Abstract
The sarcin–ricin loop (SRL) of 23S rRNA in the large ribosomal subunit is a factor-binding site that is essential for GTP-catalyzed steps in translation, but its precise functional role is thus far unknown. Here, we replaced the 15-nucleotide SRL with a GAAA tetraloop and affinity purified the mutant 50S subunits for functional and structural analysis in vitro. The SRL deletion caused defects in elongation-factor-dependent steps of translation and, unexpectedly, loss of EF-Tu-independent A-site tRNA binding. Detailed chemical probing analysis showed disruption of a network of rRNA tertiary interactions that hold together the 23S rRNA elements of the functional core of the 50S subunit, accompanied by loss of ribosomal protein L16. Our results reveal an influence of the SRL on the higher-order structure of the 50S subunit, with implications for its role in translation.
Keywords: sarcin–ricin loop, 23S rRNA, ribosome assembly, A site
INTRODUCTION
The sarcin–ricin loop (SRL) in helix 95 of 23S rRNA (nucleotides 2653–2667, Escherichia coli numbering) contains one of the longest universally conserved rRNA sequences (Gutell et al. 1992) and is a primary component of the ribosomal elongation-cycle machinery. Cleavage of the G2661–A2662 phosphodiester bond by α-sarcin (Endo and Wool 1982) or depurination of A2660 by ricin (Endo et al. 1987) inhibits translation by disrupting the interaction of ribosomes with elongation factors EF-Tu (EF-1 in eukaryotes) and EF-G (EF-2) (Hausner et al. 1987; Moazed et al. 1988; Brigotti et al. 1989). Short oligonucleotides containing the SRL sequence are recognized and cleaved by α-sarcin and ricin (Gluck et al. 1994) and have been shown to bind EF-G (Munishkin and Wool 1997). Crystal structures show that the isolated SRL adopts the same conformation (Correll et al. 1999) as found in the intact 50S subunit (Ban et al. 2000), where it projects from the interface in the 70S ribosome (Yusupov et al. 2001).
It is now known that at least five different GTPase factors of seemingly diverse functions interact with the SRL. EF-Tu catalyzes A-site tRNA binding, and EF-G accelerates translocation of tRNAs from the A and P sites to the P and E sites, respectively (Wintermeyer et al. 2004). Initiation factor IF2 stimulates fMet-tRNAfMet binding to the P site (Mazumder et al. 1969), whereas release factor RF3 catalyzes dissociation of RF1 or RF2 from the A site following peptide release (Freistroffer et al. 1997), and Tet(O) catalyzes dissociation of the antibiotic tetracycline (Trieber et al. 1998). Cryo-EM reconstructions of ribosomal complexes containing EF-G (Valle et al. 2003b), EF-Tu·GDP·aminoacyl-tRNA (Stark et al. 2002; Valle et al. 2003a), IF2 (Allen et al. 2005; Myasnikov et al. 2005), RF3 (Klaholz et al. 2004; Gao et al. 2007), and Tet(O) (Spahn et al. 2001) show that it is the GTP-binding region of each factor that interacts with the SRL.
Although their activities seem unrelated, there may be an underlying commonality to the molecular mechanisms of the GTP-dependent translation factors. Their G domains interact with the SRL, and each factor appears to act by overcoming kinetic barriers through mechanisms that likely involve structural rearrangements in their respective ribosomal functional complexes. A central question is whether the SRL serves merely as a static factor-docking site or whether its interactions with these factors trigger conformational changes in the ribosome that facilitate GTP-catalyzed steps of translation (Wool et al. 1992).
Crystal structures show that the SRL interacts with the body of the 50S subunit via the C-terminal domain of ribosomal protein L6 and loop–loop tertiary interactions with 23S rRNA helix 91 (h91) (Ban et al. 2000). H91, in turn, contacts h89, h90, and h92, which are connected directly to the A- and P-tRNA binding sites and the peptidyl transferase center (PTC). H91 also contacts h42, which connects to the so-called GTPase-associated center (GAC; h43 and h44), a second feature of 23S rRNA that has been shown to interact with the elongation factors (Moazed et al. 1988; Agrawal et al. 2001; Valle et al. 2003a). Thus, tertiary contacts link the SRL with several other critical regions of the 50S subunit, raising the possibility of a signaling pathway between the SRL and the functional core of the 50S subunit (Ban et al. 2000; Yusupov et al. 2001; Chan et al. 2006).
In this study, we examined the functional and structural consequences of deleting the entire 15-nucleotide (nt) SRL from 50S subunits and replacing it with a GAAA tetraloop. The mutation confers a dominant-lethal phenotype in vivo, consistent with the high phylogenetic conservation of the SRL. Affinity-purified mutant ribosomes show defects in both EF-Tu-dependent and nonenzymatic A-site tRNA binding, as well as in functional interactions with EF-G. Unexpectedly, detailed chemical probing experiments show that the SRL deletion leads to increased accessibility of more than forty 23S rRNA nucleotides, indicating disruption of numerous tertiary interactions in and around the functional core of the 50S subunit. The mutant ribosomes also show complete loss of ribosomal protein L16, whose binding site lies within the disrupted region of 23S rRNA. This assembly defect reveals a previously unobserved structural dependency between the SRL and the functional core of the 50S subunit that carries potential implications for the role of the SRL in translation.
RESULTS
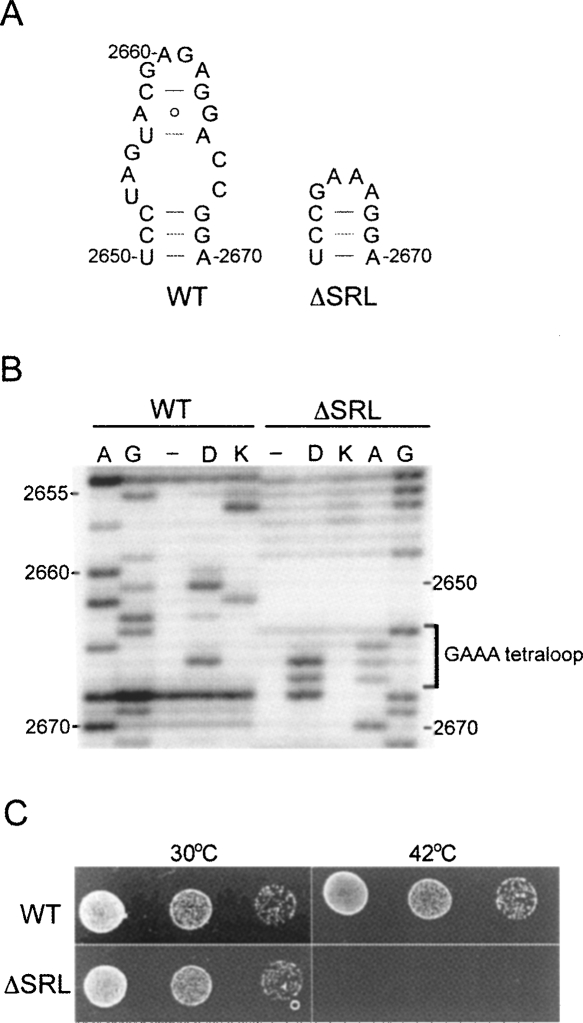
The SRL (nucleotides 2653–2667) was deleted from 23S rRNA and replaced with a GAAA tetraloop sequence (Fig. 1A) by site-directed mutagenesis, as described in Materials and Methods. The deletion mutant (ΔSRL) was constructed in plasmid pLK35.50S.MS2 (Ali et al. 2006), which contains the MS2 coat protein RNA binding sequence (LeCuyer et al. 1995) inserted into h98 of 23S rRNA to facilitate affinity purification of mutant 50S subunits (Youngman et al. 2004). Plasmid-derived ΔSRL 23S rRNA was transiently expressed from the PL promoter by inactivation of the temperature-sensitive cI857 repressor protein at 42°C (Gourse et al. 1985; Douthwaite et al. 1989). Cells expressing ΔSRL 50S subunits failed to grow in the presence of wild-type (WT) subunits (Fig. 1C, 42°C), indicating a dominant-lethal phenotype.
FIGURE 1.
Deletion of the SRL confers a dominant-lethal phenotype. (A) Wild-type (WT) sequence of 23S rRNA nucleotides 2650–2670 and that of the SRL deletion mutant (ΔSRL), in which nucleotides 2653–2667 (the SRL) were replaced with a GAAA tetraloop. (B) Primer-extension analysis of 23S rRNA from affinity-purified WT and ΔSRL 50S subunits probed with either D, dimethylsulfate, or K, kethoxal. Lanes A and G are sequencing lanes; (−) unmodified subunits. (C) E. coli strains that express either WT or ΔSRL MS2-tagged 50S subunits from plasmid, via a heat-inducible promoter, were grown at 30°C, spotted on plates, and incubated at 30°C or 42°C.
Characterization of ΔSRL ribosomes
ΔSRL and WT MS2-tagged 50S subunits were affinity purified as described in Materials and Methods. The structure of 23S rRNA in the SRL-deleted region of ΔSRL ribosomes was compared to that of WT by probing purified 50S subunits with dimethylsulfate (DMS), which methylates the N1 position of A and N3 of C, and kethoxal (KE), which modifies N1 and N2 of G (Stern et al. 1988). Probe-reactive nucleotides were detected by rRNA primer extension analysis (Fig. 1B). In WT 50S subunits G2655, A2660, G2661, A2662, and A2665 are reactive, consistent with previous probing results (Moazed et al. 1988) and the crystal structure of the SRL in the 50S subunit (Ban et al. 2000). In ΔSRL 50S subunits, all three A's of the GAAA sequence are strongly reactive, whereas the surrounding bases are unreactive, consistent with the properties of a typical tetraloop that closes off a standard helix (Heus and Pardi 1991). These results confirm the purity of the ΔSRL 50S subunits and indicate that the inserted GAAA sequence has folded as expected.
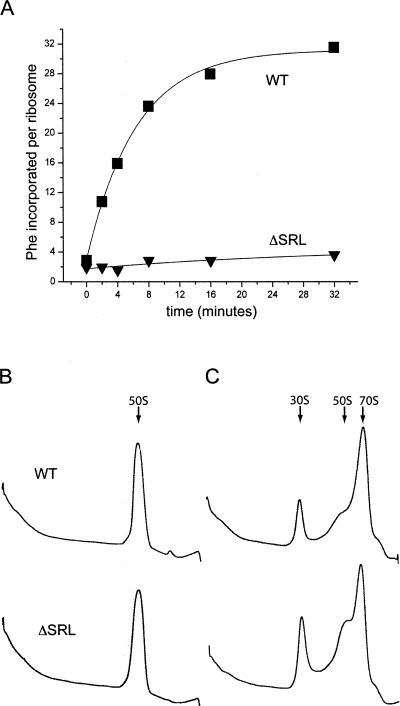
Ribosomes containing ΔSRL 50S subunits showed virtually no translational activity in vitro (Fig. 2A) when assayed by poly(U)-directed poly(Phe) synthesis (Traub et al. 1981). In order to function in this assay, 50S subunits must associate with 30S subunits, bind P-site and A-site tRNA, carry out peptidyl transfer, and interact functionally with elongation factors EF-Tu and EF-G. To determine which of these activities were affected by the SRL deletion, we performed a series of in vitro assays.
FIGURE 2.
ΔSRL 50S subunits are inactive in protein synthesis, but can associate with 30S subunits. (A) Poly-U mRNA directed [14C]-Phe incorporation was measured for ribosomes containing WT (■) or ΔSRL (▼) 50S subunits. Sucrose density-gradient sedimentation of WT and ΔSRL 50S subunits in the absence (B) or presence (C) of 30S subunits. Arrows indicate the position of 30S subunits, 50S subunits, or 70S ribosomes.
Subunit association activity was assayed by sucrose gradient centrifugation, which showed that ΔSRL 50S subunits had a sedimentation rate indistinguishable from that of WT 50S subunits (Fig. 2B), but showed a slight defect in their ability to form 70S ribosomes (Fig. 2C).
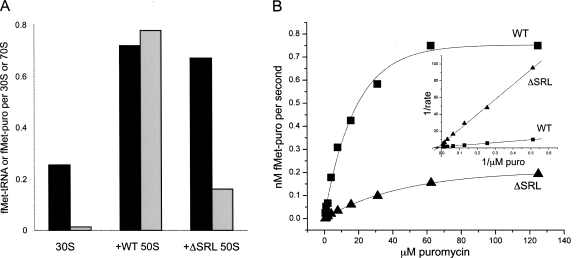
To test the function of the 50S P site, stimulation of binding of [35S]fMet-tRNAfMet to 30S subunit–mRNA complexes by 50S subunits was assayed by nitrocellulose filter binding. The results showed that both ΔSRL and WT 50S stimulated tRNA binding by about threefold (Fig. 3A, black bars), indicating that extent of P-site tRNA binding is unaffected. Peptidyl transferase (PT) activity was tested by reaction of the [35S]fMet-tRNAfMet–ribosome complex with the A-site tRNA analog puromycin. In complexes containing ΔSRL 50S, only ∼25% of the P-site-bound tRNA was puromycin reactive under conditions that are saturating for wild-type ribosomes (Fig. 3A, gray bars). This result indicates that PT activity, binding of puromycin, or both were compromised by the SRL deletion and that there is functional heterogeneity within the mutant ribosome population.
FIGURE 3.
ΔSRL 50S subunits bind P-site tRNA to WT levels, but are defective in the puromycin reaction. (A) 50S subunit stimulation of P-site tRNA binding to 30S subunits (black bars) and puromycin reactivity (gray bars) were assayed in complexes containing [35S]-fMet-tRNAfMet, mRNA, and 30S subunits formed in the presence or absence of WT or ΔSRL 50S subunits as indicated. (B) ΔSRL (▲) and WT (■) 50S subunits bound to [35S]-fMet-tRNAfMet were reacted with increasing concentrations of puromycin. The results were fit to a single exponential curve and are also shown as a double-reciprocal plot (inset).
To determine whether the puromycin-sensitive subpopulation of ΔSRL 50S subunits react with the same kinetic profile as WT 50S, we followed the PT reaction as a function of puromycin concentration (Fig. 3B) in a reaction containing [35S]fMet-tRNAfMet and methanol, which allows P-site binding in the absence of 30S subunits (Monro 1967). The Km for ΔSRL 50S was 2.7-fold higher (40 μM puromycin) than for WT (15 μM), and the ΔSRL 50S Vmax (0.23 nM/s) was about fourfold lower than that of WT (0.88 nM/s). However, if only the puromycin-reactive population of ΔSRL 50S subunits (∼25%; Fig. 3A) is taken into account, the turnover number (k cat) is about the same for ΔSRL and WT 50S (17.6 × 10−4 s−1 and 18.0 × 10−4 s−1, respectively). These results suggest that the defect in ΔSRL PT activity within the reactive subpopulation is primarily due to a reduced ability to bind puromycin to the A site, and that the majority of mutant ribosomes may not be binding puromycin at all.
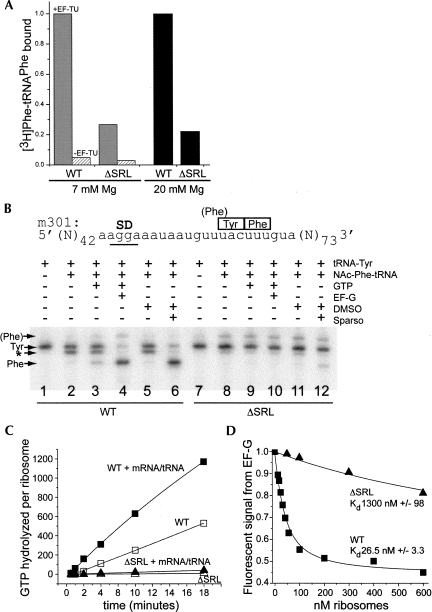
We next tested A-site tRNA binding under conditions that saturate the A site of WT ribosomes. Ribosomal complexes containing mRNA and P-site-bound fMet-tRNAfmet were assembled in 20 mM Mg++ buffer; the [Mg++] was then lowered to 7 mM to create conditions for EF-Tu-dependent aminoacyl-tRNA binding. EF-Tu·GTP-[3H]-Phe-tRNAPhe ternary complex was added, and tRNA binding was monitored by nitrocellulose filter binding. The results showed that A-site binding to ΔSRL ribosomes reached ∼25% that of WT and was dependent on the presence of EF-Tu (Fig. 4A, gray versus hatched bars). Nonenzymatic A-site binding was carried out by adding [3H]-Phe-tRNAPhe to the P-site-bound complexes and incubating in 20 mM Mg++ buffer. Under these conditions, A-site binding to ΔSRL ribosomes was also ∼25% that of WT (Fig. 4A, black bars). Similar results were obtained with complexes containing other combinations of mRNA and P- and A-site tRNAs (data not shown). These results indicate that the SRL deletion causes a general defect in binding of aminoacyl-tRNA to the 50S subunit A site and again suggest functional heterogeneity within the mutant ribosome population.
FIGURE 4.
ΔSRL 50S subunits are defective in EF-Tu-dependent and nonenzymatic A-site tRNA binding and in interaction with EF-G. (A) [3H]-Phe-tRNAPhe was bound to the A site of ribosomal complexes containing mRNA and fMet-tRNAfMet under either EF-Tu-dependent (gray and hatched bars, 7 mM Mg), or nonenzymatic (black bars, 20 mM Mg) conditions. A-site binding was normalized to the highest level obtained by WT ribosomes, which corresponds to 0.95 [3H]-Phe-tRNAPhe per ribosome (EF-Tu-dependent) and 0.8 per ribosome (nonenzymatic). (B) Toeprint analysis of tRNA binding and translocation using m301 mRNA; ribosomal complexes contain tRNA and factors as indicated. Reverse transcriptase stops (arrows) correspond to the indicated codons in the P site, and the doublet band indicates A-site-bound tRNA (*). (C) Ribosome-dependent EF-G·GTP hydrolysis. ΔSRL (triangle) or WT (square) vacant ribosomes (filled symbols) or those containing polyU mRNA and tRNAPhe (open symbols) were incubated in the presence of EF-G and [32P]-γ-GTP. (D) EF-G binding. Quenching of fluorescein-labeled EF-G was measured with increasing concentrations of ΔSRL (▲) or WT (■) ribosomal complexes containing mRNA and tRNAfMet.
We next assayed tRNA translocation catalyzed by EF-G·GTP or the antibiotic sparsomycin (Fredrick and Noller 2003) using toeprinting (Hartz et al. 1989), which monitors tRNA binding and the positioning of mRNA in the ribosome. First, complexes were formed containing tRNATyr in the P site; the position and intensity of the cDNA band corresponding to tRNATyr was similar for ΔSRL and WT ribosomes (Fig. 4B, cf. lanes 1 and 7), again indicating that the deletion did not affect P-site binding. However, when NAc-Phe-tRNAPhe was added to the A site, the doublet band indicative of A-site tRNA binding was only very faintly observed for ΔSRL ribosomes compared to WT (Fig. 4B, cf. lanes 2 and 8); instead, a weak band corresponding to an overlapping Phe P-site codon (labeled “(Phe)” in Fig. 4B) appeared in the ΔSRL ribosome complexes, likely due to partial displacement of tRNATyr by NAc-Phe-tRNAPhe. Upon addition of EF-G·GTP, a band corresponding to the translocated complex was not observed for ΔSRL ribosomes (Fig. 4B, cf. lane 4 and 10), whereas a faint band appeared in the presence of sparsomycin (Fig. 4B, cf. lanes 6 and 12). This experiment was repeated with complexes containing other combinations of mRNA and P- and A-site tRNAs. In each case, a faint doublet band indicative of A-site tRNA binding, and one corresponding to translocation by sparsomycin, but not EF-G was observed for the ΔSRL ribosomes (data not shown). These suggest that the SRL deletion does not affect tRNA translocation, but does disturb interaction with EF-G. However, interpretation of the results is complicated by the low level of A-site tRNA binding, which is required for translocation (Joseph and Noller 1998). A-site binding observed indirectly in this assay (the doublet band and that corresponding to translocation by sparsomycin) appears more defective for ΔSRL ribosomes than in the filter-binding assay (5%–10% versus 25% that of WT) under the same conditions. The reason for this discrepancy is unknown, but may reflect incomplete accommodation of A-site tRNA bound to ΔSRL ribosomes.
Functional interaction with EF-G was also assayed by monitoring ribosome-dependent EF-G·GTP hydrolysis (Rodnina et al. 1997). Ribosomes containing ΔSRL 50S subunits were inactive in this assay (Fig. 4C), even in the presence of tRNA and mRNA, which stimulated EFG·GTP hydrolysis by WT ribosomes by twofold. The Kd for EF-G binding to ΔSRL ribosomes in complex with mRNA and tRNAfMet was determined by monitoring the change in quantum yield of fluorescein-labeled EF-G·GDPNP upon binding to the ribosome (D. Ermolenko, unpubl.), based on the method of Seo et al. (2004). For ΔSRL ribosomes, the Kd was about 50-fold greater than that of WT (Fig. 4D), indicating that deletion of the SRL caused a defect in EF-G binding.
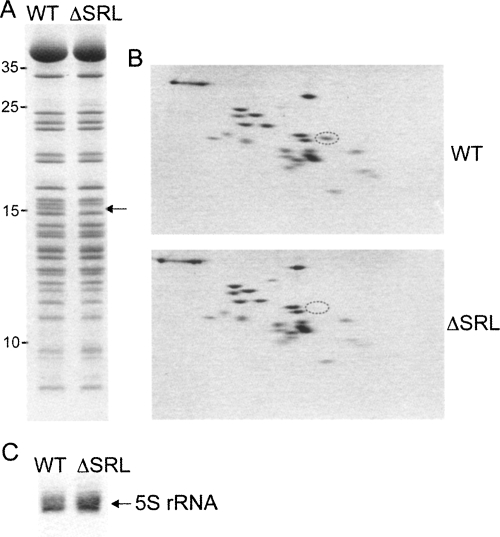
SDS gel analysis of proteins extracted from ΔSRL 50S subunits revealed the absence of a band of ∼15 kDa (Fig. 5A). The missing protein was identified by two-dimensional gel analysis (Geyl et al. 1981; Agafonov et al. 1999) as ribosomal protein L16 (Fig. 5B), which appeared to be the only protein missing from ΔSRL 50S subunits. It is known that L16 can be stripped away by treatment with 2 M LiCl (Moore et al. 1975), so we analyzed ΔSRL 50S subunits prepared from ribosomes that had not been subjected to the two standard 0.5 M NH4Cl salt washes, and found that L16 was still completely absent (data not shown). In 50S subunits, L16 contacts L25, which in turn binds 5S rRNA (Schuwirth et al. 2005). Although L16 is missing from the ΔSRL 50S subunits, both L25 and 5S rRNA (Fig. 5C) appear to be present in normal amounts.
FIGURE 5.
ΔSRL 50S subunits are missing ribosomal protein L16. (A) SDS gel of proteins extracted from affinity-purified WT and ΔSRL 50S subunits; arrow indicates the missing protein band. (B) Two-dimensional protein gel of WT and ΔSRL 50S subunits; protein L16 is circled. (C) Denaturing gel of rRNA extracted from 50S subunits shows that 5S rRNA is present in normal amounts in ΔSRL subunits.
Structural changes in the 23S rRNA of ΔSRL 50S subunits
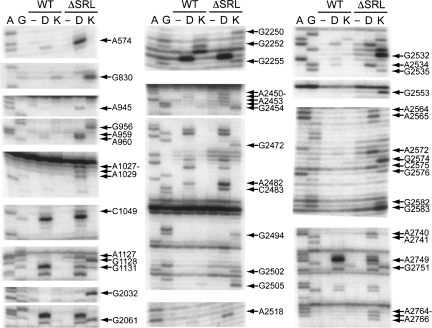
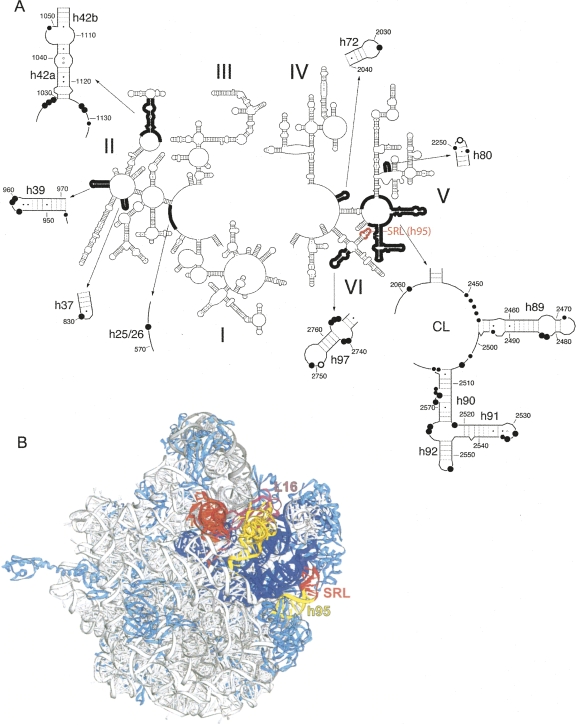
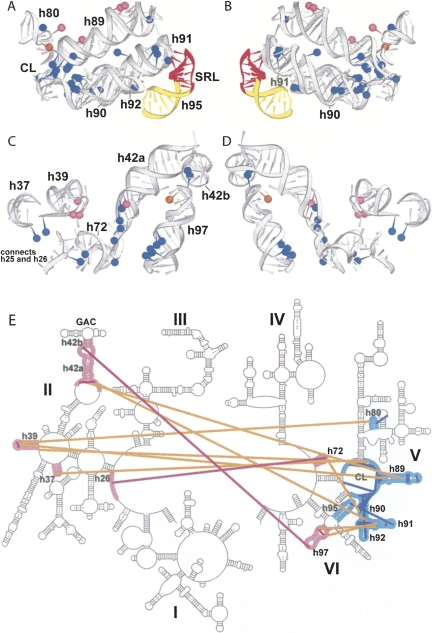
Structural changes in 23S and 5S rRNA were monitored by identifying nucleotides with altered reactivity toward base-specific structure probes in ΔSRL 50S subunits. Unexpectedly, more than 40 bases in 23S rRNA showed enhanced reactivity and 2 showed decreased reactivity toward the probes (Fig. 6, summarized in Supplemental Table 1). The affected nucleotides are widely distributed over domains II, V, and VI of 23S rRNA (Fig. 7A). Strikingly, in the E. coli ribosome crystal structure (Schuwirth et al. 2005), these nucleotides lie within features that are closely packed in three dimensions (Figs. 7B, 8). This compact region of 23S rRNA, which contains much of its functional core, can be viewed as forming essentially two layers of rRNA helices, which we will call the front and back layers (Fig. 9), according to their positions in the standard crown view of the 50S subunit (Fig. 7B). The front layer is formed primarily from domain V and the back layer from domain II, when viewed in the 23S rRNA secondary structure (Fig. 9E).
FIGURE 6.
Deletion of the SRL causes structural changes in 23S rRNA. Primer-extension analysis of 23S rRNA extracted from WT or ΔSRL 50S subunits probed with D, dimethylsufate, or K, kethoxal. A and G are sequencing lanes; (−) unmodified subunits. Arrows indicate bases with altered reactivity in ΔSRL 50S subunits as compared to WT.
FIGURE 7.
Nucleotides affected by the SRL deletion are widely distributed in the 23S rRNA secondary structure, but localized within and around the functional core of the 50S subunit. (A) Secondary structure of 23S rRNA indicating nucleotides in ΔSRL 50S subunits that show strongly enhanced (●), weakly enhanced (∙), or reduced (○) reactivity to structure probes; the SRL (h95) is indicated in red. (B) 23S rRNA regions highlighted in A (dark black line) were mapped onto the 50S subunit of the E. coli ribosome crystal structure (Schuwirth et al. 2005), and are shown in dark blue. Protein L16 (pink), 23S rRNA helix 95 (yellow), and the SRL (red) are indicated; other ribosomal proteins are light blue and 5S and 23S rRNA are gray. P-site (orange) and A-site (yellow) tRNAs were docked onto the structure using coordinates from Yusupov et al. (2001) and Korostelev et al. (2006).
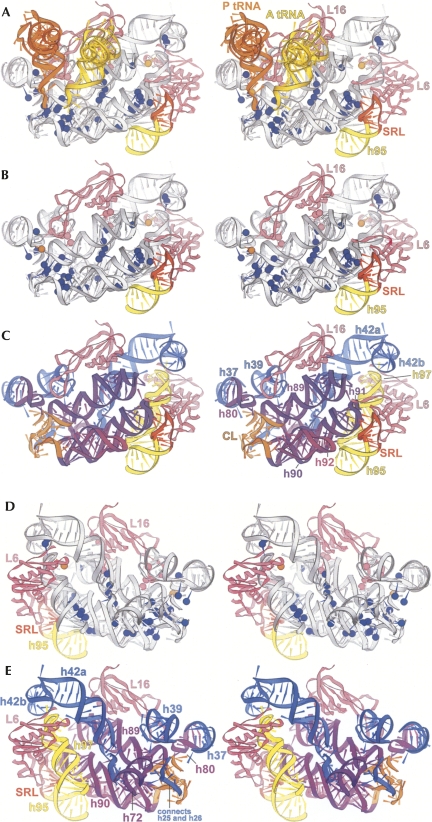
FIGURE 8.
Detailed stereo views of the ΔSRL-affected region of the 50S subunit. (A) The region of 23S rRNA indicated in dark blue in Figure 7B is shown (gray). Bases that are protected (orange) or enhanced (blue and pink) in ΔSRL 50S subunits are indicated as spheres placed at the N1 positions of A or G or the N3 of C; enhanced nucleotides that interact directly with protein L16 are shown in pink. P-tRNA (orange), A-tRNA (yellow), proteins L6 and L16 (pink), helix 95 (yellow), and the SRL (red) are indicated. (B) As in A, with tRNAs removed for clarity. (C) 23S rRNA helix numbers and domains are indicated: domain II (blue), V (purple and orange), and VI (yellow); and the SRL (red). D and E are rotated 180° around the vertical axis relative to B and C.
FIGURE 9.
Disrupted interactions within and between the front and back layers of the ΔSRL-affected region. Protected (orange) and enhanced (blue and pink) nucleotides are shown as in Figure 8; 23S rRNA (gray) helix numbers are indicated. (A) The front layer as seen in the 50S subunit crown view (Fig. 7B). (C) The corresponding back layer, with helix 95 (yellow) and the SRL (red) indicated. B and D are 180° rotations of A and C. (E) Secondary structure of 23S rRNA showing the front (cyan) and back (pink) layers; lines indicate tertiary interactions within the front (blue) or back (magenta) layers or between layers (orange) that are disrupted in the ΔSRL 50S subunits.
The front layer contains h80, h89, h90, h91, h92, and central loop (CL) (all from domain V) and the SRL (h95, domain VI) and forms the peptidyl transferase catalytic site surrounding the 3′ ends of the A- and P-tRNAs (Figs. 8A–C, 9A,B). The front layer is connected through rRNA tertiary interactions to the back layer (Figs. 8D–E, 9C,D), which is made up of elements from domain II (h37, h39, and h42a/b), domain V (h72), and domain VI (h97). H42b connects directly into the protein L11 binding site (h43 and h44), referred to as the GAC. L16, which is absent in the ΔSRL 50S subunits, straddles both layers, contacting h80, h89, h39, and h42a. L6 also traverses the layers, contacting both the SRL and h97 (Fig. 8).
The sole RNA–RNA interaction between the SRL with the rest of the 50S subunit is a limited set of tertiary contacts with 3 nt in the tip of h91. A2662 pairs with A2531, C2658–G2663–G2532 form a base triple, and the O2′ of A2665 contacts a nonbridging phosphate oxygen of U2533 (Schuwirth et al. 2005). Accordingly, in ΔSRL 50S subunits, the enhanced reactivities of G2532, A2534, and G2535 (Fig. 6) reflect the exposure of the h91 loop.
Additional bases within the front layer, several of which contact tRNA, become reactive in ΔSRL 50S subunits, indicating a disruption of their structural interactions. Enhanced bases A2572 and G2576 in the bulge region of h90 form tertiary contacts with nucleotides within the CL. A2564 and A2565, in the h90/92 connector, form consecutive Type II and Type I A-minor interactions (Nissen et al. 2001) with h91. Enhanced base G2553 in the A loop (h92) base-pairs with C75 of A-site tRNA (Kim and Green 1999; Nissen et al. 2000) and also forms a Type II A-minor-like interaction with C2507–G2582 in the CL; G2582 in turn is also enhanced. Enhanced G2472, A2482, and C2483 form tertiary interactions within the bulge and closing loop of h89, as well as backbone contacts with the T stem of A-site tRNA (Yusupov et al. 2001). In the P loop (h80), there is weak enhancement of G2255, which forms a tertiary base pair with C2275 in h81 and a backbone contact with the P-tRNA acceptor stem (Korostelev et al. 2006; Selmer et al. 2006), and protection of G2252, which pairs with P-site tRNA C74 (Samaha et al. 1995; Korostelev et al. 2006; Selmer et al. 2006). Finally, there are enhancements of several nucleotides that interact within the CL, three of which (A2450, A2451, and G2583) also contact A- or P-site tRNA (Nissen et al. 2000; Korostelev et al. 2006; Selmer et al. 2006).
Bases that form tertiary interactions within the back layer were also reactive in ΔSRL 50S subunits. These include C1049 and G2751, which form a tertiary base pair connecting h42b to h97, and G1131, at the base of h42a, which contacts the minor groove of h72. Reactive base A574 in the h25/26 connector forms a minor-groove interaction with h73.
Many nucleotides that become reactive in ΔSRL 50S subunits form tertiary interactions between the front and back layers. There are five enhanced bases in the bulge region of h97, 2764–2766 and 2740–2741, two of which (A2740 and A2741) form consecutive Type II and Type I A-minor interactions with the minor groove of h91. A2518, at the base of h91, intercalates between A1127 and G1128 in the loop below h42a, all of which are enhanced. Weakly enhanced A945 in h39 stacks on a base in the CL. G2250 in the P loop (h80) pairs with a G in the h39 stem–loop. Also in the h39 loop, enhanced bases A959 and A960 form successive Type II and Type I A-minor interactions with h89; G2494 (h89), also reactive, forms the A-minor receptor for A959. Enhanced A1028 and A1029, in the loop below h42a, also form Type II and Type I A-minors with h89. In the h72 stem–loop, G2032 contacts the minor groove of A2572 (h90) and the backbone of A2453 (CL), all of which are reactive.
Several of the nucleotides whose tertiary interactions are disrupted according to our probing data also contact protein L16 (Supplemental Table 1), which may contribute to its absence in ΔSRL 50S subunits (Fig. 5B). The major-groove sides of A959 and A960 and the backbone surface of G956, all in the h39 stem–loop, contact the 2/3 β-loop region of L16, near R81. G2250 in the P loop and the backbone of G2494 (h89) also contact the L16 2/3 β-loop. A2482 and C2483 in the h89 bulge form backbone contacts with L16 α-1, near R51. The backbone surface of A1029 at the base of h42a interacts with L16 α-2, near K118.
Figure 9E summarizes the tertiary interactions disrupted by deletion of the SRL in the context of the 23S rRNA secondary structure, the details of which are presented in Supplemental Table 1.
DISCUSSION
The SRL, which contains one of the longest universally conserved sequences in ribosomal RNA (Gutell et al. 1992), has been shown to interact with each GTPase factor involved in the ribosome translational cycle: elongation factors EF-G and EF-Tu (Moazed et al. 1988; Stark et al. 2002; Valle et al. 2003 a,b), initiation factor IF2 (La Teana et al. 2001; Allen et al. 2005; Myasnikov et al. 2005), and release factor RF3 (Klaholz et al. 2004; Gao et al. 2007). Although all of the GTPase-catalyzed steps in translation are believed to involve conformational changes in the ribosome, it is not yet known whether any of these are triggered by factor-induced changes in the structure of the SRL or its interactions with the rest of the 50S subunit (Wool et al. 1992), or whether the SRL acts solely as a static factor-docking site.
Crystal structures show that the SRL protrudes from the surface of the 50S subunit (Ban et al. 2000), making it readily accessible to factors in the interface cavity of the 70S ribosome (Yusupov et al. 2001). Contact between the SRL and the body of the 50S subunit is minimal, involving only 3 nt in the closing stem–loop of h91 and the C-terminal domain of protein L6. Surprisingly, deletion of the SRL results in extensive disruption of RNA–RNA tertiary interactions that are involved in the structural organization of the functional core of the 50S subunit. Furthermore, protein L16 is completely absent from ΔSRL 50S subunits, even though its binding site is more than 45 Å from its closest approach to the SRL.
One possible explanation for this result is that the absence of the 15-nt SRL confers a general assembly defect. This interpretation seems unlikely, because assembly of the 50S subunit is believed to occur cotranscriptionally (Nierhaus 1991), and virtually all of the 23S rRNA regions affected by the deletion (helices 37, 39, 42, 72, 80, 89, 90, 91, and 92; Fig 7A) occur upstream of the SRL (h95), and so would seem unlikely to become misfolded due to a downstream structural defect. In addition, all or part of the binding sites for proteins L3, L6, L13, L14, L17, and L19 are formed by elements of 23S rRNA downstream of the deletion (Schuwirth et al. 2005), and all of these proteins are present in normal amounts in ΔSRL 50S subunits (Fig. 5B), suggesting that this region of the subunit is assembled correctly.
Our results suggest instead that the presence of the SRL specifically influences the formation of 23S rRNA tertiary interactions that are required for correct assembly of the functional core of the 50S subunit. A plausible scenario is that these effects originate in the positioning of h91 by tertiary interactions between its closing loop and the SRL. Consistent with this model are chemical probing results (Fig. 6) indicating that several key tertiary interactions between h91 and elements of the core, involving three different domains of 23S rRNA, fail to form when the SRL is deleted (Fig. 9E). These include the interactions between h91and h97, h91 and h42, which lead directly into the GAC, and h91 and h92 (the A loop), which interact with A-site tRNA (Nissen et al. 2000; Yusupov et al. 2001). Loss of these contacts could then, in turn, explain disruption of tertiary interactions between the A loop and the CL, h97–h42, and h42–h89, which make several backbone–backbone contacts with the T stem of A-site tRNA (Yusupov et al. 2001). Incorrect positioning of h89 could then account for loss of interactions between h89 and h39 and h39 and h80 (the P loop). Nucleotides involved in the disturbed tertiary interactions between h42 and h89, h89 and h39, and h39 and h80 also help to form the binding site for protein L16, providing an explanation for its absence in ΔSRL 50S subunits.
Interaction of the SRL with protein L6 likely plays a critical role in establishing tertiary contacts between h91 and h97 during 50S subunit assembly. L6 cannot be incorporated until its main contact site, h97 (Klein et al. 2004), is transcribed, suggesting the following order of events. First, α-1 in the N-terminal domain of L6 binds the h97 minor groove; then its C-terminal domain wraps around and forms a β-sheet contact with the minor groove of the SRL (h95), whose tertiary interactions with the tip of h91 can then position it relative to h97. This could facilitate correct assembly of the rest of 23S rRNA domain V (the front layer, Fig. 9A,B) and its connection through rRNA tertiary interactions to elements of domain II (the back layer, Fig. 9C,D), thus forming the L16 binding site. Consistent with this scenario is the presence of L6 at normal levels in ΔSRL 50S subunits and the concurrent disruption of interactions between h91 and h97, as well as the absence of L16, which is thought to bind late in the 50S assembly pathway (Franceschi and Nierhaus 1990). Another intriguing possibility is that GTPases involved in 50S subunit assembly (for review, see Karbstein 2007) may interact with the SRL, for example, Obg/CgtAE, which has been shown to be important for incorporation of L16, L33, and L34 (Jiang et al. 2006).
The base reactivity changes we observe in the ΔSRL 50S subunits show extensive overlap with those in a previous study in which a G-C base pair was inserted at position 1030–1124, at the base of h42, which connects to the GAC (Sergiev et al. 2005). This mutation was predicted to lengthen and rotate h42, moving the GAC to a “downward” position. The site of the insertion mutation is ∼55 Å from the GAC, in a part of 23S rRNA that helps form the binding site for L16, the protein that straddles the two layers of helices that make up the functional core and is coincidentally the sole protein that is missing from the SRL deleted ribosomes. Thus, an alternative interpretation of the effects seen by Sergiev et al. is that, rather than being caused by repositioning of the GAC, the insertion resulted in distortion of the L16 binding site. Consistent with this interpretation, the insertion mutation resulted in enhanced reactivity of nucleotides that are in direct contact with L16 while forming tertiary interactions between h42 and h89, h89 and h39, and h39 and h80, whereas no changes in base reactivity were observed in the GAC. Thus, it is possible that the reactivity changes caused by the insertion are due to disruption of assembly of the core rather than reflecting a link between the GAC, PTC, and SRL, as proposed. In contrast, our deletion is in the SRL itself, making interpretation of the effects relatively straightforward. The only missing protein is L16, whose binding site is remote from the SRL and whose absence must therefore be an indirect effect of the mutation, which also causes disruption of tertiary RNA–RNA contacts in the core spanning the region between the SRL and L16. Thus, contacts with the SRL must be critical for establishing tertiary interactions within the core, including the L16 binding site.
Interactions between the translation factors and the SRL are unlikely to produce the sort of seismic event that we observe, some of which are clearly the result of the absence of L16. However, the profound structural effects of deleting the SRL raise the possibility that its interactions with translation factors could trigger conformational changes within the functional core of the 50S subunit to facilitate steps of translation that require movement. The functional defects caused by the SRL deletion may provide clues to how this could happen.
Deletion of the SRL reduces EF-G binding affinity by ∼50-fold (Fig. 4D), consistent with its contribution to the EF-G binding site (Hausner et al. 1987; Moazed et al. 1988). However, our results suggest that EF-G-independent translocation, catalyzed by the antibiotic sparsomycin (Fredrick and Noller 2003), is unaffected by the deletion (Fig. 4B), which would indicate that the basic translocation mechanism does not require the SRL. This is consistent with recent results showing that cleavage of the SRL does not affect elongation factor-independent translation (Chan and Wool 2008)
EF-Tu also binds the SRL (Hausner et al. 1987; Moazed et al. 1988), consistent with the defect in EF-Tu-dependent binding of aminoacyl-tRNA to the A site in ΔSRL ribosomes (Fig. 4A). However, this does not explain the observed loss of nonenzymatic A-site binding (Fig. 4A,B), which is most likely due to disruption of tertiary interactions in 23S rRNA in and around the 50S A site. Molecular dynamics simulations suggest that EF-Tu promotes entry of tRNA into the 50S subunit A site by opening an “accommodation corridor” that is otherwise blocked by interactions between conserved elements of 23S rRNA helices 43, 71, 89, 90, 92, and 93, the CL, and the SRL (Sanbonmatsu et al. 2005), which may account for the kinetic barrier to nonenzymatic A-site binding. Interestingly, deletion of the SRL causes several nucleotides that are known to contact A-site tRNA (G2472, A2482, C2483, G2553, and G2583) to become hyperreactive to chemical probes (Fig. 6). It is thus possible that interaction of EF-Tu with the SRL could transiently alter the conformation of the 50S A site, facilitating accommodation of aminoacyl-tRNA from the A/T to the A/A state. By analogy, structural changes in the functional core induced by interaction of EF-G with the SRL could help to promote movement of peptidyl-tRNA between the A/A and A/P states following peptidyl transfer.
Among the conformational changes in the 50S subunit structure caused by deletion of the SRL are numerous examples of ones involving apparent disruption of A-minor interactions (Supplemental Table 1), a widespread structural motif in rRNA (Nissen et al. 2001). It has been suggested that the properties of A-minor interactions make them particularly well suited for participation in RNA conformational switches (Noller 2005), and indeed, they are known to play a role in such diverse ribosomal functions as tRNA selection (Ogle et al. 2001), positioning of the acceptor ends of tRNA in the peptidyl transferase center (Nissen et al. 2000), formation of intersubunit bridges (Schuwirth et al. 2005), and discrimination of initiator tRNA (Lancaster and Noller 2005). Deletion of the SRL leads to disruption of a number of interconnected A-minor interactions of potential functional importance between SRL-h91, h91-h97, h91-h92, h92-CL, h89-h42, and h89-h39. Binding of translation factors has not so far been observed to cause disruptions of the kind reported here. It is nevertheless possible that their interactions with the SRL could produce more subtle and transient structural perturbation in the functional core of the subunit that would not easily be detected in experiments on static ribosomes. Such possibilities can be tested by mutational alteration of the bases that are implicated in these potential A-minor switches. A first step in this direction is the finding that mutation of A2531, which is adjacent to the A-minor interaction connecting the SRL with h91, results in partial loss of nonenzymatic A-site tRNA binding in vitro (Chan et al. 2006). It would not be surprising to find that the intricate tertiary structural networks found in rRNA serve to enable coordinated systems of long-range ribosome structural dynamics, such as the ones suggested by our findings.
MATERIALS AND METHODS
Buffer A is 50 mM KHepes (pH 7.5), 100 mM NH4Cl, 5 mM βME or 1 mM DTT, and the indicated concentration of MgCl2; for example, buffer A(10) contains 10 mM MgCl2. Buffer B is 50 mM Tris-HCl (pH 7.5), 70 mM NH4Cl, 30 mM KCl, 7 mM MgCl2, and 1 mM DTT.
tRNAfMet and tRNAPhe (Sigma) were aminoacylated, extracted, and purified as described (Moazed and Noller 1989; Lancaster and Noller 2005).
Salt-washed ribosomes were prepared from strain MRE600 (Moazed and Noller 1989), dissociated into 30S and 50S subunits, and purified from sucrose gradients (Moazed and Noller 1986).
EF-G-591-IAF (provided by R. Hickerson) is 6(His)-EF-G containing a single cysteine at position 591 (Wilson and Noller 1998) that is labeled with 5-iodoacetamidofluorescein (IAF) (Molecular Probes).
Construction of SRL deletion mutant and 50S subunit affinity purification
Site-directed mutagenesis (Kunkel 1985) was used to delete 23S rRNA nucleotides 2653–2667 and replace with sequence GAAA, using oligo 5′-CCAGTGATGCGTCCACTCCTTTCGGAGCAGCCCCCCTCAGTTC-3′, as described by Ali et al. (2006), to generate plasmid pLK35.50S.MS2.ΔSRL.
MS2-tagged 50S subunits were expressed in E. coli strain MOPOP (Ali et al. 2006); cells were lysed, and salt washed ribosomes were prepared as described (Moazed and Noller 1989). Ribosomes were dissociated into subunits prior to affinity purification over a 5 mL GST-trap FF column (Amersham) that had been prebound with MS2-GST-fusion protein as described (Lancaster and Noller 2005).
In vitro assays
All 30S and 50S subunits were heat activated at 42°C for 10 min in buffer A(20) before they were used in each in vitro assay described below.
Poly-U directed poly-Phe synthesis was carried out as described (Lancaster and Noller 2005).
For sedimentation analysis, 20 pmol 50S subunits with or without 25 pmol 30S subunits were incubated at 37°C for 10 min in Buffer A(20) prior to loading on 10%–35% sucrose gradients in buffer A(20), which were centrifuged in an SW41 rotor (Beckman) at 28,000 rpm for 13.5 h at 4°C.
To assay P-site binding, 20 pmol 30S subunits, 25 pmol [35S]-fMet-tRNAfMet (2500 cpm/pmol), and 40 pmol mRNA m291 (Fredrick and Noller 2002) were combined with or without 30 pmol 50S subunits in a total volume of 44 μL in buffer A(15) and incubated at 37°C for 20 min. Ten microliters were spotted on nitrocellulose HA filters (Millipore), which were washed three times with 5 mL ice-cold buffer A(15), then dried, dissolved in scintillation cocktail, and counted. Ten microliters of the reaction were added to 1 μL of 10 mM puromycin (Sigma) in buffer A(15) and incubated at room temperature for 25 min. One volume of MgSO4-saturated 0.3 M NaOAc (pH 5.3) was added, then [35S]-fMet-puromycin was extracted with 1 mL of ethyl acetate.
For the puromycin titration, 5 μL of a 50-μL reaction containing 30 pmol 50S subunits and 30 pmol [35S]-fMet-tRNAfMet, 50 mM Tris-HCl (pH 7.5), 400 mM KOAc, and 20 mM MgCl2 were added to 1 μL of puromycin (in the same buffer) to obtain 0 to 125 μM (final concentration) and stored on ice. The reaction was started by addition of methanol to 33%, then incubated on ice for 10 min; [35S]-fMet-puromycin was extracted as described above.
For nonenzymatic A-site tRNA binding, 12.5 pmol [3H]-Phe-tRNAPhe (6000 cpm/pmol) were added to 5 pmol of 70S-mRNA-fMet-tRNAfMet complex in a total volume of 10 μL of buffer A(20) and incubated at 37°C for 30 min. Four microliters were spotted on nitrocellulose filters as described above. For EF-Tu-dependent binding, 6His-tagged-EF-Tu (Boon et al. 1992) (5 μM) was incubated with 1 mM GTP, 5 mM phosphoenolpyruvate, and 20 μg/mL pyruvate kinase (Roche) in buffer A(10) for 15 min at 37°C, before we added [3H]-Phe-tRNAPhe and incubated it another 5 min. The Mg++ was adjusted to 7 mM by addition of buffer A(0). Ten picomoles of ternary complex were added to 5 pmol of 70S-mRNA-fMet-tRNAfMet complex that had been also adjusted to 7 mM Mg++, in a total volume of 22 μL of buffer A(7) This was incubated for 5 min at 37°C, then 9 μL were spotted on filters, which were washed in buffer A(7) as described above.
Toeprinting analysis was carried out as described by Fredrick and Noller (2003).
GTP hydrolysis was measured by combining 6.4 pmol ribosomes (either vacant or preassociated with 20 pmol tRNAPhe and 15 A26oU/mL poly-U mRNA) and 6.4 pmol of EF-G in buffer B supplemented with 2 mM GTP and trace amounts of [γ32P]-GTP (MP Biomedical, 800 Ci/mmol) in a total volume of 32 μL. The reaction was incubated at 37°C, and 4-μL aliquots were withdrawn at each time point and quenched in 4 μL of 60 mM acetic acid, then analyzed by thin layer chromatography (PEI cellulose) developed in 0.5 M KH2PO4.
For EF-G binding, ribosomal complexes containing mRNA and tRNAfMet were titrated from 0 to 600 nM in reactions that contained 20 nM EF-G-591-IAF, 0.5 mM GDPNP, 0.015% Nikkol, 1.6 mM DTT, 10 mM MgCl2, 100 mM NH4Cl, 50 mM Tris-HCl (pH 7.0), in a total volume of 30 μL, and incubated for 10 min at 37°C. Fluorescence spectra (500–600 nm) were aquired using a Cary Eclipse fluorescence spectrophotometer (Varian, Inc.) at 22°C with excitation at 485 nm, in a 10-μL cuvette (Starna Cells). The quench in fluorescent signal upon EF-G binding to the ribosome was normalized to that of EF-G alone; the Kd for EF-G binding was determined by fitting the data to the following equation:
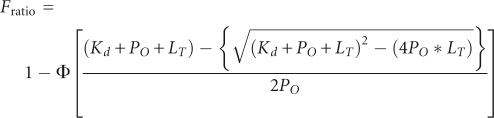 |
(Roy 2004).
Two-dimensional protein gel and rRNA structural probing
Proteins from 30 μg of purified 50S subunits were extracted with acetic acid/MgCl2 and acetone precipitated (Siegmann and Thomas 1987), then dissolved in 20 μL sample loading buffer (10 mM Bis-Tris-HAc at pH 4.0, 6 M urea, 10 mM DTT, and basic fuchsin dye), and loaded onto the first-dimension 4% acrylamide gel (1-mm spacer slab gel; Biorad Protean II), which was run in the cold room at 14 mA for 7 h: The gel and buffer system were prepared as described (Geyl et al. 1981). Each lane was cut from the first-dimension gel and briefly soaked in 100 mM Tris-HCL (pH 8.3), 100 mM tricine, 0.1% SDS, and 2% (v/v) βME, then sandwiched into glass plates for the second-dimension slab gel, which was assembled with 1-mm spacers (Biorad Protean II). A 16% acrylamide tricine/SDS gel (Schägger and von Jagow 1987) was polymerized below the sandwiched lane, then a 4% acrylamide stacking gel was polymerized around the lane, embedding it. The second-dimension gel was run at room temperature, 90 V for 2 h as described (Schägger and von Jagow 1987) and stained with Coomassie blue.
For rRNA structural probing, 20 pmol 50S subunits in 50 μL 80 mM KHepes (pH 7.5), 100 mM NH4Cl, and 20 mM MgCl2 were reacted with 1 μL of 37 mg/mL KE or 2 μL of DMS (diluted 1:14 in 95% ethanol) at 37°C for 8 min. Reactions were precipitated by addition of 2.5 V 95% ethanol; rRNA extraction and primer extension with reverse transcriptase were as described (Stern et al. 1988).
Structure figures were drawn using the program Ribbons (Carson 1997), and data were plotted using Origin 7.5 (Originlab).
SUPPLEMENTAL DATA
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank D. Ermolenko for advice regarding the EF-G binding assay and unpublished results, R. Hickerson for providing labeled EF-G protein, and I. Ali for providing plasmids that were used in the mutagenesis procedure. This work was supported by grants from the NSF and NIH (to H.F.N).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1202108.
REFERENCES
- Agafonov, D.E., Kolb, V.A., Nazimov, I.V., Spirin, A.S. A protein residing at the subunit interface of the bacterial ribosome. Proc. Natl. Acad. Sci. 1999;96:12345–12349. doi: 10.1073/pnas.96.22.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal, R.K., Linde, J., Sengupta, J., Nierhaus, K.H., Frank, J. Localization of L11 protein on the ribosome and elucidation of its involvement in EF-G-dependent translocation. J. Mol. Biol. 2001;311:777–787. doi: 10.1006/jmbi.2001.4907. [DOI] [PubMed] [Google Scholar]
- Ali, I.K., Lancaster, L., Feinberg, J., Joseph, S., Noller, H.F. Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol. Cell. 2006;23:865–874. doi: 10.1016/j.molcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Allen, G.S., Zavialov, A., Gursky, R., Ehrenberg, M., Frank, J. The cryo-EM structure of a translation initiation complex from Escherichia coli . Cell. 2005;121:703–712. doi: 10.1016/j.cell.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., Steitz, T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Boon, K., Vijgenboom, E., Madsen, L.V., Talens, A., Kraal, B., Bosch, L. Isolation and functional analysis of histidine-tagged elongation factor Tu. Eur. J. Biochem. 1992;210:177–183. doi: 10.1111/j.1432-1033.1992.tb17406.x. [DOI] [PubMed] [Google Scholar]
- Brigotti, M., Rambelli, F., Zamboni, M., Montanaro, L., Sperti, S. Effect of α-sarcin and ribosome-inactivating proteins on the interaction of elongation factors with ribosomes. Biochem. J. 1989;257:723–727. doi: 10.1042/bj2570723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson, M. Ribbons. Methods Enzymol. 1997;277B:493–505. [PubMed] [Google Scholar]
- Chan, Y.L., Wool, I.G. The integrity of the sarcin/ricin domain of 23 S ribosomal RNA is not required for elongation factor-independent peptide synthesis. J. Mol. Biol. 2008;378:12–19. doi: 10.1016/j.jmb.2008.02.016. [DOI] [PubMed] [Google Scholar]
- Chan, Y.L., Dresios, J., Wool, I.G. A pathway for the transmission of allosteric signals in the ribosome through a network of RNA tertiary interactions. J. Mol. Biol. 2006;355:1014–1025. doi: 10.1016/j.jmb.2005.11.037. [DOI] [PubMed] [Google Scholar]
- Correll, C.C., Wool, I.G., Munishkin, A. The two faces of the Escherichia coli 23 S rRNA sarcin/ricin domain: The structure at 1.11 Å resolution. J. Mol. Biol. 1999;292:275–287. doi: 10.1006/jmbi.1999.3072. [DOI] [PubMed] [Google Scholar]
- Douthwaite, S., Powers, T., Lee, J.Y., Noller, H.F. Defining the structural requirements for a helix in 23 S ribosomal RNA that confers erythromycin resistance. J. Mol. Biol. 1989;209:655–665. doi: 10.1016/0022-2836(89)93000-3. [DOI] [PubMed] [Google Scholar]
- Endo, Y., Wool, I.G. The site of action of α-sarcin on eukaryotic ribosomes. The sequence at the α-sarcin cleavage site in 28 S ribosomal ribonucleic acid. J. Biol. Chem. 1982;257:9054–9060. [PubMed] [Google Scholar]
- Endo, Y., Mitsui, K., Motizuki, M., Tsurugi, K. The mechanism of action of ricin and related toxic lectins on eukaryotic ribosomes. The site and the characteristics of the modification in 28 S ribosomal RNA caused by the toxins. J. Biol. Chem. 1987;262:5908–5912. [PubMed] [Google Scholar]
- Franceschi, F.J., Nierhaus, K.H. Ribosomal proteins L15 and L16 are mere late assembly proteins of the large ribosomal subunit. J. Biol. Chem. 1990;265:16676–16682. [PubMed] [Google Scholar]
- Fredrick, K., Noller, H.F. Accurate translocation of mRNA by the ribosome requires a peptidyl group or its analog on the tRNA moving into the 30S P site. Mol. Cell. 2002;9:1125–1131. doi: 10.1016/s1097-2765(02)00523-3. [DOI] [PubMed] [Google Scholar]
- Fredrick, K., Noller, H.F. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300:1159–1162. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
- Freistroffer, D.V., Pavlov, M.Y., MacDougall, J., Buckingham, R.H., Ehrenberg, M. Release factor RF3 in E.coli accelerates the dissociation of release factors RF1 and RF2 from the ribosome in a GTP-dependent manner. EMBO J. 1997;16:4126–4133. doi: 10.1093/emboj/16.13.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, H., Zhou, Z., Rawat, U., Huang, C., Bouakaz, L., Wang, C., Cheng, Z., Liu, Y., Zavialov, A., Gursky, R., et al. RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell. 2007;129:929–941. doi: 10.1016/j.cell.2007.03.050. [DOI] [PubMed] [Google Scholar]
- Geyl, D., Bock, A., Isono, K. An improved method for two-dimensional gel-electrophoresis: Analysis of mutationally altered ribosomal proteins of Escherichia coli . Mol. Gen. Genet. 1981;181:309–312. doi: 10.1007/BF00425603. [DOI] [PubMed] [Google Scholar]
- Gluck, A., Endo, Y., Wool, I.G. The ribosomal RNA identity elements for ricin and for α-sarcin: Mutations in the putative CG pair that closes a GAGA tetraloop. Nucleic Acids Res. 1994;22:321–324. doi: 10.1093/nar/22.3.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse, R.L., Takebe, Y., Sharrock, R.A., Nomura, M. Feedback regulation of rRNA and tRNA synthesis and accumulation of free ribosomes after conditional expression of rRNA genes. Proc. Natl. Acad. Sci. 1985;82:1069–1073. doi: 10.1073/pnas.82.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell, R.R., Schnare, M.N., Gray, M.W. A compilation of large subunit (23S- and 23S-like) ribosomal RNA structures. Nucleic Acids Res. 1992;20(Suppl):2095–2109. doi: 10.1093/nar/20.suppl.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartz, D., McPheeters, D.S., Gold, L. Selection of the initiator tRNA by Escherichia coli initiation factors. Genes & Dev. 1989;3:1899–1912. doi: 10.1101/gad.3.12a.1899. [DOI] [PubMed] [Google Scholar]
- Hausner, T.P., Atmadja, J., Nierhaus, K.H. Evidence that the G2661 region of 23S rRNA is located at the ribosomal binding sites of both elongation factors. Biochimie. 1987;69:911–923. doi: 10.1016/0300-9084(87)90225-2. [DOI] [PubMed] [Google Scholar]
- Heus, H.A., Pardi, A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991;253:191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- Jiang, M., Datta, K., Walker, A., Strahler, J., Bagamasbad, P., Andrews, P.C., Maddock, J.R. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J. Bacteriol. 2006;188:6757–6770. doi: 10.1128/JB.00444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph, S., Noller, H.F. EF-G-catalyzed translocation of anticodon stem-loop analogs of transfer RNA in the ribosome. EMBO J. 1998;17:3478–3483. doi: 10.1093/emboj/17.12.3478. [published erratum appears in EMBO J 1998 Sep 15;17(18):5519]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbstein, K. Role of GTPases in ribosome assembly. Biopolymers. 2007;87:1–11. doi: 10.1002/bip.20762. [DOI] [PubMed] [Google Scholar]
- Kim, D.F., Green, R. Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol. Cell. 1999;4:859–864. doi: 10.1016/s1097-2765(00)80395-0. [DOI] [PubMed] [Google Scholar]
- Klaholz, B.P., Myasnikov, A.G., Van Heel, M. Visualization of release factor 3 on the ribosome during termination of protein synthesis. Nature. 2004;427:862–865. doi: 10.1038/nature02332. [DOI] [PubMed] [Google Scholar]
- Klein, D.J., Moore, P.B., Steitz, T.A. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- Korostelev, A., Trakhanov, S., Laurberg, M., Noller, H.F. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Kunkel, T.A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Teana, A., Gualerzi, C.O., Dahlberg, A.E. Initiation factor IF 2 binds to the α-sarcin loop and helix 89 of Escherichia coli 23S ribosomal RNA. RNA. 2001;7:1173–1179. doi: 10.1017/s1355838201010366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster, L., Noller, H.F. Involvement of 16S rRNA nucleotides G1338 and A1339 in discrimination of initiator tRNA. Mol. Cell. 2005;20:623–632. doi: 10.1016/j.molcel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- LeCuyer, K.A., Behlen, L.S., Uhlenbeck, O.C. Mutants of the bacteriophage MS2 coat protein that alter its cooperative binding to RNA. Biochemistry. 1995;34:10600–10606. doi: 10.1021/bi00033a035. [DOI] [PubMed] [Google Scholar]
- Mazumder, R., Chae, Y.B., Ochoa, S. Polypeptide chain initiation in E. coli: Sulfhydryl groups and the function of initiation factor F2. Proc. Natl. Acad. Sci. 1969;63:98–103. doi: 10.1073/pnas.63.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed, D., Noller, H.F. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986;47:985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- Moazed, D., Noller, H.F. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989;57:585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- Moazed, D., Robertson, J.M., Noller, H.F. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature. 1988;334:362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- Monro, R.E. Catalysis of peptide bond formation by 50 S ribosomal subunits from Escherichia coli . J. Mol. Biol. 1967;26:147–151. doi: 10.1016/0022-2836(67)90271-9. [DOI] [PubMed] [Google Scholar]
- Moore, V.G., Atchison, R.E., Thomas, G., Moran, M., Noller, H.F. Identification of a ribosomal protein essential for peptidyl transferase activity. Proc. Natl. Acad. Sci. 1975;72:844–848. doi: 10.1073/pnas.72.3.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munishkin, A., Wool, I.G. The ribosome-in-pieces: Binding of elongation factor EF-G to oligoribonucleotides that mimic the sarcin/ricin and thiostrepton domains of 23S ribosomal RNA. Proc. Natl. Acad. Sci. 1997;94:12280–12284. doi: 10.1073/pnas.94.23.12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myasnikov, A.G., Marzi, S., Simonetti, A., Giuliodori, A.M., Gualerzi, C.O., Yusupova, G., Yusupov, M., Klaholz, B.P. Conformational transition of initiation factor 2 from the GTP- to GDP-bound state visualized on the ribosome. Nat. Struct. Mol. Biol. 2005;12:1145–1149. doi: 10.1038/nsmb1012. [DOI] [PubMed] [Google Scholar]
- Nierhaus, K.H. The assembly of prokaryotic ribosomes. Biochimie. 1991;73:739–755. doi: 10.1016/0300-9084(91)90054-5. [DOI] [PubMed] [Google Scholar]
- Nissen, P., Hansen, J., Ban, N., Moore, P.B., Steitz, T.A. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- Nissen, P., Ippolito, J.A., Ban, N., Moore, P.B., Steitz, T.A. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl. Acad. Sci. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller, H.F. RNA structure: Reading the ribosome. Science. 2005;309:1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- Ogle, J.M., Brodersen, D.E., Clemons, W.M., Tarry, M.J., Carter, A.P., Ramakrishnan, V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- Rodnina, M.V., Savelsbergh, A., Katunin, V.I., Wintermeyer, W. Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature. 1997;385:37–41. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- Roy, S. Fluorescence quenching methods to study protein-nucleic acid interactions. Methods Enzymol. 2004;379:175–187. doi: 10.1016/S0076-6879(04)79010-2. [DOI] [PubMed] [Google Scholar]
- Samaha, R.R., Green, R., Noller, H.F. A base pair between tRNA and 23S rRNA in the peptidyl transferase centre of the ribosome. Nature. 1995;377:309–314. doi: 10.1038/377309a0. [published erratum appears in Nature 1995 Nov 23;378(6555):419]. [DOI] [PubMed] [Google Scholar]
- Sanbonmatsu, K.Y., Joseph, S., Tung, C.S. Simulating movement of tRNA into the ribosome during decoding. Proc. Natl. Acad. Sci. 2005;102:15854–15859. doi: 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger, H., von Jagow, G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Schuwirth, B.S., Borovinskaya, M.A., Hau, C.W., Zhang, W., Vila-Sanjurjo, A., Holton, J.M., Cate, J.H. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Selmer, M., Dunham, C.M., Murphy, F.V.t., Weixlbaumer, A., Petry, S., Kelley, A.C., Weir, J.R., Ramakrishnan, V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Seo, H.S., Kiel, M., Pan, D., Raj, V.S., Kaji, A., Cooperman, B.S. Kinetics and thermodynamics of RRF, EF-G, and thiostrepton interaction on the Escherichia coli ribosome. Biochemistry. 2004;43:12728–12740. doi: 10.1021/bi048927p. [DOI] [PubMed] [Google Scholar]
- Sergiev, P.V., Lesnyak, D.V., Burakovsky, D.E., Kiparisov, S.V., Leonov, A.A., Bogdanov, A.A., Brimacombe, R., Dontsova, O.A. Alteration in location of a conserved GTPase-associated center of the ribosome induced by mutagenesis influences the structure of peptidyltransferase center and activity of elongation factor G. J. Biol. Chem. 2005;280:31882–31889. doi: 10.1074/jbc.M505670200. [DOI] [PubMed] [Google Scholar]
- Siegmann, M., Thomas, G. Separation of multiple phosphorylated forms of 40 S ribosomal protein S6 by two-dimensional polyacrylamide gel electrophoresis. Methods Enzymol. 1987;146:362–369. doi: 10.1016/s0076-6879(87)46037-0. [DOI] [PubMed] [Google Scholar]
- Spahn, C.M., Blaha, G., Agrawal, R.K., Penczek, P., Grassucci, R.A., Trieber, C.A., Connell, S.R., Taylor, D.E., Nierhaus, K.H., Frank, J. Localization of the ribosomal protection protein Tet(O) on the ribosome and the mechanism of tetracycline resistance. Mol. Cell. 2001;7:1037–1045. doi: 10.1016/s1097-2765(01)00238-6. [DOI] [PubMed] [Google Scholar]
- Stark, H., Rodnina, M.V., Wieden, H.J., Zemlin, F., Wintermeyer, W., van Heel, M. Ribosome interactions of aminoacyl-tRNA and elongation factor Tu in the codon-recognition complex. Nat. Struct. Biol. 2002;9:849–854. doi: 10.1038/nsb859. [DOI] [PubMed] [Google Scholar]
- Stern, S., Moazed, D., Noller, H.F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- Traub, P., Mizushima, S., Lowry, C.V., Nomura, M. Reconstitution of ribosomes from subribosomal components. In: Moldave K., editor. RNA and protein synthesis. Academic Press; New York: 1981. pp. 521–539. [Google Scholar]
- Trieber, C.A., Burkhardt, N., Nierhaus, K.H., Taylor, D.E. Ribosomal protection from tetracycline mediated by Tet(O): Tet(O) interaction with ribosomes is GTP-dependent. Biol. Chem. 1998;379:847–855. doi: 10.1515/bchm.1998.379.7.847. [DOI] [PubMed] [Google Scholar]
- Valle, M., Zavialov, A., Li, W., Stagg, S.M., Sengupta, J., Nielsen, R.C., Nissen, P., Harvey, S.C., Ehrenberg, M., Frank, J. Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat. Struct. Biol. 2003a;10:899–906. doi: 10.1038/nsb1003. [DOI] [PubMed] [Google Scholar]
- Valle, M., Zavialov, A., Sengupta, J., Rawat, U., Ehrenberg, M., Frank, J. Locking and unlocking of ribosomal motions. Cell. 2003b;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- Wilson, K.S., Noller, H.F. Mapping the position of translational elongation factor EF-G in the ribosome by directed hydroxyl radical probing. Cell. 1998;92:131–139. doi: 10.1016/s0092-8674(00)80905-8. [DOI] [PubMed] [Google Scholar]
- Wintermeyer, W., Peske, F., Beringer, M., Gromadski, K.B., Savelsbergh, A., Rodnina, M.V. Mechanisms of elongation on the ribosome: Dynamics of a macromolecular machine. Biochem. Soc. Trans. 2004;32:733–737. doi: 10.1042/BST0320733. [DOI] [PubMed] [Google Scholar]
- Wool, I.G., Gluck, A., Endo, Y. Ribotoxin recognition of ribosomal RNA and a proposal for the mechanism of translocation. Trends Biochem. Sci. 1992;17:266–269. doi: 10.1016/0968-0004(92)90407-z. [DOI] [PubMed] [Google Scholar]
- Youngman, E.M., Brunelle, J.L., Kochaniak, A.B., Green, R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H., Noller, H.F. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]