Abstract
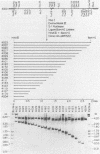
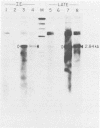
The minimum size fragment of human cytomegalovirus AD169 required to initiate transformation was determined by transfection of primary rat embryo cells with deletion fragments constructed by digestion of a cloned fragment containing the transforming region (pCM4000) with exonuclease III and S1 nuclease. The results indicate that the left-hand boundary of the minimum size sequence for transformation must reside between 490 and 318 bases from the HindIII site of pCM4000. The right-hand boundary was defined by the EcoRI site which is 20 bases from the HindIII site. The nucleotide sequence of the transforming fragment of human cytomegalovirus was determined by the chemical degradation technique of Maxam and Gilbert and with the dideoxynucleoside triphosphate chain termination method. The sequence of pCM4000 comprises 2,848 base pairs and has an A X T composition of 59.5%. Reading frame analysis of the sequence indicated the longest open reading frame was 118 amino acids in length. Northern blot analysis of polyadenylated and non-polyadenylated RNA extracted from cells at immediate early and late times after infection with human cytomegalovirus indicate that one 5.0-kilobase RNA species hybridized to pCM4000 (Jahn et al., J. Virol, in press). The direction of transcription was determined by hybridization of this transcript with M13 single-stranded probes, and S1 analysis of this RNA did not detect introns.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albrecht T., Rapp F. Malignant transformation of hamster embryo fibroblasts following exposure to ultraviolet-irradiated human cytomegalovirus. Virology. 1973 Sep;55(1):53–61. doi: 10.1016/s0042-6822(73)81007-4. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Boyer H. W., Helling R. B. Construction of biologically functional bacterial plasmids in vitro. Proc Natl Acad Sci U S A. 1973 Nov;70(11):3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarchi J. M. Human cytomegalovirus DNA: restriction enzyme cleavage maps and map locations for immediate-early, early, and late RNAs. Virology. 1981 Oct 15;114(1):23–38. doi: 10.1016/0042-6822(81)90249-x. [DOI] [PubMed] [Google Scholar]
- Fleckenstein B., Müller I., Collins J. Cloning of the complete human cytomegalovirus genome in cosmids. Gene. 1982 Apr;18(1):39–46. doi: 10.1016/0378-1119(82)90054-3. [DOI] [PubMed] [Google Scholar]
- Geelen J. L., Walig C., Wertheim P., van der Noordaa J. Human cytomegalovirus DNA. I. Molecular weight and infectivity. J Virol. 1978 Jun;26(3):813–816. doi: 10.1128/jvi.26.3.813-816.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubin G., Goldman D. S., Luce J., Neiman P. E., Cooper G. M. Molecular cloning and nucleotide sequence of a transforming gene detected by transfection of chicken B-cell lymphoma DNA. Nature. 1983 Mar 10;302(5904):114–119. doi: 10.1038/302114a0. [DOI] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Hayward W. S., Neel B. G., Astrin S. M. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981 Apr 9;290(5806):475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- Huang E. S., Kilpatrick B. A., Huang Y. T., Pagano J. S. Detection of human cytomegalovirus and analysis of strain variation. Yale J Biol Med. 1976 Mar;49(1):29–43. [PMC free article] [PubMed] [Google Scholar]
- Kuff E. L., Feenstra A., Lueders K., Rechavi G., Givol D., Canaani E. Homology between an endogenous viral LTR and sequences inserted in an activated cellular oncogene. Nature. 1983 Apr 7;302(5908):547–548. doi: 10.1038/302547a0. [DOI] [PubMed] [Google Scholar]
- Kunkel G. R., Martinson H. G. Nucleosomes will not form on double-stranded RNa or over poly(dA).poly(dT) tracts in recombinant DNA. Nucleic Acids Res. 1981 Dec 21;9(24):6869–6888. doi: 10.1093/nar/9.24.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakeman A. D., Osborn J. E. Size of infectious DNA from human and murine cytomegaloviruses. J Virol. 1979 Apr;30(1):414–416. doi: 10.1128/jvi.30.1.414-416.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marengo C., Mbikay M., Weber J., Thirion J. P. Adenovirus-induced mutations at the hypoxanthine phosphoribosyltransferase locus of Chinese hamster cells. J Virol. 1981 Apr;38(1):184–190. doi: 10.1128/jvi.38.1.184-190.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McDonough S. H., Spector D. H. Transcription in human fibroblasts permissively infected by human cytomegalovirus strain AD169. Virology. 1983 Feb;125(1):31–46. doi: 10.1016/0042-6822(83)90061-2. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Nelson J. A., Fleckenstein B., Galloway D. A., McDougall J. K. Transformation of NIH 3T3 cells with cloned fragments of human cytomegalovirus strain AD169. J Virol. 1982 Jul;43(1):83–91. doi: 10.1128/jvi.43.1.83-91.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima Y., Suzuki Y. Cloning of the silk fibroin gene and its flanking sequences. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5363–5367. doi: 10.1073/pnas.74.12.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskeva C., Roberts C., Biggs P., Gallimore P. H. Human adenovirus type 2 but not adenovirus type 12 is mutagenic at the hypoxanthine phosphoribosyltransferase locus of cloned rat liver epithelial cells. J Virol. 1983 Apr;46(1):131–136. doi: 10.1128/jvi.46.1.131-136.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne G. S., Courtneidge S. A., Crittenden L. B., Fadly A. M., Bishop J. M., Varmus H. E. Analysis of avian leukosis virus DNA and RNA in bursal tumours: viral gene expression is not required for maintenance of the tumor state. Cell. 1981 Feb;23(2):311–322. doi: 10.1016/0092-8674(81)90127-6. [DOI] [PubMed] [Google Scholar]
- Pritchett R. F. DNA nucleotide sequence heterogeneity between the Towne and AD169 strains of cytomegalovirus. J Virol. 1980 Oct;36(1):152–161. doi: 10.1128/jvi.36.1.152-161.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechavi G., Givol D., Canaani E. Activation of a cellular oncogene by DNA rearrangement: possible involvement of an IS-like element. Nature. 1982 Dec 16;300(5893):607–611. doi: 10.1038/300607a0. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975 May 25;94(3):441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- Schlehofer J. R., Hausen J. Z. Induction of mutations within the host cell genome by partially inactivated herpes simplex virus type 1. Virology. 1982 Oct 30;122(2):471–475. doi: 10.1016/0042-6822(82)90247-1. [DOI] [PubMed] [Google Scholar]
- Schreier P. H., Cortese R. A fast and simple method for sequencing DNA cloned in the single-stranded bacteriophage M13. J Mol Biol. 1979 Mar 25;129(1):169–172. doi: 10.1016/0022-2836(79)90068-8. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spector D. H., Hock L., Tamashiro J. C. Cleavage maps for human cytomegalovirus DNA strain AD169 for restriction endonucleases EcoRI, BglII, and HindIII. J Virol. 1982 May;42(2):558–582. doi: 10.1128/jvi.42.2.558-582.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinski M. F., Thomsen D. R., Stenberg R. M., Goldstein L. C. Organization and expression of the immediate early genes of human cytomegalovirus. J Virol. 1983 Apr;46(1):1–14. doi: 10.1128/jvi.46.1.1-14.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak H. F., Flavell R. A. A method for the recovery of DNA from agarose gels. Nucleic Acids Res. 1978 Jul;5(7):2321–2332. doi: 10.1093/nar/5.7.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathen M. W., Stinski M. F. Temporal patterns of human cytomegalovirus transcription: mapping the viral RNAs synthesized at immediate early, early, and late times after infection. J Virol. 1982 Feb;41(2):462–477. doi: 10.1128/jvi.41.2.462-477.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976 Sep 3;193(4256):848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]
- Weiss B., Live T. R., Richardson C. C. Enzymatic breakage and joining of deoxyribonucleic acid. V. End group labeling and analysis of deoxyribonucleic acid containing single straned breaks. J Biol Chem. 1968 Sep 10;243(17):4530–4542. [PubMed] [Google Scholar]
- Weststrate M. W., Geelen J. L., van der Noordaa J. Human cytomegalovirus DNA: physical maps for restriction endonucleases BglII, hindIII and XbaI. J Gen Virol. 1980 Jul;49(1):1–21. doi: 10.1099/0022-1317-49-1-1. [DOI] [PubMed] [Google Scholar]
- Wigler M., Silverstein S., Lee L. S., Pellicer A., Cheng Y. c., Axel R. Transfer of purified herpes virus thymidine kinase gene to cultured mouse cells. Cell. 1977 May;11(1):223–232. doi: 10.1016/0092-8674(77)90333-6. [DOI] [PubMed] [Google Scholar]