Abstract
CWH41, a gene involved in the assembly of cell wall β-1,6-glucan, has recently been shown to be the structural gene for Saccharomyces cerevisiae glucosidase I that is responsible for initiating the trimming of terminal α-1,2-glucose residue in the N-glycan processing pathway. To distinguish between a direct or indirect role of Cwh41p in the biosynthesis of β-1,6-glucan, we constructed a double mutant, alg5Δ (lacking dolichol-P-glucose synthase) cwh41Δ, and found that it has the same phenotype as the alg5Δ single mutant. It contains wild-type levels of cell wall β-1,6-glucan, shows moderate underglycosylation of N-linked glycoproteins, and grows at concentrations of Calcofluor White (which interferes with cell wall assembly) that are lethal to cwh41Δ single mutant. The strong genetic interactions of CWH41 with KRE6 and KRE1, two other genes involved in the β-1,6-glucan biosynthetic pathway, disappear in the absence of dolichol-P-glucose synthase (alg5Δ). The triple mutant alg5Δcwh41Δkre6Δ is viable, whereas the double mutant cwh41Δkre6Δ in the same genetic background is not. The severe slow growth phenotype and 75% reduction in cell wall β-1,6-glucan, characteristic of the cwh41Δkre1Δ double mutant, are not observed in the triple mutant alg5Δcwh41Δkre1Δ. Kre6p, a putative Golgi glucan synthase, is unstable in cwh41Δ strains, and its overexpression renders these cells Calcofluor White resistant. These results demonstrate that the role of glucosidase I (Cwh41p) in the biosynthesis of cell wall β-1,6-glucan is indirect and that dolichol-P-glucose is not an intermediate in this pathway.
INTRODUCTION
The cell wall of Saccharomyces cerevisiae preserves osmotic integrity and determines cell shape during growth and development. It is dynamic and constitutes 15–30% of the dry weight of the vegetative cell. Major components are mannoproteins (40% by weight) and β-glucans (50–60% by weight), with chitin representing a minor constituent found mostly in the division septum (Bulawa, 1993; Klis, 1994). β-Glucans are composed mainly of β-1,3–linked polymers averaging 1500 residues in length and smaller highly branched β-1,6-glucans with a degree of polymerization of ∼150–200 residues (Fleet and Manners, 1976).
Mutants defective in the biosynthesis of β-1,6-glucan may be selected using the S. cerevisiae K1 killer toxin, a protein secreted by killer yeast strains, which kills sensitive (nonkiller) cells. The toxin displays a lectin-like affinity for linear β-1,6-glucans and must bind to the wall of sensitive strains before initiating the killing process (Bussey et al., 1979). Selection of mutants resistant to this toxin has defined a series of KRE (killer-resistant) genes, including several required for β-1,6-glucan synthesis (Boone et al., 1990; Brown et al., 1993).
The KRE5 gene encodes a large endoplasmic reticulum (ER)1 luminal protein, which is involved in the earliest known step in the pathway for β-1,6-glucan assembly in S. cerevisiae. Kre5p shares extensive sequence homology, size, and subcellular location with the UDP-glucose:glycoprotein glucosyltransferase from Drosophila (Parker, et al., 1995) and Schizosaccharomyces pombe (Fernandez et al., 1996) responsible for the transient reglucosylation of oligosaccharides of misfolded proteins in the ER lumen. KRE5 deletion strains do not have any detectable β-1,6-glucan polymer, show aberrant morphology, and are extremely compromised in growth (Meaden et al., 1990). This is in agreement with the recently proposed hypothesis that the β-1,6-glucan is the central molecule or “glue” that holds together the other components of the cell wall, β-1,3-glucan, mannoproteins, and a portion of cellular chitin (Kollar et al., 1997). The specific function of Kre5p is not known. Another protein of the ER, Cwh41p, also has a role in β-1,6-glucan synthesis. This is an N-glycosylated integral membrane protein, and null mutants synthesize approximately half the normal amounts of β-1,6-glucan (Jiang et al., 1996).
Further along the secretory pathway, other proteins have also been implicated in β-1,6-glucan synthesis. KRE6 and SKN1 are two highly homologous genes encoding 80- and 87-kDa integral membrane glycoproteins likely to be localized in the Golgi apparatus. These genes have been proposed to function independently and early in the assembly of the β-1,6 polymer, possibly as glucan synthases (Roemer et al., 1994). KRE1 gene encodes a secreted, O-glycosylated, threonine- and serine-rich agglutinin-like protein necessary for the addition of β-1,6–linked outer chains to a core glucan structure; it can be found mainly at the cell wall (Boone et al., 1990; Roemer and Bussey 1995). The CWH41 gene displays strong genetic interactions with KRE1 and KRE6. The cwh41Δkre6Δ double mutant is nonviable, whereas the cwh41Δkre1Δ double mutant displays strong synthetic defects, such as a severe slow growth phenotype and a 75% reduction in β-1,6-glucan (Jiang et al. 1996).
Very recently, CWH41 has been shown to be the structural gene for S. cerevisiae glucosidase I (Romero et al., 1997). This enzyme initiates the pathway of N-glycan processing by removing the terminal α-1,2-glucose residue from Glc3Man9GlcNAc2 oligosaccharide chains attached to newly synthesized proteins in the ER. Glucosidase II then removes the two remaining α-1,3–linked glucoses to continue the trimming process. This unexpected finding gives rise to a puzzling question: what is the role of glucosidase I (Cwh41p), a very well-characterized enzyme of the N-glycosylation pathway, in the biosynthesis of cell wall β-1,6-glucan?
The first possibility is a direct involvement of glucosidase I (Cwh41p) as a synthetic component together with (or after) Kre5p action in the initial assembly of β-1,6-glucan polymer in the lumen of the rough ER (Jiang et al., 1996). The biochemical reactions leading to the initiation of β-1,6-glucan polymer have not been deciphered yet. If a transitory protein primer were implicated, Kre5p and Cwh41p (glucosidase I) could be involved in glucose addition and removal from the hypothetical primer. Perhaps the covalent linkage between the glycosylphosphatidylinositol anchor of mannoproteins and β-1,6-glucan existent in the cell wall (Kollar et al., 1997) could be preassembled during their concomitant synthesis in the lumen of the ER and be mediated by Cwh41p and Kre5p. Glucosidase I (Cwh41p) could also be hypothesized to have transglucosidase activity and to transfer the glucose it removes in the trimming of N-glycans (originated from dol-P-glucose) to the nascent β-1,6-glucan polymer in the ER lumen.
The second possibility is an indirect role of glucosidase I (Cwh41p) in the biosynthesis of cell wall β-1,6-glucan chains (Romero et al., 1997). Mammalian studies have demonstrated that blocking glucose removal with glucosidase inhibitors such as 1-deoxynojirimycin or castanospermine causes accumulation and in some cases degradation of some glycoproteins in the ER (Datema et al., 1987; Elbein, 1991). This in turn may decrease the amount of certain glycoproteins at the cell surface or in different compartments of the secretory pathway. The failure to correctly process N-linked glycoproteins involved in β-1,6-glucan synthesis could reduce or abolish their ability to function properly through instability, mislocalization, or reduced enzymatic activity.
We took a combined genetic and biochemical approach to distinguish between a direct or indirect role of glucosidase I (Cwh41p) in the biosynthesis of cell wall β-1,6-glucan. We reasoned that if the effect were indirect, i.e., due to the abnormal presence of the three glucose residues in the N-linked chains of glycoproteins, it would be abolished in a mutant that did not have glucose added to N-linked oligosaccharides. The alg5 mutant, which is defective in the synthesis of dol-P-glucose, accumulates Man9GlcNAc2-PP-dolichol as oligosaccharide donor. In vivo this nonglucosylated oligosaccharide is transferred to proteins with reduced efficiency, leading to a moderate underglycosylation; nevertheless the alg5 mutant grows normally under laboratory conditions (Huffaker and Robbins, 1983; Runge et al., 1984; te Heesen et al., 1994). On the other hand, if the role of Cwh41p in β-1,6-glucan biosynthesis were direct, an alg5Δcwh41Δ double mutant would still have reduced levels of β-1,6-glucan and should have an even more severe phenotype if dolichol-P-glucose were required for β-1,6-glucan biosynthesis.
We constructed a null allele of the ALG5 gene in the same genetic background (SEY6210) where the cell wall phenotype and genetic interactions between cwh41 and kre mutants have been described (Jiang et al., 1996). We found that in the absence of dolichol-P-glucose (alg5Δ), wild-type levels of cell wall β-1,6-glucans are made. The double mutant alg5Δcwh41Δ exhibited the same phenotype as alg5Δ alone, and the strong genetic interaction displayed by the CWH41 gene with KRE6 and KRE1 genes disappeared in an alg5 deletion background. The triple mutant alg5Δcwh41Δkre6Δ is viable in the same genetic background, whereas a double mutant cwh41Δkre6Δ is not (Jiang et al., 1996). Kre6p, a putative Golgi glucan synthase, was found to be selectively unstable in the cwh41Δ strain, and its overexpression renders this strain Calcofluor White (CFW) resistant. These results therefore demonstrate that glucosidase I (Cwh41p) has an indirect role in the biosynthesis of cell wall β-1,6-glucan because of the nonphysiological retention of the three terminal glucoses on the N-linked oligosaccharides, which are trimmed in wild-type cells.
MATERIALS AND METHODS
Strains and Growth Conditions
S. cerevisiae strains were grown in YPD or SD medium supplemented with the required amino acids (Sherman et al., 1986). Cells were grown at 24°C. Solid medium was made by adding 2% agar to the liquid stock. Standard procedures were used for genetic crosses, sporulation of diploids, and dissection of tetrads (Sherman et al., 1986). Strains used for this study are isogenic to SEY6210 and are listed in Table 1. Because no differential auxotrophic markers were available for the selection of diploids, zygotes were visually identified under the dissecting microscope and dragged with the aid of a micromanipulator 4–6 h after complementary haploids were mated on filter. Null alleles were identified by whole yeast cell PCR and/or resistance to S. cerevisiae K1 killer toxin (Brown et al., 1994). Yeast and bacterial transformation were done by electroporation as described (Abeijon et al., 1996). Escherichia coli DH5α was used for plasmid propagation and grown in Luria-Bertani medium, containing ampicillin (100 μg/ml) when needed (Maniatis et al., 1982).
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| SEY 6210 | MATα leu2-3, 112 ura3-52 his3-Δ200 lys2-80 trp1-Δ901 suc2-Δ9 | H. Bussey |
| HAB 251-15B | Mata/MATα leu2,3,112/leu2,3,112 ura3-52/ura3-52 his3-Δ200/his3-Δ200 lys2-80/lys2-80 trp1-Δ901/trp1-Δ901 suc2-Δ9/suc2-Δ9 | H. Bussey |
| HAB 855 | MATα cwh41Δ∷HIS3 in SEY 6210 | H. Bussey |
| HAB 635 | MATa kre1Δ∷HIS3 in SEY 6210 | H. Bussey |
| TR 92 | MATa kre6Δ∷HIS3 in SEY 6210 | H. Bussey |
| LCY 15 | MATa/MATα ALG5/alg5Δ∷HIS3 in HAB 251-15B | This study |
| LCY 20 | MATa/MATα ALG5/alg5Δ∷HIS3 CWH41/cwh41Δ∷HIS3 in HAB 251-15B | This study |
| LCY 21 | MATα SEY 6210 | This study |
| LCY 22 | MATα alg5Δ∷HIS3 in SEY 6210 | This study |
| LCY 23 | MATa alg5Δ∷HIS3 cwh41Δ∷HIS3 in SEY 6210 | This study |
| LCY 24 | MATa cwh41Δ∷HIS3 in SEY 6210 | This study |
| LCY 30 | MATa/MATα ALG5/alg5Δ∷HIS3 KRE6/kre6∷HIS3 in HAB 251-15B | This study |
| LCY 33 | MATα alg5Δ∷HIS3 kre6Δ∷HIS3 in SEY 6210 | This study |
| LCY 45 | MATa/MATα alg5Δ∷HIS3/alg5Δ∷HIS3 KRE6/kre6Δ∷HIS3 CWH41/cwh41Δ∷HIS3 in HAB 251-15B | This study |
| LCY 47 | MATa kre6Δ∷HIS3 cwh41Δ∷HIS3 alg5Δ∷HIS3 in SEY 6210 | This study |
| LCY 55 | MATa/MATα KRE1/kre1Δ∷HIS3 ALG5/alg5Δ∷HIS3 in HAB 251-15B | This study |
| LCY 58 | MATα kre1Δ∷HIS3 alg5Δ∷HIS3 in SEY 6210 | This study |
| LCY 65 | MATa/MATα alg5Δ∷HIS3/alg5Δ∷HIS3 KRE1/kre1Δ∷HIS3 CWH41/cwh41Δ∷HIS3 in HAB 251-15B | This study |
| LCY 66 | MATa alg5Δ∷HIS3 kre1Δ∷HIS3 cwh41Δ∷HIS3 in SEY 6210 | This study |
| LCY 80 | MATa/MATα KRE1/kre1Δ∷HIS3 CWH41/cwh41Δ∷HIS3 in HAB 251-15B | This study |
| LCY 81 | MATα kre1Δ∷HIS3 cwh41Δ∷HIS3 in SEY 6210 | This study |
Disruption of the ALG5 Locus
The heterozygous alg5Δ::HIS3/ALG5 strain LCY16 was made via one-step gene replacement (Rothstein, 1983) by transforming diploid strain HAB251-15B (obtained by mating type switching of SEY6210; Roemer and Bussey, 1991) with a 2.8-kb BamHI–HpaI linear fragment, from pALG5Δ (Spe–Bgl) HIS (te Heesen et al., 1994) to histidine prototrophy. In the deletion construct, a 1.2-kb SpeI-BglI fragment containing the promoter and more than two-thirds of the ALG5 coding region were replaced by a 1.8-kb fragment containing the S. cerevisiae HIS3 locus transcribing in the same direction as the ALG5 open reading frame (te Heesen et al., 1994). Sporulation yielded haploid alg5Δ::HIS3 strains LCY17 and LCY18, which were mated appropriately to originate some of the strains listed in Table 1. Transformants and haploid progeny were checked for correct homologous recombination by whole yeast cell PCR as described bellow. Forward primer Alg5-F (5′-GCACAAAGGACCATAGTCACTGTG-3′) starting at position −137 (ATG = +1) and reverse primer Alg5-R (5′-AGCAAATGCCCTTGAGCGAG-3′) ending at position 1109 give rise to a 1246-bp PCR product from the wild-type ALG5 locus and no product from the alg5Δ::HIS3 locus. A forward primer his3-F (5′-CGTGCGTGGAGTAAAAAGGTRTTG-3′) starting at position 330 (ATG = 1) of the HIS3 locus yielded a 1230-bp PCR product with the Alg5-R primer from the disrupted allele and no product from the wild-type ALG5 gene, indicating that the disruption was at the ALG5 locus.
Identification of the Allele at the CWH41 Locus by Whole-Cell PCR
The null allele cwh41Δ::HIS3 used throughout this study originates in strain HAB855 and was described by Jiang et al. (1996). Briefly, an internal 1152-bp BamHI–XbaI fragment was deleted from the coding region of the CWH41 gene and replaced with a 1.8-kb BamHI fragment containing the HIS3 gene transcribing in the same direction. We used forward primer cwh41-F (5′-TGGTTGGGAAGTGTATGATCCAAG-3′) starting at position 297 (ATG = 1) and reverse primer cwh41-R (5′-TCGTGGAGCAAGCCAGGATTG-3′) starting at position 1616, which gave rise to a 1319-bp PCR product from the wild-type CWH41 gene and a 2-kb product from the null allele cwh41Δ::HIS3. Also the His3-F primer described in the previous section and the cwh41-R primer originated a 1-kb PCR product from the null allele and no product from the wild-type CWH41 gene. These combinations of primers allowed identification of the allele present at the CWH41 locus in the strains constructed for this study in a whole-cell PCR reaction. Approximately 5 × 105 yeast were included in a 25-μl PCR reaction mix (10 mM Tris-HCl, pH 8.3, 50 mM KCl, 2.5 mM MgCl2, 0.2 mM dNTP, 1 μM primers). The mixture was overlaid with a drop of mineral oil and heated to 94°C for 5 min (hot start), then 1 U of Taq DNA polymerase (Promega, Madison, WI) was added, and PCR was started: 30 cycles of 30 s at 94°C, 30 s at 50°C, and 90 s at 72°C.
Seeded Plate Assay for Killer Resistance
The assay was done as described by Brown et al. (1994). Briefly, yeast strains were grown to stationary phase in liquid media, and 50 μl of this culture were used to inoculate 10 ml of 1% agar, 1× Halvorson’s buffered YPD (pH 4.7), 0.001% methylene blue at 45°C. The agar was promptly poured onto 60- × 15-mm Petri dishes and allowed to cool. Five microliters of a fresh culture of K1 killer toxin secreting strain 514a/T158C (Bussey et al., 1983) were spotted, and the plates were incubated at 18°C overnight, followed by a 24°C incubation of 48 h.
Western Analysis of Wbp1p
Approximately 2.5 OD600 units of yeast cells were grown in YPD and harvested in log phase, and a total protein extract was prepared with 20% trichloroacetic acid. Cells harboring plasmids were grown in SD medium to an OD600 of 1.5. Proteins ( of each extract) were resolved by SDS-PAGE gels and transferred to a polyvinylidene difluoride membrane (Bio-Rad, Hercules, CA). Blots were incubated for 1 h at room temperature in Tris-buffered saline (20 mM Tris-HCl, pH 7.5, 150 mM NaCl) containing 0.05% Tween 20 and 2% nonfat dried milk. The incubation with antibodies against Wbp1p (a generous gift of Stephen te Heesen, University of Zurich, Zurich, Switzerland) was done overnight at 4°C. mAb HA.11 (Babco, Richmond, CA) was diluted 1:1000. Incubations were for 2 h at room temperature. Horseradish peroxidase-conjugated secondary antibody (Promega) was visualized using enhanced chemiluminescence (Western Blot Chemiluminescence Plus, New England Nuclear, Boston, MA). Quantification by volume integration was done with a Fluoro-S multi-imaging system and Multi-Analyst software from Bio-Rad.
of each extract) were resolved by SDS-PAGE gels and transferred to a polyvinylidene difluoride membrane (Bio-Rad, Hercules, CA). Blots were incubated for 1 h at room temperature in Tris-buffered saline (20 mM Tris-HCl, pH 7.5, 150 mM NaCl) containing 0.05% Tween 20 and 2% nonfat dried milk. The incubation with antibodies against Wbp1p (a generous gift of Stephen te Heesen, University of Zurich, Zurich, Switzerland) was done overnight at 4°C. mAb HA.11 (Babco, Richmond, CA) was diluted 1:1000. Incubations were for 2 h at room temperature. Horseradish peroxidase-conjugated secondary antibody (Promega) was visualized using enhanced chemiluminescence (Western Blot Chemiluminescence Plus, New England Nuclear, Boston, MA). Quantification by volume integration was done with a Fluoro-S multi-imaging system and Multi-Analyst software from Bio-Rad.
Plasmids
YEp 24-KRE6 was constructed as a 4.6-kb KRE6 BamHI-SalI fragment in the multicopy plasmid YEp24. The same KRE6 insert was subcloned into the centromeric plasmid pRS315, and nucleotides encoding the influenza virus hemagglutinin sequence were inserted in frame, giving rise to a functional, epitope-tagged pRS315-KRE6-HA (Roemer et al., 1994).
Isolation and Analysis of Cell Walls
Cell walls were isolated from 100 ml of fresh stationary phase cultures. After breaking the cells with glass beads on ice, cold 50 mM Tris-HCl (pH 7.5) with protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1.5 μg/ml leupeptin, 3.0 μg/ml pepstatin A), the cell wall fraction was collected by centrifugation at 1000 × g for 5 min and washed three times with water. β-1,6-Glucan content of isolated cell walls was determined as described by Brown et al. (1994). Briefly, alkali-insoluble glucans were extracted from the cell wall material, and after β-1,3-glucanase (Zymolyase 100T, ICN Pharmaceuticals, Costa Mesa, CA) digestion and dialysis, the β-1,6-glucan was collected and quantified as hexose by the phenol-sulfuric acid method (McKelvy and Lee, 1969).
RESULTS
The Double Mutant alg5Δcwh41Δ Is Not Hypersensitive to CFW
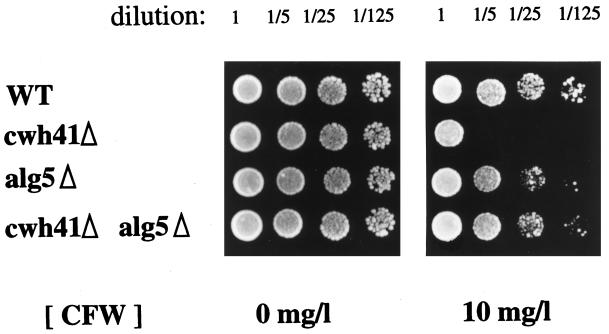
Phenotypic analysis of the double mutant alg5Δcwh41Δ is pivotal in determining whether glucosidase I (Cwh41p) has a direct or indirect role in the biosynthesis of cell wall β-1,6-glucan. We constructed the isogenic LCY20 strain, which is heterozygous for the cwh41Δ/CWH41 and alg5Δ/ALG5 locus, and analyzed the phenotype of tetratype tetrads obtained after sporulation. We initially looked at the effect of CFW, a negatively charged fluorescent dye that does not enter cells but interferes with the extracellular assembly of yeast cell wall, amplifying the consequences of cell wall mutations (Elorza et al., 1983; Murgui et al., 1985). Wild-type cells are resistant to the presence of 10 mg/l CFW in the growth medium, whereas glucosidase I-deficient cells (cwh41Δ) are not (Ram et al., 1994) (Figure 1). The double mutant cwh41Δalg5Δ grows well at 10 mg/l CFW, like the alg5Δ single mutant and the wild type (Figure 1). Because cells with a weakened cell wall are not able to tolerate additional disturbances of the cell wall caused by CFW, this result indicates that the cell wall of the double mutant cwh41Δalg5Δ is stronger than that of the single mutant cwh41Δ.
Figure 1.
Calcofluor sensitivity test. Yeast cells were resuspended at 106 cells/ml in sterile H2O. Serial dilutions of 1-, 5-, 25-, and 125-fold were made in sterile water, and 4 μl of each dilution were spotted on YPD plates containing the indicated concentration of CFW. The plates were incubated for 3 d at 24°C before photography. Strains LCY21 (SEY6210, WT), LCY24 (cwh41Δ), LCY22 (alg5Δ), and LCY23 (alg5Δcwh41Δ) represent a tetrad from strain LCY 20.
N-Glycosylation of the ER Resident Protein Wbp1p in the cwh41Δalg5Δ Double Mutant Is The Same as in the alg5Δ Single Mutant
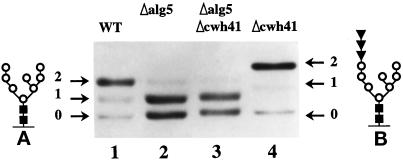
The cwh41Δ mutant has no glucosidase I activity in vitro, and all the N-linked oligosaccharides remain fully glucosylated in vivo (Romero et al., 1997). We decided to study the glycosylation of Wbp1p (a subunit of the oligosaccharyltransferase) as a model for ER resident proteins, which contain only core oligosaccharides. There is a reduction in electrophoretic mobility with respect to wild type caused by the retention of three glucoses per carbohydrate chain in cwh41Δ strains, which is clearly detected (Figure 2, lane 4 vs. 1). Similar decrease in electrophoretic mobility was seen by Esmon et al. (1984) for core glycosylated invertase and carboxypeptidase Y in gls1-1 mutants deficient in glucosidase I activity. Recently Simons et al. (1998) demonstrated that the CWH41 gene can complement the gls1-1 mutation. We observed moderate underglycosylation of Wbp1p in the alg5Δ strain (Figure 2, lane 2 vs. 1), as previously reported for this protein (Karaoglu et al., 1995, their Figure 5C, lane g vs. lane a) and for carboxypeptidase Y (te Heesen et al., 1994) in alg5Δ mutants. The isogenic double mutant cwh41Δalg5Δ showed underglycosylation of Wbp1p to the same extent as the alg5Δ single mutant (Figure 2, lane 3 vs. 2) indicating that in the absence of Dol-P-glucose synthase activity (alg5Δ), the presence or absence of glucosidase I (Cwh41p) is irrelevant. In wild-type cells where Glc3Man9GlcNAc2 is transferred to proteins, glucosidase I (Cwh41p) initiates the trimming process by removing the outermost α-1,3–linked glucose followed by glucosidase II, which removes the two remaining α-1,2–linked glucoses. Our finding in the double mutant cwh41Δalg5Δ is as expected, because in alg5Δ strains, the suboptimal substrate for oligosaccharyl transferase Man9GlcNAc2-PP-Dol is used as an oligosaccharide donor and transferred to proteins, eliminating the need for trimming.
Figure 2.
N-glycosylation of Wbp1p. Total cell extracts from strains LCY21 (lane 1), LCY22 (lane2), LCY23 (lane 3), and LCY24 (lane 4) were analyzed by 8% SDS/PAGE and Western blot. Arrows indicate the number of glycosylation sites used. (A and B) Structure of the oligosaccharides, N-acetylglucosamine (closed squares), mannose (open circles), and glucose (closed inverted triangles).
Cell Wall β-1,6-Glucan and Resistance to K1 Killer Toxin in the cwh41Δalg5Δ Double Mutant
Disruption of the CWH41 gene leads to a K1 killer toxin-resistant phenotype and a 50% reduction in the amount of cell wall β-1,6-glucan (Jiang et al., 1996). When we prepared the cell wall, removed mannoproteins, enzymatically digested the β-1,3-glucan, and determined the level of this polymer and the susceptibility to K1 killer toxin of the double mutant cwh41Δalg5Δ, we found both parameters to be indistinguishable from those of the alg5Δ single mutant (Table 2). We measured on average 15% less β-1,6-glucan polymer per milligram of cell wall in alg5Δ mutants compared with wild type (Table 2). Because of dispersion of the data, the variation in the level of β-1,6-glucan between wild type and alg5Δ mutants was found to be statistically nonsignificant using the Student’s t test (p > 0.05). Alg5Δ mutants were found to be slightly resistant to K1 killer toxin (Table 2). Some mutants with no apparent alterations in β-1,6-glucan, such as glycosylation-defective gda1 or mnt1/kre2 mutants, were also found to be partially or totally resistant to K1 toxin (Häusler et al., 1992; Hill et al., 1992; Abeijon, et al., 1993).
Table 2.
Interaction between CWH41 and ALG5 genes
| Strain | Genotype | Killer toxin size (mm)a | Cell wall β-1, 6-glucan (μg/mg of cell wall dry weight)b |
|---|---|---|---|
| LCY21 | CWH41ALG5 | 13.0 ± 0.5 | 148 ± 17 |
| LCY24 | cwh41Δ | 7.0 ± 0.3 | 69 ± 12 |
| LCY22 | alg5Δ | 10.3 ± 0.5 | 129 ± 19 |
| LCY23 | cwh41Δalg5Δ | 10.5 ± 0.3 | 124 ± 23 |
Results represent the mean ± 1 SD of five independent determinations.
Sensitivity to K1 killer toxin was determined by seeded plate tests as described in MATERIALS AND METHODS.
Total cell walls were first isolated from stationary phase cells, and then the major cell wall polymers were fractionated and measured as described by Brown et al. (1994).
The Triple Mutant cwh41Δkre6Δalg5Δ Is Viable and Has the Same Amount of Cell Wall β-1,6-glucan as kre6Δ Strains
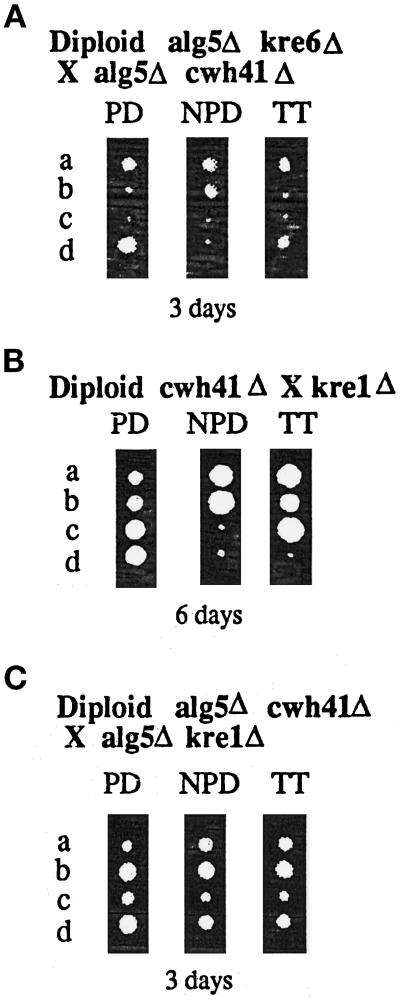
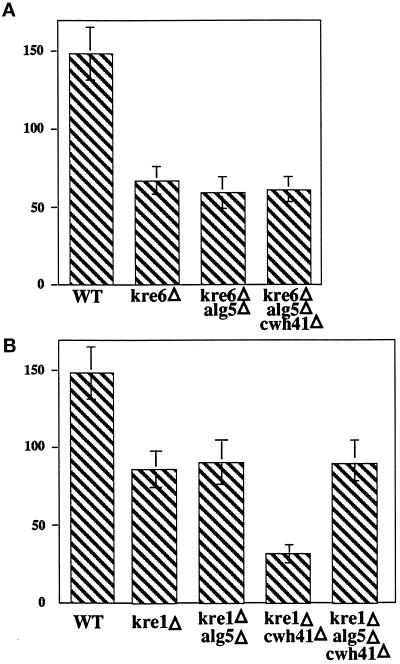
To further assess the role of CWH41 in the synthesis of β-1,6-glucan, we wanted to reexamine its interactions with KRE6 and KRE1 genes, this time in an alg5Δ background. We constructed the isogenic LCY45 strain, which is homozygous for alg5Δ/alg5Δ and heterozygous for the cwh41Δ/CWH41 and kre6Δ/KRE6 locus, sporulated, and did tetrad analysis. A total of 21 complete tetrads of 30 dissected were further analyzed and yielded 14 tetratypes (TT), 4 parental ditypes (PD), and 3 nonparental ditypes (NPD). On Figure 3A an example of each is shown. NPD and TT tetrads were found, indicating that the triple mutant cwh41Δkre6Δalg5Δ is viable in the same genetic background (SEY6210), whereas the double mutant cwh41Δkre6Δ is nonviable (Jiang et al., 1996). Spores carrying the kre6::HIS3 allele were identified by the small size of the colonies and by the total resistance to K1 killer toxin. The cwh41Δ::HIS3 allele was identified by whole-cell PCR as described in MATERIALS AND METHODS. No synthetic phenotype was observed when both null alleles (kre6 and cwh41) were present in the same spore. KRE6 encodes an integral membrane Golgi glycoprotein, probably a β-glucan synthase. Null mutants exhibit a 50% reduction in cell wall β-1,6-glucan (Roemer and Bussey, 1991). As seen in Figure 4A, the triple mutant kre6Δcwh41Δalg5Δ has the same level of cell wall β-1,6-glucan as kre6Δ mutants and alg5Δkre6Δ, showing that in the absence of dolichol-P-glucose synthase (alg5Δ), the presence or absence of glucosidase I (Cwh41p) does not have influence on β-1,6-glucan synthesis.
Figure 3.
Germination and growth of tetrads. Diploid strains were sporulated and dissected onto YPD plates and incubated at 24°C for the amount of time indicated. Tetrad types shown are parental ditype (PD), nonparental ditype (NPD), and tetratype (TT). The four spore progeny derived from each tetrad are indicated by the letters a–d to the left of each panel. (A) Diploid strain LCY45 (alg5Δ/alg5Δ, kre6Δ/KRE6, cwh41Δ/CWH41). (B) Diploid strain LCY80 (kre1Δ/KRE1, cwh41Δ/CWH41). (C) Diploid strain LCY65 (alg5Δ/alg5Δ, kre1Δ/KRE1, cwh41Δ/CWH41).
Figure 4.
Levels of cell wall β-1,6-glucan. Alkali-insoluble β-1,6-glucan was extracted from cell wall preparations of various strains and quantified in micrograms per milligram of cell wall dry weight as described in MATERIALS AND METHODS. The data represent the mean ± SD of five independent experiments. Strains A: SEY6210 (wild type), TR92 (kre6Δ), LCY33(kre6Δalg5Δ), LCY47 (kre6Δcwh41Δalg5Δ); strains on B: HAB635(kre1Δ), LCY58 (kre1Δalg5Δ), LCY81 (kre1Δcwh41Δ), LCY66 (alg5Δkre1Δcwh41Δ).
Genetic Interactions between CWH41 and KRE1 Genes
The cwh41Δkre1Δ double mutation has been shown to result in strong synergistic defects with a severely slow growth phenotype and a drastic (75%) reduction in cell wall β-1,6-glucan level (Jiang et al., 1996). To find out whether the role of glucosidase I (Cwh41p) in β-1,6-glucan synthesis is direct or indirect, we determined whether these strong genetic interactions persisted in an alg5Δ background. We constructed the isogenic strains LCY65, which is alg5Δ/alg5Δ cwh41Δ/CWH41 kre1Δ/KRE1, and LCY80 (cwh41Δ/CWH41 kre1Δ/KRE1), and after sporulation tetrad analysis was performed. As seen in Figure 3B, the cwh41Δkre1Δ double mutant displayed an extremely slow growth phenotype, giving tiny colonies even after 6 d of incubation at 24°C and had a large reduction (70%) in cell wall β-1,6-glucan (Figure 4B) as previously observed (Jiang et al., 1996). Both phenotypes were absent in the alg5Δcwh41Δkre1Δ triple mutant. Kre1Δ segregants, which were identified by its complete resistance to K1 killer toxin, formed slightly smaller colonies upon spore germination independently of the allele at the CWH41 locus in the alg5Δ background (Figure 3C, spores A and C). The level of β-1,6-glucan in the alg5Δcwh41Δkre1Δ triple mutant is the same as in the kre1Δ single mutant (Figure 4B), showing once more that the strong synergistic interaction between CWH41 and KRE1 genes is abolished in an alg5Δ background.
Kre6p Is Selectively Unstable in cwh41Δ Strains
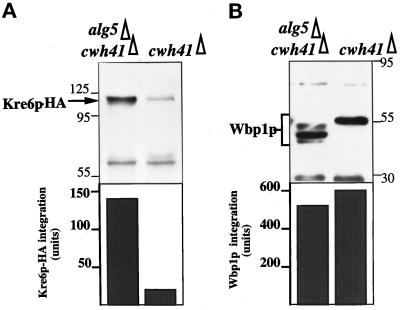
To understand why the lack of glucosidase I activity can lead to a defect in β-1,6-glucan synthesis, we looked at the stability of Kre6p in cwh41Δ strains because this protein is likely to be a glucan synthase and is a glycoprotein (Roemer et al., 1994). An 86% reduction in Kre6p in cell extracts of cwh41Δ mutant cells was found, compared with the double mutant alg5Δcwh41Δ in which nonglucosylated oligosaccharides are present (Figure 5A). This instability was selective because the ER glycoprotein Wbp1p was not degraded in cwh41Δ mutant cells (Figure 5B). Although the glycoforms of Wbp1p in cwh41Δ and alg5Δcwh41Δ mutants are different (Figures 2 and 5B), the total amount of this glycoprotein is not decreased (Figure 5B).
Figure 5.
Stability of Kre6p in the cwh41Δ mutant. Total cell extracts from strains LCY24 (cwh41Δ) and LCY23 (alg5Δcwh41Δ) were subject to 10% SDS-PAGE and Western blots, followed by quantification of the indicated proteins (see MATERIALS AND METHODS). Integration is expressed in arbitrary units. The same membrane was probed successively with antiserum against the hemagglutinin epitope tag of Kre6p (A) and Wbp1p (B).
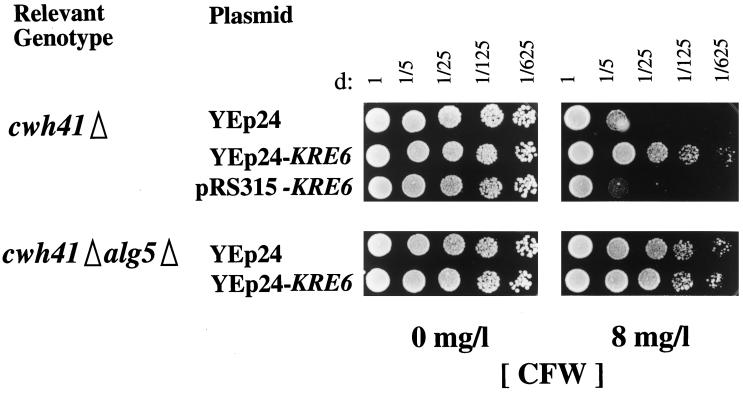
We reasoned that if a reduction in the amount of Kre6p available in glucosidase I mutants was a critical factor for the synthesis of β-1,6-glucan and for cell wall stability, overexpression of this protein should alleviate this phenotype. As seen in Figure 6, overexpression of Kre6p almost completely abolished the hypersensitivity of cwh41Δ strains to CFW, whereas it had no effect on the double mutant alg5Δcwh41Δ.
Figure 6.
Kre6p overexpression and CFW sensitivity of cwh41Δ mutant. The sensitivity test was done as described in the legend of Figure 1. Concentrations of CFW are indicated. Strains LCY24 (cwh41Δ) and LCY23 (alg5Δcwh41Δ) were transformed with either multicopy (YEp24) or centromeric (pRS315) plasmids containing KRE6 or KRE6-HA.
DISCUSSION
Our results indicate that glucose trimming by glucosidase I (Cwh41p) is indirectly required for the biosynthesis of cell wall β-1,6-glucan in S. cerevisiae. When the nonglucosylated oligosaccharide Man9GlcNAc2 is transferred to N-linked glycoproteins as in the alg5Δ strains, the amount of β-1,6-glucan in the cell wall is not influenced by the presence or absence of glucosidase I. This implies that there is no other function for glucosidase I (Cwh41p) in the biosynthesis of cell wall components, besides its established role in glucose trimming during N-linked glycoprotein processing. Because ALG5 is the single gene encoding dolichol-P-glucose synthase in S. cerevisiae (te Heesen et al., 1994), our results also demonstrate that dolichol-P-glucose does not serve as a glucose donor in the biosynthesis of β-1,6-glucan.
How can the retention of three glucoses per N-linked carbohydrate chain have profound effects on the ability of gls1/cwh41 mutant cells to maintain wild-type levels of cell wall β-1,6-glucan even though growth rate, mannan outer chain addition to glyco-proteins, and secretion are normal? A possible explanation is our observation of a severe and selective instability of a glycoprotein Kre6p, required for β-1,6-glucan synthesis in glucosidase I mutants. To our knowledge, this is the first time such a phenomenon has been found in yeast, even though it is well documented in mammalian cells (for reviews, see Datema et al., 1987; Elbein, 1991). Several glycoproteins encoded by KRE genes, responsible for β-1,6-glucan synthesis, reside in organelles along the secretory pathway and in the periplasmic space; others may be affected too. The lack of glucose trimming has also been shown to have physiological consequences in lower eukaryotes, where disruption of the glucosidase II gene in Dictyostelium discoideum results in a developmental phenotype with mutants growing well but forming abnormal fruiting bodies (Freeze et al., 1997).
Not much is known about the mechanism of biosynthesis of β-1,6-glucan. The site of initiation of the polymer chains is very likely the ER lumen, where Kre5p resides. Although this protein is essential for β-1,6-glucan synthesis (Meaden et al., 1990) and has some homology with glucosyltransferases (Parker et al., 1995; Fernandez et al., 1996), no enzymatic activity has yet been assigned to it. Our work demonstrates that dolichol-P-glucose is not the glucose donor in the biosynthesis of β-1,6-glucan. Consequently, UDP-glucose is very likely the sugar donor in this process. This raises a topographical problem, because nucleotide sugars are synthesized in the cytosol and require specific transporter proteins located on the organellar membranes, such as ER and Golgi, to become available in the lumen as substrates for macromolecular synthesis (Abeijon et al., 1997). We have detected transport of UDP-glucose into microsomal vesicles from S. cerevisiae (Abeijon, unpublished data), and further work will address the issue of whether UDP-glucose transport is a prerequisite for the biosynthesis of β-1,6-glucan. A very drastic and specific reduction in the levels of this polymer was found when mutations in either CWH41/GLS1 or GLS2 genes were combined with mutation in KAR2, the yeast homologue of the ER molecular chaperone Bip (Simons et al., 1998).
Further along the secretory pathway, there are other glycoproteins that participate in the biosynthesis of β-1,6-glucan. Kre6p and Skn1p are type II integral membrane proteins in the Golgi apparatus likely to be glucan synthases (Roemer and Bussey, 1991; Roemer et al., 1993, 1994). Kre1p is a heavily O-glycosylated, agglutinin-like protein that localizes mainly to the periplasmic space and is necessary for the addition of β-1,6-linked outer chains to a core glucan structure (Boone et al., 1990; Roemer and Bussey, 1995).
Our conclusion that the role of glucosidase I/Cwh41p in the biosynthesis of β-1,6-glucan is indirect is strengthened by the observations indicating that the triple mutant cwh41Δalg5Δkre6Δ is viable, whereas the double mutant cwh41Δkre6Δ in the same genetic background is not (Jiang et al., 1996), and it contains the same amount of β-1,6-glucan in the cell wall as the single kre6Δ mutant. Because the alg5Δcwh41Δ double mutant had normal levels of cell wall β-1,6-glucan, the interactions between CWH41, KRE6, and KRE1 genes should not be present in an alg5 deletion background. Synergistic defects detected in the cwh41Δkre1Δ double mutant (Jiang et al., 1996) are not present in the triple mutant alg5Δcwh41Δkre1Δ. Clearly the trimming of N-linked oligosaccharides affects the biosynthesis of cell wall β-1,6-glucan only indirectly, by selectively reducing the stability of Kre6p and possibly other glycoproteins involved in the pathway.
The cellular location where β-1,6-glucan becomes covalently attached to cell wall mannoproteins is still unknown and could be intracellular. It is only in the periplasmic space that all the components of the yeast cell wall get together. Available evidence indicates that chitin (Bulawa, 1993) and β-1,3-glucan (Drgonova et al., 1996) are synthesized at the plasma membrane with simultaneous secretion into the periplasmic space. On the other hand, mannoproteins and probably β-1,6-glucan are synthesized in the ER and modified during their transport through the secretory pathway and end up anchored in the external leaflet of the plasma membrane or in the periplasmic space. As the covalent linkage between β-1,6-glucan and mannoproteins in the cell wall is through a portion of the glycosylphosphatidylinositol anchor (Kollar et al., 1997), it is possible that subcomplexes between both polymers may be preassembled during their intracellular synthesis. Linkages have been demonstrated between mannoproteins, β-1,6-glucan, and β-1,3-glucan (Kapteyn et al., 1996) and between the latter and chitin (Kollar et al., 1995). The central molecule that appears to keep together all components of the yeast cell wall is β-1,6-glucan (Kollar, et al., 1997); therefore, understanding its biosynthesis is of critical importance.
ACKNOWLEDGMENTS
We are grateful to Howard Bussey for generosity providing many strains and plasmids used in this study. We also thank Peter Dijkgraaf for a detailed protocol on how to measure β-1,6-glucan, Stephen te Heesen and Markus Aebi for the pALG5Δ(Spe–Bgl) HIS plasmid and the antibody against Wbp1p, and Duane Jenness and Reid Gilmore for helpful discussions. This work was supported by National Institutes of Health grant GM 30365 to C. Hirschberg and a Small Grant Program award from University of Massachusetts Medical Center (to C.A.).
Abbreviations used:
- CFW
Calcofluor White
- ER
endoplasmic reticulum
REFERENCES
- Abeijon C, Mandon E, Hirschberg CB. Transport of nucleotide sugars, nucleotide sulfate and ATP into the Golgi apparatus. Trends Biochem Sci. 1997;22:203–207. doi: 10.1016/s0968-0004(97)01053-0. [DOI] [PubMed] [Google Scholar]
- Abeijon C, Robbins PW, Hirschberg CB. Molecular cloning of the Golgi apparatus uridine diphosphate N-acetylglucosamine transporter by phenotypic correction of a yeast mutant. Proc Natl Acad Sci USA. 1996;93:5963–5972. doi: 10.1073/pnas.93.12.5963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeijon C, Yanagisawa K, Mandon EC, Hausler A, Moremen K, Hirschberg CB, Robbins PW. Guanosine diphosphatase is required for protein and sphingolipid glycosylation in the Golgi lumen of Saccharomyces cerevisiae. J Cell Biol. 1993;122:307–323. doi: 10.1083/jcb.122.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone C, Sommer SS, Hensel A, Bussey H. Yeast KRE genes provide evidence for a pathway of cell wall β-glucan assembly. J Cell Biol. 1990;110:1833–1843. doi: 10.1083/jcb.110.5.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL, Kossaczka Z, Jiang B, Bussey H. A mutational analysis of killer toxin resistance in Saccharomyces cerevisiae identifies new genes involved in cell wall β 1,6-glucan synthesis. Genetics. 1993;133:837–849. doi: 10.1093/genetics/133.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL, Roemer T, Lussier M, Sdicu A-M, Bussey H. The K1 killer toxin: molecular and genetic applications to secretion and cell surface assembly. In: Johnston JR, editor. Molecular Genetics of Yeast. A Practical Approach. Oxford, United Kingdom: IRL Press; 1994. pp. 217–232. [Google Scholar]
- Bulawa CE. Genetics and molecular biology of chitin synthesis in fungi. Annu Rev Microbiol. 1993;47:505–534. doi: 10.1146/annurev.mi.47.100193.002445. [DOI] [PubMed] [Google Scholar]
- Bussey H, Saville D, Hutchins K, Palfree RGE. Binding of yeast killer toxin to a cell wall receptor on sensitive Saccharomyces cerevisiae. J Bacteriol. 1979;140:888–892. doi: 10.1128/jb.140.3.888-892.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussey H, Steinmetz O, Saville D. Protein secretion in yeast: two chromosomal mutants that oversecrete killer toxin in Saccharomyces cerevisiae. Curr Genet. 1983;7:449–456. doi: 10.1007/BF00377610. [DOI] [PubMed] [Google Scholar]
- Datema R, Olofson S, Romero PA. Inhibitors of protein glycosylation and glycoprotein processing in viral systems. Pharmacol Ther. 1987;33:221–286. doi: 10.1016/0163-7258(87)90066-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drgonová J, Drgon T, Tanaka K, Kollar R, Chen G-C, Ford RA, Chan CSM, Takai Y, Cabib E. Rholp, a yeast protein at the interface between cell polarization and morphogenesis. Science. 1996;272:277–279. doi: 10.1126/science.272.5259.277. [DOI] [PubMed] [Google Scholar]
- Elbein AD. Glycosidase inhibitors as antiviral and/or antitumor agents. Semin Cell Biol. 1991;2:309–317. [PubMed] [Google Scholar]
- Elorza MV, Rico H, Sentandreu R. Calcofluor White alters the assembly of chitin fibrils in Saccharomyces cerevisiae and Candida albicans cells. J Gen Microbiol. 1983;129:1577–1582. doi: 10.1099/00221287-129-5-1577. [DOI] [PubMed] [Google Scholar]
- Esmon B, Esmon PC, Schekman R. Early steps in processing of yeast glycoproteins. J Biol Chem. 1984;259:10322–10327. [PubMed] [Google Scholar]
- Fernandez F, Jannatipour M, Hellman U, Rokeach LA, Parodi A. A new stress protein: synthesis of Schizosaccharomyces pombe UDP-Glc:glycoprotein glucosyltransferase mRNA is induced by stress conditions but the enzyme is not essential for viability. EMBO J. 1996;15:705–713. [PMC free article] [PubMed] [Google Scholar]
- Fleet GH, Manners DJ. Isolation and composition of an alkali-soluble glucan from the cell walls of Saccharomyces cerevisiae. J Gen Microbiol. 1976;94:180–192. doi: 10.1099/00221287-94-1-180. [DOI] [PubMed] [Google Scholar]
- Freeze HH, Lammertz M, Iranfar N, Fuller D, Panneerselvam K, Loomis WF. Consequences of disrupting the gene that encodes α-glucosidase II in the N-linked oligosaccharide biosynthesis pathway of Dictyostelium discoideum. Dev Genet. 1997;21:177–186. doi: 10.1002/(SICI)1520-6408(1997)21:3<177::AID-DVG1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Häusler A, Ballou L, Ballou CE, Robbins PW. Yeast glycoprotein biosynthesis: MNTI encodes an α-1,2-mannosyltransferase involved in O-glycosylation. Proc Natl Acad Sci USA. 1992;89:6846–6850. doi: 10.1073/pnas.89.15.6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K, Boone C, Goebl M, Puccia R, Sdicu A-M, Bussey H. Yeast KRE2 defines a new gene family encoding probable secretory proteins, and is required for the correct N-glycosylation of proteins. Genetics. 1992;130:273–283. doi: 10.1093/genetics/130.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker TC, Robbins PW. Yeast mutants deficient in protein glycosylation. Proc Natl Acad Sci USA. 1983;80:7466–7470. doi: 10.1073/pnas.80.24.7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B, Sheraton J, Ram AFJ, Dijkgraaf GJP, Klis FM, Bussey H. CWH41 encodes a novel endoplasmic reticulum membrane N-glycoprotein involved in β 1,6-glucan assembly. J Bacteriol. 1996;178:1162–1171. doi: 10.1128/jb.178.4.1162-1171.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapteyn JC, Montijn RC, Vink E, de la Cruz J, Llobell A, Shimoi H, Lipke P, Klis FM. Retention of Saccharomyces cerevisiae cell wall proteins through a phosphodiester linked β-1,3/β-1,6 glucan heteropolymer. Glycobiology. 1996;6:337–345. doi: 10.1093/glycob/6.3.337. [DOI] [PubMed] [Google Scholar]
- Karaoglu D, Kelleher DJ, Gilmore R. Functional Characterization of Ost3p. Loss of the 34kD subunit of the Saccharomyces cerevisiae oligosaccharyltransferase results in biased underglycosylation of acceptor substrates. J Cell Biol. 1995;130:567–577. doi: 10.1083/jcb.130.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klis FM. Cell wall assembly in yeast. Yeast. 1994;10:851–869. doi: 10.1002/yea.320100702. [DOI] [PubMed] [Google Scholar]
- Kollar R, Petrakova E, Ashwell G, Robbins PW, Cabib E. Architecture of the yeast cell wall. The linkage between chitin and β (1→3) glucan. J Biol Chem, 1995;270:1170–1178. doi: 10.1074/jbc.270.3.1170. [DOI] [PubMed] [Google Scholar]
- Kollar R, Reinolds BB, Petrakova E, Yeh HJC, Ashwell G, Drgonova J, Kapteyn JC, Klis FM, Cabib E. Architecture of the yeast cell wall. β (1→6)-Glucan interconnects mannoprotein, β (1→3)-glucan, and chitin. J Biol Chem. 1997;272:17762–17775. doi: 10.1074/jbc.272.28.17762. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- McKelvy J, Lee YC. Microheterogeneity of the carbohydrate group of Aspergillus oryzae α-amylase. Arch Biochem Biophys. 1969;132:99–110. doi: 10.1016/0003-9861(69)90341-5. [DOI] [PubMed] [Google Scholar]
- Meaden P, Hill K, Wagner J, Slipetz D, Sommer SS, Bussey H. The yeast KRE5 gene encodes a probable endoplasmic reticulum protein required for (1–6)-β-d-glucan synthesis and normal cell growth. Mol Cell Biol. 1990;10:3013–3019. doi: 10.1128/mcb.10.6.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgui A, Elorza MV, Sentandreu R. Effect of papulacandin B and Calcofluor White on the incorporation of mannoproteins in the wall of Candida albicans blastopores. Biochim Biophys Acta. 1985;841:215–222. doi: 10.1016/0304-4165(85)90024-8. [DOI] [PubMed] [Google Scholar]
- Parker CG, Fessler LI, Nelson RE, Fessler JH. Drosophila UDP-glucose:glycoprotein glucosyltransferase: sequence and characterization of an enzyme that distinguishes between denatured and native proteins. EMBO J. 1995;14:1294–1303. doi: 10.1002/j.1460-2075.1995.tb07115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram AFJ, Walters A, Hooper RT, Klis FM. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to Calcofluor White. Yeast. 1994;10:1019–1030. doi: 10.1002/yea.320100804. [DOI] [PubMed] [Google Scholar]
- Roemer T, Bussey H. Yeast β-glucan synthesis: KRE6 encodes a predicted type II membrane protein required for glucan synthesis in vivo and for glucan synthase activity in vitro. Proc Natl Acad Sci USA. 1991;88:11295–11299. doi: 10.1073/pnas.88.24.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roemer T, Bussey H. Yeast Kre1p is a cell surface O-glycoprotein. Mol Gen Genet. 1995;249:209–216. doi: 10.1007/BF00290368. [DOI] [PubMed] [Google Scholar]
- Roemer T, Paravicine G, Payton MA, Bussey H. Characterization of the yeast (1–6)-β-glucan biosynthetic components, Kre6p and Skn1p, and genetic interactions between the PKC1 pathway and extracellular matrix assembly. J Cell Biol. 1994;127:567–579. doi: 10.1083/jcb.127.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero PA, Dijkgraaf GJP, Shahinian S, Herscovics A, Bussey H. The yeast CWH41 gene encodes glucosidase. Glycobiology. 1997;7:997–1004. doi: 10.1093/glycob/7.7.997. [DOI] [PubMed] [Google Scholar]
- Rothstein RJ. One step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Runge KW, Huffaker TC, Robbins PW. Two yeast mutations in glucosylation steps of the asparagine glycosylation pathway. J Biol Chem. 1984;259:412–417. [PubMed] [Google Scholar]
- Sherman G, Fink GR, Hicks JB. Methods in Yeast Genetics, A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Simons JF, Ebersold M, Helenius A. Cell wall 1,6-β-glucan synthesis in S. cerevisiae depends on ER glucosidases I and II, and molecular chaperone Bip/Kar2p. EMBO J. 1998;17:396–405. doi: 10.1093/emboj/17.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Heesen S, Lehle L, Weissmann A, Aebi M. Isolation of the ALG5 locus encoding the UDP-glucose: dolichol-phosphate glucosyltransferase from Saccharomyces cerevisiae. Eur J Biochem. 1994;224:71–79. doi: 10.1111/j.1432-1033.1994.tb19996.x. [DOI] [PubMed] [Google Scholar]