Abstract
Ciliates are unicellular eukaryotic organisms containing two types of nuclei: macronuclei and micronuclei. After the sexual pathway takes place, a new macronucleus is formed from a zygote nucleus, whereas the old macronucleus is degraded and resorbed. In the course of macronuclear differentiation, polytene chromosomes are synthesized that become degraded again after some hours. Most of the DNA is eliminated, and the remaining DNA is fragmented into small DNA molecules that are amplified to a high copy number in the new macronucleus. The protein Pdd1p (programmed DNA degradation protein 1) from Tetrahymena has been shown to be present in macronuclear anlagen in the DNA degradation stage and also in the old macronuclei, which are resorbed during the formation of the new macronucleus. In this study the identification and localization of a Pdd1p homologous protein in Stylonychia (Spdd1p) is described. Spdd1p is localized in the precursor nuclei in the DNA elimination stage and in the old macronuclei during their degradation, but also in macronuclei and micronuclei of starved cells. In all of these nuclei, apoptotic-like DNA breakdown was detected. These data suggest that Spdd1p is a general factor involved in programmed DNA degradation in Stylonychia.
INTRODUCTION
Ciliates contain several nuclei with different structural and functional characteristics. Micronuclei are diploid and transcriptionally inactive, whereas DNA-rich macronuclei contain fragmented DNA molecules that are transcribed during vegetative life cycle (for review, see Prescott, 1994). During sexual reproduction, meiotic products of the micronucleus are exchanged between conjugating partners and fuse with the stationary haploid micronucleus, resulting in a diploid zygote nucleus. After the mitotic division of this syncaryon, one of the daughter nuclei differentiates into a new macronucleus, and the other differentiates into new micronuclei. During this process, the old macronucleus is degraded and its contents are resorbed (Grell, 1973).
In the hypotrichous ciliate Stylonychia lemnae, the above process takes ∼100 h, and the precursor nuclei can be distinguished by light microscopy (Ammermann et al., 1974). Macronuclear development is accompanied by extensive elimination, rearrangement, and amplification of DNA. Some micronuclear chromosomes are completely eliminated, before the other chromosomes undergo several rounds of DNA replication to form polytene chromosomes. These chromosomes are degraded again, and most of the DNA is eliminated. The remaining DNA is fragmented into small DNA molecules that are specifically amplified in a second round of replication, which results in the vegetative macronucleus. These macronuclear DNA molecules have sizes of 0.5–30 kb and normally contain only one gene and its regulatory regions (for review, see Prescott, 1994; Lipps and Eder, 1996).
In Stylonychia, ∼98% of the micronuclear genome is eliminated during macronuclear differentiation; only 2% of the genome therefore contain all of the information required for vegetative growth of the cell. A few of these eliminated sequences of hypotrichs have been characterized. Typically these belong to middle repetitive sequences such as, for example, transposon-like elements in Euplotes crassus, telomere-bearing elements in Oxytricha nova, or a repetitive element in Stylonychia lemnae (for review, see Klobutcher and Jahn, 1991; Maercker et al., 1997). In addition, many unique micronuclear sequences are eliminated during macronuclear development. Some of those sequences have been characterized as flanking sequences of precursor genes, spacer sequences between precursor genes, or short sequences that are integrated in precursor genes (internal eliminated sequences), but get eliminated during nuclear differentiation (Prescott, 1994). In addition to the elimination process in the macronuclear anlagen, the whole DNA from the old macronucleus also becomes degraded.
Very little is known about the molecular mechanisms involved in these DNA elimination processes in hypotrichous ciliates, but it has to be assumed that elimination of the various sequence elements requires different protein-encoding enzymatic machineries. The resorption of whole micronuclear chromosomes takes place soon after the formation of the zygote nucleus, and the chromosomes become localized at the periphery of the nucleus before their degradation takes place (Ammermann et al., 1974). The elimination of transposon-like elements starts early during macronuclear development and takes a few hours (Frels et al., 1996). The excision of internal eliminated sequences starts early, during formation of the polytene chromosomes, and is finished only in the stage of fully developed polytene chromosomes, and it is performed in a specific order (Tausta et al., 1991; Wen et al., 1996). Transposon-like elements and also the larger internal eliminated sequences can be detected as circles during their elimination (Jaraczewski and Jahn, 1993; Klobutcher et al., 1993; Williams et al., 1993). In addition, a dramatic reorganization of chromatin is found during macronuclear development. DNA sequences to be eliminated are organized in 30-nm loops, which are eventually released from the axis and degraded (Meyer and Lipps, 1980). Also, in the holotrichous ciliate Tetrahymena, the processing of a gene has been shown to be due to high-frequency intragenic recombination during macronuclear development (Deak and Doerder, 1998). Collectively, the observations make it very likely that recombination events are involved in DNA rearrangement during macronuclear differentiation and that these events are catalyzed by stage-specific proteins.
To date, no enzyme involved in the programmed DNA elimination has been identified. Transposase- or integrase-like motifs have been detected in the ORFs of transposon-like elements (Doak et al., 1994), but transcripts derived from these repetitive DNA sequences could only be detected in very low abundance (Jaraczewski et al., 1994); however, a number of genes and proteins active during macronuclear maturation of ciliates have been isolated recently. The CON ZA7 gene of Euplotes crassus possibly codes for a DNA-binding protein that might function as a transcription factor (Ling et al., 1997). The Tetrahymena CNJ C gene might encode an RNA polymerase subunit (Martindale, 1990).
The first conjugation-specific proteins isolated from ciliates that might be involved in DNA elimination are a small family of programmed DNA degradation proteins (Pddps) (Madireddi et al., 1994). Two such proteins, Pdd1p and Pdd2p, have been characterized in some detail (Madireddi et al., 1996; Smothers et al., 1997a,b). Interestingly, both polypeptides localize to the Tetrahymena macronuclear anlagen during the time of DNA degradation and also to the old macronucleus during its resorption. More importantly, cross-linking and fluorescent in situ hybridization analyses demonstrate that Pdd1p closely associates with DNA segments that are destined for elimination. Analysis of the PDD1 gene indicates that the protein contains three chromodomains (Callebaut et al., 1997) that are known to be able to bind to heterochromatic DNA or silenced euchromatin (for review, see Elgin, 1996), and so far, Pddps localize to specialized heterochromatic structures in developing macronuclei (Smothers et al., 1997b).
Here we describe the identification of a Pdd1p-homologous protein in Stylonychia lemnae (Spdd1p) by immunochemical methods. Spdd1p has approximately the same apparent mass as Tetrahymena Pdd1p. It seems to be involved not only in DNA degradation in the old macronuclei and macronuclear anlagen during nuclear differentiation but also in limited DNA degradation in micronuclei and macronuclei of starved cells. DNA breakdown in nuclei of exconjugants and of starved cells expressing Spdd1p was also demonstrated with the TUNEL assay. These results indicate that similar mechanisms might be involved in the apoptotic-like DNA degradation in exconjugants and starved Stylonychia cells.
MATERIALS AND METHODS
Cultivation of Cells and Isolation of Nuclei
Cultivation and mating of Stylonychia and isolation of nuclei were performed as described by Ammermann et al. (1974).
Western Blot Analysis
Vegetative cells or exconjugant cells during different stages of macronuclear development were harvested and micronuclei, macronuclei, and macronuclear anlagen were separated as described previously (Ammermann et al., 1974), using 10-, 20-, 25-, 30-, and 40-μm gazes (Heidland, Gütersloh, Germany). The nuclei were lysed in 2 × SDS sample buffer before the nuclear proteins of ∼104 cells per lane were separated on 12.5% polyacrylamide gels (SDS-PAGE) according to Laemmli (1970). By transferring the separated proteins on a supported nitrocellulose membrane (Hybond-C super, Amersham, Buckinghamshire, UK) at 1–2 mA/cm2 for 2 h, immunodetection was possible according to the DIG Detection Kit protocol (Boehringer Mannheim, Mannheim, Germany) at room temperature with the following modifications. The membrane was blocked in blocking solution (1% [wt/vol] blocking reagent in maleic acid buffer: 0.15 M maleic acid, 0.15 M NaCl, pH 7.5), incubated for 2 h with Tetrahymena Pdd1p antiserum (Madireddi et al., 1994), diluted 1:1000 in blocking solution, washed three times for 10 min each in maleic acid buffer, and incubated for 1 h in goat anti-rabbit IgG (H+L) AP conjugate (0.6 mg/ml, Dianova, Hamburg, Germany), diluted 1:10,000 in blocking solution. The washing and staining procedure was performed according to the Boehringer Mannheim protocol.
Blocking Experiment
Blocking solution (500 μl) including 4 μl of Pdd1p antiserum was preincubated without or together with different amounts of HPLC-purified Tetrahymena Pdd1p (Madireddi et al., 1994, 1996) at room temperature on a shaker for 1 h. Then 7.5 ml of blocking solution were added, leading to the 1:2000 final dilution of the antiserum. After that, Western blot analysis was continued as described above.
Immunofluorescence Analysis
Whole cells were fixed in ethanol:acetic acid (3:1) for 3 min, dropped onto slides, and air-dried. Single polytene chromosomes were obtained by overnight fixation of isolated nuclei in methanol:acetic acid (3:1), centrifugation (1000 rpm, 1–3 min, Heraeus Megafuge, Heraeus, Osterode, Germany), and resuspension in 45% acetic acid and then were squashed on a slide. For immunodetection, incubation with Pdd1p antiserum (1:100) in PBS (137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4.7H20, 1.4 mM KH2P04) was followed by three washes in PBS for 2 min each, incubation in anti-rabbit IgG Cy2 conjugate (Dianova; 1.5 mg/ml, diluted 1:100) for 1 h at 37°C, washing three times in PBS for 10 min each, and DAPI staining (0.1 μg/ml PBS, 10 min). Finally, the slides were rinsed with H20 and analyzed by light microscopy (phase-contrast, UV light for DAPI stain, 470 nm for the Cy2 label; Leitz DM RB microscope, Leica, Wetzlar, Germany objective 63× Plan Apochromat, magnification 630×). Photographs were taken with Kodak (Rochester, NY) 100 ASA positive film or Kodak Tri-X pan 400 ASA film.
TUNEL Assay
The TUNEL assay was performed as described previously (Gavrieli et al., 1992; Madireddi et al., 1996) with slight modifications. Slides with ethanol:acetic acid (3:1)-fixed cells were washed in H2O for 15 min and air-dried. The TUNEL reaction containing components of the Boehringer Mannheim DIG oligonucleotide tailing kit (4 μl 5 × reaction buffer, 4 μl 5 × CoCl2 solution, 1 μl DIG-dUTP, 1 μl TdT (50 U), 10 μl H2O per field) was performed at 37°C for 1 h. The slide was washed three times in PBS for 2 min each and then incubated in anti-DIG Fab fragments fluorescein conjugate (Boehringer Mannheim; 200 μg/ml, 1:100 dilution) at 37°C for 1 h and washed in PBS for 2 min. DAPI staining and microscopy were performed as described above.
RESULTS
Localization of a Protein Homologous to Tetrahymena Pdd1p (Spdd1p) in Nuclei of Stylonychia Exconjugants
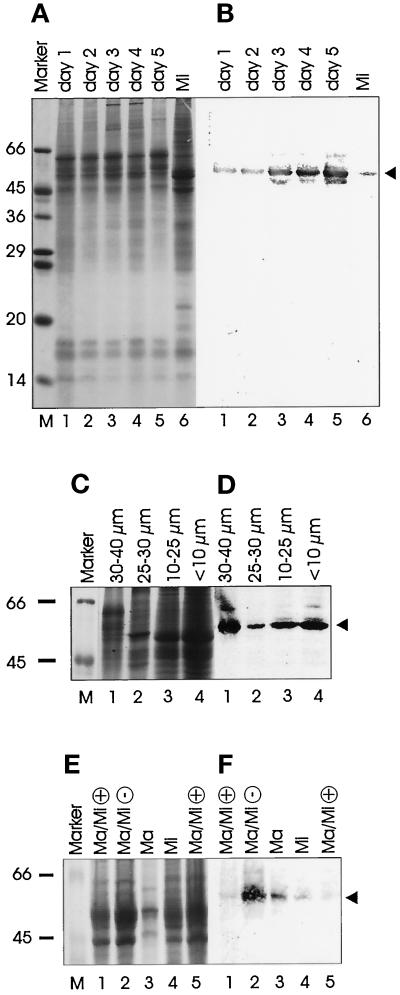
Pdd1p and Pdd2p were recently biochemically identified in Tetrahymena as being enriched in the developing macronuclei and in old macronuclei during nuclear differentiation, indicating that they might be involved in programmed DNA degradation processes (Madireddi et al., 1994; 1996; Smothers et al., 1997b). To test whether proteins homologous to Tetrahymena Pdd1p and Pdd2p also exist in the hypotrichous ciliate Stylonychia lemnae, antisera against both proteins (Madireddi et al., 1994; Smothers et al., 1997b) were used for immunoblot analysis of Stylonychia nuclear proteins. Although no specific signal was obtained with the Pdd2p antiserum (our unpublished results), the experiments with the Pdd1p antiserum revealed very reliable results after Western blotting. Two different strains of opposite mating types were mixed, and the nuclei were prepared from exconjugant cells at different time points after mating. One to five days after pairing, total nuclear protein from nuclei >10 μm (macronuclei, macronuclear anlagen) and micronuclear proteins of vegetative cells (from nuclei <10 μm) were resolved by SDS-PAGE (Figure 1a). When the proteins were blotted and probed with the Pdd1p antiserum, a band corresponding to 53–54 kDa was detected, similar to the size estimated for Tetrahymena Pdd1p (Figure 1b) (Madireddi et al., 1996), suggesting that a protein homologous to Tetrahymena Pdd1p also exists in Stylonychia (hereafter referred to as Spdd1p). The intensity of the band was very weak at days 1 and 2 and became stronger at day 3 after mating, the time period when fragmentation of the polytene chromosomes and programmed DNA degradation occurs. Western analysis of micronuclear proteins of vegetative cells with the Pdd1p antiserum revealed a weak signal (Figure 1b, lane 6). To localize Spdd1p in nuclei of exconjugants in more detail, a crude separation and enrichment of exconjugant nuclei were made at day 2 after mating as follows. Micronuclei (<10 μm), macronuclei, including old macronuclei to be resorbed during differentiation (10–25 μm), large, polytene chromosomes containing nuclei (30–40 μm), and small macronuclear anlagen (25–30 μm), representing precursor nuclei before or after the polytene chromosome stage, were separated. The most prominent signal was found in the large macronuclear anlagen (30–40 μm) (Figure 1d); however, a reaction could also be detected in all other nuclear fractions, suggesting that a low level of Spdd1p is present in all kinds of nuclei from exconjugant cells or in single nuclei enriched in the different nuclear fractions. The Pdd1p expression in micronuclei (<10 μm) was remarkably strong in this experiment (Figure 1d, lane 4), in contrast to the low-level expression in micronuclei of vegetative cells (Figure 1b, lane 6; 1f, lane 4). The reason for this phenomenon might be that the degradation of old micronuclei in the exconjugants preferentially takes place at day 2 after conjugation. Therefore, Spdd1p may also be present in some micronuclei, an observation that was not obvious in initial studies in Tetrahymena (Madireddi et al., 1996) but now also appears to be the case (Coyne, Nikiforov, Smothers, Allis, and Yao, unpublished observations).
Figure 1.
Detection of a protein homologous to Tetrahymena Pdd1p (Spdd1p) in Stylonychia nuclear extracts by immunoblot analyses. Nuclear proteins with the same concentrations were separated by SDS-PAGE, stained with Coomassie Blue (a, c, e), blotted, and incubated with the Pdd1p antiserum (b, d, f). (a) Whole nuclear extracts from exconjugants (days 1–5 after mating). (b) Anti-Pdd1p Western blot of the proteins shown in a. (c) Nuclear extracts of exconjugant nuclei (day 2 after mating), which were separated by size. Lane 1, 30–40 μm; lane 2, 25–30 μm; lane 3, 10 -25 μm; lane 4, <10 μm. (d) Anti-Pdd1p Western blot analysis of the extracts shown in c. (e) Whole nuclear extracts from vegetative growing cells (+, lane 1 and lane 5 with the double concentration), from starved cells (3 d without feeding, −, lane 2), from isolated macronuclei (diameter 10–25 μm, lane 3), and from isolated micronuclei (<10 μm, lane 4). (f) Anti-Pdd1p (dilution of antiserum 1:1000) Western analysis of the extracts shown in e. Spdd1p is labeled by an arrow. Sizes are given in kilodaltons (Sigma molecular weight marker).
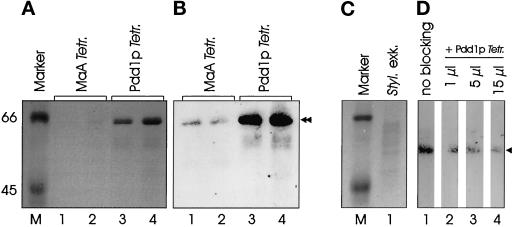
To exclude the possibility that the band appearing with the Tetrahymena Pdd1p antiserum is only due to cross-reactivity with a nonhomologous protein in Stylonychia, a blocking experiment using Tetrahymena Pdd1p was performed. The antiserum was preincubated with purified Tetrahymena Pdd1p in different concentrations before continuing with the Western blot analysis. By increasing the concentration of Tetrahymena Pdd1p in the assay, an ever-increasing inhibition of the Western reaction was observed (Figure 2). This is a very strong indication that the Stylonychia protein reacting with the Pdd1p antiserum is a homologue of Tetrahymena Pdd1p, although Spdd1p seems to be slightly smaller than Tetrahymena Pdd1p (Figure 2a).
Figure 2.
Tetrahymena Pdd1p inhibits binding of Pdd1p antiserum to Spdd1p. (a) Whole Tetrahymena macronuclear anlagen (MaA) nuclear lysates (lane 1, 10 μl; lane 2, 30 μl) and HPLC-purified Tetrahymena Pdd1p (lane 3, 10 μl; lane 4, 30 μl), separated by SDS-PAGE. (b) Anti-Pdd1p Western analysis (dilution of antiserum 1:2000) of the extracts shown in a (control for d). (c) Stylonychia exconjugants nuclear extracts (day 4) after pairing, separated by SDS-PAGE (lane 1). (d) The same amount of protein as in c, lane 1, was separated by SDS-PAGE, blotted, and incubated with the Pdd1p antiserum (dilution 1:2000) after preincubation without Tetrahymena Pdd1p (lane 1) or after preincubation with 1 μl (lane 2), 5 μl (lane 3), or 15 μl (lane 4) Tetrahymena Pdd1p (purified protein fraction as shown in a and b, lanes 3 and 4). The staining reaction was performed for 4 h with each membrane strip. Sizes are given in kilodaltons (Sigma molecular weight marker). Arrowhead, Spdd1p; double arrowhead, Tetrahymena Pdd1p.
The immunoblot analysis shown in Figure 1 only distinguishes between the size of the different nuclei, but in some cases it is not possible to distinguish between different kinds of nuclei by this parameter. For example, small macronuclear anlagen before or after DNA elimination and old macronuclei overlap considerably in size. In addition, conjugation is not completely synchronous, so that at any individual time point, not all macronuclear anlagen are exactly in the same developmental stage, and a small fraction of the cells (10–20%) represent nonconjugants.
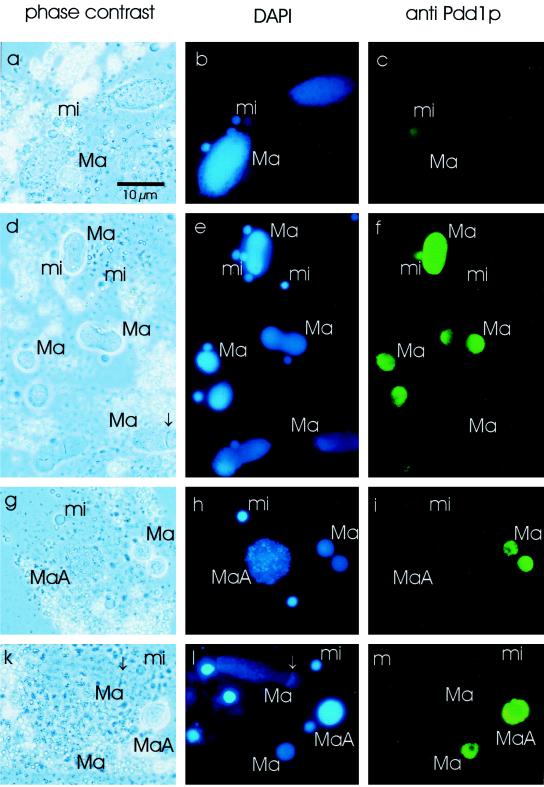
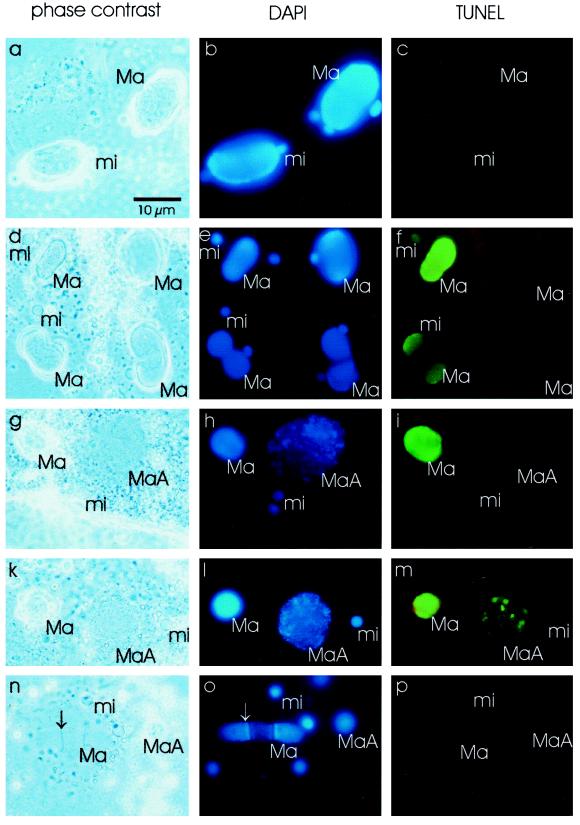
For these reasons, immunofluorescence analyses were performed to distinguish between the nuclei in single cells and anti-Pdd1p signals therein. Normal vegetative cells and exconjugants were fixed and incubated with Pdd1p antiserum. The DNA was counterstained with DAPI, and the slides were analyzed by phase-contrast and fluorescence microscopy. Nuclei of fast growing vegetative cells as a control did not show a signal with Pdd1p antiserum (Figure 3c). In exconjugants, during the polytene chromosome stage (day 2 after mating), the old macronucleus, which is degraded during development of the new macronucleus, was stained by Pdd1p antiserum (Figure 3i). At a later time point during macronuclear development (day 4 after mating), when DNA degradation in old macronuclei and late macronuclear anlagen occurs, staining of macronuclear anlagen as well as old macronuclei was observed. In new macronuclei in which DNA was replicated (replication band, marked by an arrow in Figure 3, k and l), no Pdd1p staining was detected (Figure 3m).
Figure 3.
Immunofluorescence analysis of vegetatively growing, starved, and exconjugant cells. Vegetative growing cells (a–c), starved cells (2 d without feeding, d–f), and exconjugant cells during polytene chromosome stage (g–i) and at a later stage (k–m) were fixed and incubated with the Pdd1p antiserum (secondary anti-rabbit Cy2-labeled antibody), and the DNA was stained with DAPI. (a, d, g, k) Phase-contrast microscopy, (b, e, h, l) DAPI stain (UV light), (c, f, i, m) anti-Pdd1p stain (blue light). Ma, macronucleus; MaA, macronuclear anlagen; mi, micronucleus. The replication band is labeled by an arrow. Bar, 10 μm.
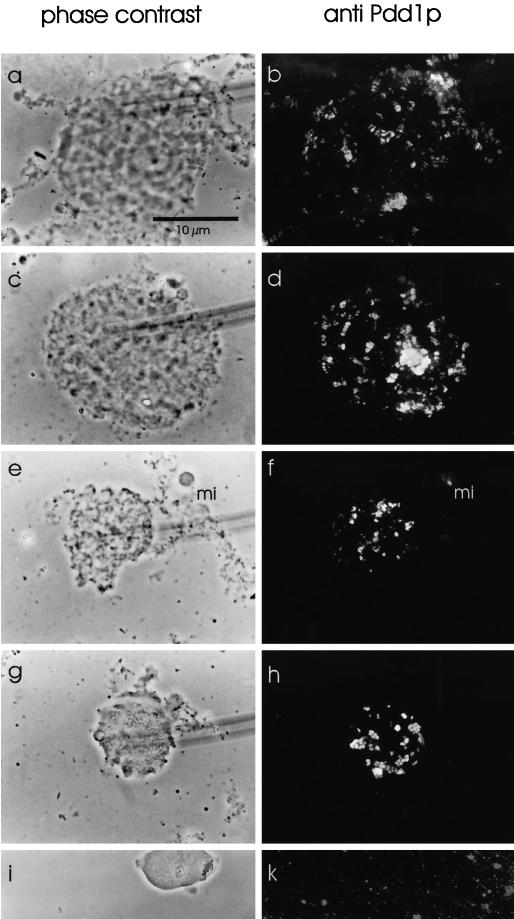
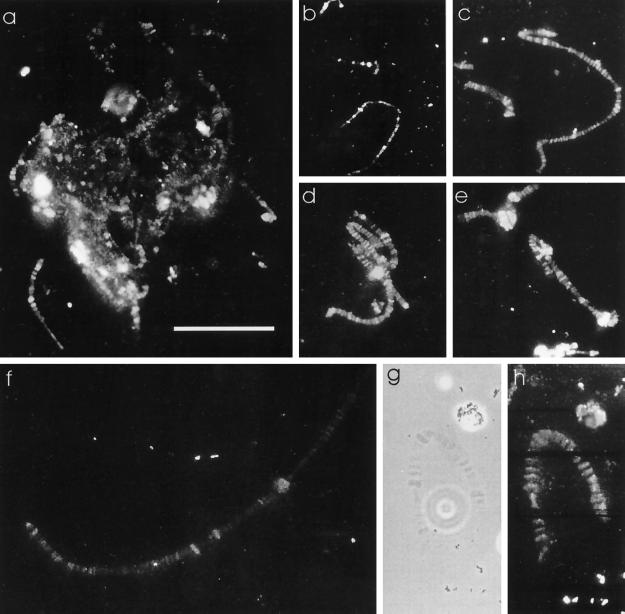
The observation that the polytene chromosome nucleus did not react with Pdd1p antiserum (Figure 3i) was in contrast to the strong signal resulting from macronuclear anlagen nuclei in Western analysis (Figure 1). One explanation could be that because whole cells were used for the experiments shown in Figure 3, Spdd1p in some macronuclear anlagen may not have been accessible for the antiserum. Therefore, macronuclear anlagen in the polytene chromosome stage and in later stages were squashed in 45% acetic acid. With this preparation method, staining of Spdd1p on polytene chromosomes was obtained (Figure 4, a and b) as well as in later anlagen stages, when DNA fragmentation starts (Figure 4c–f) and during DNA degradation in the vesicle stage (Figure 4, g and h), whereas macronuclei of nonconjugating cells did not stain (Figure 4k). To investigate how Spdd1p localized during different maturation steps of the polytene chromosomes, nuclei in different developmental stages were squashed and analyzed by immunofluorescence microscopy. These experiments revealed that Spdd1p associated with the banded regions of the fully developed polytene chromosomes (Figure 5, g and h) and also with heterochromatic regions of the chromosomes (Figure 5, d and e). Even in an early stage, when the polytene chromosomes have not yet reached their full size, Spdd1p already binds to the chromatin (Figure 5b).
Figure 4.
Spdd1p expression in late Stylonychia macronuclear anlagen. Macronuclear anlagen during polytene chromosome stage (a, b), at the beginning of DNA fragmentation (c–f), and in the vesicle stage during DNA degradation (g, h) were fixed, squashed, and stained with the Pdd1p antiserum (Cy2-stained anti-rabbit secondary antibody). (i, k) Macronucleus of nonconjugating cells (negative control). (a, c, e, g, i) Phase-contrast microscopy. (b, d, f, h, k) Anti-Pdd1p immunofluorescence (blue light). mi, micronucleus. Bar, 10 μm.
Figure 5.
Localization of Spdd1p to the bands of polytene chromosomes. Macronuclear anlagen were fixed, squashed, incubated with the Pdd1p antiserum, and analyzed by immunofluorescence microscopy. (a–f, h) Anti-Pdd1p immunofluorescence stain (blue light); (g) phase-contrast image of h. Bar, 10 μm.
Expression of Pdd1p in Starved Stylonychia Cells
Because the Pdd1p antiserum showed a reaction with nuclear proteins of vegetative cells in several experiments (for example, Figure 1b, lane 6), we sought to determine whether Spdd1p expression in vegetative Stylonychia cells is in any way dependent on the physiological status of the cells. Therefore, total nuclear extracts from actively growing cells and from starved cells (3 d without feeding), as well as from isolated macronuclei and micronuclei from a growing culture fed every day, were prepared for immunoblot analysis. The anti-Pdd1p signal was very weak with nuclear extracts of rapidly growing vegetative cells (Figure 1f, lanes 1 and 5). Both separated macronuclei and micronuclei of cells from a continuously growing culture contained detectable levels of Spdd1p; also the macronuclear signal was stronger than the micronuclear signal (Figure 1f, lanes 3 and 4). The whole nuclei of starved cells not fed for 3 d, however, revealed a very strong signal (Figure 1f, lane 2).
The above results were confirmed by immunofluorescence analysis. In starved cells not fed for 2 d, single macronuclei displayed a strong reaction to the antibodies, whereas others did not (Figure 3F). In nondividing cells, smaller macronuclei typically stained intensely, in contrast to macronuclei of nonstarved cells where the macronuclear DNA is being replicated, as indicated by the presence of a replication band (arrow in Figure 3d). Some micronuclei of starved cells also showed an anti-Pdd1p reaction (Figure 3f).
Because a signal with Pdd1p antiserum was obtained with nuclei of starved cells, the question was raised concerning whether DNA degradation may occur in these cells. As a first experiment, DAPI images of macronuclei of ∼30 normal-growing as well as the same number of nuclei of starved cells expressing Spdd1p were scanned with a densitometer. Their size and the absorption, reflecting the DNA content, were quantitatively estimated with densitometric analyses. The average size of macronuclei of growing cells was more than twice the size of macronuclei of starved cells. At the same time, the absorption per micrometers squared was approximately the same with nuclei from starved and growing cells. The average DNA content of starved cells was only ∼35% of the DNA content of macronuclei of growing cells (our unpublished results). Because Pdd1p in Tetrahymena has been localized exclusively to nuclei in which DNA degradation takes place (Madireddi et al., 1996), we expected that in Stylonychia Spdd1p is present in macronuclei and in some cases in micronuclei of starved cells, where DNA degradation may occur.
Apoptotic-like DNA Degradation in Stylonychia Exconjugants and Starved Cells
Programmed DNA degradation in macronuclear anlagen and old macronuclei from Tetrahymena exconjugants was shown to be an apoptotic-like DNA degradation process by the TUNEL assay (Madireddi et al., 1996; Mpoke and Wolfe, 1996). We applied the same technique to exconjugant and starved Stylonychia cells. Nicked DNA on slides with fixed whole cells was labeled with DIG dUTP, which could be detected by anti-DIG dUTP immunofluorescence microscopy. As expected, nicked DNA could be detected in macronuclear anlagen during fragmentation of the polytene chromosomes and DNA degradation (Figure 6m) as well as in the old macronuclei resorbed during macronuclear differentiation (Figure 6, i and m). Small macronuclei from nondividing cells showed a strong TUNEL reaction, often preferentially at the border of the macronuclei (Figure 6f), because it was seen with macronuclei in starved cells by anti-Pdd1p immunofluorescence experiments (Figure 3f). In addition, some micronuclei in starved and exconjugant cells were labeled by DIG dUTP (Figure 6c) (not shown for exconjugants; our unpublished results). These results confirm the assumption that apoptotic-like DNA degradation takes place in exconjugants as well as in starved cells. Because the TUNEL reaction obviously colocalizes with Spdd1p, the protein might be involved in this process in both types of cells.
Figure 6.
TUNEL assay with starved and exconjugant cells. Vegetatively growing cells (a–c), starved cells (d–f), exconjugants during polytene chromosome stage (g–i), exconjugants during beginning of DNA fragmentation (k–m), and cells with very late macronuclear anlagen and a newly synthesized macronucleus (n–p) were fixed for the TUNEL assay. In addition, nuclear DNA was stained with DAPI. (a, d, g, k, n) Phase contrast microscopy; (b, e, h, l, o) DAPI stain; (c, f, i, m, p) TUNEL assay (FITC-labeled secondary anti-mouse antibody). Ma, macronucleus; MaA, macronuclear anlagen; mi, micronucleus. Arrows, replication band. Bar, 10 μm.
DISCUSSION
Nuclear differentiation in ciliates is accompanied by extensive DNA rearrangement and elimination processes. Pdd1p, the first protein most likely involved in these processes, was identified as being an abundant, stage-specific polypeptide from Tetrahymena (Madireddi et al., 1994; 1996; Smothers et al., 1997b). Pdd1p belongs to the group of chromodomain-containing proteins such as heterochromatin protein 1 (HP1), which is most likely a component of heterochromatin or otherwise condensed chromatin and also might mediate telomere behavior (Elgin, 1996; Fanti et al., 1998; Wakimoto, 1998). In general, no direct DNA binding activity is associated with proteins in this class. As expected from other chromodomain proteins, Pdd1p contains multiple phosphorylation sites, which are specifically phosphorylated during nuclear differentiation (Madireddi et al., 1996; Smothers et al., 1997b). We used an antiserum raised against Pdd1p to test whether a homologous protein can be found in the hypotrichous ciliate Stylonychia lemnae. Stylonychia nuclear extracts from different developmental stages were tested for the presence of this protein by Western blot analysis. Because Pdd1p antiserum detected a stage-specific polypeptide with a size of ∼54 kDa (Spdd1p, Figures 1 and 2), which is similar to the size of Tetrahymena Pdd1p, the localization and expression pattern of Spdd1p in Stylonychia during sexual reproduction and vegetative growth was further analyzed.
Tetrahymena Pdd1p is expressed in exconjugant nuclei where DNA degradation takes place, both in the late macronuclear anlagen and in the old macronuclei, which are resorbed during the formation of the new macronucleus (Madireddi et al., 1996). To localize Pdd1p-like proteins in ciliates in more detail, Stylonychia has been chosen as a model system, because in this ciliate, DNA degradation is extreme—approximately 98% of the micronuclear genome is eliminated during macronuclear development—and a very prominent polytene chromosome stage precedes DNA fragmentation and amplification of the macronuclear DNA molecules (Prescott, 1994).
Spdd1p Is a Homologue to Tetrahymena Pdd1p
Immunoblot analyses of Stylonychia nuclear extracts from old macronuclei and macronuclear anlagen in exconjugants revealed a very distinct band with a size of ∼54 kDa with the Tetrahymena Pdd1p antiserum (Figures 1 and 2). This led to the assumption that Stylonychia also contains a protein homologous to Tetrahymena Pdd1p. This was confirmed by the blocking experiments (Figure 2). There seem to be some differences between Tetrahymena Pdd1p and Spdd1p: Tetrahymena Pdd1p behaves slightly differently on SDS gels, running more slowly than Spdd1p (Figure 2, a and b). Moreover, with a Tetrahymena PDD1 gene probe (Madireddi et al., 1996) only a very weak hybridization signal was obtained with Stylonychia DNA (our unpublished results). Therefore, it seems likely that the Stylonychia protein might be homologous to epitopes in common with the Tetrahymena protein but that other regions are more different and that it only exhibits a low degree of conservation at the DNA level. Cloning and sequencing of the Stylonychia gene will be necessary to compare the two genes completely.
DNA Elimination Takes Place in Exconjugant and Starved Stylonychia Cells
In agreement with Tetrahymena studies, Spdd1p was found not only in macronuclear anlagen and old macronuclei but also in some micronuclei of Stylonychia exconjugants (Figure 1f, lane 4). We assume that these micronuclei represent “old” nuclei that are resorbed when the new micronuclei are formed. In Tetrahymena, DNA breakdown has occasionally been observed in micronuclei identified as degenerating haploid products of meiosis (Mpoke and Wolfe, 1996). In our Stylonychia experiments, some micronuclei of exconjugants showed a TUNEL reaction (our unpublished results). Therefore, we assume that this DNA breakdown in micronuclei of ciliates during sexual reproduction is accompanied by Pddp1 in the same nuclei.
The binding of the protein to polytene chromosomes has been suggested by immunofluorescence microscopic studies. The protein seems to be associated with nearly every band of the polytene chromosomes (Figure 5). This raises the question of whether most, if not all, of the bands contain micronucleus-specific DNA to be eliminated after chromosomal fragmentation. This hypothesis is also supported by results from electron microscopy, showing that DNA from these bands is eliminated after fragmentation of the polytene chromosomes (Kloetzel, 1970; Meyer and Lipps, 1980) and by in situ hybridizations performed with micronucleus-specific repetitive sequences, which exclusively hybridize to polytene chromosome bands (Maercker et al., 1997).
By Western blot as well as by immunofluorescence microscopic analyses, Spdd1p expression has been demonstrated in starved Stylonychia cells. Nuclei from starved cells that contain Spdd1p are smaller than macronuclei from growing cells not expressing Spdd1p (Figure 3f). As shown by our densitometric measurements, the smaller size of the macronuclei during starvation is not simply due to chromatin condensation, but instead these nuclei exhibit a reduced DNA content compared with actively growing vegetative cells (our unpublished results). Because the macronuclear DNA content in starved cells is even less than one-half of the DNA content in growing cells, the possibility that the result is only due to a prolonged G1 stage during starvation can be excluded; instead the DNA content must be reduced in all starving cells. This relationship of the DNA contents in both types of cells is in agreement with studies in Paramecium where the DNA content of the macronucleus is closely coupled with growth rate (Berger and Ching, 1989; for review, see Adl and Berger, 1996). The result that Spdd1p is also expressed in macronuclei of starved Stylonychia cells could suggest not only that DNA replication and gene expression are reduced but that active DNA degradation also takes place in starved cells. The fact that the TUNEL assays were not only positive in exconjugant cells in the DNA degradation stage as expected from the Tetrahymena results (Figure 6, i and m) (Davis et al., 1992; Madireddi et al., 1996; Mpoke and Wolfe, 1996), but also in starved cells (Figure 6f), suggested that in both starved and exconjugant cells apoptotic-like DNA degradation might be responsible for a programmed degradation of DNA. Whether active DNA degradation takes place in starved Tetrahymena cells is not known; however, at least a reduction in macronuclear volume and an increase in the size of electron-dense chromatin bodies takes place, probably with the help of H1/HP1-like protein, which becomes hyperphosphorylated in these cells (Huang et al., 1999). Because recent results show that Pdd1p also is present in starved Tetrahymena cells (Coyne, Nikiforov, Smothers, Allis, and Yao, unpublished observations), it also might be involved in this chromatin condensation process.
Still not clear to us is why DNA degradation seems to happen in micronuclei of starved Stylonychia cells, because these micronuclei are not supposed to contain multicopy DNA. The question remains whether some micronuclei might be completely resorbed during starvation of Stylonychia for some unknown reason.
Spdd1p Is Possibly a General Factor Involved in Programmed DNA Degradation
Recently a chromodomain protein (H1/HP1-like protein) has been identified in Tetrahymena that is exclusively localized in macronuclei and seems to be involved in the formation or maintenance of the heterochromatic bodies, the silent chromatin of the Tetrahymena macronuclear genome (Huang et al., 1998). In contrast to H1/HP1-like protein, which seems to bind to any silenced chromatin in the macronuclei, Spdd1p only seems to be associated with any DNA that is eventually degraded during macronuclear differentiation and after starvation of Stylonychia cells. It binds not only to heterochromatin but also to the bands of polytene chromosomes and to macronuclear chromatin. This suggests that Spdd1p is not only a heterochromatin specific protein but that it also binds to repressed euchromatin similar to other chromodomain proteins (Elgin, 1996).
At present we can only speculate about the possible function of Spdd1p in Stylonychia. It seems likely that Spdd1p is a part of a protein complex whose binding properties to DNA are not yet clear. The fact that Spdd1p is most often enriched in nuclei when DNA is degraded suggests an important but poorly understood role in DNA degradation. The abundant nature of Pdd1p in Tetrahymena and Spdd1p in Stylonychia suggests a structural role, leaving other proteins responsible for the initiation of the DNA degradation process. Different factors binding to the chromatin in different nuclei might have binding sites for Spdd1p, which itself could be a general structural factor that somehow facilitates specific DNA degradation. The Drosophila chromodomain protein Su(var)3–7 has been shown to be associated with heterochromatic regions of Drosophila polytene chromosomes, being part of a protein complex containing other proteins, including the chromodomain protein HP1 (Cléard et al., 1997). Preliminary cross-linking experiments revealed that Spdd1p is associated not only with DNA but also with at least two other proteins with sizes of ∼48 and 60 kDa. This suggests an association of a whole protein complex containing Spdd1p with DNA, although the DNA interaction seems to be weak (our unpublished results). Spdd1p and the 48- and 60-kDa proteins do not show a direct DNA binding activity in the Southwestern analysis (our unpublished results). We have not found core histones to be associated with Spdd1p as it was detected for Tetrahymena Pdd1p (Smothers et al., 1997b), but this association cannot be excluded, because we never applied sensitive methods such as radiolabeling of proteins.
The results of the TUNEL assay raise the question regarding whether similar enzymes are involved in apoptotic-like DNA degradation in different situations. It is not clear yet whether there are homologies between ciliate proteins and proteins involved in apoptotic DNA degradation in mammalian cells. We never obtained specific anti-Pdd1p signals by Western blot analysis with proteins from apoptotic hamster and human cells in cell culture, although a homologous protein still might be expressed in low amounts at certain time points (our unpublished results). Because the DNA fragmentation in Tetrahymena is Ca2+ and Zn2+ insensitive (Mpoke and Wolfe, 1996), the enzymes involved in this process may be different in ciliates from those involved in apoptosis in mammalian cells (for review, see Montague and Cidlowski, 1996; Walker and Sikorska, 1997). The regulation of DNA degradation in exconjugants and starved cells may differ, because the old macronucleus in exconjugants is completely resorbed, whereas only a partial DNA degradation, leading to a reduced metabolism, takes place in starved cells.
How far the modifications of Spdd1p play a role in these processes—possibly they are different in exconjugant and starved cells—is not known. On low percentage SDS gels, an electrophoretic heterogeneity of Tetrahymena Pdd1p in exconjugant nuclear extracts, caused by phosphorylation, could be eliminated by bacterial alkaline phosphatase (Madireddi et al., 1994). A detailed phosphorylation analysis has not yet been performed for Stylonychia. It seems likely that the complete identification and characterization of Spp1p-associated polypeptides and possibly other factors will allow further insight into the DNA degradation processes in the different nuclei during nuclear differentiation or reduced nutrient level and how these processes are regulated.
ACKNOWLEDGMENTS
We are very grateful to Sabine Feiler for the cultivation of the Stylonychia cells and to Sabine Feiler and Berthold Wilden for the preparation of nuclei. We also acknowledge Dr. Andreas Eger for valuable tips about cross-linking experiments, Dr. Franziska Jönsson for critically reading this manuscript, and Boris Pasek and Christian P. Fetzer for their help with the graphics. This work was supported by the Alfried Krupp von Bohlen and Hahlbach Foundation, the Fonds der Chemischen Industrie, and the Deutsche Forschungsgemeinschaft.
Abbreviations used:
- Pdd1p
programmed degradation protein 1
- Spdd1p
Stylonychia Pdd1p
- TUNEL
terminal deoxynucleotidyl transferase (TdT)-mediated dUTP-digoxygenin nick end labeling assay
REFERENCES
- Adl SM, Berger JD. Commitment to division in ciliate cell cycles. J Eukaryot Microbiol. 1996;43:77–86. doi: 10.1111/j.1550-7408.1996.tb04484.x. [DOI] [PubMed] [Google Scholar]
- Ammermann D, Steinbrück G, von Berger L, Hennig W. The development of the macronucleus in the ciliated protozoan Stylonychia mytilus. Chromosoma. 1974;45:401–429. doi: 10.1007/BF00283386. [DOI] [PubMed] [Google Scholar]
- Berger JD, Ching AS. Commitment to division in Paramecium: effect of nutrient level on the macronuclear DNA increment. Exp Cell Res. 1989;182:90–104. doi: 10.1016/0014-4827(89)90282-6. [DOI] [PubMed] [Google Scholar]
- Callebaut I, Courvalin J-C, Wormann HJ, Mornon J-P. Hydrophobic cluster analysis reveals a third chromodomain in the Tetrahymena Pdd1p protein of the chromo superfamily. Biochem Biophys Res Commun. 1997;235:103–107. doi: 10.1006/bbrc.1997.6748. [DOI] [PubMed] [Google Scholar]
- Cléard F, Delattre M, Spierer P. SU(VAR)3–7, a Drosophila heterochromatin-associated protein and companion of HP1 in the genomic silencing of position-effect variegation. EMBO J. 1997;16:5280–5288. doi: 10.1093/emboj/16.17.5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MC, Ward G, Herrick JG, Allis CD. Programmed nuclear death: apoptotic-like degradation of specific nuclei in conjugating Tetrahymena. Dev Biol. 1992;154:419–432. doi: 10.1016/0012-1606(92)90080-z. [DOI] [PubMed] [Google Scholar]
- Deak JC, Doerder FP. High frequency intragenic recombination during macronuclear development in Tetrahymena thermophila restores the wild-type SerH1 gene. Genetics. 1998;148:1109–1115. doi: 10.1093/genetics/148.3.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doak TG, Doerder FP, Jahn CL, Herrick G. A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common “D35E” motif. Proc Natl Acad Sci USA. 1994;91:942–946. doi: 10.1073/pnas.91.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin SCR. Heterochromatin and gene regulation in Drosophila. Curr Opin Genet Dev. 1996;6:193–202. doi: 10.1016/s0959-437x(96)80050-5. [DOI] [PubMed] [Google Scholar]
- Fanti L, Giovinazzo G, Berloco M, Pimpinelli S. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol Cell. 1998;2:527–538. doi: 10.1016/s1097-2765(00)80152-5. [DOI] [PubMed] [Google Scholar]
- Frels JS, Tebeau CM, Doktor SZ, Jahn CL. Differential replication and DNA elimination in the polytene chromosomes of Euplotes crassus. Mol Biol Cell. 1996;7:755–768. doi: 10.1091/mbc.7.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrieli Y, Sherman Y, Ben-Sasson SA. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grell KG. Protozoology. Berlin: Springer-Verlag; 1973. [Google Scholar]
- Huang H, Wiley EA, Lending CR, Allis CD. An HP1-like protein is missing from transcriptionally silent micronuclei of Tetrahymena. Proc Natl Acad Sci USA. 1998;95:13624–13629. doi: 10.1073/pnas.95.23.13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Smothers JF, Wiley EA, Allis CD. A nonessential HP1-like protein affects starvation-induced assembly of condensed chromatin and gene expression in macronuclei of Tetrahymena thermophila. Mol Cell Biol. 1999;19:3624–3634. doi: 10.1128/mcb.19.5.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaraczewski JW, Frels JS, Jahn CL. Developmentally regulated, low abundance Tec element transcripts in Euplotes crassus: implications for DNA elimination and transposition. Nucleic Acids Res. 1994;22:4535–4542. doi: 10.1093/nar/22.21.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaraczewski JW, Jahn CL. Elimination of Tec elements involves a novel excision process. Genes Dev. 1993;7:95–105. doi: 10.1101/gad.7.1.95. [DOI] [PubMed] [Google Scholar]
- Klobutcher LA, Jahn CL. Developmentally controlled genomic rearrangements in ciliated protozoa. Curr Opin Genet Dev. 1991;1:397–403. doi: 10.1016/s0959-437x(05)80306-5. [DOI] [PubMed] [Google Scholar]
- Klobutcher LA, Turner LR, LaPlante J. Circular forms of developmentally excised DNA in Euplotes crassus have a heteroduplex junction. Genes Dev. 1993;7:84–94. doi: 10.1101/gad.7.1.84. [DOI] [PubMed] [Google Scholar]
- Kloetzel JA. Compartmentalization of the developing macronucleus following conjugation in Stylonychia and Euplotes. J Cell Biol. 1970;47:395–407. doi: 10.1083/jcb.47.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Ling Z, Ghosh S, Jacobs ME, Klobutcher LA. Conjugation-specific genes in the ciliate Euplotes crassus: gene expression from the old macronucleus. J Eukaryot Microbiol. 1997;44:1–11. doi: 10.1111/j.1550-7408.1997.tb05682.x. [DOI] [PubMed] [Google Scholar]
- Lipps HJ, Eder C. Macronucleus structure and macronucleus development in hypotrichous ciliates. Int J Dev Biol. 1996;40:141–147. [PubMed] [Google Scholar]
- Madireddi MT, Coyne RS, Smothers JF, Mickey KM, Yao M-C, Allis CD. Pdd1p, a novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination in Tetrahymena. Cell. 1996;87:75–84. doi: 10.1016/s0092-8674(00)81324-0. [DOI] [PubMed] [Google Scholar]
- Madireddi MT, Davis MC, Allis CD. Identification of a novel polypeptide involved in the formation of DNA-containing vesicles during macronuclear development in Tetrahymena. Dev Biol. 1994;165:418–431. doi: 10.1006/dbio.1994.1264. [DOI] [PubMed] [Google Scholar]
- Maercker C, Stoll S, Rosenkranz K, Becker E-M, Lipps HJ. Characterization of a family of repetitive sequences that is eliminated late during macronuclear development of the hypotrichous ciliate Stylonychia lemnae. Dev Genet. 1997;21:201–211. doi: 10.1002/(SICI)1520-6408(1997)21:3<201::AID-DVG3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Martindale DW. A conjugation-specific gene (cnjC) from Tetrahymena encodes a protein homologous to yeast RNA polymerase subunits (RPB3, RPC40) and similar to a portion of the prokaryotic RNA polymerase alpha subunit (rpoA) Nucleic Acids Res. 1990;18:2953–2960. doi: 10.1093/nar/18.10.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer GF, Lipps HJ. Chromatin elimination in the hypotricous ciliate Stylonychia mytilus. Chromosoma. 1980;77:285–297. doi: 10.1007/BF00286054. [DOI] [PubMed] [Google Scholar]
- Montague JW, Cidlowski JA. Cellular catabolism in apoptosis: DNA degradation and endonuclease activation. Experientia. 1996;52:957–962. doi: 10.1007/BF01920104. [DOI] [PubMed] [Google Scholar]
- Mpoke S, Wolfe J. DNA digestion and chromatin condensation during nuclear death in Tetrahymena. Exp Cell Res. 1996;125:357–365. doi: 10.1006/excr.1996.0186. [DOI] [PubMed] [Google Scholar]
- Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–266. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smothers JF, Madireddi MT, Warner FD, Allis CD. Programmed DNA elimination and nucleolar biogenesis occur in distinct organelles during macronuclear development in Tetrahymena. J Eukaryot Microbiol. 1997a;44:79–88. doi: 10.1111/j.1550-7408.1997.tb05942.x. [DOI] [PubMed] [Google Scholar]
- Smothers JF, Mizzen CA, Tubbert MM, Cook RG, Allis CD. Pdd1p associates with germline-restricted chromatin and a second novel anlagen-enriched protein in developmentally programmed DNA elimination structures. Development. 1997b;124:4537–4545. doi: 10.1242/dev.124.22.4537. [DOI] [PubMed] [Google Scholar]
- Tausta SL, Turner LR, Buckley LK, Klobutcher LA. High fidelity developmental excision of Tec1 transposons and internal eliminated sequences in Euplotes crassus. Nucleic Acids Res. 1991;19:3229–3236. doi: 10.1093/nar/19.12.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakimoto BT. Beyond the nucleosome: epigenetic aspects of position-effect variegation in Drosophila. Cell. 1998;93:321–324. doi: 10.1016/s0092-8674(00)81159-9. [DOI] [PubMed] [Google Scholar]
- Walker PR, Sikorska M. New aspects of the mechanism of DNA fragmentation in apoptosis. Biochem Cell Biol. 1997;75:287–299. [PubMed] [Google Scholar]
- Wen J, Maercker C, Lipps HJ. Sequential excision of internal eliminated DNA sequences in the differentiating macronucleus of the hypotrichous ciliate Stylonychia lemnae. Nucleic Acids Res. 1996;24:4415–4419. doi: 10.1093/nar/24.22.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Doak TG, Herrick G. Developmental precise excision of Oxytricha trifallax telomere-bearing elements and formation of circles closed by a copy of the flanking target duplication. EMBO J. 1993;12:4593–4601. doi: 10.1002/j.1460-2075.1993.tb06148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]