Abstract
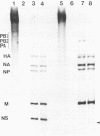
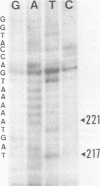
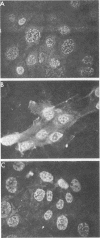
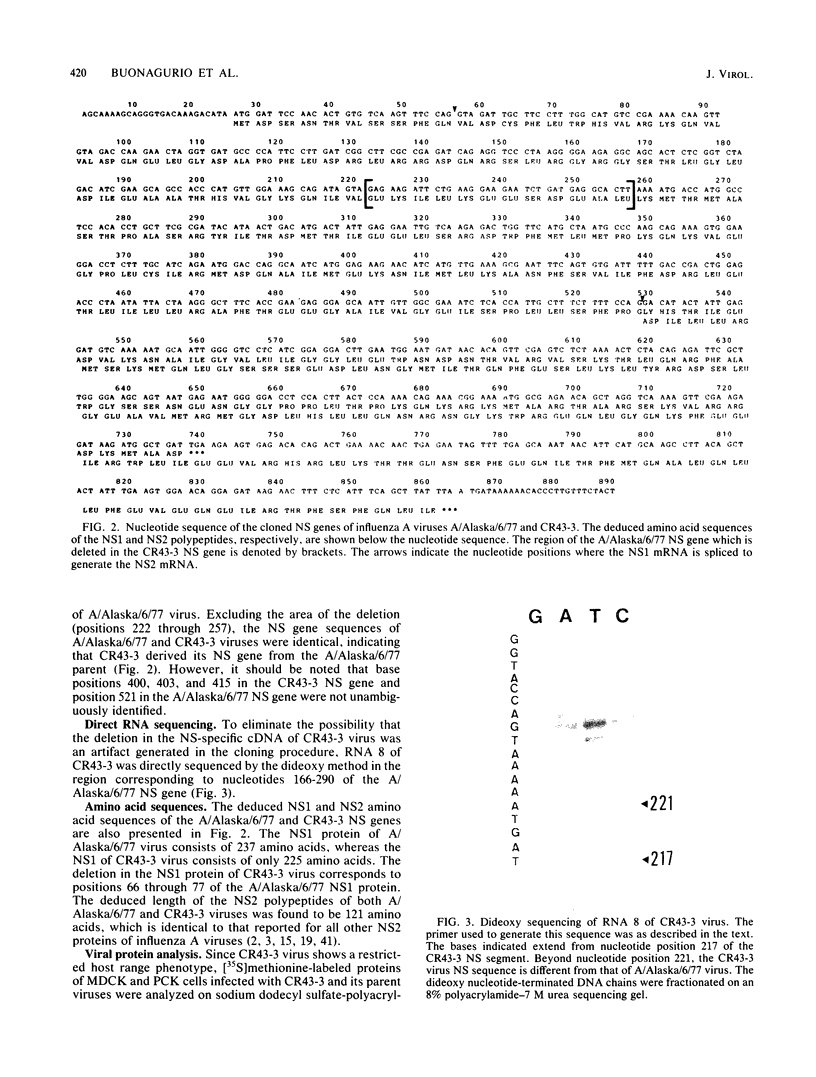
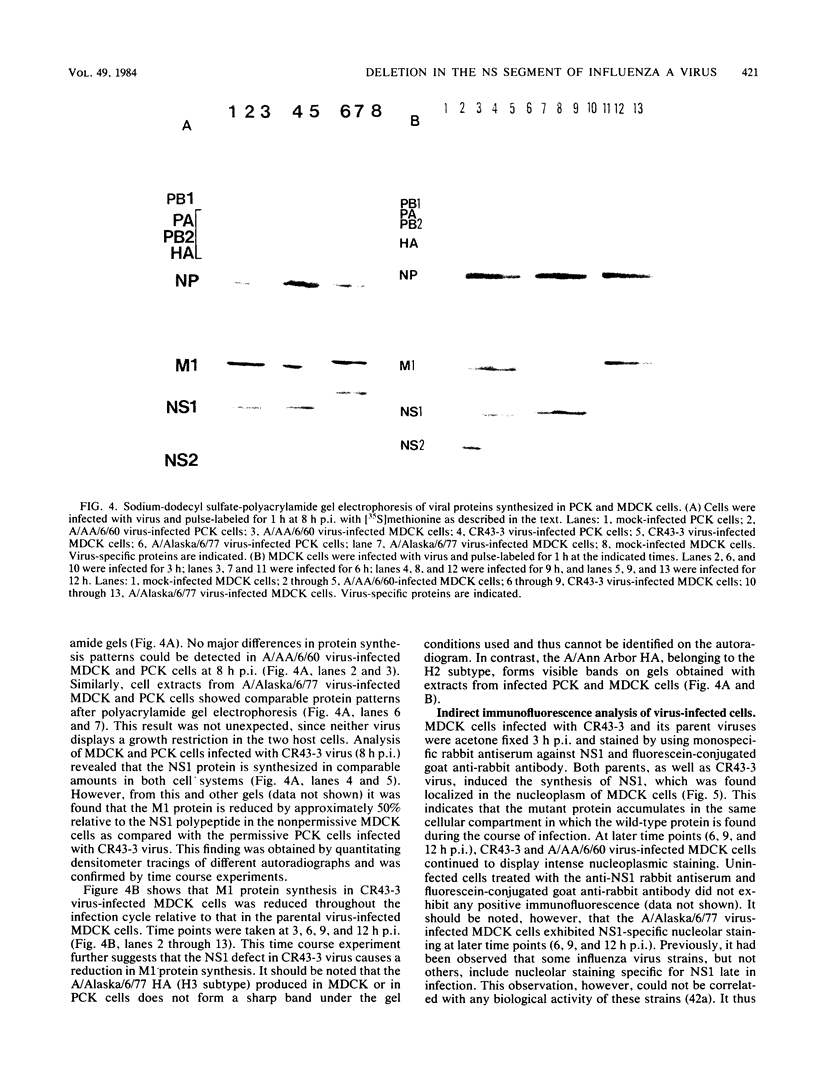
The influenza virus host range mutant CR43-3, derived by recombination from the A/Alaska/6/77 and the cold-adapted and temperature-sensitive A/Ann Arbor/6/60 viruses, has previously been shown to possess a defect in the NS gene. To characterize this defect, nucleotide sequence data were obtained from cloned cDNAs. The CR43-3 NS gene was found to be 854 nucleotides long and to derive from the NS gene of the A/Alaska/6/77 parent virus by an internal deletion of 36 nucleotides. Direct sequencing of RNA 8 of CR43-3 virus confirmed that the deletion in the NS1-coding region was not an artifact that was generated during the cloning procedure. Protein analysis indicated that the NS1 protein of CR43-3 virus was synthesized in equal amounts in the restrictive (MDCK) cells as well as in the permissive (PCK) host cells. Also, indirect immunofluorescence studies of virus-infected cells showed that the NS1 protein of CR43-3 virus, like that of the parent viruses, accumulates in the nuclei of both cell systems. Although no differences in synthesis or localization of the NS1 protein could be detected, a consistent reduction in M1 protein was noted in CR43-3 virus-infected, nonpermissive cells as compared with that of the permissive host. Since analysis of the CR43-3 virus required us to obtain the NS nucleotide sequence of the 1977 isolate A/Alaska/6/77, we were able to compare this sequence with those of corresponding genes of earlier strains. The result of this analysis supports the idea of a common lineage of human influenza A viruses isolated over a 43-year period.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Almond J. W., McGeoch D., Barry R. D. Method for assigning temperature-sensitive mutations of influenza viruses to individual segments of the genome. Virology. 1977 Aug;81(1):62–73. doi: 10.1016/0042-6822(77)90058-7. [DOI] [PubMed] [Google Scholar]
- Baez M., Taussig R., Zazra J. J., Young J. F., Palese P., Reisfeld A., Skalka A. M. Complete nucleotide sequence of the influenza A/PR/8/34 virus NS gene and comparison with the NS genes of the A/Udorn/72 and A/FPV/Rostock/34 strains. Nucleic Acids Res. 1980 Dec 11;8(23):5845–5858. doi: 10.1093/nar/8.23.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baez M., Zazra J. J., Elliott R. M., Young J. F., Palese P. Nucleotide sequence of the influenza A/duck/Alberta/60/76 virus NS RNA: conservation of the NS1/NS2 overlapping gene structure in a divergent influenza virus RNA segment. Virology. 1981 Aug;113(1):397–402. doi: 10.1016/0042-6822(81)90166-5. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blok J., Air G. M. Block deletions in the neuraminidase genes from some influenza A viruses of the N1 subtype. Virology. 1982 Apr 15;118(1):229–234. doi: 10.1016/0042-6822(82)90337-3. [DOI] [PubMed] [Google Scholar]
- Caton A. J., Brownlee G. G., Yewdell J. W., Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell. 1982 Dec;31(2 Pt 1):417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- Desselberger U., Palese P. Molecular weights of RNA segments of influenza A and B viruses. Virology. 1978 Jul 15;88(2):394–399. doi: 10.1016/0042-6822(78)90297-0. [DOI] [PubMed] [Google Scholar]
- Dimmock N. J. New virus-specific antigens in cells infected with influenza virus. Virology. 1969 Oct;39(2):224–234. doi: 10.1016/0042-6822(69)90042-7. [DOI] [PubMed] [Google Scholar]
- Fields S., Winter G. Nucleotide sequences of influenza virus segments 1 and 3 reveal mosaic structure of a small viral RNA segment. Cell. 1982 Feb;28(2):303–313. doi: 10.1016/0092-8674(82)90348-8. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis S. C., Barrett T., Brown C. M., Almond J. W. The smallest genome RNA segment of influenza virus contains two genes that may overlap. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3790–3794. doi: 10.1073/pnas.76.8.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koennecke I., Boschek C. B., Scholtissek C. Isolation and properties of a temperature-sensitive mutant (ts 412) of an influenza A virus recombinant with a ts lesion in the gene coding for the nonstructural protein. Virology. 1981 Apr 15;110(1):16–25. doi: 10.1016/0042-6822(81)90003-9. [DOI] [PubMed] [Google Scholar]
- Krug R. M., Soeiro R. Studies on the intranuclear localization of influenza virus-specific proteins. Virology. 1975 Apr;64(2):378–387. doi: 10.1016/0042-6822(75)90114-2. [DOI] [PubMed] [Google Scholar]
- Krystal M., Buonagurio D., Young J. F., Palese P. Sequential mutations in the NS genes of influenza virus field strains. J Virol. 1983 Feb;45(2):547–554. doi: 10.1128/jvi.45.2.547-554.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystal M., Young J. F., Palese P., Wilson I. A., Skehel J. J., Wiley D. C. Sequential mutations in hemagglutinins of influenza B virus isolates: definition of antigenic domains. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4527–4531. doi: 10.1073/pnas.80.14.4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R. A., Choppin P. W. Segment 8 of the influenza virus genome is unique in coding for two polypeptides. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4908–4912. doi: 10.1073/pnas.76.10.4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R. A., Lai C. J. Sequence of interrupted and uninterrupted mRNAs and cloned DNA coding for the two overlapping nonstructural proteins of influenza virus. Cell. 1980 Sep;21(2):475–485. doi: 10.1016/0092-8674(80)90484-5. [DOI] [PubMed] [Google Scholar]
- Lazarowitz S. G., Compans R. W., Choppin P. W. Influenza virus structural and nonstructural proteins in infected cells and their plasma membranes. Virology. 1971 Dec;46(3):830–843. doi: 10.1016/0042-6822(71)90084-5. [DOI] [PubMed] [Google Scholar]
- Maassab H. F. Adaptation and growth characteristics of influenza virus at 25 degrees c. Nature. 1967 Feb 11;213(5076):612–614. doi: 10.1038/213612a0. [DOI] [PubMed] [Google Scholar]
- Maassab H. F., DeBorde D. C. Characterization of an influenza A host range mutant. Virology. 1983 Oct 30;130(2):342–350. doi: 10.1016/0042-6822(83)90088-0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odagiri T., DeBorde D. C., Maassab H. F. Cold-adapted recombinants of influenza A virus in MDCK cells. I. Development and characterization of A/Ann Arbor/6/60 X A/Alaska/6/77 recombinant viruses. Virology. 1982 May;119(1):82–95. doi: 10.1016/0042-6822(82)90067-8. [DOI] [PubMed] [Google Scholar]
- Palese P., Young J. F. Variation of influenza A, B, and C viruses. Science. 1982 Mar 19;215(4539):1468–1474. doi: 10.1126/science.7038875. [DOI] [PubMed] [Google Scholar]
- Parvin J. D., Young J. F., Palese P. Nonsense mutations affecting the lengths of the NS1 nonstructural proteins of influenza A virus isolates. Virology. 1983 Jul 30;128(2):512–517. doi: 10.1016/0042-6822(83)90280-5. [DOI] [PubMed] [Google Scholar]
- Ritchey M. B., Palese P., Schulman J. L. Mapping of the influenza virus genome. III. Identification of genes coding for nucleoprotein, membrane protein, and nonstructural protein. J Virol. 1976 Oct;20(1):307–313. doi: 10.1128/jvi.20.1.307-313.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J. S., Robertson E., Roditi I., Almond J. W., Inglis S. C. Sequence analysis of fowl plague virus mutant ts47 reveals a nonsense mutation in the NS1 gene. Virology. 1983 Apr 15;126(1):391–394. doi: 10.1016/0042-6822(83)90489-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtissek C., Spring S. B. Extragenic suppression of temperature-sensitive mutations in RNA segment 8 by replacement of different rna segments with those of other influenza A virus prototype strains. Virology. 1982 Apr 15;118(1):28–34. doi: 10.1016/0042-6822(82)90316-6. [DOI] [PubMed] [Google Scholar]
- Shaw M. W., Lamon E. W., Compans R. W. Surface expression of a nonstructural antigen on influenza A virus-infected cells. Infect Immun. 1981 Dec;34(3):1065–1067. doi: 10.1128/iai.34.3.1065-1067.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K., Mullinix M. G., Chanock R. M., Murphy B. R. Temperature-sensitive mutants of influenza A/Udorn/72 (H3N2) virus. I. Isolation of temperature-sensitive mutants some of which exhibit host-dependent temperature sensitivity. Virology. 1982 Feb;117(1):38–44. doi: 10.1016/0042-6822(82)90505-0. [DOI] [PubMed] [Google Scholar]
- Shimizu K., Mullinix M. G., Chanock R. M., Murphy B. R. Temperature-sensitive mutants of influenza A/Udorn/72 (H3N2) virus. II. Genetic analysis and demonstration of intrasegmental complementation. Virology. 1982 Feb;117(1):45–61. doi: 10.1016/0042-6822(82)90506-2. [DOI] [PubMed] [Google Scholar]
- Shimizu K., Mullinix M. G., Chanock R. M., Murphy B. R. Temperature-sensitive mutants of influenza A/Udorn/72 (H3N2) virus. III. Genetic analysis of temperature-dependent host range mutants. Virology. 1983 Jan 15;124(1):35–44. doi: 10.1016/0042-6822(83)90288-x. [DOI] [PubMed] [Google Scholar]
- Staden R. A strategy of DNA sequencing employing computer programs. Nucleic Acids Res. 1979 Jun 11;6(7):2601–2610. doi: 10.1093/nar/6.7.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Further procedures for sequence analysis by computer. Nucleic Acids Res. 1978 Mar;5(3):1013–1016. doi: 10.1093/nar/5.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Sequence data handling by computer. Nucleic Acids Res. 1977 Nov;4(11):4037–4051. doi: 10.1093/nar/4.11.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. M., Hampson A. W., Layton J. E., White D. O. The polypeptides of influenza virus. IV. An analysis of nuclear accumulation. Virology. 1970 Nov;42(3):744–752. doi: 10.1016/0042-6822(70)90320-x. [DOI] [PubMed] [Google Scholar]
- Winter G., Fields S., Gait M. J., Brownlee G. G. The use of synthetic oligodeoxynucleotide primers in cloning and sequencing segment of 8 influenza virus (A/PR/8/34). Nucleic Acids Res. 1981 Jan 24;9(2):237–245. doi: 10.1093/nar/9.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolstenholme A. J., Barrett T., Nichol S. T., Mahy B. W. Influenza virus-specific RNA and protein syntheses in cells infected with temperature-sensitive mutants defective in the genome segment encoding nonstructural proteins. J Virol. 1980 Jul;35(1):1–7. doi: 10.1128/jvi.35.1.1-7.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. F., Desselberger U., Palese P., Ferguson B., Shatzman A. R., Rosenberg M. Efficient expression of influenza virus NS1 nonstructural proteins in Escherichia coli. Proc Natl Acad Sci U S A. 1983 Oct;80(19):6105–6109. doi: 10.1073/pnas.80.19.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. F., Palese P. Evolution of human influenza A viruses in nature: recombination contributes to genetic variation of H1N1 strains. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6547–6551. doi: 10.1073/pnas.76.12.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]