaux and Van Hoof 2006) is shown on top. The 5′-fragment of ribozyme mediated cleavage of the transcript encoded by this gene construct is the unadenylated Protein A mRNA whose half-life is measured in this experiment. Cells were grown to log phase in galactose medium to express the reporter mRNA, whose transcription is then shut off by shifting cells to glucose medium. Following this, RNA was made from the cells at different time points and subjected to Northern analysis followed by PhosphorImager quantitation of the bands to determine the rate of disappearance of the Protein A mRNA. Level of this mRNA in each sample was normalized for that of 7S RNA of SRP (to serve as loading control), which was determined by reprobing the blot for that RNA. Strains used for the experiment and the half-lives determined are shown on the left and right of the Northern blot phosphorimages.
DISCUSSION
The highly conserved Lsm1p-7p-pat1p complex is a key activator of decapping in the 5′ to 3′ mRNA decay pathway of eukaryotes, wherein oligoadenylated but not polyadenylated mRNAs are selectively decapped. Consistently, the purified Lsm1p-7p-Pat1p complex has a strong binding preference for oligoadenylated RNA over unadenylated and polyadenylated RNA in vitro (Chowdhury et al. 2007). The studies on the lsm1-9 and lsm1-14 mutants presented here reveal that loss of such binding preference of the Lsm1p-7p-Pat1p complex can result in inhibition of mRNA decay in vivo. Mutant complexes made in the lsm1-9 and lsm1-14 cells fail to show enhanced affinity for oligoadenylated RNAs over unadenylated or polyadenylated RNAs but are similar to the wild-type complex with regard to several RNA binding properties like binding near the 3′-end of the RNA, having higher affinity for unadenylated RNAs that carry U-tracts near the 3′-end over those that do not, and exhibiting similar affinities for unadenylated and polyadenylated RNAs. Yet these mutants are clearly defective in mRNA decay in vivo. These observations suggest that the oligo(A) tail-mediated augmentation of Lsm1p-7p-Pat1p complex interaction with the mRNA is important for efficient targeting of the mRNA by the 5′ to 3′ pathway. Consistent with such an idea, the in vivo decay of unadenylated mRNA (generated by ribozyme cleavage) is much less efficient via the 5′ to 3′ pathway than the 3′ to 5′ pathway.
Although the lsm1-9 and lsm1-14 alleles bear lesions in the predicted RNA binding residues of Lsm1p, the mutant complexes made in lsm1-9 and lsm1-14 cells have not completely lost their RNA binding ability and have retained many of the RNA binding properties of the wild-type complex. This could be due to two reasons. First, in each of these alleles only a subset of the predicted RNA binding residues of Lsm1p is changed (Tharun et al. 2005). Second, the subunits other than Lsm1p (Lsm2p through Lsm7p and Pat1p) could also play a substantial role in binding RNA. In fact, structural studies on the Sm-like protein complexes Hfq and AF-Sm1 show that adjacent nucleotide bases of the RNA contact different subunits of the complex (Toro et al. 2001; Schumacher et al. 2002) such that RNA binding is determined by the collective interactions of multiple subunits. In any case, consistent with the mutations affecting the predicted RNA binding residues of Lsm1p in these mutants, the overall RNA binding ability of the mutant complexes is lower than that of the wild-type complex (Figs. 2–5).
Our in vitro studies clearly show that the inability of the mutant complexes to recognize oligoadenylated RNAs is due to their decreased affinity (compared to the wild-type complex) for such RNAs and not because their affinity for unadenylated and polyadenylated RNAs is increased (compared to the wild-type complex) to a level that they are bound as efficiently as the oligoadenylated RNAs (Figs. 2–5). Since oligoadenylated mRNAs are the main substrates for decapping in vivo, this implies that the less efficient binding of oligoadenylated mRNAs by the mutant Lsm1p-7p-Pat1p complexes is likely to be at least one of the reasons for the impairment of mRNA decay observed in lsm1-9 and lsm1-14 cells.
Our studies show that addition of a 3′-A5 tail to the RNA stimulates RNA binding by the wild-type complex but not RNA binding by the mutant complexes purified from lsm1-9 and lsm1-14 cells. Although in the case of MFA2 RNA, addition of 3′-A5 tail results in inhibition of binding by the mutant complexes (while it leads to stimulation of binding by the wild-type complex), such inhibition is also observed upon adding 5-nt-long non-oligo(A) sequence, CAGAC to the 3′-end of MFA2 RNA. These results suggest that the mutant complexes are impaired in their ability to specifically recognize the oligo(A) tail. Given this, the lower affinity of the mutant complexes for the MFA2-A5 and MFA2-CAGAC RNAs (compared to MFA2 RNA) could be because these RNAs are perceived by the mutant complexes as substrates that carry the U-tract farther from the 3′-end. Because we have shown earlier that while the presence of U-tract near the 3′-end stimulates RNA binding by the Lsm1p-7p-Pat1p complex, such stimulation is impaired if the U-tract is moved away from the 3′-end (Chowdhury et al. 2007). Consistent with this, although addition of 3′-A5 tail to MFA2 RNA stimulates its binding by the wild-type complex, addition of the CAGAC sequence at the 3′-end of this RNA results in impairment of binding by the wild-type complex (Fig. 4). Finally, the U-tract is 4 nt away from the 3′-end in MFA2 RNA but is at the very 3′-end or just 1 nt away from the 3′-end in the PGK1 and RPL41A RNAs, respectively. Therefore, this and other primary structural differences between the MFA2 and the latter two RNAs could be responsible for the differential effects of 3′-A5 tail addition to these RNAs on the binding by the mutant complexes.
Our results argue that the Lsm1p subunit is crucial for the unique ability of the Lsm1p-7p-Pat1p complex to preferentially bind to oligoadenylated RNAs (Chowdhury et al. 2007). This is consistent with the fact that Lsm1p is a key subunit that distinguishes this complex from the related Lsm2p-8p complex. However, this does not rule out a role for the Pat1p subunit in this function since Pat1p also associates specifically with the Lsm1p-7p complex but not Lsm2p-8p complex (Bouveret et al. 2000; Tharun et al. 2000). Further, Pat1p by itself can bind poly(U) (Pilkington and Parker 2008). Therefore, both Lsm1p and Pat1p are likely to be involved in the recognition of oligo(A) tail and U-tracts at the 3′-end.
We showed earlier that in both lsm1-9 and lsm1-14 mutants, Lsm2p, Lsm7p, and (mutant) Lsm1p localize to the P-bodies just as in wild-type cells (Tharun et al. 2005). Our results presented here show that the RNA binding ability of the mutant complexes made in these cells is not completely abolished. These observations together suggest that in the lsm1-9 and lsm1-14 mutants, transport of mRNP to P-bodies may occur normally and decay could be blocked at any of the later step(s) involving the activation of the decapping enzyme.
Our studies on the decay of unadenylated mRNA (generated by ribozyme cleavage) reveal that such mRNA is poorly targeted by the Lsm1p-mediated 5′ to 3′ decay pathway in vivo and therefore decays mainly via the 3′ to 5′ pathway. This is in contrast to the known fact that oligoadenylated mRNAs mainly decay via the Lsm1p-mediated 5′ to 3′ pathway in yeast. These results together with the observations that mRNA decay in vivo is impaired in lsm1-9 and lsm1-14 mutants and that the mutant complexes made in these cells do not have enhanced affinity for the oligoadenylated RNAs over unadenylated RNAs imply that presence of 3′-oligo(A) tail on the mRNA may be very important for efficient destruction of the mRNA through the Lsm1p-mediated 5′ to 3′ decay pathway. Thus, while the poly(A) tail plays a key role in facilitating translation, the oligo(A) tail of the mRNA could play an important role in promoting the 5′ to 3′ decay of the mRNA.
Interestingly, in prokaryotes 3′-oligo(A) tail addition facilitates RNA degradation via 3′ to 5′ exonucleolytic attack by polynucleotide phosphorylase (PNPase) and RNase-II (Dreyfus and Regnier 2002; Kushner 2002). Similarly, in eukaryotic nucleus, 3′-oligo(A) tail addition by the TRAMP complex facilitates 3′-processing and degradation of different RNAs by the exosome (Anderson 2005; Egecioglu et al. 2006). This is presumably because the oligo(A) tail provides a single-stranded site for the 3′ to 5′ exonuclease to hold on to the RNA stably so that it can work through the upstream secondary structures. Our studies presented here reveal the significance of the 3′-oligo(A) tail in facilitating decay from the 5′-end. Thus oligo(A) tailed 3′-end of RNA could serve as a recruiting site for factors facilitating decay from both 3′- and 5′-ends of the RNA in different cellular contexts.
MATERIALS AND METHODS
Yeast strains and plasmids
Strains used in this study are in the genetic background of yRP841 (Hatfield et al. 1996). The LSM1, lsm1-9, and lsm1-14 strains used for the purification of the Lsm1p-7p-Pat1p complex were made by transforming yST247 (Chowdhury et al. 2007) (MAT α, leu2, ura3, lys2, trp1, cup1Δ∷LEU2(PM), lsm1Δ∷TRP1, LSM5-6xHis-NEOr) with the plasmids pST17 (Chowdhury et al. 2007), pST66, and pST68, respectively. The lsm1Δ (yRP1365), ski2Δ (yRP1192), and lsm1Δ ski2Δ (yRP1424) strains have been previously described (He and Parker 2001).
Mutations of lsm1-9 and lsm1-14 were introduced by the Quikchange mutagenesis procedure (Stratagene) using pST17 (Chowdhury et al. 2007) as the template to generate pST66 and pST68. The FLAG-lsm1 inserts in these plasmids carried the native flanking sequences (including the promoter and UTR) of LSM1. Plasmid construct coding for ribozyme cleaved mRNA (pAV226) has been described previously (Meaux and Van Hoof 2006).
Protein purification and RNA analyses
Purification of mutant and wild-type complexes was carried out as described before (Chowdhury et al. 2007). Radiolabeled RNA substrates for in vitro experiments were made by in vitro transcription using T7 RNA polymerase and α-32P-labeled UTP as described earlier (Chowdhury et al. 2007). MFA2(u) and MFA2(u)A55 RNAs were made using plasmid template (Chowdhury et al. 2007), while all the other RNAs were made using oligonucleotide templates with the help of the Mirvana kit (Ambion, Inc.). Purification of the substrate RNAs, RNA binding reactions, and gel shift analyses were done as described (Chowdhury et al. 2007). Concentration of the radiolabeled RNA substrate in the binding reaction was 0.33 nM for the experiment in Figure 5A. In all the other experiments, it was 0.05 nM so that the molar concentration of the radiolabeled RNA substrate is several-fold lower than the lowest molar concentration of the purified complex used in the experiment. In all the experiments, substrate RNA was heat denatured before it was included in the binding reaction. RNA substrates whose binding affinities are compared in an experiment [e.g., comparison of MFA2, MFA2-A5, and MFA2-6xUΔ RNAs; comparison of PGK1 and PGK1-A5 RNAs; comparison of MFA2(u) and MFA2(u)A55 RNAs] were always prepared and tested together. BSA was used at final 147 nM concentration in the control reactions. Oligonucleotide inhibition experiments and RNase-H reactions were carried out as described (Chowdhury et al. 2007). Northern analyses and determination of mRNA half-lives were also done as described before (Tharun and Parker 1999; Tharun et al. 2005).
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Roy Parker for helpful comments on the manuscript. We thank Ambro van Hoof's laboratory for the plasmid construct encoding ribozyme cleaved mRNA and Roy Parker's laboratory for ski2Δ strains. This work was supported by USUHS intramural grant (C071HJ) and NIH grant (GM072718) to S.T.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1094208.
REFERENCES
- Achsel, T., Brahms, H., Kastner, B., Bachi, A., Wilm, M., Luhrmann, R. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 1999;18:5789–5802. doi: 10.1093/emboj/18.20.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani, N., Ghosh, S., Mangus, D.A., Jacobson, A. Translation factors promote the formation of two states of the closed-loop mRNP. Nature. 2008;453:1276–1280. doi: 10.1038/nature06974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, J.T. RNA turnover: Unexpected consequences of being tailed. Curr. Biol. 2005;15:R635–R638. doi: 10.1016/j.cub.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Beelman, C.A., Stevens, A., Caponigro, G., LaGrandeur, T.E., Hatfield, L., Fortner, D.M., Parker, R. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;382:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- Boeck, R., Lapeyre, B., Brown, C.E., Sachs, A.B. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol. Cell. Biol. 1998;18:5062–5072. doi: 10.1128/mcb.18.9.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnerot, C., Boeck, R., Lapeyre, B. The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol. Cell. Biol. 2000;20:5939–5946. doi: 10.1128/mcb.20.16.5939-5946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret, E., Rigaut, G., Shevchenko, A., Wilm, M., Seraphin, B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro, G., Parker, R. Multiple functions for the poly(A)-binding protein in mRNA decapping and deadenylation in yeast. Genes & Dev. 1995;9:2421–2432. doi: 10.1101/gad.9.19.2421. [DOI] [PubMed] [Google Scholar]
- Chowdhury, A., Mukhopadhyay, J., Tharun, S. The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA. 2007;13:998–1016. doi: 10.1261/rna.502507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller, J., Parker, R. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- Coller, J.M., Gray, N.K., Wickens, M.P. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes & Dev. 1998;12:3226–3235. doi: 10.1101/gad.12.20.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller, J.M., Tucker, M., Sheth, U., Valencia-Sanchez, M.A., Parker, R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker, C.J., Parker, R. A turnover pathway for both stable and unstable mRNAs in yeast: Evidence for a requirement for deadenylation. Genes & Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Dreyfus, M., Regnier, P. The poly(A) tail of mRNAs: Bodyguard in eukaryotes, scavenger in bacteria. Cell. 2002;111:611–613. doi: 10.1016/s0092-8674(02)01137-6. [DOI] [PubMed] [Google Scholar]
- Dunckley, T., Parker, R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley, T., Tucker, M., Parker, R. Two related proteins, Edc1p and Edc2p, stimulate mRNA decapping in Saccharomyces cerevisiae . Genetics. 2001;157:27–37. doi: 10.1093/genetics/157.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egecioglu, D.E., Henras, A.K., Chanfreau, G.F. Contributions of Trf4p- and Trf5p-dependent polyadenylation to the processing and degradative functions of the yeast nuclear exosome. RNA. 2006;12:26–32. doi: 10.1261/rna.2207206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, N., Weis, K. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J. 2002;21:2788–2797. doi: 10.1093/emboj/21.11.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfield, L., Beelman, C.A., Stevens, A., Parker, R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae . Mol. Cell. Biol. 1996;16:5830–5838. doi: 10.1128/mcb.16.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, W., Parker, R. The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3′ termini from partial degradation. Genetics. 2001;158:1445–1455. doi: 10.1093/genetics/158.4.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, C.L., Stevens, A. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelfinger, D., Arndt-Jovin, D.J., Luhrmann, R., Achsel, T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Khanna, R., Kiledjian, M. Poly(A)-binding-protein-mediated regulation of hDcp2 decapping in vitro. EMBO J. 2004;23:1968–1976. doi: 10.1038/sj.emboj.7600213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kshirsagar, M., Parker, R. Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae . Genetics. 2004;166:729–739. doi: 10.1093/genetics/166.2.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner, S.R. mRNA decay in Escherichia coli comes of age. J. Bacteriol. 2002;184:4658–4665. doi: 10.1128/JB.184.17.4658-4665.2002. discussion 4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus, D.A., Evans, M.C., Jacobson, A. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayes, A.E., Verdone, L., Legrain, P., Beggs, J.D. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 1999;18:4321–4331. doi: 10.1093/emboj/18.15.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaux, S., Van Hoof, A. Yeast transcripts cleaved by an internal ribozyme provide new insight into the role of the cap and poly(A) tail in translation and mRNA decay. RNA. 2006;12:1323–1337. doi: 10.1261/rna.46306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, S., Temme, C., Wahle, E. Messenger RNA turnover in eukaryotes: Pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- Muhlrad, D., Parker, R. The yeast EDC1 mRNA undergoes deadenylation-independent decapping stimulated by Not2p, Not4p, and Not5p. EMBO J. 2005;24:1033–1045. doi: 10.1038/sj.emboj.7600560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad, D., Decker, C.J., Parker, R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes & Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- Parker, R., Song, H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Pilkington, G.R., Parker, R. Pat1 contains distinct functional domains that promote P-body assembly and activation of decapping. Mol. Cell. Biol. 2008;28:1298–1312. doi: 10.1128/MCB.00936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez, C.V., Vilela, C., Berthelot, K., McCarthy, J.E. Modulation of eukaryotic mRNA stability via the cap-binding translation complex eIF4F. J. Mol. Biol. 2002;318:951–962. doi: 10.1016/S0022-2836(02)00162-6. [DOI] [PubMed] [Google Scholar]
- Salgado-Garrido, J., Bragado-Nilsson, E., Kandels-Lewis, S., Seraphin, B. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 1999;18:3451–3462. doi: 10.1093/emboj/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher, M.A., Pearson, R.F., Moller, T., Valentin-Hansen, P., Brennan, R.G. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: A bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, D.C., Parker, R. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae . Mol. Cell. Biol. 1999;19:5247–5256. doi: 10.1128/mcb.19.8.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth, U., Parker, R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun S.Z., Jr, Sachs, A.B. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Teixeira, D., Sheth, U., Valencia-Sanchez, M.A., Brengues, M., Parker, R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun, S., Parker, R. Analysis of mutations in the yeast mRNA decapping enzyme. Genetics. 1999;151:1273–1285. doi: 10.1093/genetics/151.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun, S., Parker, R. Targeting an mRNA for decapping: Displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol. Cell. 2001;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- Tharun, S., He, W., Mayes, A.E., Lennertz, P., Beggs, J.D., Parker, R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- Tharun, S., Muhlrad, D., Chowdhury, A., Parker, R. Mutations in the Saccharomyces cerevisiae LSM1 gene that affect mRNA decapping and 3′-end protection. Genetics. 2005;170:33–46. doi: 10.1534/genetics.104.034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro, I., Thore, S., Mayer, C., Basquin, J., Seraphin, B., Suck, D. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 2001;20:2293–2303. doi: 10.1093/emboj/20.9.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, M., Valencia-Sanchez, M.A., Staples, R.R., Chen, J., Denis, C.L., Parker, R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae . Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- Wells, S.E., Hillner, P.E., Vale, R.D., Sachs, A.B. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- Wilusz, C.J., Gao, M., Jones, C.L., Wilusz, J., Peltz, S.W. Poly(A)-binding proteins regulate both mRNA deadenylation and decapping in yeast cytoplasmic extracts. RNA. 2001;7:1416–1424. [PMC free article] [PubMed] [Google Scholar]
- Yamashita, A., Chang, T.C., Yamashita, Y., Zhu, W., Zhong, Z., Chen, C.Y., Shyu, A.B. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]