Abstract
Purified Golgi membranes were mixed with cytosol and microtubules (MTs) and observed by video enhanced light microscopy. Initially, the membranes appeared as vesicles that moved along MTs. As time progressed, vesicles formed aggregates from which membrane tubules emerged, traveled along MTs, and eventually generated extensive reticular networks. Membrane motility required ATP, occurred mainly toward MT plus ends, and was inhibited almost completely by the H1 monoclonal antibody to kinesin heavy chain, 5′-adenylylimidodiphosphate, and 100 μM but not 20 μM vanadate. Motility was also blocked by GTPγS or AlF4− but was insensitive to AlCl3, NaF, staurosporin, or okadaic acid. The targets for GTPγS and AlF4− were evidently of cytosolic origin, did not include kinesin or MTs, and were insensitive to several probes for trimeric G proteins. Transport of Golgi membranes along MTs mediated by a kinesin has thus been reconstituted in vitro. The motility is regulated by one or more cytosolic GTPases but not by protein kinases or phosphatases that are inhibited by staurosporin or okadaic acid, respectively. The pertinent GTPases are likely to be small G proteins or possibly dynamin. The in vitro motility may correspond to Golgi-to-ER or Golgi-to-cell surface transport in vivo.
INTRODUCTION
The Golgi complex in interphase animal cells is often located near a site where the minus ends of microtubules (MTs)1 are clustered. In fibroblasts, for example, radially arranged MTs emanate from a perinuclear MT organizing center, where their minus ends are attached and near which the Golgi apparatus is centered (Kreis, 1990). By contrast, both MT minus ends and the Golgi reside near the apical surface of many polarized epithelial cells, including differentiated MDCK cells in culture (Bacallao et al., 1989), hepatocytes (Porter and Bonneville, 1973; Ihrke et al., 1993; Marks et al., 1994), and Sertoli cells (Redenbach and Boekelheide, 1994) in vivo.
The structural integrity and intracellular location of the Golgi reflect a balance between two opposing forms of tubulovesicular membrane flow along MTs. Outward flow involves transport intermediates that bud off of the Golgi and are delivered to the cell surface and endoplasmic reticulum (ER), among other locations. In contrast, transport intermediates that move inward from peripheral sites fuse with the Golgi when they reach the MT ends near which the Golgi complex is located. Hence, the maintenance and localization of an intact Golgi result from the coordinated action of a set of uniformly polarized MTs, motor proteins that move membranes along MTs toward and away from the Golgi, and factors that promote membrane vesiculation and fusion.
Several recent studies have shed light on the molecular mechanisms that underlie Golgi membrane dynamics. For example, methods have been developed for converting isolated Golgi stacks into small vesicles and tubules using mitotic cytosol (Misteli and Warren, 1994) and then reassembling the resulting Golgi fragments into cisterna and stacks using interphase cytosol (Rabouille et al., 1995) or buffer alone (Rabouille et al., 1995). These in vitro phenomena closely parallel the breakup and reformation of the Golgi, which occur respectively near the onset and completion of mitosis to ensure equal partitioning of Golgi membranes between daughter cells (Lucocq and Warren, 1987). Further analysis of these reconstituted systems indicated a requirement of COP-I proteins for production of mitotic Golgi fragments (Misteli and Warren, 1994) and the involvement of several other proteins, including NSF, α-SNAP, γ-SNAP, p115, p97, and syntaxin 5, in reformation of Golgi cisternae (Rabouille et al., 1995, 1998). A parallel set of studies has focused on the reformation of dispersed Golgi stacks and cisternae in perforated, ilimaquinone-treated cells. Here again, requirements for NSF, the SNAPs, and p97 were indicated for reassembly of Golgi cisternae (Acharya et al., 1995a,b). Furthermore, activation of a trimeric G protein was shown to trigger Golgi vesiculation in the absence of ilimaquinone, through release of free βγ subunit complex (Jamora et al., 1997).
The exact roles played by MTs and MT motor proteins in Golgi dynamics are also gradually being revealed. Evidently, membrane transport both toward and away from the Golgi normally takes place along MTs, even though transport may eventually become efficient in cells exposed to MT-depolymerizing drugs (Lippincott-Schwartz et al., 1990; Cole et al., 1996; Bloom and Goldstein, 1998). Recently, we found that the MT motor protein kinesin is present on intermediate compartment membranes that cycle constitutively between the ER and Golgi. Furthermore, we obtained evidence that in cells with radially arranged MTs emanating from a perinuclear MT organizing center, kinesin is the motor for Golgi-to-ER transport, but is inactive for ER-to-Golgi motility (Lippincott-Schwartz et al., 1995). Thus, it appears that the activity of kinesin is regulated during cyclic membrane transport between the ER and Golgi, and that ER-to-Golgi transport depends on another MT motor, probably dynein (Burkhardt et al., 1997; Presley et al., 1997), whose activity is also regulated.
To begin the task of determining how MT-based transport of secretory pathway membranes is regulated, we have reconstituted vigorous motility in vitro using Golgi membranes and cytosol isolated from rat liver and purified bovine brain tubulin assembled into MTs using Taxol. Motility in this reconstituted system occurs predominantly toward MT plus ends and is driven primarily by a kinesin. It is insensitive to staurosporin or okadaic acid but can be blocked almost completely by low concentrations of GTPγS or AlF4−. The molecular targets of GTPγS and AlF4− are of cytosolic origin, do not include conventional kinesin or MTs, and are insensitive to several probes for trimeric G proteins. These results imply that Golgi-derived membrane transport along MTs is regulated by at least one type of GTPase, such as a low-molecular-weight G protein or dynamin, but not by any of the protein kinases or phosphatases that are inhibited by staurosporin or okadaic acid, respectively. The regulatory mechanisms for Golgi transport along MTs must be distinct from the okadaic acid-stimulated mechanism which controls MT-based motility of ER membranes (Allan, 1995). The collective properties of the new motility system raise the possibility that it is an in vitro equivalent of transport from the Golgi to the ER or plasma membrane in vivo.
MATERIALS AND METHODS
Materials
GTPγS and 5′-adenylylimidodiphosphate (AMP-PNP) were purchased from Boehringer Mannheim (Indianapolis, IN). Taxol was generously supplied by Nancita R. Lomax (National Cancer Institute, Bethesda, MD). The monoclonal mouse antibody H1, which is specific for kinesin heavy chain (Pfister et al., 1989), was purified from ascites fluids using Affi-Gel protein A-agarose (Bio-Rad, Hercules, CA) according to the vendor’s instructions. The following polyclonal rabbit antibodies were kindly provided by colleagues, as indicated: anti-galactosyltransferase, Dr. Eric G. Berger (University of Zurich, Zurich, Switzerland) (Berger et al., 1987); anti-BiP, Dr. Linda M. Hendershot (St. Jude Children’s Research Hospital, Memphis, TN) (Hendershot et al., 1995); anti-transferrin receptor, Dr. Elizabeth A. Rutledge (University of Washington, Seattle, WA) and Dr. Caroline A. Enns (Oregon Health Science Center, Portland, OR) (Williams and Enns, 1991); anti-cathepsin D, Dr. William J. Brown (Cornell University, Ithaca, NY) (Park et al., 1991); and anti-rabkinesin-6, Dr. Bruno Goud (Institut Curie, Paris, France) (Echard et al., 1998). As a positive control for immunoblotting, Dr. Goud also provided cytosol isolated from HeLa cells that overexpressed rabkinesin-6 by transfection. HRP-labeled goat anti-mouse immunoglobulin G (IgG) and HRP-labeled goat anti-rabbit IgG were purchased from Jackson ImmunoResearch (West Grove, PA). Anti-HIPYR, a rabbit polyclonal anti-pan-kinesin antibody, was purchased from Babco (Richmond, CA). Purified, recombinant, full-length, his6-tagged Drosophila melanogaster kinesin heavy chain (Hancock and Howard, 1998) was kindly donated by Drs. William O. Hancock and Jonathan Howard (University of Washington). Luminol enhancer and stable peroxide solutions for chemiluminescent immunoblotting and Coomassie Plus protein assay reagent were purchased from Pierce (Rockford, IL). Immobilon-P membranes for Western blotting were obtained from Millipore (Bedford, MA). Diethylaminoethyl (DEAE)-Sephadex and DEAE-Toyopearl chromatography media were purchased from Pharmacia (Piscataway, NJ) and Supelco (Bellefonte, PA), respectively. Contrad 70 was obtained from Curtin Matheson (Houston, TX). Recombinant Go, Gs, and Gi3 (Lee et al., 1994) and β1γ2 (Iñiguez-Lluhi et al., 1992) subunits of trimeric G proteins were generously provided Drs. Hsin Chieh Lin and Alfred G. Gilman (University of Texas Southwestern Medical Center, Dallas, TX). 32P-NAD was purchased from Amersham (Arlington Heights, IL). Sigma (St. Louis, MO) was the source of all other biochemical reagents, including pertussis toxin (catalog number P0317) and cholera toxin A subunit (catalog number C8180).
Isolation of Cytosol, Crude Microsomes, and Golgi Membranes from Rat Liver
Freshly dissected rat liver was minced into small pieces with a razor blade and was then homogenized using an Omni 500 tissue grinder (Omni International, Gainesville, VA) at a ratio of 1 g of tissue/ml of acetate buffer (100 mM potassium acetate, 3 mM magnesium acetate, 5 mM EGTA, 1 mM DTT, 10 mM HEPES, pH 7.1) supplemented with a 10 μg/ml concentration of each of the following protease inhibitors: pepstatin A, leupeptin, and chymostatin. For preparation of microsomes or Golgi membranes, the buffer also contained 0.25 M sucrose. All subsequent steps were performed at 4°C.
Cytosol was prepared by centrifuging homogenates at 50,000 rpm (135,240 × gmax) for 30 min at 4°C in a Beckman Instruments (Fullerton, CA) 100.3 rotor and discarding the pellets and floating lipid layers. Golgi membranes and crude microsomes were purified from liver homogenates using a modified version of previously published procedures (Leelavathi et al., 1970; Allan and Vale, 1991). First, the homogenate was spun at 2500 rpm (907 × gmax) for 10 min at 4°C in a Sorvall RC-5C centrifuge using an SA-600 rotor. The floating fat layer and the pellet were then removed and discarded, yielding a suspension of crude microsomal membranes in cytosol. Microsomes were prepared by centrifuging the suspension at 36,000 rpm (150,668 × gmax) for 1 h at 4°C in a Beckman 45Ti rotor and resuspending the pellet in a small volume of buffer.
Golgi membranes were purified from the suspension of crude microsomes in cytosol by the following consecutive steps. Acetate buffer containing a 1 μg/ml concentration of each protease inhibitor plus variable sucrose concentrations (see below) was used throughout the procedure. 1) Centrifuge tubes for a Beckman 45Ti rotor were filled with 15 ml of membrane suspension layered on top of 40 ml of 1.4 M sucrose and were then spun at 36,000 rpm (150,668 × gmax) for 1 h at 4°C. 2) Material that collected at the 0.25/1.4 M sucrose interface was harvested and diluted into acetate buffer containing 1.25 M sucrose at a ratio of 1 ml of interface material/5 ml of 1.25 M sucrose. The refractive index of the resulting suspension was then adjusted to ηD = 1.3939 (equivalent to 1.255 M sucrose), as measured by a refractometer. 3) Centrifuge tubes for a Beckman SW 28 rotor were filled with the following sucrose step gradient: 10 ml of 1.4 M, 15 ml of membrane suspension at 1.255 M, 10 ml of 1.1 M, and 3 ml of 0.25 M. The tubes were then centrifuged at 22,000 rpm (87,275 × gmax) for 1.5 h. 4) Golgi membranes were collected from the 0.25/1.1 M sucrose interface, and protease inhibitors were added to a final concentration of 10 μg/ml each.
Cytosol, Golgi membranes, and microsomes were divided into small aliquots, snap frozen in liquid nitrogen, and stored at −80°C. Thawed aliquots of cytosol and Golgi membranes were used only on the day of thawing.
Purification of Tubulin
MT protein was purified from bovine brain cytosol by one to three cycles of GTP-dependent assembly at 37°C and cold-induced disassembly. Tubulin was then separated from MT-associated proteins by DEAE-Sephadex or DEAE-Toyopearl chromatography, divided into small aliquots, snap frozen in liquid nitrogen, and stored at −80°C (Bloom et al., 1988).
Isolation of Flagellar Axonemes from Chlamydomonas reinhardtii
C. reinhardtii were generously supplied by the laboratory of our departmental colleague, Dr. William Snell. Flagellae were isolated from dibucaine-treated cells and subsequently were demembranated using the nonionic detergent Nonidet P-40 (Witman, 1986). The membrane-free axonemes were then extracted with salt to remove flagellar dyneins (King et al., 1986) and stored at −20°C in Tris-KCl (50 mM KCl, 5 mM MgSO4, 0.5 mM EDTA, 20 mM Tris-Cl, pH 7.6) containing 50% glycerol.
Quantitative Immunoblotting
A Bio-Rad Mini-Protean II cell was used for both SDS-PAGE (Laemmli, 1970) and Western blotting (Towbin et al., 1979) onto Immobilon-P membranes. The originally described transfer buffer (Towbin et al., 1979) was modified by the inclusion of SDS to 0.1%, and samples that were blotted included crude microsomes and the Golgi membranes that were purified from them. Secondary antibodies were labeled with HRP, and immunoreactive proteins were visualized by a chemiluminescent assay using Luminol enhancer and stable peroxide solutions at 50% the concentration recommended by the vendor (Pierce).
To determine the degree of enrichment or deenrichment of each marker protein in the purified Golgi fractions, compared with the microsomes, the protein concentration of each was measured using a Coomassie dye binding assay (Bradford, 1976), and several volumes of each sample were loaded onto gels. After the blotting procedure was completed for each antibody, the chemiluminescent blots were scanned using a scanning laser Personal Densitometer (Molecular Dynamics, Sunnyvale, CA). For each immunoreactive protein, volumes of Golgi membranes and microsomes that yielded immunoreactive bands of equivalent integrated intensities were thereby determined. For each marker protein, comparison of the total protein contents of Golgi and microsome aliquots that contained equivalent immunoreactivity indicated the relative enrichment or depletion of the marker in the purified Golgi fraction.
Electron Microscopy
Purified Golgi membranes were pelleted by centrifugation and fixed in buffer (50 mM sodium cacodylate, pH 7.4) containing 2% ρ-formaldehyde plus 2% glutaraldehyde for 1 h and then in buffer containing 2% glutaraldehyde plus 5 mM CaCl2 for 1 h, followed by three rinses of 5 min each. Samples were then post-fixed for 1.5 h in 2% OsO4 plus 0.8% K3Fe(CN)6, rinsed three times in deionized water, stained en bloc for 1 h in 2% uranyl acetate, and washed three additional times in deinonized water. Finally, the fixed membranes were dehydrated through a graded ethanol series and embedded in 100% Epon after passage through a graded series of Epon in ethanol. Thin (silver) sections were examined and photographed using a JEOL 1200 electron microscope.
Membrane Motility Assays
All steps were performed at room temperature. Each motility assay was performed in an ∼20-μl specimen chamber formed by a pair of number 0 thickness washed coverslips separated by a pair of thin spacers (Brady et al., 1985). The coverslips were washed by the following procedure. 1) Sonicate coverslips in a 2% solution of Contrad 70 cleaning agent for 45 min in a Cole-Parmer (Niles, IL) ultrasonic cleaner. 2) Rinse 10 times in hot tap water. 3) Sonicate 30 min in hot tap water. 4) Rinse 10 times in deionized water. 5) Sonicate 30 min in deionized water. 6) Rinse several times in 100% ethanol. 7) Sonicate 30 min in 100% ethanol. 8) Store coverslips in a clean jar in 100% ethanol. 9) Immediately before use, spin coverslips at 1000 rpm in a clinical centrifuge to remove residual ethanol.
To begin each experiment, cytosol diluted in motility buffer (acetate buffer containing 150 mM sucrose) to 5–10 mg/ml was introduced into a specimen chamber. After a 10-min incubation, the chamber was flushed with motility buffer to remove unadsorbed protein, and MTs were then added to the chamber. The MTs were polymerized from 10–20 μM (1–2 mg/ml) purified tubulin and 50% equimolar Taxol for 45 min and were typically longer than 20 μm. MTs became immobilized on the upper and lower surfaces of the specimen chamber by binding to adsorbed cytosolic proteins during a 10- to 45-min incubation period. Unattached MTs were then removed by flushing the chamber with additional motility buffer. Finally, a mixture of Golgi membranes, cytosol, and an ATP-regenerating system (5 mM ATP, 37.5 mM creatine phosphate, 0.2 U/μl creatine phosphokinase) was added to the chamber. Typically, both the membranes and the cytosol were diluted 10-fold into motility buffer to yield final protein concentrations of ∼0.25 and ∼5 mg/ml, respectively.
Immediately after the final components of the motility system were placed in the specimen chamber (Brady et al., 1985), the chamber was mounted on either a Zeiss (Thornwood, NY) IM-35 microscope fitted with a 63×, 1.4 numerical aperture (NA) plan-apochromatic objective or a Zeiss Axiomat microscope fitted with a 100×, 1.3 NA plan-apochromatic objective. Illumination for both microscopes was provided by a 100-W mercury arc lamp connected to a 1.4 NA condenser by a fiber optic light scrambler. Differential interference contrast images were collected and processed for noise subtraction and contrast enhancement using either of the following Hamamatsu (Bridgewater, NJ) video systems: a Chalnicon camera head and C1966 AVEC image processor or a C5985 cooled, charge-coupled device and Argus-20 image processor. Video sequences were recorded in real time using a Panasonic Super-VHS videocassette recorder. Velocities of motile membrane vesicles or tubules were made using a Hamamatsu C2117 Videomanipulator. The public domain program NIH Image (http://rsb.info.nih.gov/nihimage/) was used to import recorded video sequences into an Apple (Cupertino, CA) Power Macintosh computer equipped with either a built-in 24-bit digitizer or an 8-bit LG-3 frame grabber card (Scion, Frederick, MD) and to generate QuickTime movies that are played back at real-time rates.
Membrane tubule formation was quantitated as follows. First, recorded video frames were imported into a Power Macintosh computer as described in the previous paragraph. Next, membrane tubule lengths were measured by either of two procedures. 1) Captured images were printed. A ruler was then used to measure the sum of the lengths (in millimeters) of all tubules within an individual print, after which the sum was divided by the length (in millimeters) of a 5-μm scale bar that was superimposed on the printed micrograph. 2) Alternatively, NIH Image software was used to measure tubule lengths directly on images displayed on the computer monitor.
Time course data, as in Figure 5, were obtained by applying this procedure to images captured from multiple time points of a single field of view of an individual experiment and plotting total tubule length versus time. To quantitate the effects on tubule formation of experimental additives, such as GTPγS ± GTP, all samples that were mutually compared were prepared simultaneously and observed and recorded in parallel using the same aliquots of cytosol, membranes, and MTs. The total length of membrane tubules formed in five randomly chosen fields of view were then measured in each sample 1–2 h after the start of an experiment. In any single experiment, the total tubule length for the control (no additive) sample was defined as 100%. Finally, the results were plotted as bar graphs, as in Figures 8 and 9.
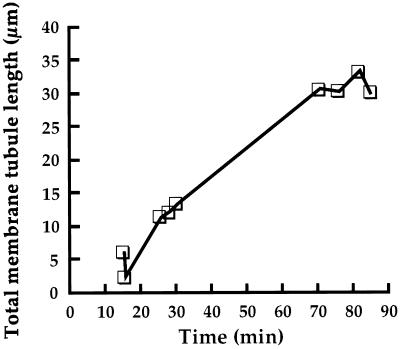
Figure 5.
Kinetics of membrane tubule elongation. The sum of the lengths of all membrane tubules present in a single field of view was measured at various time points of a control experiment, and the lengths were then plotted versus time.
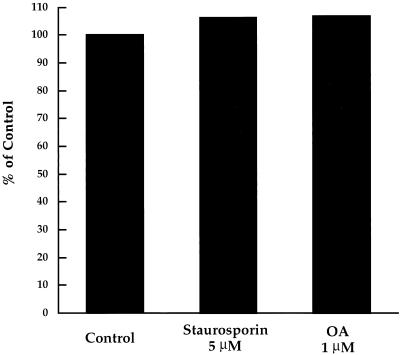
Figure 8.
Motility is not affected by inhibitors of protein kinases or phosphatases. Motility assays were performed using samples containing no additives (Control), 5 μM staurosporin, or 1 μM okadaic acid (OA). After 60–75 min, the extent of membrane tubule formation was measured as decribed in MATERIALS AND METHODS. Each compound was compared with a control in a minimum of three separate experiments. We consistently observed that membrane tubule formation in samples containing staurosporin or OA, which inhibit several protein kinases or phosphatases, respectively, were nearly indistinguishable from controls. Illustrated here are results from one representative experiment.
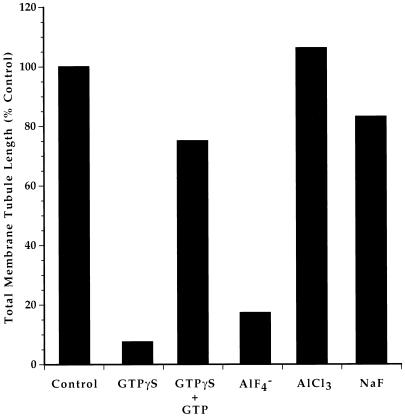
Figure 9.
Motility is potently inhibited by GTPγS or AlF4−. Motility assays were performed using samples containing no additives (Control), 1 μM GTPγS ± 100 μM GTP, AlF4− (20 μM AlCl3 + 3 mM NaF), 20 μM AlCl3, or 3 mM NaF. After 60–90 min, the extent of membrane tubule formation was measured as decribed in MATERIALS AND METHODS. Each experimental condition was compared with a control in a minimum of three separate experiments. We consistently observed that membrane tubule formation was nearly abolished by GTPγS or AlF4−, that the effects of GTPγS were minimized by addition of a 100-fold molar excess of GTP, and that neither AlCl3 nor NaF alone was inhibitory. Illustrated here are merged results from two representative experiments, one of which examined GTPγS ± GTP and the other of which tested AlF4−, AlCl3, and NaF. GTPγS and AlF4− also nearly abolished motility of small Golgi fragments at early stages of the experiment, but neither agent prevented formation of membrane aggregates.
Perturbations of Trimeric G Proteins
Samples containing standard concentrations of cytosol, Golgi membranes, and components of the ATP-regenerating system (see Membrane Motility Assays) were mixed with 25 μM NAD plus 1 μg/ml pertussis toxin or cholera toxin for 1 h at 37°C or 4 h at room temperature. The samples were then added to microscope chambers containing preadsorbed MTs, and membrane motility was assayed as described above. To verify that the toxins were enzymatically active, Golgi membranes were incubated with 32P-NAD and cholera toxin or pertussis toxin for 1–4 h at 0°C or 7.5–60 min at 20 or 30°C. Final protein concentrations were 1.76 mg/ml for the proteins and 0.8 μg/ml for the toxins. Radiolabeled proteins were observed by SDS-PAGE autoradiography at all incubation times and temperatures, indicating that the toxins were, indeed, active.
Membrane motility assays were also performed in the presence of recombinant Gα or βγ subunits (see Materials). The Gα subunits were initially saturated with GTPγS or GDP, after which a desalting column was used to separate free nucleotide from complexes of Gα–GTPγS or Gα–GDP. Immediately before motility assays began, Gα-guanine nucleotide complexes or free βγ subunits were added directly to samples containing standard amounts of cytosol, Golgi membranes, and components of the ATP-regenerating system.
MT Gliding Assays
Unless stated otherwise, all steps were performed at room temperature. A specimen chamber (Brady et al., 1985) was exposed sequentially for 5 min each to 1) BRB80 buffer (80 mM piperazine-N,N′-bis(2-ethanesulfonic acid), pH 6.8, 1 mM MgCl2, 1 mM GTP) supplemented with 0.2 mg/ml casein and 1 mM ATP (BBR80CA); 2) recombinant kinesin heavy chain (see Materials) in BRB80CA; and 3) MTs in BRB80CA supplemented with 10 μM Taxol. The MTs were assembled in advance in BRB80 by incubating 20 μl of 50 μM (5 mg/ml) purified bovine brain tubulin with 1 μM Taxol for 5 min at 37°C and then adding an additional 20 μl of 50 μM tubulin and incubating at 37°C for 10 more minutes (Paschal and Vallee, 1993). MTs prepared in this manner typically ranged in length from 2 to 8 μm and were diluted to a final concentration of 5 μM assembled tubulin. Kinesin-mediated MT gliding (Vale et al., 1985) was observed and recorded as described above for membrane motility assays. These experiments were performed using the Zeiss Axiomat microscope and Hamamatsu Argus-20 image processor and, for measurements of MT gliding, velocities, the Hamamatsu C2117 Videomanipulator.
RESULTS
Characterization of Purified Golgi Membranes
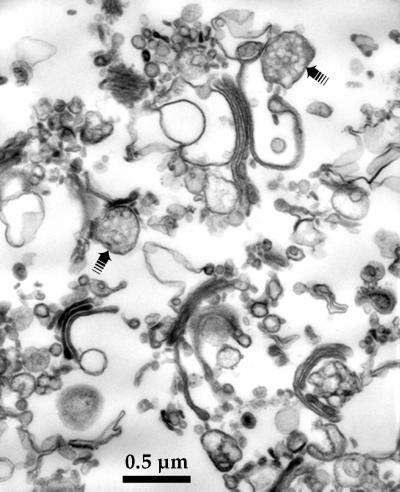
The purified membranes that were used for motility assays were extensively characterized ultrastructurally and biochemically. As shown by thin-section electron microscopy (EM) in Figure 1, membranes with the typical appearance of Golgi cisternae and stacks were abundant in purified membrane fractions and were most often seen as cross-sectionally cut profiles. The Golgi cisternae were variable in size, but most were <1 μm long. The purified membrane fractions also contained numerous apparent vesicles and short tubules that ranged in diameter from ∼50 to 100 nm and lesser numbers of lipoprotein particles, which are often found at the margins of Golgi cisternae in liver (Porter and Bonneville, 1973).
Figure 1.
Ultrastructural appearance of purified membranes. Pelleted membranes were processed for thin-section EM. Note the abundance of Golgi cisternae and stacks and apparent vesicles and short membrane tubules. Also visible in this micrograph are a few examples of lipoprotein particles (➟), which are characteristically associated with the margins of liver Golgi cisternae (Porter and Bonneville, 1973). Bar, 200 nm.
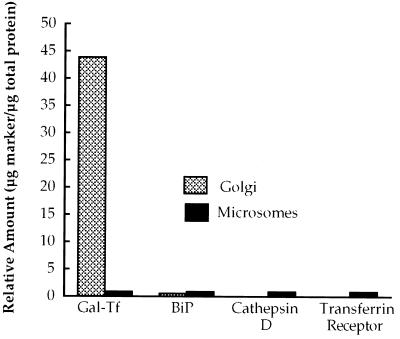
To gauge the extent of contamination of Golgi-enriched fractions by non-Golgi membranes, both purified fractions and the crude microsomes from which they were obtained were analyzed by quantitative immunoblotting, as described in MATERIALS AND METHODS. Representative results are summarized in Figure 2. Compared with the microsomes, the Golgi membranes were enriched >40-fold for the Golgi enzyme galactosyltransferase and were deenriched by ∼50% for the ER marker BiP. When assayed for cathepsin D, the microsomes were found to contain two mature lysosomal forms of the enzyme, as well as a higher-molecular-weight precursor. In contrast, only the prelysosomal form of cathepsin D was present in the Golgi fraction. The transferrin receptor, a marker for plasma membrane and early endosomes, was detectable in the microsomes but not in the purified Golgi fraction. Based on the combined EM and immunoblotting results, we concluded that the purified membrane fraction was highly enriched in Golgi stacks, was minimally contaminated by ER, and did not contain detectable levels of lysosomal, endosomal, or plasma membrane markers.
Figure 2.
The purified membranes are highly enriched for a Golgi marker enzyme and deenriched for marker proteins of the ER, lysosomes, endosomes, and plasma membrane. Golgi-enriched membranes and the crude microsomes from which they were purified were analyzed by quantitative immunoblotting, as described in MATERIALS AND METHODS. Compared with the microsomes, the purified membranes were enriched ∼44-fold for the Golgi enzyme galactosyltransferase and were deenriched by ∼50% for the ER luminal protein BiP. Both the mature form of cathepsin D, a lysosomal enzyme, and the transferrin receptor, which is a marker for endosomes and the plasma membrane, were readily detectable in the microsomes but could not be detected in the purified membrane fraction.
Motility of Purified Golgi Membranes Along MTs
Video-enhanced differential interference contrast microscopy (Allen et al., 1981) was used to determine whether the purified Golgi membranes were capable of moving along MTs in the presence of rat liver cytosol and an ATP-regenerating system. Membranes and MTs that were freely suspended throughout the specimen chamber underwent constant and rapid Brownian motion and thus were impossible to keep in focus and analyze carefully. Fortunately, MTs and associated membranes became loosely bound to the inner surface of the lower coverslip and remained in focus near that plane for extended periods. Accordingly, most observations were made at or slightly above the lower surface of the specimen chamber.
When initially observed, most Golgi membranes appeared as small, vesicle-like structures. Within minutes of being placed in a specimen chamber, vesicles were seen to move vigorously along MTs (Figure 3, top). Among the vesicles that were visible near the lower surface of the chamber, the proportion that were motile varied among different experiments but reached a level as high as ∼50%. During continuous movement, vesicle velocities were consistently 1.4–1.5 μm/s. Individual vesicles commonly moved without obvious interruption for several micrometers, but discontinuous excursions of variable length were also often seen. Motility was predominantly unidirectional, but on rare occasions, vesicles were observed to change direction while moving along a single MT.
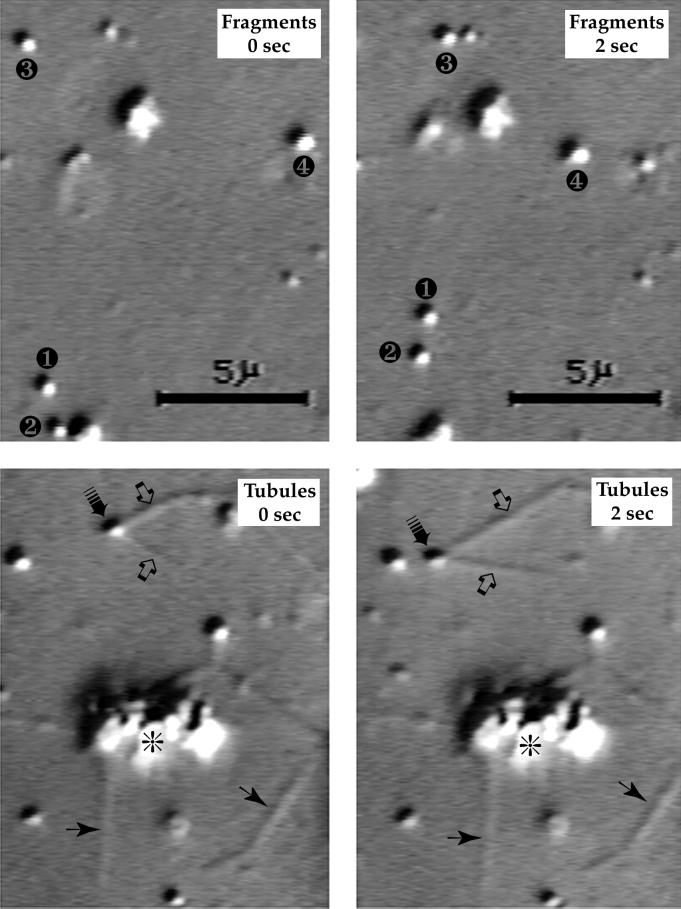
Figure 3.
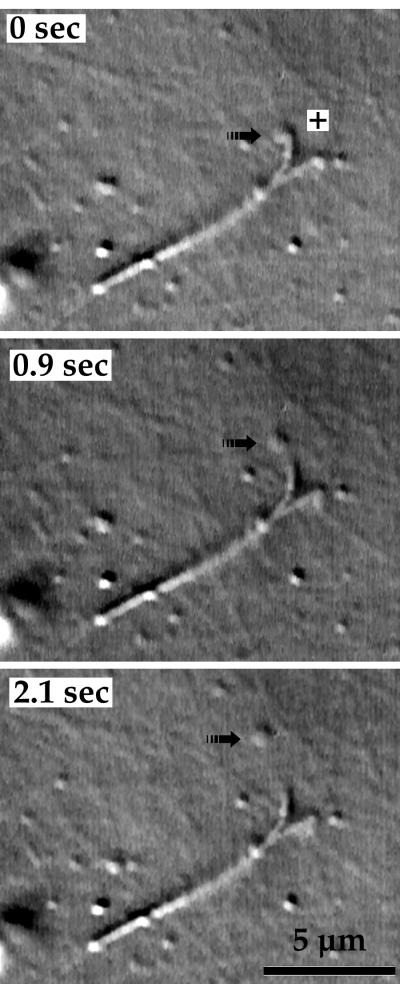
The purified Golgi membranes move along MTs in vitro. Top, small Golgi fragments are illustrated in two video frames captured at a 2-s interval during the early stage of a control assay. Four of the fragments in this field of view moved. Their instantaneous velocities were, 1.4–1.5 μm/s, but because not all of them were continuously motile during the time span shown here, their average velocities were generally slower (➊, 1.37; ➋, 1.29; ➌, 1.13; and ➍, 1.45 μm/s). Bottom, at a later stage of a different control assay, fragments of Golgi membranes merged to form large aggregates (*), out of which membrane tubules grew. As shown in this sequence of two video frames captured at a 2-s interval, a motile site (➟) on one sharply bent tubule (➩), which was attached to an aggregate outside the field of view, moved along an MT for 2.74 μm at an average velocity of 1.37 μm/s. Additional membrane tubules are also highlighted (➛). The 5-μm bars apply to all four images. MTs are only faintly visible in these micrographs because they were below the planes of focus.
Within 20–30 min after the start of typical control experiments, membranous structures much larger than the vesicle-sized Golgi fragments became abundant throughout the specimen chambers. These variably shaped structures, the longest dimensions of which occasionally exceeded 10 μm, were not commonly seen at any focal plane immediately after samples were introduced into the chambers and appeared regardless of whether MTs were also present. They thus represented aggregates of the much smaller Golgi fragments and their formation occurred in a time-dependent manner. The membrane aggregates formed independently of MT-based motility, because agents that inhibited motility did not necessarily prevent vesicle aggregation (see Figure 6). Although we did not conduct a detailed characterization of the aggregation process and do not know whether it included membrane fusion, the process was at least superficially reminiscent of the assembly of Golgi cisternae and stacks from isolated mitotic Golgi fragments placed in interphase cytosol (Rabouille et al., 1995) or buffer (Rabouille et al., 1995).
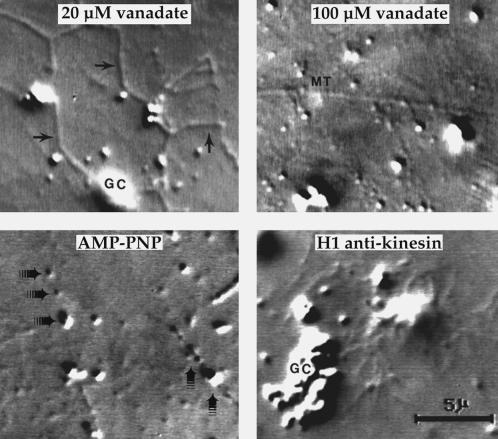
Figure 6.
Motility is inhibited by vanadate, AMP-PNP, or a monoclonal anti-kinesin. Video frames from the late stages of experiments are illustrated here. Membrane tubulation along MTs was inhibited almost completely by 100 μM vanadate, 5 mM AMP-PNP (equimolar to the exogenous ATP), or 0.3 mg/ml purified H1, a monoclonal antibody to kinesin heavy chain (Hirokawa et al., 1989; Pfister et al., 1989). Transport of small Golgi fragments at early stages of experiments was also abolished by all three agents, but 20 μM vanadate had no effect on the motility of either small Golgi fragments or membrane tubules (➛), as shown here. Aggregation of Golgi fragments into larger complexes (GC) was inhibited by 100 μM vanadate and 5 mM AMP-PNP, perhaps because they interfered with ATPases, such as NSF and p97, which are required for fusion of Golgi fragments (Acharya et al., 1995a; Rabouille et al., 1995). In contrast, H1 anti-kinesin did not prevent membrane aggregation, even though it inhibited the motility of small Golgi fragments and elongation of membrane tubules. Note the accumulation of small Golgi fragments along MTs in the presence of AMP-PNP (➟). Bar, 5 μm.
The membrane aggregates did not move along MTs, presumably because their large surface areas ensured that they became attached to the underlying glass surface. Occasionally, however, a localized region on the surface of a membrane aggregate became bound to an MT, moved outward along the MT, and thus formed a membrane tubule that steadily lengthened while remaining attached to the aggregate (Figure 3, bottom). Membrane tubules elongated at 1.4–1.5 μm/s during uninterrupted motility, the same velocity observed for motility of small Golgi fragments along MTs. Formation and elongation of membrane tubules proceeded for >1 h after the start of typical experiments. Moving tubules commonly switched direction as they jumped from MT to MT and often appeared to merge with other tubules whose paths they had crossed. In addition, new membrane tubules frequently grew out of preexisting tubules. The net result of this action was the formation of a reticular network of membrane tubules that occupied large expanses of the specimen chamber (Figure 4).
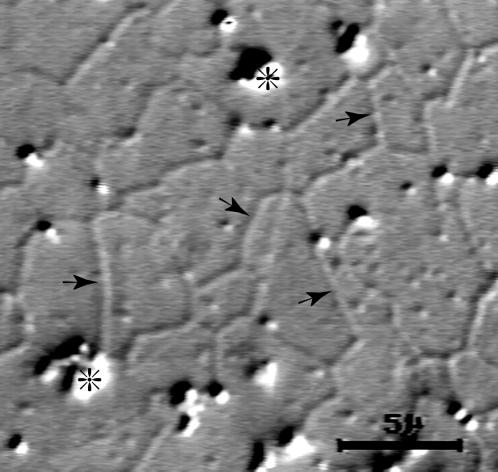
Figure 4.
Membrane tubule networks. Shown here is a stable network of Golgi membrane tubules (➛) and aggregates (*) photographed >4 h after the start of a control assay. Bar, 5 μm.
During the scores of hours that membrane motility was directly observed, MTs were never seen to glide along glass coverslips. Thus, it is unlikely that any of the Golgi motility can be explained by motile MTs simply dragging along membranes that were bound to them in a rigor manner. Instead, most or all of the Golgi motility that was detected must have been due to membranes being transported along the surfaces of MTs that were stationary relative to their underlying substrates.
Motility of Golgi membranes along MTs was analyzed quantitatively by measuring the total lengths of membrane tubules in an individual field of view at several time points during an experiment. Results of such an analysis from one typical experiment are illustrated in Figure 5. Network formation was first evident at ∼15 min and expanded at a roughly linear rate for another 45 min. In the example shown in Figure 5, ∼35 μm of total membrane tubule length was formed within a single 36.3- × 36.3-μm field of view within 1 h of the start of the experiment.
A Kinesin Is the Predominant Motor for Motility
To identify the MT motor(s) responsible for Golgi membrane motility in the reconstituted system, motility was studied in the presence of several potential inhibitors, and the directionality of transport relative to MT polarity was analyzed. In the presence of a starting concentration of 5 mM exogenous ATP, network formation (Figure 6) and vesicle transport (our unpublished results) were almost completely prevented by 5 mM AMP-PNP, or by 100 μM, but not 20 μM vanadate. These effects of AMP-PNP and vanadate are consistent with a kinesin but not a dynein, being the principal motor for motility (Kuznetsov and Gelfand, 1986; Paschal and Vallee, 1987; Porter et al., 1987; Shpetner et al., 1988; Cohn et al., 1989; Wagner et al., 1989). Further implicating a kinesin was the observation that purified H1, an IgG1 monoclonal antibody to kinesin heavy chain (Pfister et al., 1989), inhibited vesicle motility (our unpublished results) and network formation (Figure 6), as well. Concentrations of H1 as low as 0.3 mg/ml were maximally inhibitory, whereas a control monoclonal antibody had no effects at concentrations as high as 1 mg/ml. It is noteworthy that H1 also potently inhibited Golgi-to-ER but not ER-to-Golgi membrane transport when microinjected into live cultured cells (Lippincott-Schwartz et al., 1995).
If a kinesin were responsible for most Golgi membrane motility, transport should have occurred most frequently toward MT plus ends, the direction in which most kinesins, but no known dyneins move their cargo along MTs (Hirokawa, 1998). To assay the direction of transport in the in vitro system, we analyzed Golgi membranes moving along MTs growing out of demembranated, salt-extracted axonemes isolated from C. reinhardtii. The plus and minus ends of the axonemes are morphologically distinct (Paschal and Vallee, 1987), allowing unambiguous determination of the direction in which Golgi membranes moved (Figure 7).
Figure 7.
Motility occurs mainly toward MT plus ends. Shown here is a small Golgi fragment (➟) moving along an MT that grew out of the frayed or plus (+) end of a detergent-extracted, salt-washed axoneme isolated from C. reinhardtii. Note that the Golgi fragment moved toward the MT plus end. Of a total of 16 motile membranes whose directionality was unambiguously determined in this manner, 13 moved exclusively toward MT plus ends, 1 moved solely toward a MT minus end, and 2 moved bidirectionally.
A total of 16 membranes were observed to move along polarity marked MTs. Thirteen of the membranes moved exclusively in the MT plus end direction, whereas only one moved solely toward a MT minus end. The other two membranes initially moved in one direction, then stopped, and finally resumed moving in the opposite direction. In each of the two cases in which bidirectional motility was observed, all motility appeared to occur along a single MT, because the refractility of the transport fiber was as weak as the refractility of isolated, individual MTs that were commonly seen throughout the specimen chamber. Thus, of a total of 18 motile events involving 16 membranes, 15 events were toward an MT plus end and 3 were toward an MT minus end.
Simple inspection of these data implies that a MT plus end-directed motor was predominant. Nevertheless, χ2 analysis was used to test this idea formally. Based on the hypothesis that plus and minus end–directed motility were equally likely, the χ2 value for the data was calculated to equal 8.00, and at 1 df the probability that fifteen 15 of 18 motile events occurred toward an MT plus end is <0.005. The hypothesis that motility toward MT plus and minus ends was equally probable is thus very unlikely to be correct. Instead, the directionality results (Figure 7) represent statistically significant evidence that the preferred direction of Golgi membrane motility was toward MT plus ends. This conclusion is fully consistent with pharmacological data (Figure 6) that implicated a member of the kinesin superfamily as being responsible for most Golgi membrane transport in the in vitro motility system. In light of the fact that the H1 monoclonal antibody to conventional kinesin heavy chain (Pfister et al., 1989) potently inhibited motility (Figure 6), conventional kinesin is a strong candidate for the motor in question.
GTPγS and AlF4− Inhibit Motility
Motility in the in vitro system closely resembled intracellular Golgi-to-ER transport, which we had shown earlier to be inhibited by monoclonal H1 anti-kinesin and to be regulated by a mechanism that remains to be established (Lippincott-Schwartz et al., 1995). To attempt to shed light on the in vivo regulatory mechanism, various probes for potential regulatory factors were introduced into the in vitro system. Motility was not affected by 5 μM staurosporin (Tamaoki et al., 1986) or 1 μM okadaic acid (Cohen et al., 1989), broad-spectrum inhibitors of serine/threonine protein kinases or phosphatases, respectively (Figure 8). The biological activities of the staurosporin and okadaic acid were verified by phosphorylation assays for the axonal MT-associated protein τ in bovine brain cytosol and in primary mouse brain cell cultures (our unpublished results).
In contrast to staurosporin and okadaic acid, two compounds that bind to many GTP-binding proteins, GTPγS and AlF4−, dramatically inhibited transport of both vesicles and membrane tubules. In the presence of 1 μM GTPγS, vesicle aggregation was unimpaired, but vesicle transport along MTs was rarely observed, and the formation of tubular membrane networks was inhibited by >90% (Figure 9). Less potent but noticeable inhibition was observed at GTPγS concentrations as low as 200 nm (our unpublished results). Network formation was inhibited by only 25% in the presence of 1 μM GTPγS plus 100 μM GTP (Figure 9), demonstrating that GTPγS acts in this system as a poorly hydrolyzable substrate for one or more pertinent regulatory GTPases. The effects of GTPγS were mimicked by AlF4− (Figure 9), which activates both trimeric (Gilman, 1987) and low-molecular-weight (Mittal et al., 1996; Reza et al., 1997; Hoffman et al., 1998) G proteins. AlF4− was formed by addition of AlCl3 and NaF to final concentrations of 20 μM and 3 mM, respectively. When used alone at those concentrations, neither AlCl3 nor NaF alone had any demonstrable effect on motility (Figure 9).
The Targets of GTPγS and AlF4− Are Cytosolic and Include Neither Kinesin nor MTs
The relevant targets of GTPγS and AlF4− could have been membrane associated or cytosolic and could have included either of the two most basic components of the motile machinery, the kinesin or the MTs. To test whether the targets were membrane associated, purified Golgi membranes were incubated in the absence or presence of rat liver cytosol at 4°C for as long as 30 min with GTPγS at a concentration as high as 10 μM or with 20 μM AlF4− (20 μM AlCl3 plus 3 mM NaF). The membranes were then centrifuged, resuspended in buffer lacking GTPγS and AlF4−, and tested for motile capacity. The rationale for including cytosol in some samples was the possibility that GTPγS or AlF4− acted by binding to a cytosolic protein, which as a result became tightly associated with the membranes. A well-characterized example of such a phenomenon is the GTPγS-stimulated tight binding of cytosolic ARF and COP I to Golgi membranes (Kreis et al., 1995). In the present case, however, transient exposure of Golgi membranes to GTPγS or AlF4− in either the absence or presence of cytosol had no detectable effect of the ability of the membranes to move along MTs and to form reticular networks (our unpublished results). Taken at face value, these results imply that the inhibiton of motility by GTPγS and AlF4− did not involve binding of either compound to an integral membrane protein or to a protein that is soluble in a GDP-bound state but tightly associated with membranes when bound to GTPγS (or GTP). By process of elimination, the results also imply that the targets of GTPγS and AlF4− were cytosolic proteins that associated with membranes either weakly or not at all.
The identification of cytosol as the origin of the GTPγS-sensitive and AlF4−-sensitive factors raised the possibility that a kinesin or MTs were the relevant factors. Although ATP is its preferred nucleotide substrate, conventional kinesin can hydrolyze GTP (Kuznetsov and Gelfand, 1986) and couple the free energy released by hydrolysis to force generation (Porter et al., 1987). Likewise, tubulin has long been known as a GTPase that polymerizes into MTs by a GTP-stimulated mechanism, hydrolyzes GTP after the assembly step, and promotes MT lability when present in the polymer in a GDP-bound state (Gelfand and Bershadsky, 1991). Even though MTs were supplied to our in vitro motility system as preassembled bovine brain tubulin, the tubulin was of cytosolic origin and thus qualifies as a potential target of GTPγS or AlF4−.
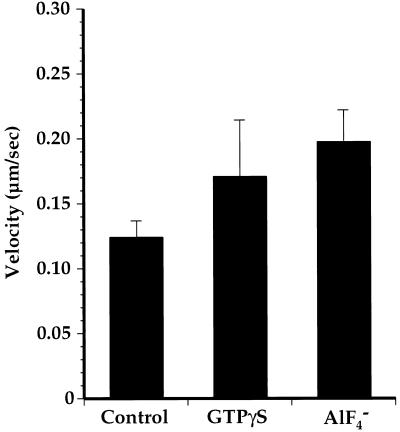
An MT gliding assay (Vale et al., 1985) was used to test the hypothesis that GTPγS or AlF4− inhibited Golgi membrane motility by directly interfering with a kinesin or MTs. The only proteins present were recombinant D. melanogaster kinesin heavy chain (Hancock and Howard, 1998), which was used to coat glass coverslips, polymerized bovine brain tubulin, and casein, which was used for blocking protein binding sites on the inner glass surface of the specimen chambers. In the absence of GTPγS or AlF4−, nearly all MTs in contact with the substrata moved at a velocity of 0.124 ± 0.013 μm/s. When 1 μM GTPγS or 20 μM AlF4− were present, transport velocities were 0.170 ± 0.042 and 0.197 ± 0.024 μm/s, respectively, and again, nearly all MTs moved (Figure 10). Although these velocities were slower than those that have been reported for the same recombinant kinesin (Hancock and Howard, 1998), it is noteworthy that GTPγS and AlF4− did not inhibit motility but instead may have stimulated the velocity of MT transport to a modest degree. We therefore conclude that neither conventional kinesin nor MTs were targets of GTPγS or AlF4− in the Golgi membrane motility assays.
Figure 10.
GTPγS and AlF4− do not inhibit MT gliding mediated by conventional kinesin. Video-enhanced light microscopy was used to assay MT gliding in the presence of 1 mM ATP on glass coverslips coated with recombinant conventional kinesin heavy chain. One micromolar ATP was present in all samples; controls contained no additions; and final concentrations of GTPγS and AlF4− were 1 and 20 μM, respectively. Nearly all MTs in all samples moved, and shown here are the mean velocities ± SDs for five velocity measurements for each condition. Note that motility was not inhibited by either GTPγS or AlF4−.
Probes for Trimeric G Proteins Do Not Inhibit Membrane Motility
In light of reports that several subunit polypeptides of trimeric G proteins, including Gαi2 (Montmayeur and Borrelli, 1994), Gαi3 (Ercolani et al., 1990; Wilson et al., 1994; Denker et al., 1996), Gαq/11 (Denker et al., 1996), and Gαs (Denker et al., 1996), are localized on the Golgi, two sets of experiments were performed to assess whether a trimeric G protein might be a pertinent target of GTPγS or AlF4−. The first set made use of bacterial toxins that catalyze transfer of ADP-ribose from NAD to Gα subunits, which thereby become locked in either an activate or inactive state. Neither cholera toxin, which activates Gαs (Gill and Meren, 1978), nor pertussis toxin, which inactivates Gαi isoforms and G0 (Moss et al., 1983), had any detectable effect on membrane motility (Table 1). It is also noteworthy that pertussis toxin treatment did not confer subsequent protection against GTPγS, implying that activation of G0 or a Gαi by GTPγS does not explain the inhibition of membrane motility by GTPγS (our unpublished results).
Table 1.
Membrane motility assayed in the presence of specific probes for trimeric G poteins
| Probe | Level of motility |
|---|---|
| Cholera toxin | Equal to control |
| Pertussis toxin | Equal to control |
| Gα0-GTPγS | Equal to control |
| Gαi3-GTPγS | Equal to control |
| Gαs-GTPγS | Equal to control |
| βγ subunits | Equal to control |
The second set of experiments directed at trimeric G proteins was based on the fact that GTPγS and AlF4− exert dual effects on those proteins: activation of Gα and generation of free Gα and βγ subunits (Gilman, 1987). We reasoned that if the inhibitory factor for membrane motility in GTPγS-containing samples was either a Gα–GTPγS complex or free βγ, it should be possible to prevent motility by simply adding the appropriate trimeric G protein subunit to the complete motility system. An identical stratgey was recently used to demonstrate a role for trimeric G proteins in regulating Golgi structure in vitro (Jamora et al., 1997). To test this hypothesis, stable complexes (Gilman, 1987) of GTPγS bound to Gα0, Gαs, or Gαi3 (see Perturbations of Trimeric G Proteins) were added to the motility system to final concentrations as high as 440 nM, or free βγ was added to 1 μM. In all cases, motility was equivalent to that observed for controls. The most straightforward interpretation of these results is that the inhibitory effects of GTPγS on membrane motility were not due to generation of free βγ or complexes of GTPγS bound to any of the Gα subunits that were tested.
DISCUSSION
In a previous report, we presented evidence that kinesin is localized on membranes that move bidirectionally along MTs between the ER and the Golgi complex but appears to be active only for the Golgi-to-ER arm of the motility cycle (Lippincott-Schwartz et al., 1995). The cell therefore regulates the membrane transport activities of both kinesin and another MT motor protein, which evidently is a dynein (Burkhardt et al., 1997; Presley et al., 1997) and is responsible for ER-to-Golgi transport. To attempt to uncover mechanisms by which MT-based membrane transport in the secretory pathway is regulated, we have now developed an in vitro system in which purified Golgi membranes move vigorously along MTs. Individual bursts of motility lasted from less than a second to several seconds and involved small, vesicle-like Golgi fragments, as well as membrane tubules that emanated from large Golgi aggregates or other tubules (Figure 3). Membrane tubules gradually intersected with one another to form stable reticular networks (Figure 4), and quantitative estimates of total motility were made by summing the lengths of all membrane tubules in randomly sampled fields of view (Figures 5, 8, and 9).
Several results consistently point to a kinesin as being the motor for most of the observed transport. The most compelling evidence is that motility occurred predominantly toward MT plus ends (Figure 7), the direction that most kinesins move along MTs, but the opposite direction of movement for dyneins (Hirokawa, 1998). In addition, AMP-PNP potently inhibited motility when present at a level equimolar to the exogenous ATP (Figure 6). Under such conditions, AMP-PNP has been shown to inhibit conventional kinesin (Cohn et al., 1987; Porter et al., 1987; Wagner et al., 1989) but not dyneins (Lye et al., 1987; Paschal and Vallee, 1987). Finally, the observation that 100 μM but not 20 μM vanadate inhibited transport (Figure 6) is consistent with the idea that a kinesin but not a dynein was the most active motor in our system (Lye et al., 1987; Paschal and Vallee, 1987; Shpetner et al., 1988; Wagner et al., 1989).
Conventional kinesin is a leading candidate for the pertinent motor for a number of reasons. First, the H1 monoclonal antibody to conventional kinesin heavy chain (Pfister et al., 1989) was a potent inhibitor of motility (Figure 6). Next, a band that comigrated with conventional kinesin heavy chain was the most immunoreactive protein recognized in the motility system by anti-HIPYR (our unpublished results), an antibody that was made against a peptide sequence that is highly conserved among kinesins and that reacts with several known members of the kinesin superfamily (Swain et al., 1992). Finally, by immunoblotting, H1 did not recognize rabkinesin-6 (our unpublished results), a recently discovered kinesin that is localized on the Golgi and has been suggested to be a motor for Golgi-to-ER transport (Echard et al., 1998).
Despite these points, additional considerations lead us to keep an open mind about the identity of the predominant kinesin in the in vitro motility system. As striking as the results with H1 may be, we are mindful that antibodies can inhibit function by both direct and indirect mechanisms. For example, we found that H2, another monoclonal antibody to conventional kinesin heavy chain (Pfister et al., 1989), inhibited organelle transport toward MT plus and minus ends in squid axoplasm (Brady et al., 1990). The effect of H2 on minus end-directed transport must have been indirect, because kinesin is a plus end-directed motor. We are thus forced to consider the possibility that H1 inhibited Golgi membrane motility along MTs in the present case by indirectly interfering with a protein other than conventional kinesin. We also note that the instantaneous velocity for Golgi membrane motility along MTs was ∼1.5 μm/s, which is much higher than reported velocities for transport of MTs along glass coverslips coated with conventional kinesin (Vale et al., 1985; Hancock and Howard, 1998) or of conventional kinesin-coated microshperes moving along MTs (Svoboda et al., 1993). Although we suspect that conventional kinesin acts as a slower motor when attached to nonphysiological substrata as opposed to biological membranes, the speed with which Golgi membranes moved in our in vitro system leaves open the possibility that a different kinesin was responsible for the observed motility. On balance, therefore, we conclude that the most active motor in our system must be a kinesin and favor the idea that it is conventional kinesin itself.
Two agents that activate G proteins, GTPγS and AlF4−, inhibited transport almost completely (Figure 9). GTPγS was maximally effective at 1 μM in the presence of a robust ATP regenerating system that initially supplied 5 mM ATP. In contrast, 1 μM GTPγS inhibited transport by only ∼25% when a 100-fold molar excess of GTP was also present. The effects of GTPγS were therefore guanine nucleotide specific and prompt us to speculate that each burst of Golgi membrane motility along MTs is regulated by a cycle of GTP binding and hydrolysis by a G protein. One potential explanation of our data is that motility begins when GTP is hydrolyzed by a G protein and persists until a membrane-associated kinesin loses its grip on the MT. Reinitiation of motility would then require another round of GTP binding and hydrolysis, and motility would not be possible if the nucleotide binding site on G protein were occupied by a nonhydrolyzeable pseudosubstrate, such as GTPγS or GDP plus AlF4−.
Although the G protein in question remains unknown, several proteins can be eliminated as likely candidates. Conventional kinesin and tubulin, both of which bind and hydrolyze GTP (Kuznetsov and Gelfand, 1986; Porter et al., 1987; Gelfand and Bershadsky, 1991), can be ruled out as the direct targets of GTPγS and AlF4−, because neither inhibitor of Golgi membrane motility inhibited MT gliding mediated by conventional kinesin (Figure 10). For several reasons it is also doubtful that the target corresponds to Gα0 or any of the following Golgi-associated Gα species: Gαi2 (Montmayeur and Borrelli, 1994), Gαi3 (Ercolani et al., 1990; Wilson et al., 1994; Denker et al., 1996), Gαq/11 (Denker et al., 1996), and Gαs (Denker et al., 1996). First, motility was unaffected by cholera toxin (Table 1), which like GTPγS, activates Gαs (Gill and Meren, 1978) and, with less efficiency, several other Gα species as well (Gilman, 1987). Next, motility was also insensitive to pertussis toxin (Table 1), which permanently inactivates Gα0, Gαi2, and Gαi3 (Moss et al., 1983). If activation of any of those proteins by GTPγS or AlF4− prevented membrane motility, pertussis toxin should have conferred protection against the inhibitory compounds, but no such protection was observed (Table 1). Furthermore, if sustained motility required cycles of GTP hydrolysis by Gα0, Gαi2, or Gαi3, motility should have been prevented by pertussis toxin. Finally, motility was also unaffected by excess levels of Gα0, Gαs, and Gαi3 stably complexed with GTPγS (Table 1). It is also important to note that free βγ subunits did not inhibit motility (Table 1), implying that their dissociation from Gα subunits induced by GTPγS or AlF4− did not account for the inhibition of motility caused by either compound. Taken together, the results summarized here favor the hypothesis that the inhibition of Golgi motility caused by GTPγS or AlF4− did not involve trimeric G proteins.
What, then, might be the relevant targets of GTPγS and AlF4−? Because AlF4− was long known to be an activator of Gαs (Sternweiss and Gilman, 1982) but was shown to be inert toward numerous purified small G proteins (Kahn, 1991), we initially considered the latter group of proteins to be poor candidates. Recently, however, several groups have demonstrated formation of stable complexes of small G proteins bound simultaneously to AlF4− and GTPase-activating proteins (Mittal et al., 1996; Reza et al., 1997; Hoffman et al., 1998). Presumably, the small G protein in such a complex behaves as if it were in the activated, or GTP-bound, state. Considering that GTPase-activating proteins must have been present in the unfractionated cytosol that was a major component of our complete motility system, it is very likely that AlF4− was able to form stable complexes with the GTPase-activating proteins and small G proteins. We thus regard small G proteins, such as members of the Ras, Rab, and ARF families, and especially the Rho family, to be potential relevant targets for GTPγS and AlF4− in our Golgi membrane motility system. Activated forms of the Rho family members, RhoA, Rac1, and Cdc42, have been shown to interact with a putative kinesin receptor kinectin (Toyoshima et al., 1992) in a yeast two-hybrid screen (Hotta et al., 1996). Using the same approach, the protein kinases MLK2 and MLK3 were found to interact with activated Rac and Cdc42, as well as with the kinesin superfamily member KIF3 (Nagata et al., 1998). Another candidate protein is dynamin, a form of which has been localized to the Golgi (Henley and McNiven, 1996), and the type II isoform of which has been implicated in the formation of transport vesicles from the trans-Golgi network (Jones et al., 1998).
Using a variety of distinct, but related approaches, several groups have reconstituted MT-dependent formation of reticular membrane networks from isolated cellular components. Despite some similarities to the networks that have been studied in other laboratories, the Golgi networks described here are distinguished by a number of unique traits. Networks were first observed in mixtures of cytosol and crude microsomes isolated from chick embryo fibroblasts (Dabora and Sheetz, 1988) and in samples of partially purified squid kinesin, which were contaminated with membranous material of unknown identity (Vale and Hotani, 1988). Xenopus oocyte cytosol has been shown to drive formation of networks from crude oocyte microsomes and ER- and Golgi-enriched membranes isolated from rat liver (Allan and Vale, 1991; Allan and Vale, 1994; Allan, 1995; Niclas et al., 1996). The networks that formed from crude oocyte microsomes were predominantly ER (Allan, 1995) and dynein dependent (Niclas et al., 1996). Furthermore, network formation in interphase cytosol was stimulated by levels of okadaic acid that inhibit protein phosphatase 1 (Allan, 1995), whereas metaphase cytosol supported substantially less transport because of phosphorylation of a dynein subunit polypeptide and concomitant dissociation of dynein from membrane surfaces (Niclas et al., 1996). In contrast to the networks formed from Xenopus oocyte microsomes, the networks described here were formed from highly enriched Golgi membranes, were dependent on a kinesin, were not affected by okadaic acid, and were potently inhibited by GTPγS or AlF4−. These contrasting sets of results emphasize the likelihood that dynein-dependent ER motility and Golgi transport mediated by a kinesin are regulated by distinct mechanisms. In further support of the idea that motor- and organelle-specific mechanisms exist for regulating organelle transport along MTs, we found earlier that GTPγS, but not AlF4−, inhibited fast axonal transport in squid giant axons (Bloom et al., 1993). GTPγS inhibited transport bidirectionally in this case, although we cannot yet explain why AlF4− suppressed MT-based transport of Golgi but not axonal membranes.
The collective properties of our motility system raise the possibility that it represents an in vitro equivalent of an in vivo transport pathway that recycles resident ER components that escape to the Golgi. Using cultured cells with Golgi complexes that were localized near MT minus ends and the cell center, we obtained evidence that Golgi-to-ER transport occurs in a regulated, kinesin-dependent manner toward MT plus ends (Lippincott-Schwartz et al., 1995). In the present study, we used cytosol and Golgi membranes isolated from liver. Approximately 80% of the cells in liver are hepatocytes (Fawcett, 1986), in which the Golgi complex is localized near MT minus ends at the apical surface (Fawcett, 1986; Ihrke et al., 1993). Thus, in hepatocytes, the source of most of the cytosol and Golgi membranes in our reconstituted system, Golgi-to-ER transport must occur toward MT plus ends, just as it did in the cultured cells that we studied earlier (Lippincott-Schwartz et al., 1995). As in the cultured cells, transport in the reconstituted system occurred away from the Golgi, toward MT plus ends, by a regulated method mediated by a kinesin. There are thus striking parallels between Golgi-to-ER transport in live cells and Golgi transport along MTs in our reconstituted system. In the absence of further evidence, however, we cannot exclude the possibility that the in vitro motility more closely resembles transport from the Golgi to other organelles, such as prelysosomal structures, or to the cell surface. Regarding the latter possibility, it is worth noting that the velocity with which membranes moved in our in vitro system is similar to the velocity with which vesicles have been reported to move from the Golgi to the plasma membrane in living cells (Hirschberg et al., 1997). Irrespective of which intracellular transport steps correspond to the in vitro motility described here, further investigations aimed at identifying the GTPases that regulate motility in the reconstituted system and characterizing their mechanisms of action are bound to shed light on how Golgi membrane transport along MTs is regulated in vivo.
ACKNOWLEDGMENTS
We thank Karen Barker and David Mena for technical assistance and Drs. Bill Brown, Eric Berger, Caroline Enns, Al Gilman, Bruno Goud, Will Hancock, Linda Hendershot, Joe Howard, Calvin Lin, Libby Rutledge, and Bill Snell for providing antibodies and other reagents that were essential for this study. We also express our appreciation to Dr. Clare Waterman-Storer for advice about how to clean coverslips. This work was supported by grants from the American Cancer Society (CB-58E), the National Institutes of Health (NS30485 and DK52395) and the Robert A. Welch Foundation (I-1236) to G.S.B.
Abbreviations used:
- AMP-PNP
5′-adenylylimidodiphosphate
- DEAE
diethylaminoethyl
- EM
electron microscopy
- ER
endoplasmic reticulum
- Ig
immunoglobulin
- MT
microtubule
Footnotes
REFERENCES
- Acharya U, Jacobs R, Peters J-M, Watson N, Farquhar MG, Malhotra V. The formation of Golgi stacks from vesiculated Golgi membranes requires two distinct fusion events. Cell. 1995a;82:895–904. doi: 10.1016/0092-8674(95)90269-4. [DOI] [PubMed] [Google Scholar]
- Acharya U, McCaffery JM, Jacobs R, Malhotra V. Reconstitution of vesiculated Golgi membranes into stacks of cisternae: requirement of NSF in stack formation. J Cell Biol. 1995b;129:577–590. doi: 10.1083/jcb.129.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan V. Protein phosphatase 1 regulates the cytoplasmic dynein-driven formation of endoplasmic reticulum networks in vitro. J Cell Biol. 1995;128:879–891. doi: 10.1083/jcb.128.5.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan V, Vale R. Movement of membrane tubules along microtubules in vitro: evidence for specialised sites of motor attachment. J Cell Sci. 1994;107:1885–1897. doi: 10.1242/jcs.107.7.1885. [DOI] [PubMed] [Google Scholar]
- Allan VJ, Vale RD. Cell cycle control of microtubule-based membrane transport and tubule formation in vitro. J Cell Biol. 1991;113:347–359. doi: 10.1083/jcb.113.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen RD, Allen NS, Travis JL. Video-enhanced contrast, differential interference contrast (AVEC-DIC) microscopy: a new method capable of analyzing microtubule-related motility in the reticulopodial network of Allogromia laticollaris. Cell Motil. 1981;1:291–302. doi: 10.1002/cm.970010303. [DOI] [PubMed] [Google Scholar]
- Bacallao R, Antony C, Dotti C, Karsenti E, Stelzer EHK, Simons K. The subcellular organization of Madin-Darby canine kidney cells during the formation of a polarized epithelium. J Cell Biol. 1989;109:2817–2832. doi: 10.1083/jcb.109.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger EG, Muller U, Aegerter E, Strous GJ. Biology of galactosyltransferase: recent developments. Biochem Soc Trans. 1987;15:610–613. doi: 10.1042/bst0150610. [DOI] [PubMed] [Google Scholar]
- Bloom GS, Goldstein LSB. Cruising along microtubule highways: how membranes move through the secretory pathway. J Cell Biol. 1998;140:1277–1280. doi: 10.1083/jcb.140.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom GS, Richards BW, Leopold PL, Ritchey DM, Brady ST. GTPγS inhibits organelle transport along axonal microtubules. J Cell Biol. 1993;120:467–476. doi: 10.1083/jcb.120.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom GS, Wagner MC, Pfister KK, Brady ST. Native structure and physical properties of bovine brain kinesin and identification of the ATP-binding subunit polypeptide. Biochemistry. 1988;27:3409–3416. doi: 10.1021/bi00409a043. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brady ST, Lasek RJ, Allen RD. Video microscopy of fast axonal transport in extruded axoplasm: a new model for study of molecular mechanisms. Cell Motil. 1985;5:81–101. doi: 10.1002/cm.970050203. [DOI] [PubMed] [Google Scholar]
- Brady ST, Pfister KK, Bloom GS. A monoclonal antibody against kinesin inhibits both anterograde and retrograde fast axonal transport in squid axoplasm. Proc Natl Acad Sci USA. 1990;87:1061–1065. doi: 10.1073/pnas.87.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhardt JK, Echeverri CJ, Nilsson T, Vallee RB. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J Cell Biol. 1997;139:469–484. doi: 10.1083/jcb.139.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen P, Klummp S, Schelling DL. An improved procedure for identifying and quantifying protein phosphatases in mammalian tissues. FEBS Lett. 1989;250:596–600. doi: 10.1016/0014-5793(89)80803-8. [DOI] [PubMed] [Google Scholar]
- Cohn SA, Ingold AL, Scholey JM. Correlation between the ATPase and microtubule translocating activities of sea urchin egg kinesin. Nature. 1987;328:160–163. doi: 10.1038/328160a0. [DOI] [PubMed] [Google Scholar]
- Cohn SA, Ingold AL, Scholey JM. Quantitative analysis of sea urchin egg kinesin-driven microtubule motility. J Biol Chem. 1989;264:4290–4297. [PubMed] [Google Scholar]
- Cole NB, Sciaky N, Marotta A, Song J, Lippincott-Schwartz J. Golgi dispersal during microtubule disruption: regeneration of Golgi stacks at peripheral endoplasmic reticulum exit sites. Mol Biol Cell. 1996;7:631–650. doi: 10.1091/mbc.7.4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabora SL, Sheetz MP. The microtubule-dependent formation of a tubulovesicular network with characteristics of the ER from cultured cells. Cell. 1988;54:27–35. doi: 10.1016/0092-8674(88)90176-6. [DOI] [PubMed] [Google Scholar]
- Denker SP, McCaffery JM, Palade GE, Insel PA, Farquhar MG. Differential distribution of α subunits and βγ subunits of heterotrimeric G proteins on Golgi membranes of the exocrine pancreas. J Cell Biol. 1996;133:1027–1040. doi: 10.1083/jcb.133.5.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echard A, Jollivet F, Martinez O, Lacapère J-J, Rousselet A, Janoueix-Lerosey I, Goud B. Interaction of a Golgi-associated kinesin-like protein with rab6. Science. 1998;279:580–585. doi: 10.1126/science.279.5350.580. [DOI] [PubMed] [Google Scholar]
- Ercolani L, Stow JL, Boyle JF, Holtzman EJ, Lin H. Membrane localization of the pertussis toxin-sensitive G-protein subunits αi2 and αi3 and expression of a metallothionein-αi2 fusion gene in LLC-PK1 cells. Proc Natl Acad Sci USA. 1990;87:4635–4639. doi: 10.1073/pnas.87.12.4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett DW. A Textbook of Histology. 11th ed. Philadelphia, PA: W.B. Saunders; 1986. [Google Scholar]
- Gelfand VI, Bershadsky AD. Microtubule dynamics: mechanism, regulation and function. Annu Rev Cell Biol. 1991;7:93–116. doi: 10.1146/annurev.cb.07.110191.000521. [DOI] [PubMed] [Google Scholar]
- Gill DM, Meren R. ADP-ribosylation of membrane proteins catalyzed by cholera toxin: basis of the activation of adenylate cyclase. Proc Natl Acad Sci USA. 1978;75:3050–3054. doi: 10.1073/pnas.75.7.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman AG. G proteins: transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- Hancock WO, Howard J. Processivity of the motor protein kinesin requires two heads. J Cell Biol. 1998;140:1395–1405. doi: 10.1083/jcb.140.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot LM, Wei J, Gaut JR, Lawson B, Freiden PJ, Murti KG. In vivo expression of mammalian BiP ATPase mutants causes disruption of the endoplasmic reticulum. Mol Biol Cell. 1995;6:283–296. doi: 10.1091/mbc.6.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henley JR, McNiven MA. Association of a dynamin-like protein with the Golgi apparatus in mammalian cells. J Cell Biol. 1996;133:761–775. doi: 10.1083/jcb.133.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Pfister KK, Yorifuji H, Wagner MC, Brady ST, Bloom GS. Submolecular domains of bovine brain kinesin identified by electron microscopy and monoclonal antibody decoration. Cell. 1989;56:867–878. doi: 10.1016/0092-8674(89)90691-0. [DOI] [PubMed] [Google Scholar]
- Hirschberg K, Presley J, Cole N, Lippincott-Schwartz J. Golgi to plasma membrane trafficking in living cells: characterization of post-Golgi intermediates. Mol Biol Cell. 1997;8:194a. (abstract). [Google Scholar]
- Hoffman GR, Nassar N, Oswald RE, Cerione RA. Fluoride activation of the rho family GTP-binding protein Cdc42Hs. J Biol Chem. 1998;273:4392–4399. doi: 10.1074/jbc.273.8.4392. [DOI] [PubMed] [Google Scholar]
- Hotta K, Tanaka K, Mino A, Kohno H, Takai Y. Interaction of the Rho family small G proteins with kinectin, an anchoring protein of kinesin motor. Biochem Biophys Res Commun. 1996;225:69–74. doi: 10.1006/bbrc.1996.1132. [DOI] [PubMed] [Google Scholar]
- Ihrke G, Neufeld EB, Meads T, Shanks MR, Cassio D, Laurent M, Schroer TA, Pagano RE, Hubbard AL. WIF-B cells: an in vitro model for studies of hepatocyte polarity. J Cell Biol. 1993;123:1761–1775. doi: 10.1083/jcb.123.6.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iñiguez-Lluhi JA, Simon MI, Robishaw JD, Gilman AG. G protein βγ subunits synthesized in Sf9 cells: functional characterization and the significance of prenylation of γ. J Biol Chem. 1992;267:23409–23417. [PubMed] [Google Scholar]
- Jamora C, Takizawa PA, Zaarour RF, Denesvre C, Faulkner DJ, Malhotra V. Regulation of Golgi structure through trimeric G proteins. Cell. 1997;91:617–626. doi: 10.1016/s0092-8674(00)80449-3. [DOI] [PubMed] [Google Scholar]
- Jones SM, Howell KE, Henley JR, Cao H, McNiven MA. Role of dynamin in the formation of transport vesicles from the trans-Golgi network. Science. 1998;279:573–577. doi: 10.1126/science.279.5350.573. [DOI] [PubMed] [Google Scholar]
- Kahn RA. Fluoride is not an activator of the smaller (20–25 kDa) GTP-binding proteins. J Biol Chem. 1991;266:15595–15597. [PubMed] [Google Scholar]
- King SM, Otter T, Witman GB. Purification and characterization of Chlamydomonas flagellar dyneins. Methods Enzymol. 1986;134:291–306. doi: 10.1016/0076-6879(86)34097-7. [DOI] [PubMed] [Google Scholar]
- Kreis TE. Role of microtubules in the organization of the Golgi apparatus. Cell Motil Cytoskel. 1990;15:67–70. doi: 10.1002/cm.970150202. [DOI] [PubMed] [Google Scholar]
- Kreis TE, Lowe M, Pepperkok R. COPs regulating membrane traffic. Annu Rev Cell Dev Biol. 1995;11:677–706. doi: 10.1146/annurev.cb.11.110195.003333. [DOI] [PubMed] [Google Scholar]
- Kuznetsov SA, Gelfand VI. Bovine brain kinesin is a microtubule-activated ATPase. Proc Natl Acad Sci USA. 1986;83:8530–8534. doi: 10.1073/pnas.83.22.8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee E, Linder ML, Gilman AG. Expression of G-protein α subunits in Escherichia coli. Methods Enzymol. 1994;237:146–164. doi: 10.1016/s0076-6879(94)37059-1. [DOI] [PubMed] [Google Scholar]
- Leelavathi DE, Estes LW, Feingold DS, Lombardi B. Isolation of a Golgi-rich fraction from rat liver. Biochim Biophys Acta. 1970;211:124–138. [Google Scholar]
- Lippincott-Schwartz J, Cole NB, Marotta A, Conrad PA, Bloom GS. Kinesin is the motor for microtubule-mediated Golgi-to-ER membrane traffic. J Cell Biol. 1995;128:293–306. doi: 10.1083/jcb.128.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Donaldson JG, Schweizer A, Berger EG, Hauri H-P, Yuan LC, Klausner RD. Microtubule-dependent retrograde transport of proteins into the ER in the presence of brefeldin A suggests an ER recycling pathway. Cell. 1990;60:821–836. doi: 10.1016/0092-8674(90)90096-w. [DOI] [PubMed] [Google Scholar]
- Lucocq JM, Warren G. Fragmentation and partitioning of the Golgi apparatus during mitosis in HeLa cells. EMBO J. 1987;6:3239–3246. doi: 10.1002/j.1460-2075.1987.tb02641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lye RJ, Porter ME, Scholey JM, McIntosh JR. Identification of a microtubule-based cytoplasmic motor in the nematode C. elegans. Cell. 1987;51:309–318. doi: 10.1016/0092-8674(87)90157-7. [DOI] [PubMed] [Google Scholar]
- Marks DL, Larkin JM, McNiven MA. Association of kinesin with the Golgi apparatus in rat hepatocytes. J Cell Sci. 1994;107:2417–2426. doi: 10.1242/jcs.107.9.2417. [DOI] [PubMed] [Google Scholar]
- Misteli T, Warren G. COP-coated vesicles are involved in the mitotic fragmentation of Golgi stacks in a cell-free system. J Cell Biol. 1994;125:269–282. doi: 10.1083/jcb.125.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal R, Ahmadian MR, Goody RS, Wittinghofer A. Formation of a transition-state analog of the Ras GTPase reaction by Ras-GDP, tetrafluoroaluminate, and GTPase-activating proteins. Science. 1996;273:115–117. doi: 10.1126/science.273.5271.115. [DOI] [PubMed] [Google Scholar]
- Montmayeur J-P, Borrelli E. Targetting of Gαi2 to the Golgi by alternative spliced carboxyl-terminal region. Science. 1994;263:95–98. doi: 10.1126/science.8272874. [DOI] [PubMed] [Google Scholar]
- Moss J, Stanley SJ, Burns DL, Hsia JA, Yost DA, Myers GA, Hewlett EL. Actiavation by thiol of the latent NAD glycohydrolase and ADP-ribosyltransferase activities of Bordetella pertussis toxin (islet activating protein) J Biol Chem. 1983;258:11879–11882. [PubMed] [Google Scholar]
- Nagata K, Puls A, Futter C, Aspenstrom P, Schaefer E, Nakata T, Hirokawa N, Hall A. The MAP kinase kinase kinase MLK2 co-localizes with activated JNK along microtubules and associates with kinesin superfamily motor KIF3. EMBO J. 1998;17:149–158. doi: 10.1093/emboj/17.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niclas J, Allan VJ, Vale RD. Cell cycle regulation of dynein association with membranes modulates microtubule-based organelle transport. J Cell Biol. 1996;133:585–593. doi: 10.1083/jcb.133.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Draper RK, Brown WJ. Biosynthesis of lysosomal enzymes in cells of the End3 complementation group conditionally defective in endosomal acidification. Somat Cell Mol Genet. 1991;17:137–150. doi: 10.1007/BF01232971. [DOI] [PubMed] [Google Scholar]
- Paschal BM, Vallee RB. Retrograde transport by the microtubule-associated protein MAP 1C. Nature. 1987;330:181–183. doi: 10.1038/330181a0. [DOI] [PubMed] [Google Scholar]
- Paschal BM, Vallee RB. Microtubule and axoneme gliding assays for force production by microtubule motor proteins. Methods Cell Biol. 1993;39:65–74. doi: 10.1016/s0091-679x(08)60161-2. [DOI] [PubMed] [Google Scholar]
- Pfister KK, Wagner MC, Stenoien DA, Brady ST, Bloom GS. Monoclonal antibodies to kinesin heavy and light chains stain vesicle-like structures, but not microtubules, in cultured cells. J Cell Biol. 1989;108:1453–1463. doi: 10.1083/jcb.108.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter KR, Bonneville MA. Fine Structure of Cells and Tissues. Philadelphia, PA: Lea and Febiger; 1973. [Google Scholar]
- Porter ME, Scholey JM, Stemple DL, Vigers GPA, Vale RD, Sheetz MP, McIntosh JR. Characterization of the microtubule movement produced by sea urchin egg kinesin. J Biol Chem. 1987;262:2794–2802. [PubMed] [Google Scholar]
- Presley JF, Cole NB, Schroer TA, Lippincott-Schwartz J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Kondo H, Newman R, Hui N, Freemont P, Warren G. Syntaxin 5 is a common component of the NSF- and p97-mediated reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro. Cell. 1998;92:603–610. doi: 10.1016/s0092-8674(00)81128-9. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Levine TP, Peters J-M, Warren G. An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell. 1995;82:905–914. doi: 10.1016/0092-8674(95)90270-8. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Misteli T, Watson R, Warren G. Reassembly of Golgi stacks from mitotic Golgi fragments in a cell-free system. J Cell Biol. 1995;129:605–618. doi: 10.1083/jcb.129.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redenbach DM, Boekelheide K. Microtubules are oriented with their minus-ends directed apically before tight junction formation in rat Sertoli cells. Eur J Cell Biol. 1994;65:246–257. [PubMed] [Google Scholar]
- Reza M, Mittal R, Hall A, Wittinghofer A. Aluminum fluoride associates with the small guanine nucleotide binding proteins. FEBS Lett. 1997;408:315–318. doi: 10.1016/s0014-5793(97)00422-5. [DOI] [PubMed] [Google Scholar]
- Shpetner HS, Paschal BM, Vallee RB. Characterization of the microtubule-activated ATPase of brain cytoplasmic dynein (MAP 1C) J Cell Biol. 1988;107:1001–1009. doi: 10.1083/jcb.107.3.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternweiss PC, Gilman AG. Aluminum: a requirement for activation of the regulatory component of adenylate cyclase by fluoride. Proc Natl Acad Sci USA. 1982;79:4888–4891. doi: 10.1073/pnas.79.16.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- Swain KE, Mitchison TJ, Wordeman LG. Evidence for kinesin-related proteins in the mitotic apparatus using peptide antibodies. J Cell Sci. 1992;101:303–313. doi: 10.1242/jcs.101.2.303. [DOI] [PubMed] [Google Scholar]
- Tamaoki T, Nomoto H, Takahashi I, Kato Y, Morimoto M, Tomita F. Staurosporine, a potent inhibitor of phospholipid/Ca++-dependent protein kinase. Biochem Biophys Res Commun. 1986;135:397–402. doi: 10.1016/0006-291x(86)90008-2. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima I, Yu H, Steuer ER, Sheetz MP. Kinectin, a major kinesin-binding protein on ER. J Cell Biol. 1992;118:1121–1131. doi: 10.1083/jcb.118.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Hotani H. Formation of membrane networks in vitro by kinesin-driven microtubule movement. J Cell Biol. 1988;107:2233–2241. doi: 10.1083/jcb.107.6.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner MC, Pfister KK, Bloom GS, Brady ST. Copurification of kinesin polypeptides with microtubule-stimulated Mg-ATPase activity and kinetic analysis of enzymatic processes. Cell Motil Cytoskel. 1989;12:195–215. doi: 10.1002/cm.970120403. [DOI] [PubMed] [Google Scholar]
- Williams AM, Enns CA. A mutated transferrin receptor lacking asparagine-linked glycosylation sites shows reduced functionality and an association with binding immunoglobulin protein. J Biol Chem. 1991;266:17648–17654. [PubMed] [Google Scholar]
- Wilson BS, Komuro M, Farquhar MG. Cellular variations in heterotrimeric G protein localization and expression in rat pituitary. Endocrinology. 1994;134:233–244. doi: 10.1210/endo.134.1.8275939. [DOI] [PubMed] [Google Scholar]
- Witman GB. Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 1986;134:280–290. doi: 10.1016/0076-6879(86)34096-5. [DOI] [PubMed] [Google Scholar]