Abstract
Quantum dots (QDs) are often cell-impermeable and require transporters to facilitate crossing over cell membranes. Here we present a simple and versatile method that utilizes enzymes, matrix metalloprotease 2 (MMP-2) and MMP-7, to modulate the cellular uptake of QDs. QD-peptide conjugates could be efficiently taken up into cells after the MMP treatment. This enzyme-modulated cellular uptake of QDs may be applied to other nanoparticles for biological imaging and selective drug delivery into tumor cells.
Quantum dots (QDs) are fluorescent semiconductor nanocrystals with unique optical properties such as high quantum yield, high molar extinction coefficients, narrow emission spectra, size-depending emission, and high photo stability, and are emerging as an attractive alternative to organic fluorophores as fluorescent, non-isotopic labels.1–8 QDs have to be water-soluble for biological applications, so they are often coated with amphiphilic ligands such as thiol-containing molecules (for example, dihydrolipoic acid and peptides) 9–11, oligomeric phosphines,12–13 amphiphilic polymers or phospholipid micelles 14–16. Water-soluble QDs are further functionalized with biomolecules such as small peptides, 17 carbohydrates, 18–19 proteins,20 and nucleic acids, 21 for in vitro and in vivo imaging studies. Such QD conjugates are typically in the range of 10 to 30 nm in diameter, and usually cell-impermeable. Several methods have been developed to facilitate the introduction of QDs into living cells including physical methods such as microinjection, electroporation and transferring or lipofectamine-mediated transfection.22–24 QDs coated with cationic peptides or polymers have also been demonsstrated to cross cell plasma membranes.25 In this work, we show that extracellular enzymes can be applied to modulate uptake of QDs into cells.
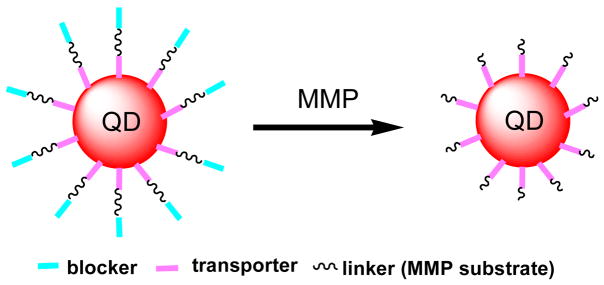
A recent study by Jiang et al. has shown that the transducing activity of cationic membrane-penetrating peptides decreases with the number of appended negatively charged glutamate residues, and can be restored after the appended glutamates are removed.26 In testing whether this approach could be applied to modulate the uptake of QDs, we designed QD conjugates shown in Scheme 1. The QDs we used here were coated with 5–10 copies of streptavidin, and conjugated with biotinylated peptide ligands via the strong biotin and streptavidin binding. The biotinylated peptide ligands consisted of 1) a transporting group (transporter) that could transport QDs into cells, 2) a blocking group (blocker) that could diminish the cellular uptake of QDs, and 3) a sensing group, sandwiched between the transporter and blocker, that could be cleaved by an extracellular enzyme. Two members of matrix metalloproteases (MMPs), gelatinase MMP-2 and stromelysin MMP-7, were selected to evaluate our design.
Scheme 1.
Schematic of QD conjugates that can be activated by MMPs for cellular uptake.
Matrix metalloproteases (MMPs) are a family of zinc-dependent secreted endopeptidases crucial for regulated degradation and processing of extracellular matrices.27–28 Malignant tumor cells need to breach extracellular matrices and invade, so MMPs are upregulated in almost every type of human cancers. Their over-expression correlates with advanced tumor stage, increased invasion and metastasis, and shortened survival.29–31 The significant role of MMPs in promoting cancer progression makes them important targets for drug development and in vivo tumor detection. Both fluorescence and magnetic resonance-based approaches have been used to detect MMPs activity.26,32–34 Reported fluorescence-based methods utilized small organic fluorophores such as near infrared dyes Cy5 and Cy5.5.26,32 Since the broad absorption spectra of QDs allow multiplexing—multiple emissions under single excitation for simultaneous detection of multiple targets, and there are at least 26 identified MMPs, a QD-based method should take advantage of this unique property of QDs and permit multiplexed detection and imaging of multiple MMPs. Here we describe this QD-based method to detect the activity of gelatinase MMP-2 and stromelysin MMP-7.
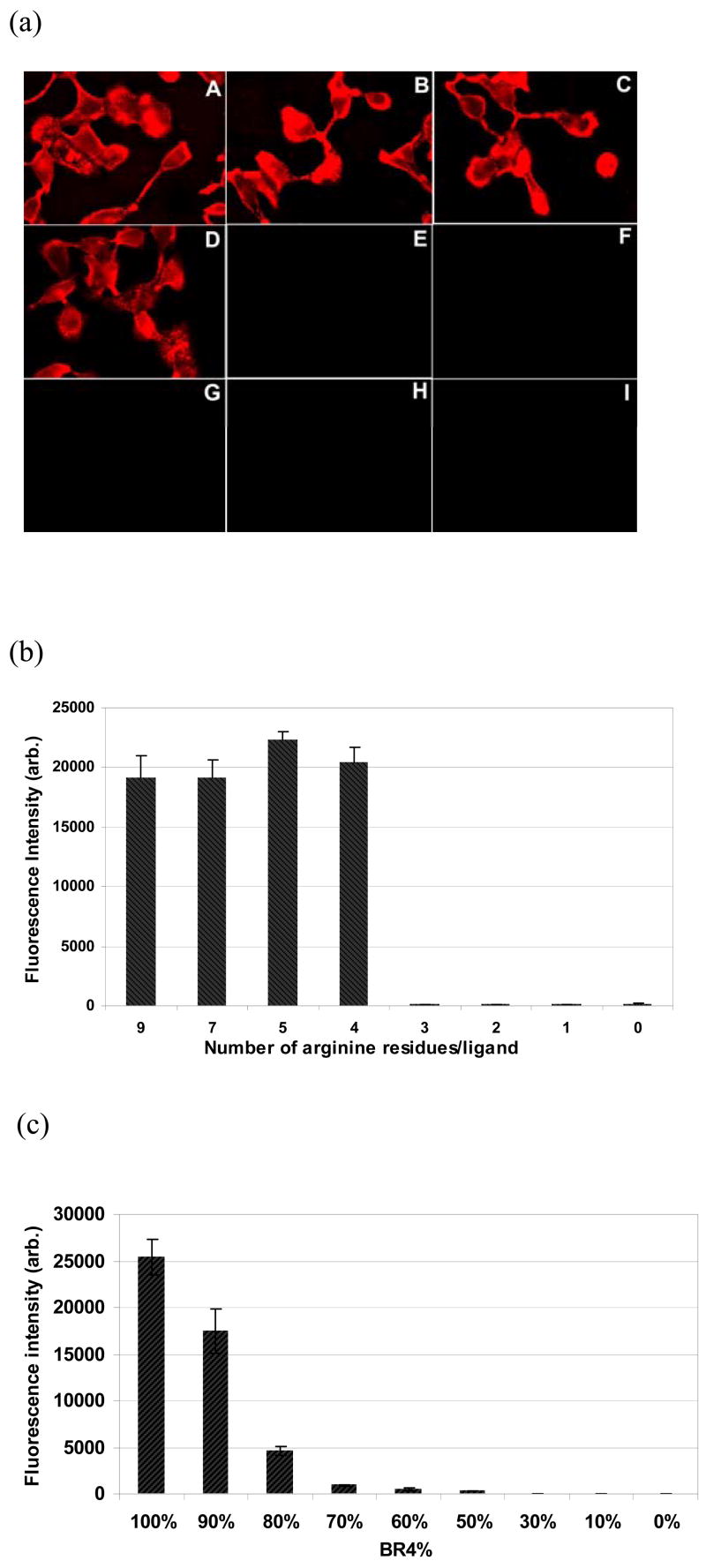
It has been reported that a biotinylated nine arginine oligomer is able to deliver streptavidin conjugated QDs into mammalian cells,25 but it was not clear whether all of 9 arginine residues were required for efficient uptake of QDs. We thus prepared a series of biotinylated oligomers with a varying number of arginine residues (from 1 to 9), and conjugated with streptavidin-coated QDs. These QD conjugates were incubated with HT-1080 (human fibrosarcoma) cells and examined under a fluorescence microscope to evaluate their uptake efficiency. As shown in Figure 1a, cells incubated with QD@BR9, QD@BR7, QD@BR5, and QD@BR4 showed strong fluorescence (average fluorescence signal was between 800–1200 after background subtraction) (Fig. 1a, A to D). After trysinization to detach from the glass surface and media washing, these cells still displayed strong QD fluorescence emission, suggesting that the QDs were indeed taken up into cells instead of just at the cell membrane. In contrast, little fluorescence could be detected in cells incubated with QD@BR3, QD@BR2 and QD@BR (average fluorescence signal was less than 10 after background subtraction). QD@BRn (n=1,2,3) appeared to possess little transducing activity like the negative control (the conjugate of biotin with QDs without arginine). The same pattern has been observed with COS-7 (monkey kidney) cells.
Figure 1.
(a) Fluorescence images of HT-1080 cells stained with QD@BRn conjugates (n = 9, 7, 5, 4, 3, 2, 1, 0 for A to H, respectively) and QD (I); (b) Fluorescence intensity of COS-7 cells in 96 microtiter plates incubated with QD@BRn (n as indicated for each column); (c) Fluorescence intensity of COS-7 cells incubated with QDs conjugated with a mixture of BR4 and BR at indicated ratios.
A microtiter plate reader was used to verify the microscope observation by quantifying the uptake of QD conjugates in a large population of cells (Fig. 1b). Consistent with the microscope imaging data, COS-7 cells stained with conjugates with >4 arginine residues (from QD@BR9 to QD@BR4) displayed around 100 times higher fluorescence intensity than those with < 4 arginine residues. No uptake was observed in cells incubated with QD@BR3. Other polycationic conjugates such as QD@BKn displayed the similar result (data not shown).
This result is very different from the reported dependence of the transducing efficiency on the number of arginine residues for a linear polyariginine,35 especially the dramatic decrease in uptake of the QD@BR3 conjugate. This observation may be related to the polymeric presence of arginine on the QD surface: each QD was coated with 5 to 10 copies of streptavidin and each streptavidin can bind four biotinylated peptides, thus there were around 20–40 biotinylated arginine oligomers on each QD. Due to this polymeric nature, a small change in the ligand from BR4 to BR3 –just one residue– was amplified to a big decrease in the total number of arginine residues on each QD conjugate, about 20–40.
To test this hypothesis, we conjugated the QDs with a mixture of BR4 and BR at a various ratio (ranging from 100% to 0), resulting in a smaller change in the total number of arginine residues on each QD conjugate. For example, from 100% BR4 to 90% BR4/10% BR, the total number of arginine residues on each QD conjugate decreased by about 6–12. Indeed, the cellular uptake of the QDs now decreased continuously with the decrease of the ratio of BR4 in the mixture (Fig. 1c). With a mixture of 70% of BR4 and 30% BR, the total number of the arginine residues on each QD was approximately the same to that of BR3, and correspondingly, little cellular uptake was observed (Fig. 1c).
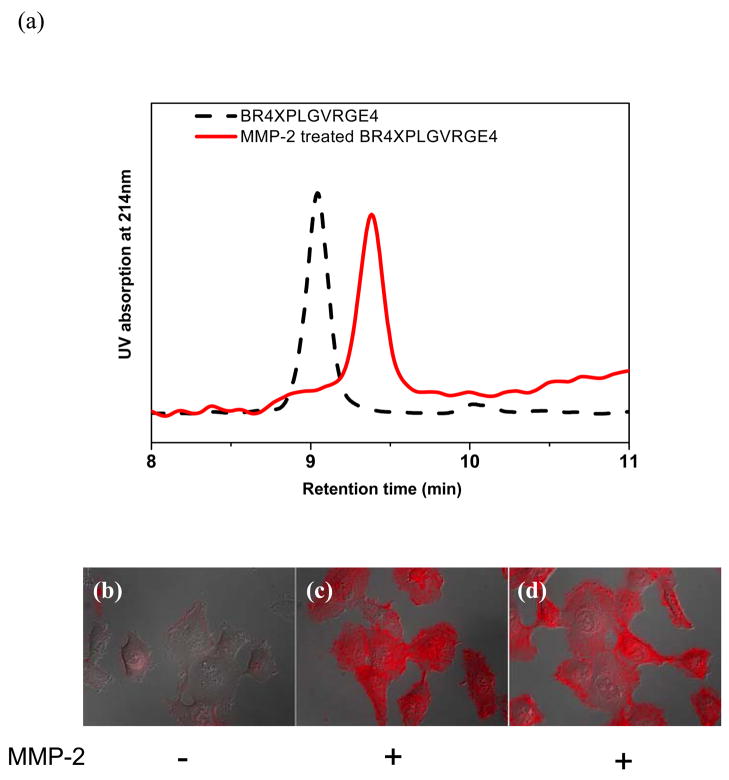
The gelatinase MMP-2 has been identified as one of the key MMPs in degrading type-IV collagen. Its preferential peptide substrate contains an amino acid sequence of PLGVR.37 We appended PLGVR to the biotinylated ligand BR4, and the resulting conjugate QD@BR4PLGVR was able to cross the plasma membranes. Based on this result, we designed the first peptide ligand R4XPLGVRGE4, where X denotes 6-aminohexanoyl as a spacer inserted to minimize non-favorable interactions with the enzyme. Peptide R4XPLGVRGE4 was synthesized and biotinylated to afford BR4XPLGVRGE4. The biotinylated peptide substrate (5 nmol) was incubated with active MMP-2 (1 μg) in borate buffer at room temperature for four hours, and its cleavage reaction by MMP-2 was analyzed by the HPLC which indicated a new peak with the retention time at 9.4 min different from the original substrate peak at 9.0 min (Fig. 2a). MMP-2 cleaves the amide bond between the glycine and valine residues, affording two cleaved products BR4XPLG and VRGE4. Analysis of the new peak at 9.4 min by the MALDI-MS revealed a mass corresponding to the cleaved product BR4XPLG, confirming that BR4XPLGVRGE4 can be efficiently cleaved by MMP-2 (Supporting Information).
Figure 2.
(a) HPLC analysis of the MMP-2 catalyzed cleavage of the substrate BR4XPLGVRGE4; (b–d) Overlaid fluorescence and differential interference contrast (DIC) images of HT-1080 cells stained with the conjugate QD@BR4XPLGVRGE4 (b), QD@BR4XPLG (formed with MMP-2 cleaved BR4XPLGVRGE4) (c), and MMP-2 treated conjugate QD@BR4XPLGVRGE4 (d), respectively. Display scale: 0–600.
The biotinylated peptide substrate was then conjugated with the QDs for cellular uptake. The conjugate QD@BR4XPLGVRGE4 was first incubated with MMP-2 (1 μg) for 4 hours, and subsequently incubated with HT-1080 cells in HBSS buffer (at 5 nM). Fluorescence microscopic imaging showed strong fluorescence signals nearly from all cells (Fig. 2d). When the biotinylated peptide was treated first with MMP-2 before conjugation with QDs, the resulting QD conjugates produced fluorescence intensity in incubated HT-1080 cells comparable to that in Fig. 2D (Fig. 2c). In contrast, a negative control containing the conjugate but without MMP-2 treatment showed little uptake with an average intensity of <10% of that treated MMP-2 (Fig. 2b). These results indicate that MMP-2 can efficiently cleave the peptide substrate immobilized on QDs and induce cellular uptake of QD conjugates.
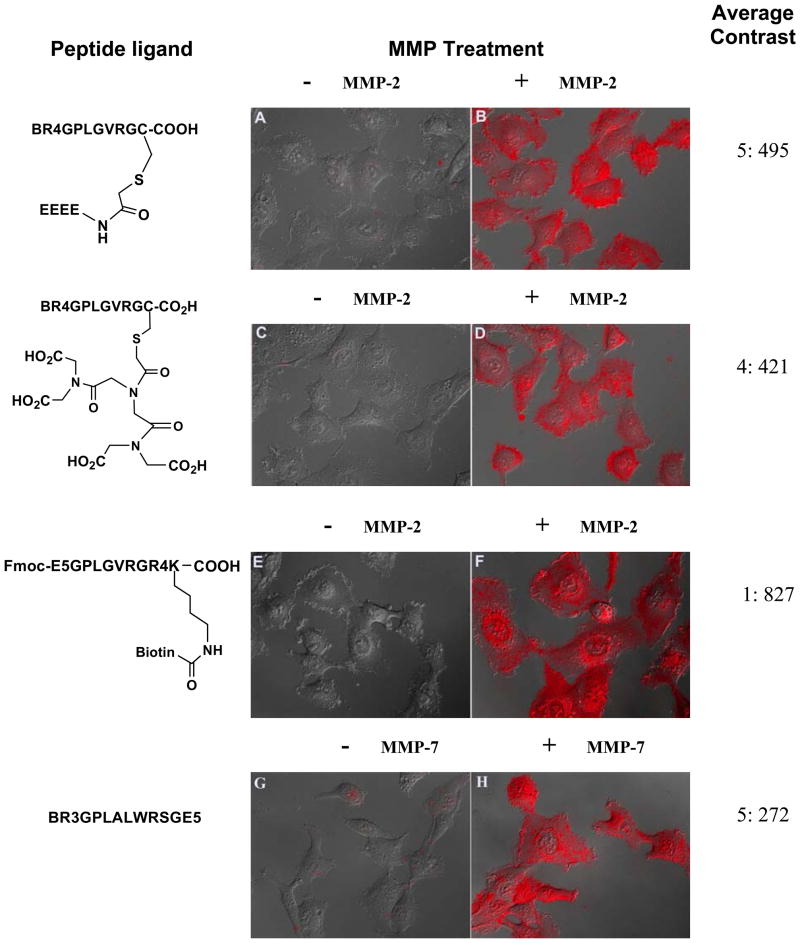
We were wondering whether the anionic glutamate residues could block the transducing activity if located on the side chain of the peptide substrate. We introduced a cysteine residue to the C-terminus of the substrate, and alkylation of the thiol side chain of cysteine by an iodoacetylated glutamate tetramer afforded the branched peptide BR4GPLGVRGC(E4). Cells incubated with this QD conjugate displayed no detectable fluorescence signal under the microscope, confirming the efficient blocking of the transducing activity by the glutamate residues on the side chain (A in Fig. 3). Cells incubated with MMP-2 treated conjugate showed a 100-fold increase in the average fluorescence intensity, and almost 2-fold higher than cells incubated with the MMP-2 activated QD@BR4XPLGVRGE4 conjugate (B in Fig. 3). Interestingly, when the linear glutamate tetramer in BR4GPLGVRGC(E4) was replaced by a molecule branched with four carboxylic acid groups, iminodiacetyl diiminodiacetic acid (IDA(IDA)2), cellular uptake experiment of its QD conjugate showed that the four branched carboxylate groups efficiently blocked the transduction of the conjugate (C in Fig. 3), and overnight incubation with MMP-2 could activate the transduction potential of the conjugates and induce the uptake in HT-1080 cells (D in Fig. 3).
Figure 3.
Overlaid fluorescence and DIC images of HT-1080 cells incubated with QD conjugates with indicated peptides with and without MMP-2 treatment (A–F), and MMP-7 (G–H). Display scale: 0–1000 for A–D; 0–1200 for E, F and 0–500 for G, H.
Next, we tested whether the arginine oligomer and the glutamate oligomer can exchange their positions in the design. We designed a peptide Fmoc-E5GPLGVRGR4K (Biotin) where the arginine oligomer was at the C-terminal side and the glutamate residues at the N-terminal side. The biotin group was coupled to the amino group on the side chain of the C-terminal lysine to afford Fmoc-E5GPLGVRGR4K(Biotin). QDs conjugated with this peptide turned out to be an excellent activatable cell imaging probe. The blocking efficiency of E5 on the cellular uptake of QD conjugates was quite high (E in Fig. 3, no fluorescent cells could be detected under microscope), and the QD conjugates could be activated within 4 hours by MMP-2, resulting in the highest cellular uptake of QDs among all peptide ligands tested under the same conditions (F in Fig. 3).
Finally, we asked whether this strategy could be applied to other MMPs. Stromelysin MMP-7 cleaves a peptide substrate RPLALWRS efficiently between the alanine and leucine residues.38 We similarly prepared a biotinylated peptide BR3GRPLALWRSGE5 that contained an arginine trimer at the N-terminus and a glutamate pentamer at the C-terminus. HT-1080 cells incubated with this peptide and QD conjugate did not show any significant fluorescence signal, but MMP-7 treated conjugates produced bright fluorescence after incubation (Images G and H in Fig. 3). This result confirms the generality of this strategy in modulating cellular uptake of QDs by MMP enzymes.
In summary, we report here an example of applying enzymes to modulate the cellular uptake of QD conjugates. We have found out that polycationic peptide-mediated uptake of QD conjugates can be blocked by the simultaneous presence of negatively charged groups on the QD conjugates. The negatively charged blocking groups can be presented linearly or branched on the peptide ligands. Proteases like MMP-2 and MMP-7 can remove the negatively charged groups by enzymatic hydrolysis and lead to the uptake of the QD conjugates into live cells. Since the matrix metalloproteases are greatly involved with tumor formation and progress in many types of human cancers, this strategy may be applied to image MMPs in vivo. Modulation of uptake of nanoparticles into cells with tumor-specific enzymes could lead to selective accumulation of nanoparticles in tumor cells and find wide applications in nanomedicine.
Supplementary Material
Syntheses and characterizations of the biotinylated MMP-2 peptide ligands, and experimental procedures for fluorescence microscopic imaging and microplate assay of cellular uptake. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
This work was supported by the Burroughs Wellcome Fund and the National Cancer Institute Centers of Cancer Nanotechnology Excellence (CCNE). Y.Z. acknowledges the Dean’s Fellowship from the Stanford School of Medicine.
References
- 1.Bruchez M, Moronne M, Gin P, Weiss S, Alivisatos AP. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 2.Chan WCW, Nie SM. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 3.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Nat Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 5.Jaiswal JK, Simon SM. Trends in Cell Biology. 2004;14:497–504. doi: 10.1016/j.tcb.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 6.Smith AM, Nie SM. Analyst. 2004;129:672–677. doi: 10.1039/b404498n. [DOI] [PubMed] [Google Scholar]
- 7.Dabbousi BO, RodriguezViejo J, Mikulec FV, Heine JR, Mattoussi H, Ober R, Jensen KF, Bawendi MG. J Phys Chem B. 1997;101:9463–9475. [Google Scholar]
- 8.Alivisatos P. Nat Biotechnol. 2004;22:47–52. doi: 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- 9.Uyeda HT, Medintz IL, Jaiswal JK, Simon SM, Mattoussi H. J Am Chem Soc. 2005;127:3870–3878. doi: 10.1021/ja044031w. [DOI] [PubMed] [Google Scholar]
- 10.Pinaud F, King D, Moore H-P, Weiss S. J Am Chem Soc. 2004;126:6115–6123. doi: 10.1021/ja031691c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mattoussi H, Mauro JM, Goldman ER, Anderson GP, Sundar VC, Mikulec FV, Bawendi MG. J Am Chem Soc. 2000;122:12142–12150. [Google Scholar]
- 12.Kim S, Bawendi MG. J Am Chem Soc. 2003;125:14652–14653. doi: 10.1021/ja0368094. [DOI] [PubMed] [Google Scholar]
- 13.Kim S, Lim YT, Soltesz EG, De Grand AM, Lee J, Nakayama A, Parker JA, Mihaljevic T, Laurence RG, Dor DM, Cohn LH, Bawendi MG, Frangioni JV. Nat Biotechnol. 2004;22:93–97. doi: 10.1038/nbt920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao XH, Cui YY, Levenson RM, Chung LWK, Nie SM. Nat Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 15.Wu XY, Liu HJ, Liu JQ, Haley KN, Treadway JA, Larson JP, Ge NF, Peale F, Bruchez MP. Nat Biotechnol. 2003;21:452–452. doi: 10.1038/nbt764. [DOI] [PubMed] [Google Scholar]
- 16.Dubertret B, Skourides P, Norris DJ, Noireaux V, Brivanlou AH, Libchaber A. Science. 2002;298:1759–1762. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]
- 17.Akerman ME, Chan WCW, Laakkonen P, Bhatia SN, Ruoslahti E. Proc Natl Acd Sci USA. 2002;99:12617–12621. doi: 10.1073/pnas.152463399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun XL, Cui W, Haller C, Chaikof EL. ChemBioChem. 2004;5:1593–1596. doi: 10.1002/cbic.200400137. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Ji T, Rosenzweig Z. Nano Lett. 2003;3:581–584. [Google Scholar]
- 20.So MK, Xu C, Loening AM, Gambhir SS, Rao J. Nat Biotechnol. 2006;22:339–343. doi: 10.1038/nbt1188. [DOI] [PubMed] [Google Scholar]
- 21.Charkrabarti R, Klibanov AM. J Am Chem Soc. 2003;125:12531–12540. doi: 10.1021/ja035399g. [DOI] [PubMed] [Google Scholar]
- 22.Chen FQ, Gerion D. Nano Lett. 2004;4:1827–1832. [Google Scholar]
- 23.Derfus AM, Chan WCW, Bhatia SN. Adv Mater. 2004;16:961–966. [Google Scholar]
- 24.Voura EB, Jaiswal JK, Mattoussi H, Simon SM. Nat Medicine. 2004;10:993–998. doi: 10.1038/nm1096. [DOI] [PubMed] [Google Scholar]
- 25.Lagerholm BC, Wang MM, Ernst LA, Ly DH, Liu HJ, Bruchez MP, Waggoner AS. Nano Lett. 2004;4:2019–2022. [Google Scholar]
- 26.Jiang T, Olson ES, Nguyen QT, Roy M, Jennings PA, Tsien RY. Proc Natl Acd Sci USA. 2004;101:17867–17872. doi: 10.1073/pnas.0408191101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brinckerhoff CE, Matrisian LM. Nat Rev Mol Cell Biol. 2002;3:207–214. doi: 10.1038/nrm763. [DOI] [PubMed] [Google Scholar]
- 28.Egeblad M, Werb Z. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 29.Giambernardi TA, Grant GM, Taylor GP, Hay RJ, Maher VM, McCormick JJ, Klebe RJ. Matrix Biol. 1998;16:483–496. doi: 10.1016/s0945-053x(98)90019-1. [DOI] [PubMed] [Google Scholar]
- 30.Stearns ME, Wang M. Cancer Res. 1993;53:878–883. [PubMed] [Google Scholar]
- 31.Davies B, Waxman J, Wasan H, Abel P, Williams G, Krausz T, Neal D, Thomas D, Hanby A, Balkwill F. Cancer Res. 1993;53:5365–5369. [PubMed] [Google Scholar]
- 32.Bremer C, Bredow S, Mahmood U, Weissleder R, Tung CH. Radiology. 2001;221:523–529. doi: 10.1148/radiol.2212010368. [DOI] [PubMed] [Google Scholar]
- 33.McIntyre JO, Fingleton B, Wells KS, Piston DW, Lynch CC, Gautam S, Matrisian LM. Biochem J. 2004;377:617–628. doi: 10.1042/BJ20030582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao M, Josephson L, Tang Y, Weissleder R. Angew Chem Int ed. 2003;42:1375–1378. doi: 10.1002/anie.200390352. [DOI] [PubMed] [Google Scholar]
- 35.Rothbard JB, Kreider E, VanDeusen CL, Wright L, Wylie BL, Wender PA. J Med Chem. 2002;45:3612–3618. doi: 10.1021/jm0105676. [DOI] [PubMed] [Google Scholar]
- 36.Mitchell DJ, Kim DT, Steinman L, Fathman CG, Rothbard JB. J Peptide Res. 2000;56:318–325. doi: 10.1034/j.1399-3011.2000.00723.x. [DOI] [PubMed] [Google Scholar]
- 37.Seltzer JL, Akers KT, Weingarten H, Grant GA, McCourt DW, Eisen AZ. J Biol Chem. 1990;265:20409–20413. [PubMed] [Google Scholar]
- 38.Welch AR, Holman CM, Browner MF, Gehring MR, Kan CC, Vanwart HE. Arch Biochem Biophys. 1995;324:59–64. doi: 10.1006/abbi.1995.9929. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Syntheses and characterizations of the biotinylated MMP-2 peptide ligands, and experimental procedures for fluorescence microscopic imaging and microplate assay of cellular uptake. This material is available free of charge via the Internet at http://pubs.acs.org.