Abstract
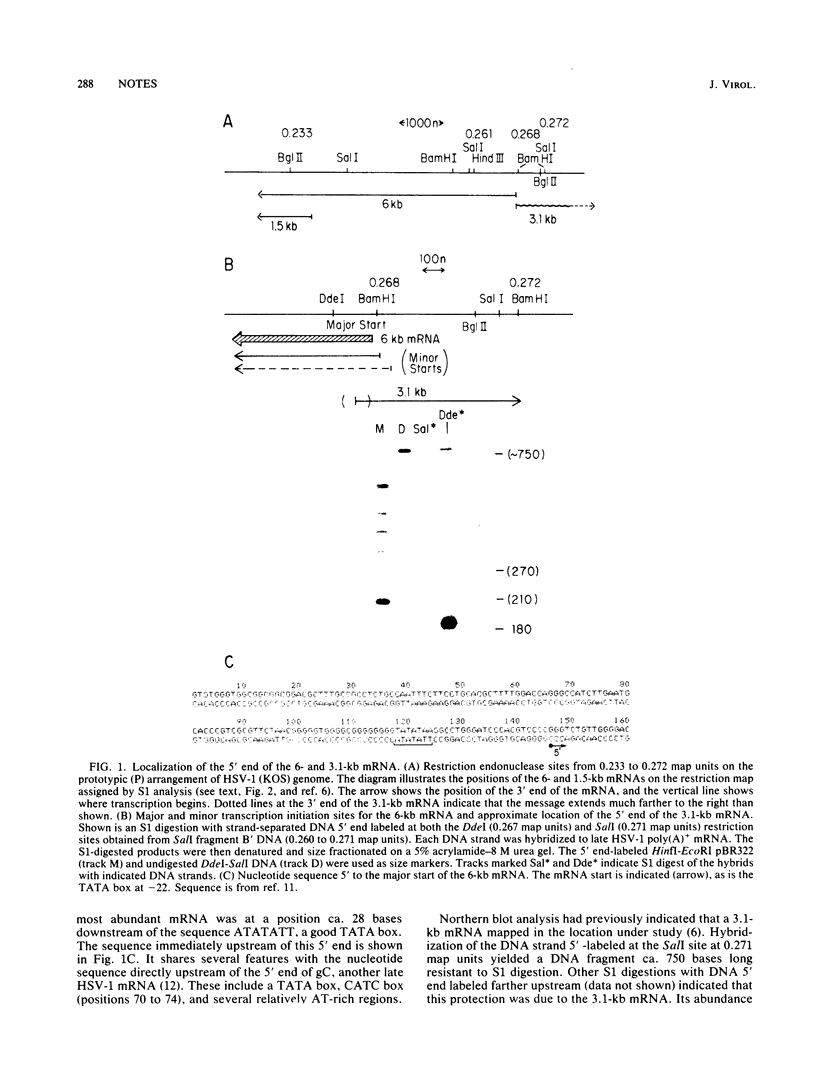
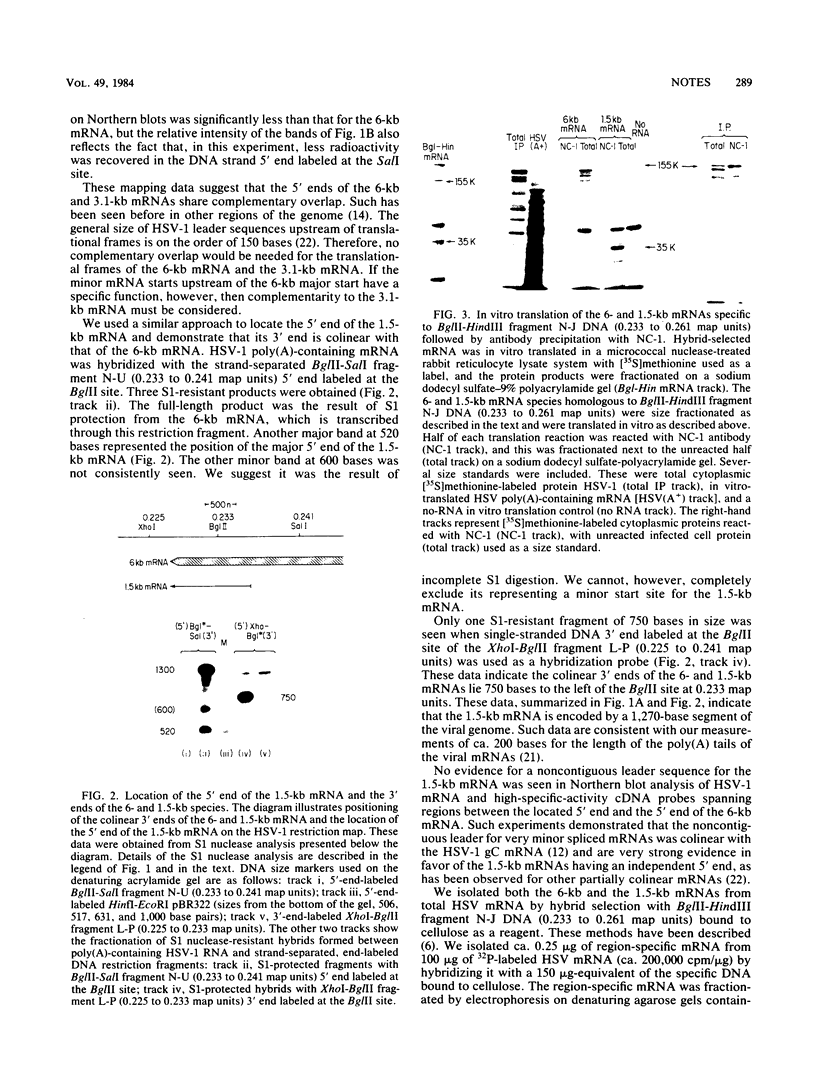
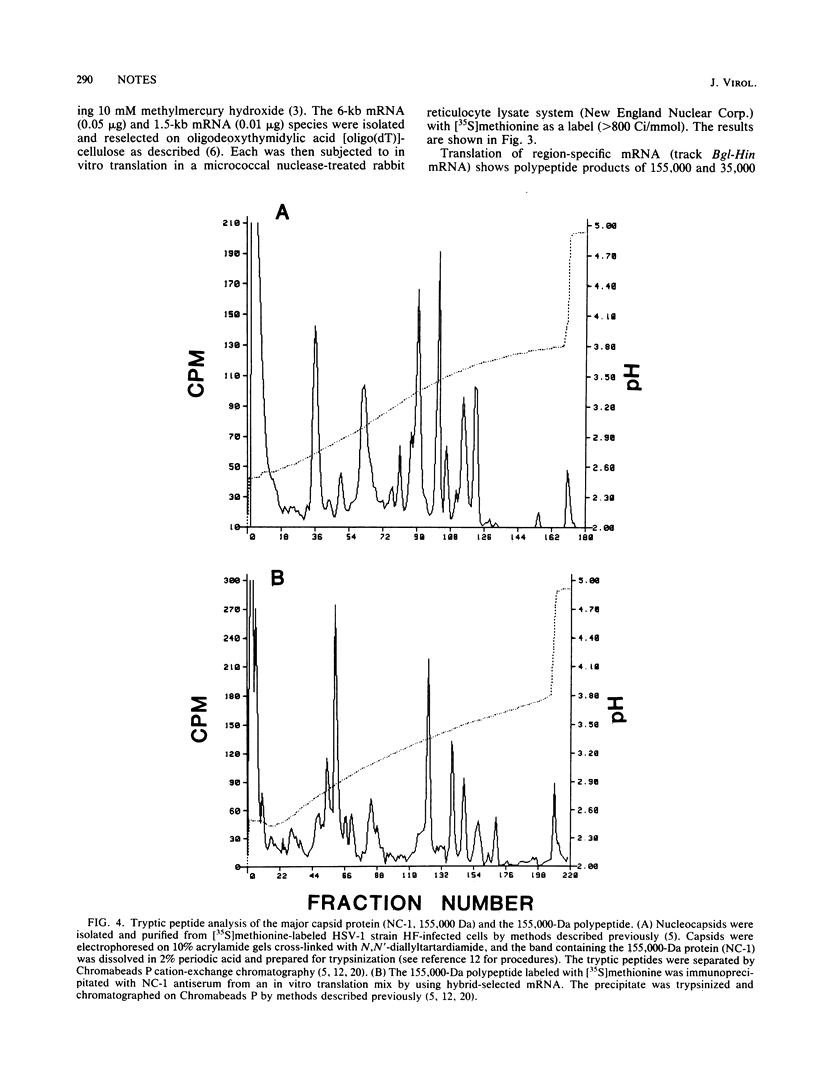
The two partially colinear 6-kilobase (kb) and 1.5-kb mRNAs mapping between 0.23 and 0.27 map units on the herpes simplex virus type 1 genome were precisely located. The 5' end of the 6-kb mRNA was located 28 bases downstream of the sequence ATATATT and was 10 bases to the left of the BamHI site at 0.268. This position is ca. 90 bases to the left of our earlier reported sequence (R. J. Frink, K. G. Draper, and E. K. Wagner, Proc. Natl. Acad. Sci. U.S.A. 78:6139-6143, 1981). We used a polyclonal antibody made against purified herpes simplex virus type 1 VP5 to demonstrate that the 155,000-dalton translation product of the 6-kb mRNA is this capsid protein. The antibody did not react with the 35,000-dalton translation product of the 1.5-kb mRNA. We also confirmed our identification of VP5 as the translation product of the 6-kb mRNA by comparison of tryptic peptides of the in vitro-translated protein and authentic VP5.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson K. P., Frink R. J., Devi G. B., Gaylord B. H., Costa R. H., Wagner E. K. Detailed characterization of the mRNA mapping in the HindIII fragment K region of the herpes simplex virus type 1 genome. J Virol. 1981 Mar;37(3):1011–1027. doi: 10.1128/jvi.37.3.1011-1027.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson K. P., Stringer J. R., Holland L. E., Wagner E. K. Isolation and localization of herpes simplex virus type 1 mRNA. J Virol. 1979 Jun;30(3):805–820. doi: 10.1128/jvi.30.3.805-820.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J. M., Davidson N. Methylmercury as a reversible denaturing agent for agarose gel electrophoresis. Anal Biochem. 1976 Jan;70(1):75–85. doi: 10.1016/s0003-2697(76)80049-8. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Cohen G. H., Ponce de Leon M., Diggelmann H., Lawrence W. C., Vernon S. K., Eisenberg R. J. Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J Virol. 1980 May;34(2):521–531. doi: 10.1128/jvi.34.2.521-531.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R. H., Devi B. G., Anderson K. P., Gaylord B. H., Wagner E. K. Characterization of a major late herpes simplex virus type 1 mRNA. J Virol. 1981 May;38(2):483–496. doi: 10.1128/jvi.38.2.483-496.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R. H., Draper K. G., Banks L., Powell K. L., Cohen G., Eisenberg R., Wagner E. K. High-resolution characterization of herpes simplex virus type 1 transcripts encoding alkaline exonuclease and a 50,000-dalton protein tentatively identified as a capsid protein. J Virol. 1983 Dec;48(3):591–603. doi: 10.1128/jvi.48.3.591-603.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper K. G., Frink R. J., Wagner E. K. Detailed characterization of an apparently unspliced beta herpes simplex virus type 1 gene mapping in the interior of another. J Virol. 1982 Sep;43(3):1123–1128. doi: 10.1128/jvi.43.3.1123-1128.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutia B. M. Ribonucleotide reductase induced by herpes simplex virus has a virus-specified constituent. J Gen Virol. 1983 Mar;64(Pt 3):513–521. doi: 10.1099/0022-1317-64-3-513. [DOI] [PubMed] [Google Scholar]
- Edwards S. A., Fan H. gag-Related polyproteins of Moloney murine leukemia virus: evidence for independent synthesis of glycosylated and unglycosylated forms. J Virol. 1979 May;30(2):551–563. doi: 10.1128/jvi.30.2.551-563.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg R. J., Long D., Pereira L., Hampar B., Zweig M., Cohen G. H. Effect of monoclonal antibodies on limited proteolysis of native glycoprotein gD of herpes simplex virus type 1. J Virol. 1982 Feb;41(2):478–488. doi: 10.1128/jvi.41.2.478-488.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frink R. J., Draper K. G., Wagner E. K. Uninfected cell polymerase efficiently transcribes early but not late herpes simplex virus type 1 mRNA. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6139–6143. doi: 10.1073/pnas.78.10.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frink R. J., Eisenberg R., Cohen G., Wagner E. K. Detailed analysis of the portion of the herpes simplex virus type 1 genome encoding glycoprotein C. J Virol. 1983 Feb;45(2):634–647. doi: 10.1128/jvi.45.2.634-647.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway D. A., Goldstein L. C., Lewis J. B. Identification of proteins encoded by a fragment of herpes simplex virus type 2 DNA that has transforming activity. J Virol. 1982 May;42(2):530–537. doi: 10.1128/jvi.42.2.530-537.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L. M., Draper K. G., Frink R. J., Costa R. H., Wagner E. K. Herpes simplex virus mRNA species mapping in EcoRI fragment I. J Virol. 1982 Aug;43(2):594–607. doi: 10.1128/jvi.43.2.594-607.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huszar D., Bacchetti S. Is ribonucleotide reductase the transforming function of herpes simplex virus 2? Nature. 1983 Mar 3;302(5903):76–79. doi: 10.1038/302076a0. [DOI] [PubMed] [Google Scholar]
- Jenkins F. J., Howett M. K., Spector D. J., Rapp F. Detection by RNA blot hybridization of RNA sequences homologous to the BglII-N fragment of herpes simplex virus type 2 DNA. J Virol. 1982 Dec;44(3):1092–1096. doi: 10.1128/jvi.44.3.1092-1096.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden H. S., Stow N. D., Preston V. G., Timbury M. C., Wilkie N. M. Physical mapping of herpes simplex virus-induced polypeptides. J Virol. 1978 Nov;28(2):624–642. doi: 10.1128/jvi.28.2.624-642.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Morse L. S., Pereira L., Roizman B., Schaffer P. A. Anatomy of herpes simplex virus (HSV) DNA. X. Mapping of viral genes by analysis of polypeptides and functions specified by HSV-1 X HSV-2 recombinants. J Virol. 1978 May;26(2):389–410. doi: 10.1128/jvi.26.2.389-410.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizer L. I., Cohen G. H., Eisenberg R. J. Effect of tunicamycin on herpes simplex virus glycoproteins and infectious virus production. J Virol. 1980 Apr;34(1):142–153. doi: 10.1128/jvi.34.1.142-153.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer J. R., Holland L. E., Swanstrom R. I., Pivo K., Wagner E. K. Quantitation of herpes simplex virus type 1 RNA in infected HeLa cells. J Virol. 1977 Mar;21(3):889–901. doi: 10.1128/jvi.21.3.889-901.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]