Abstract
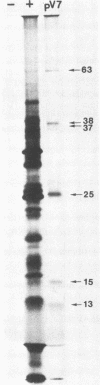
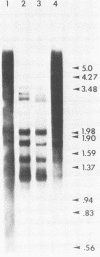
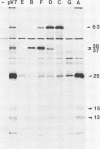
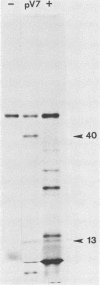
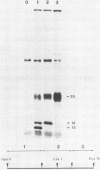
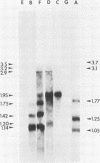
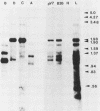
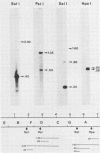
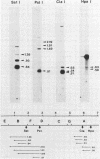
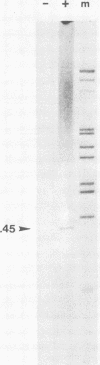
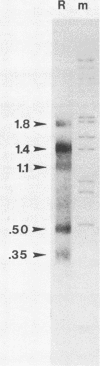
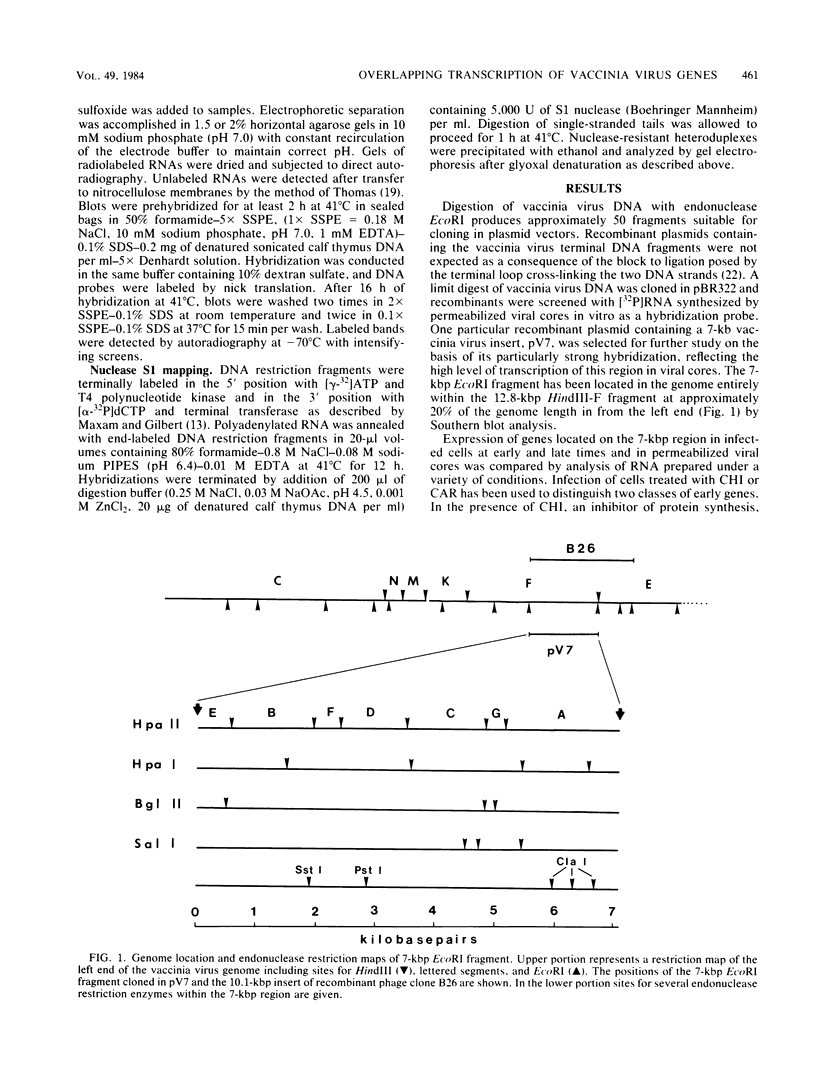
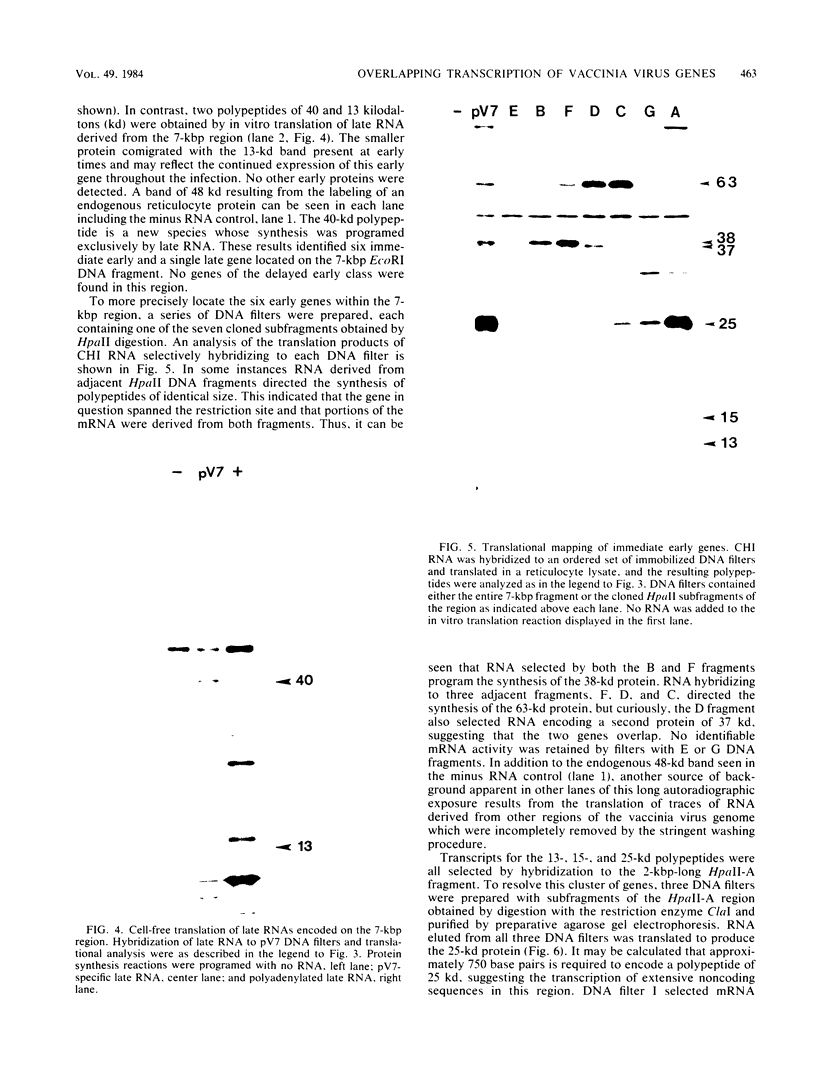
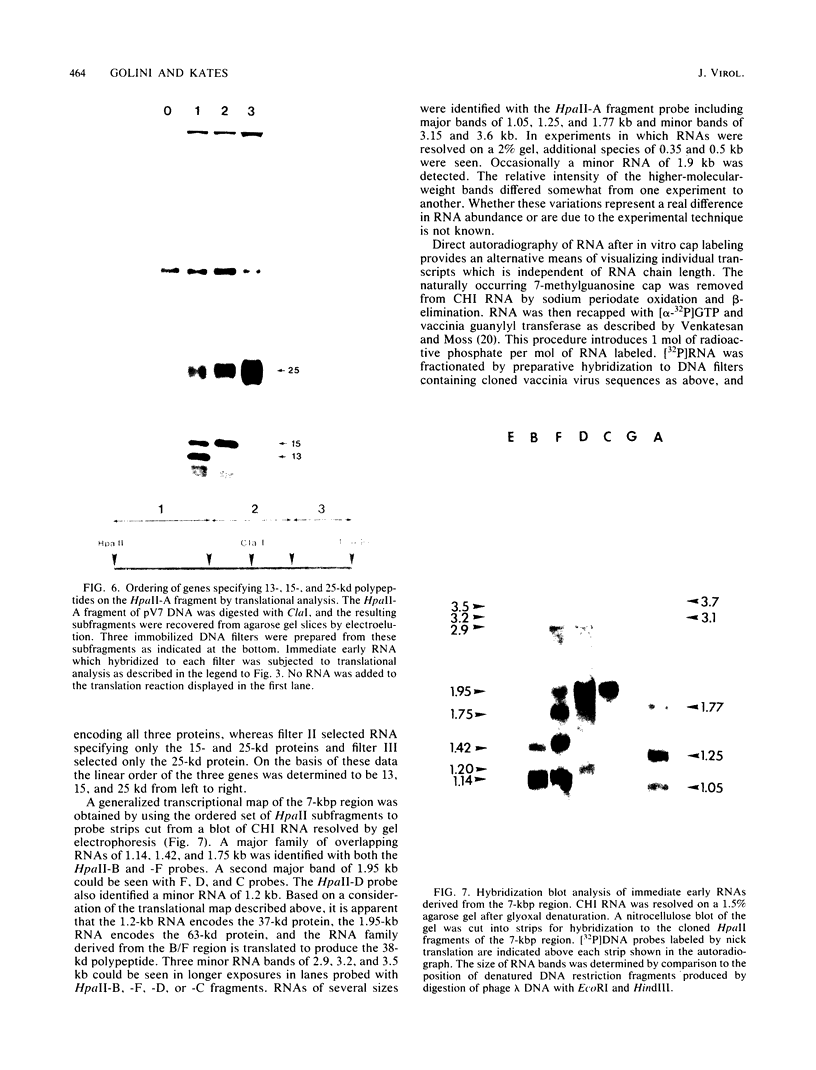
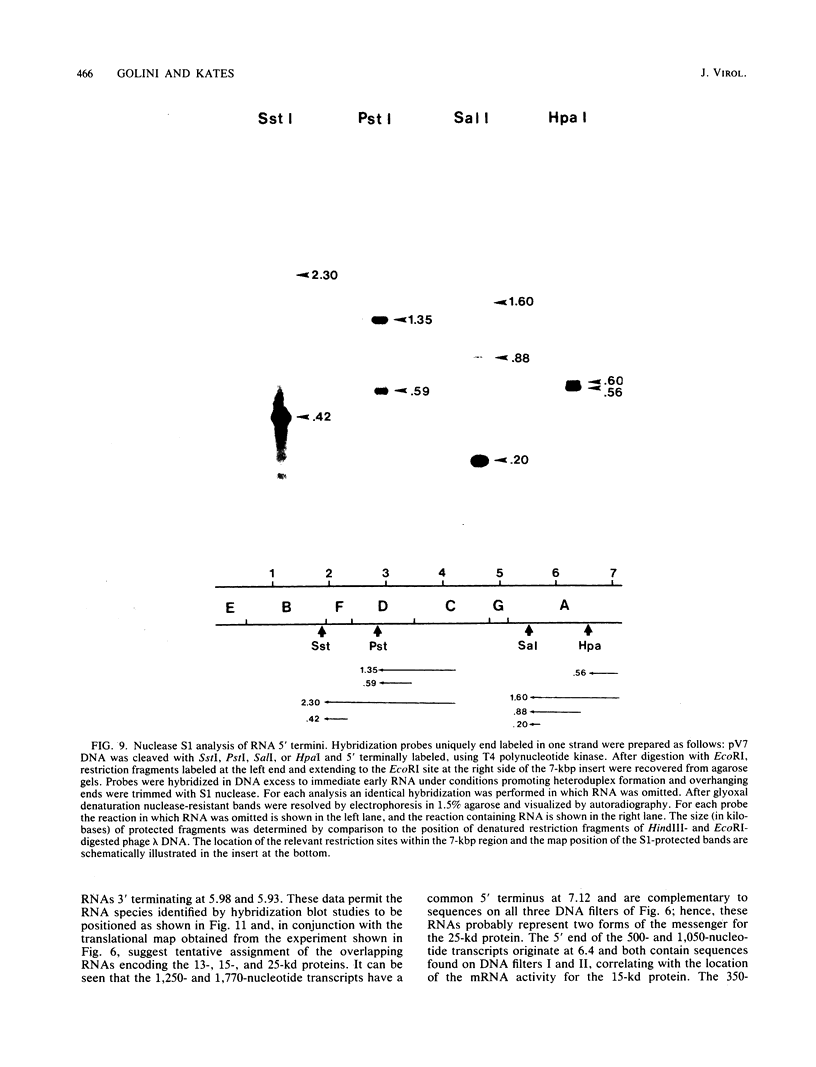
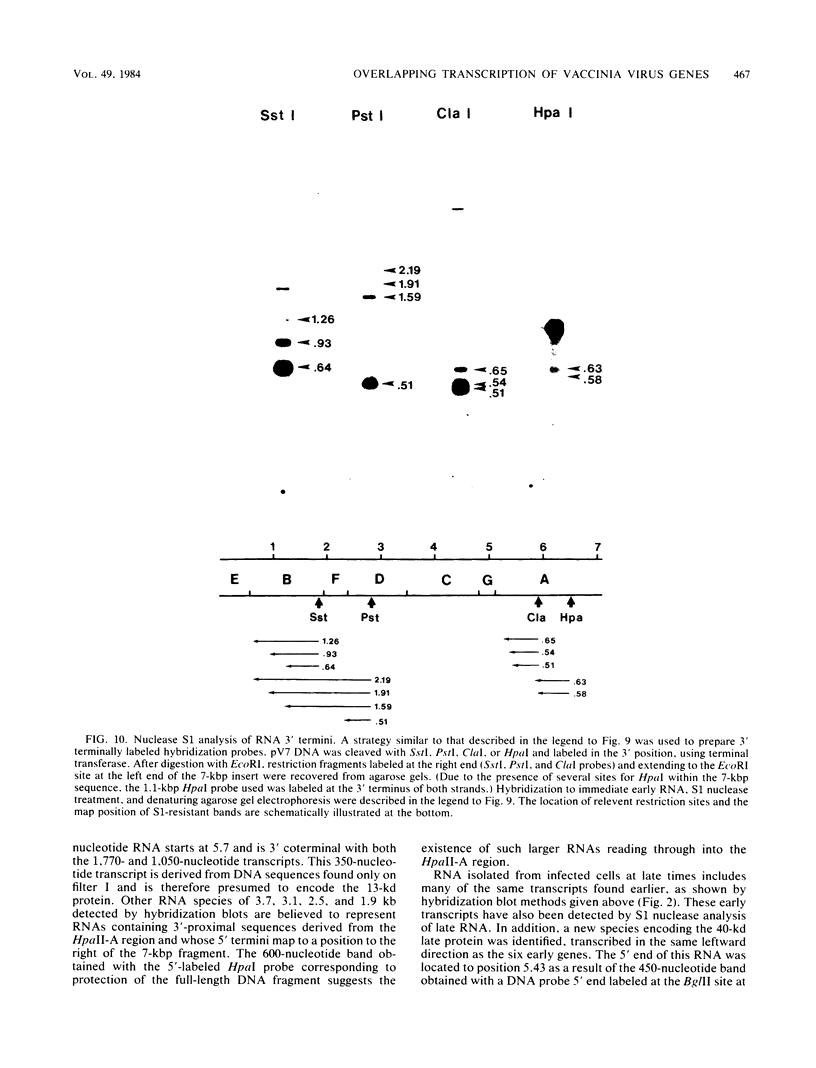
A transcriptional and translational map was obtained for a 7-kilobase pair EcoRI fragment of vaccinia virus DNA containing 4% of the viral genome. This particular region contains a cluster of early genes which are transcribed in viral cores in vitro and in infected cells before uncoating of the viral DNA. Expression of this region was characterized by using vaccinia virus DNA sequences cloned in phage and plasmid vectors. Polypeptides encoded on the 7-kilobase pair fragment were identified by cell-free translation of viral mRNA selected by hybridization to immobilized DNA fragments. Early RNA programmed the synthesis of six proteins having apparent molecular weights of 63,000, 38,000, 37,000, 25,000, 15,000, and 13,000. The same result was obtained with RNA synthesized in vitro. A new species of 40 kilodaltons was synthesized in response to late RNA. Of the six "early" polypeptides, only the 13-kilodalton component was synthesized from late mRNA. RNA derived from the 7-kilobase pair region was analyzed by a variety of methods including hybridization blot, in vitro recapping, and S1 nuclease techniques. A surprisingly complex pattern of early transcription was found, indicating the existence of families of overlapping RNA species which share common 5'-proximal sequences. In addition, larger RNAs were identified spanning two or more adjacent genes. These RNAs appear to possess a common 5' terminus with transcripts derived from the first gene and are coterminal at the 3' end with RNAs from the downstream gene. Late RNA encoding the 40-kilodalton protein was shown to be heterogeneous in size. A single 5' terminus but no unique 3' terminus was identified for this class of transcripts. RNA species synthesized by cores in vitro were of similar size to authentic transcripts isolated from infected cells at early times.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bajszár G., Wittek R., Weir J. P., Moss B. Vaccinia virus thymidine kinase and neighboring genes: mRNAs and polypeptides of wild-type virus and putative nonsense mutants. J Virol. 1983 Jan;45(1):62–72. doi: 10.1128/jvi.45.1.62-72.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Bochner B. R., Huang H. C., Schieven G. L., Ames B. N. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980 Aug;143(2):926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Backman K. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 1979;68:245–267. doi: 10.1016/0076-6879(79)68018-7. [DOI] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Cooper J. A., Wittek R., Moss B. Extension of the transcriptional and translational map of the left end of the vaccinia virus genome to 21 kilobase pairs. J Virol. 1981 Sep;39(3):733–745. doi: 10.1128/jvi.39.3.733-745.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. A., Wittek R., Moss B. Hybridization selection and cell-free translation of mRNA's encoded within the inverted terminal repetition of the vaccinia virus genome. J Virol. 1981 Jan;37(1):284–294. doi: 10.1128/jvi.37.1.284-294.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Meselson M. Plasmid screening at high colony density. Gene. 1980 Jun;10(1):63–67. doi: 10.1016/0378-1119(80)90144-4. [DOI] [PubMed] [Google Scholar]
- Hruby D. E., Guarino L. A., Kates J. R. Vaccinia virus replication. I. Requirement for the host-cell nucleus. J Virol. 1979 Feb;29(2):705–715. doi: 10.1128/jvi.29.2.705-715.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Hardison R. C., Lacy E., Lauer J., O'Connell C., Quon D., Sim G. K., Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978 Oct;15(2):687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K. I., Joklik W. K. Hybridization and sedimentation studies on "early" and "late" vaccinia messenger RNA. J Mol Biol. 1967 Aug 14;27(3):395–419. doi: 10.1016/0022-2836(67)90047-2. [DOI] [PubMed] [Google Scholar]
- Pelham H. R., Jackson R. J. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur J Biochem. 1976 Aug 1;67(1):247–256. doi: 10.1111/j.1432-1033.1976.tb10656.x. [DOI] [PubMed] [Google Scholar]
- Seed B. Diazotizable arylamine cellulose papers for the coupling and hybridization of nucleic acids. Nucleic Acids Res. 1982 Mar 11;10(5):1799–1810. doi: 10.1093/nar/10.5.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O. Recovery of DNA from gels. Methods Enzymol. 1980;65(1):371–380. doi: 10.1016/s0076-6879(80)65048-4. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan S., Moss B. In vitro transcription of the inverted terminal repetition of the vaccinia virus genome: correspondence of initiation and cap sites. J Virol. 1981 Feb;37(2):738–747. doi: 10.1128/jvi.37.2.738-747.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittek R., Barbosa E., Cooper J. A., Garon C. F., Chan H., Moss B. Inverted terminal repetition in vaccinia virus DNA encodes early mRNAs. Nature. 1980 May 1;285(5759):21–25. doi: 10.1038/285021a0. [DOI] [PubMed] [Google Scholar]
- Wittek R., Cooper J. A., Barbosa E., Moss B. Expression of the vaccinia virus genome: analysis and mapping of mRNAs encoded within the inverted terminal repetition. Cell. 1980 Sep;21(2):487–493. doi: 10.1016/0092-8674(80)90485-7. [DOI] [PubMed] [Google Scholar]