Abstract
Topoisomerase II is able to break and rejoin double-strand DNA. It controls the topological state and forms and resolves knots and catenanes. Not much is known about the relation between the chromosome segregation and condensation defects as found in yeast top2 mutants and the role of topoisomerase II in meiosis. We studied meiosis in a heat-sensitive top2 mutant of Schizosaccharomyces pombe. Topoisomerase II is not required until shortly before meiosis I. The enzyme is necessary for condensation shortly before the first meiotic division but not for early meiotic prophase condensation. DNA replication, prophase morphology, and dynamics of the linear elements are normal in the top2 mutant. The top2 cells are not able to perform meiosis I. Arrested cells have four spindle pole bodies and two spindles but only one nucleus, suggesting that the arrest is nonregulatory. Finally, we show that the arrest is partly solved in a top2 rec7 double mutant, indicating that topoisomerase II functions in the segregation of recombined chromosomes. We suggest that the inability to decatenate the replicated DNA is the primary defect in top2. This leads to a loss of chromatin condensation shortly before meiosis I, failure of sister chromatid separation, and a nonregulatory arrest.
INTRODUCTION
In meiosis one round of DNA replication is followed by two nuclear divisions. The chromosome number is halved, and four daughter cells are formed. In meiotic prophase, after DNA replication, the homologous chromosomes pair and recombine. In the following two nuclear divisions the homologous chromosomes (meiosis I) and the sister chromatids (meiosis II) are segregated. In the great majority of sexually reproducing eukaryotes a meiosis-specific tripartite structure is formed during meiotic prophase: the synaptonemal complex (for review, see von Wettstein et al., 1984). The fission yeast Schizosaccharomyces pombe is an exception; no synaptonemal complex is formed. During meiotic prophase so-called linear elements appear, which resemble the axial elements of other eukaryotes. These elements are formed discontinuously along the chromosomes and never form a tripartite structure (Olson et al., 1978; Hirata and Tanaka, 1982; Bähler et al., 1993). In meiotic prophase the nuclei become elongated and are called horse tail nuclei (Robinow, 1977). A striking nuclear movement takes place during this time, which is led by the spindle pole body with attached telomeres (Chikashige et al., 1994).
Topoisomerase II is an enzyme that is able to break and rejoin double-strand DNA molecules. In this way, it can control the topological state of the DNA and form and resolve knots and catenanes in duplex DNA. Several results suggest that topoisomerase II is required for segregation of replicated circular and linear DNA sister molecules. It has also been shown that topoisomerase II is necessary for mitotic chromosome condensation. Other roles for topoisomerase II have also been suggested (for review, see Watt and Hickson, 1994; Koshland and Strunnikov, 1996).
For mitosis, the phenotypes as seen in Saccharomyces cerevisiae and S. pombe temperature-sensitive (ts)1 top2 mutants can be explained by defects in segregation and chromosome condensation. In both yeasts topoisomerase II is essential for viability (DiNardo et al., 1984; Goto and Wang, 1984; Uemura and Yanagida, 1984). Synchronously dividing cells of ts top2 mutants become inviable, when shifted to restrictive temperature, at the time of mitotic division (Holm et al., 1985; Uemura and Yanagida, 1986). Different studies in S. cerevisiae and S. pombe suggest a role for topoisomerase II in the separation of sister chromatids. In ts top2 mutants the cells arrest at the mitotic nuclear division. In these arrested cells plasmids are found in the form of catenated dimers (DiNardo et al., 1984; Uemura and Yanagida, 1986). The mitotic arrest is nonregulatory in both yeasts. It was shown in S. cerevisiae that the lack of topoisomerase II activity leads to nondisjunction and chromosome breakage in mitosis (Holm et al., 1989; Spell and Holm, 1994). In S. pombe a mitotic spindle is formed and tries to separate the sister chromatids. Then the septum appears and cuts the nucleus across (cut phenotype; Uemura and Yanagida, 1984, 1986). The stabilization of normally unstable large circular minichromosomes in S. pombe by overexpression of topoisomerase II is also consistent with a role of topoisomerase II in the segregation of replicated DNA daughter molecules (Murakami et al., 1995). Uemura et al. (1987) showed that topoisomerase II is necessary for proper chromosome condensation in mitosis of S. pombe.
In meiosis, cold-sensitive (cs) top2 cells of S. cerevisiae arrest at the first meiotic division at restrictive temperature. Premeiotic DNA replication, chromosome condensation during prophase, and recombination are not affected. Complementary temperature shift experiments suggest that the enzyme is only required at the time of the nuclear divisions (Rose et al., 1990). The arrest has been interpreted to be regulatory. The spindle pole bodies do not separate, and no spindle is formed; the arrested cells are able to return to mitotic growth. The cs top2 cells seem to be delayed in the exit from pachytene (Rose and Holm, 1993). In a top2 rad50 double mutant, in which recombination is blocked and no synaptonemal complex is formed, the cells are able to perform the first but not the second meiotic division (Rose et al., 1990). Because of the dual phenotype of rad50, two hypotheses about the nature of the arrest were put forward. The first one is that topoisomerase II is needed for the segregation of recombined chromosomes (Rose et al., 1990). The second one is that topoisomerase II functions in the resolution of interlocks that occur during the course of homologous chromosome pairing. The synaptonemal complex might play a role in detecting the unresolved interlocks and triggering the regulatory arrest (Rose and Holm, 1993).
A number of questions still remain to be answered about the role of topoisomerase II in segregation and chromosome condensation. It is not clear whether or how the defects in chromosome segregation and condensation are related. Especially about the role of topoisomerase II in meiosis not much is known. Therefore we studied meiosis in topoisomerase II deficient fission yeast, a widely used eukaryotic model organism.
MATERIALS AND METHODS
Strains
The diploid strains used in this study are h+/h− ade6-M216/ade6-149 (Bähler et al., 1993; in this paper referred to as wild type), h+/h− top2-191/top2-191 leu1-32/leu1-32 ade6-M216/ade6-M210 (referred to as top2), and h+/h− top2-191/top2-191 rec7-102/rec7-102 leu1-32/leu1-32 ade6-M210/ade6-M210 (referred to as top2 rec7). The top2-191 allele was isolated by Uemura and Yanagida (1984). The rec7-102 allele was isolated by Ponticelli and Smith (1989). For strain construction, standard genetic methods were used (Gutz et al., 1974).
Culture Conditions and Meiotic Time Courses
The cells were cultured and shifted to meiosis-inducing medium as described by Bähler et al. (1993). Permissive and restrictive temperatures of 24 and 34°C, respectively, were used for meiotic time courses. Because after shift to meiotic medium the cells first have to perform a mitotic division (which is lethal at restrictive temperature for the top2 and top2 rec7 strains), the cultures were first incubated for 2.5 h at 24°C and then divided in two cultures, which were incubated at 24 and 34°C, respectively, for the rest of the time course (but not for the complementary temperature shift experiment; see below). At different time points after shift to meiosis-inducing conditions samples were taken and processed for spreading and 4′,6-diamino-2-phenylindole (DAPI) staining as described below. Samples for immunofluorescence were taken and processed as described below.
Complementary Temperature Shift Experiments
In the first experiment (see Figure 1A) top2 and wild-type diploid cells were shifted to meiotic conditions and incubated at restrictive temperature. At hourly intervals aliquots were removed and shifted to permissive temperature. At the same time cells were fixed for DAPI staining, and the percentage of cells that completed the first meiotic division was determined. Sporulation efficiency was determined the next day. In the second experiment (Figure 1B), after shift to meiotic conditions, cells were incubated at permissive temperature, and every hour an aliquot was removed and shifted to restrictive temperature. The rest of the experiment was performed as described for the first experiment. For both experiments, at least 100 cells were counted from every time point for determination of the sporulation efficiency and percentage of cells that performed the first meiotic division.
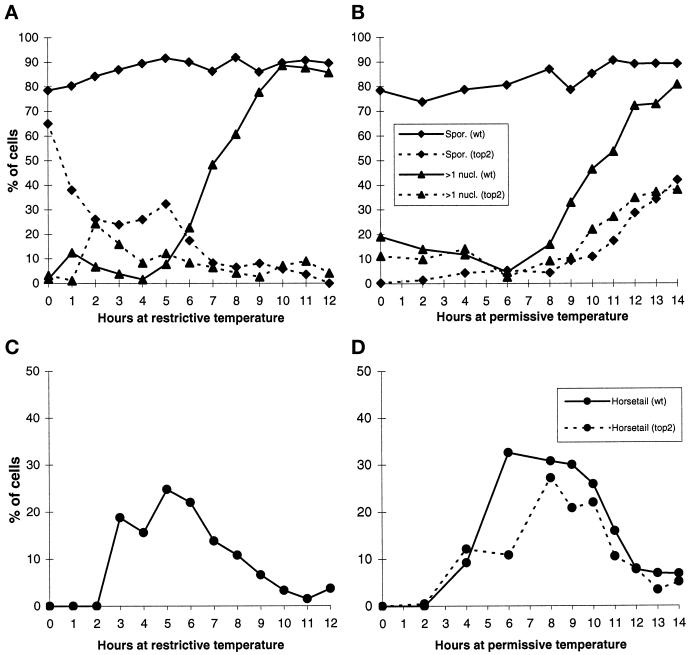
Figure 1.
Complementary temperature shift experiment. For details see MATERIALS AND METHODS. (A) In the first experiment top2 and wild-type (wt) cells were shifted to meiotic conditions and incubated at restrictive temperature. At hourly intervals aliquots were removed and shifted to permissive temperature. (B) In the second experiment, after shift to meiotic conditions, cells were incubated at permissive temperature, and every hour an aliquot was removed and shifted to restrictive temperature. (C) The percentage of horse tail nuclei for the experiment presented in A is shown for wild type. Data about horse tail nuclei in top2 could not be included, because the high abundance of mitotic cells with an elongated nucleus caused by the cut phenotype made an objective estimation impossible. (D) Data about horse tail nuclei in top2 as well as in wild type for the experiment presented in B are shown. In this experiment the first meiotic division was delayed in the top2 strain compared with wild type at permissive temperature. The top2 mutation has a slow growth phenotype at permissive temperature, probably because the cells have a problem with the mitotic division. The same problem probably delayed meiosis I in the top2 mutant at permissive temperature. Spor., sporulation efficiency; >1 nucl., percentage of cells that performed the first meiotic division.
Nuclear Spreading, DAPI Staining, and Immunofluorescence
Nuclear spreading and DAPI staining were performed as described by Bähler et al. (1993). After staining with silver nitrate and transfer to grids, nuclear spreads were examined by electron microscopy with a Philips EM300 at 60 kV (Bähler et al., 1993). For each time point at least 100 nuclei were analyzed. DAPI staining was performed as described by Bähler et al. (1993), and at least 100 cells per time point were analyzed by fluorescence microscopy. For immunofluorescence double staining the two primary antibodies were incubated simultaneously, as were the secondary antibodies. The rabbit polyclonal SAD1 antibody against the spindle pole body (Hagan and Yanagida, 1995) was isolated and generously provided by I. Hagan (University of Manchester, United Kingdom). The monoclonal rabbit TAT1 antibody against microtubules (Woods et al., 1989) was generously provided by K. Gull (University of Manchester). For immunofluorescence we used the protocol as described by Hagan and Hyams (1988), with some alterations as described by Svoboda et al. (1995). As secondary antibodies affinity-isolated goat anti-mouse immunoglobulin G TRITC conjugate (Fab specific, Sigma T-6528) and affinity-isolated goat anti-rabbit immunoglobulin G FITC conjugate (whole molecule, Sigma F-0382) were used.
Analysis of DNA Replication and Chromatin Condensation
Cultures of wild type and top2 were shifted to meiosis as described above. Cells were cultured for 2.5 h at 24°C and then shifted to 34°C.
For the analysis of DNA replication, every hour 1 ml of each meiotic culture was briefly centrifuged, resuspended in 1 ml 70% ethanol, and stored at 4°C. After staining the cells with propidium iodide, the DNA content of the cells was determined by flow cytometry (Beach et al., 1985).
For the analysis of chromatin condensation, every hour a sample was taken and prepared for DAPI staining (see above). From each time point pictures were taken with a digital charge-coupled device camera on a fluorescence microscope and stored for further analysis. Exposure settings of the camera were identical throughout the experiment. For each time point, from at least 200 cells containing one nucleus the nuclear area and the total nuclear fluorescence (giving a measure of DNA content) were determined with the University of Texas Health Science Center (San Antonio, TX) ImageTool program (http://ddsdx.uthscsa.edu/dig/itdesc.html) using the same threshold settings throughout to define the outline of the nuclei. The total fluorescence as determined with this method is roughly linear to the DNA content of the nucleus, as suggested by the fact that G2 nuclei give twice the fluorescence intensity of G1 nuclei (our unpublished results). In the top2 mutant, cells that showed the cut phenotype (see INTRODUCTION) were left out of the analysis. For each nucleus, the nuclear fluorescence was divided by the nuclear area, giving a measure of nuclear condensation.
RESULTS
Topoisomerase II Is Not Required during Meiotic Prophase but at the Time of the Meiotic Divisions
To determine at which time topoisomerase II is needed during meiosis, we performed two complementary temperature shift experiments (Rose et al., 1990; also see Materials and Methods). The first experiment revealed from which time on topoisomerase II was needed for sporulation (Figure 1A). The sporulation efficiency of the wild-type strain stayed constant over time. However, the top2 strain showed a strong decrease immediately after shift to restrictive temperature. This first decrease reflected that the majority of the cells had to perform a mitotic division, which was fatal at restrictive temperature, before they could start meiosis from G1. Accordingly, cells with two nuclei showing the cut phenotype were apparent during this time (our unpublished results). Therefore, we interpreted this decrease as a result of mitotic inviability. After 2 h, the sporulation efficiency reached a plateau. As can be seen in Figure 1C, this corresponds to an increase in the amount of horse tail nuclei, typical for meiotic prophase, in wild type. A second decrease in sporulation efficiency took place after 5 h. This coincided with an increase in the number of wild-type cells that completed the first meiotic division and a decrease in the number of horse tail nuclei. Data about top2 horse tail nuclei could not be presented in Figure 1C, because the high abundance of mitotic cells with an elongated nucleus attributable to the cut phenotype made an objective estimation impossible. The low percentage of top2 cells containing more than one nucleus at later time points (Figure 1A) suggests that the cells arrest before the first meiotic division (also see below). A repetition of this experiment gave similar results (our unpublished results). From these experiments we concluded that topoisomerase II is first needed at the premeiotic division, and again at the time of the first meiotic division, but not during meiotic prophase.
The complementary experiment (see MATERIALS AND METHODS) showed from which time on topoisomerase II was no longer needed for sporulation (Figure 1B). Again the sporulation efficiency of the wild-type strain stayed constant over time. The top2 strain showed an increase in sporulation efficiency at ∼8 h. This coincided with an increase in the number of top2 cells that performed a meiotic division and a decrease in the number of horse tail nuclei. From this experiment we concluded that topoisomerase II is not required for sporulation after the meiotic divisions.
DNA Replication and Chromatin Condensation
In S. pombe topoisomerase II is necessary for mitotic chromosome condensation (Uemura et al., 1987; see INTRODUCTION). We studied DNA replication and meiotic chromatin condensation in wild-type and top2 strains in a meiotic time course at 34°C. To compare meiotic DNA replication between wild-type and top2 cells at restrictive temperature, we determined the DNA content of the cells by flow cytometry (see MATERIALS AND METHODS). For every hour the percentage of cells in G2 was determined (Figure 2A). Beginning at 1 h there was a decrease in the percentage of G2 cells, reflecting the mitotic division before the start of meiosis (see Figure 2E). After 6 h the percentage of G2 cells increased again, reflecting meiotic DNA replication. The kinetics of DNA replication between wild type and top2 was similar. At the end of meiosis the percentage of G2 cells was somewhat lower in the top2 mutant. This was probably because the analysis of DNA replication in the top2 mutant was obscured by the contribution of arrested dead cells showing a G1 DNA content (cut phenotype; see INTRODUCTION). The DNA content of the arrested top2 cells (11 h) was similar to the mitotic G2 DNA content (0 h), indicating that DNA replication is normal in top2 (Figure 2C).
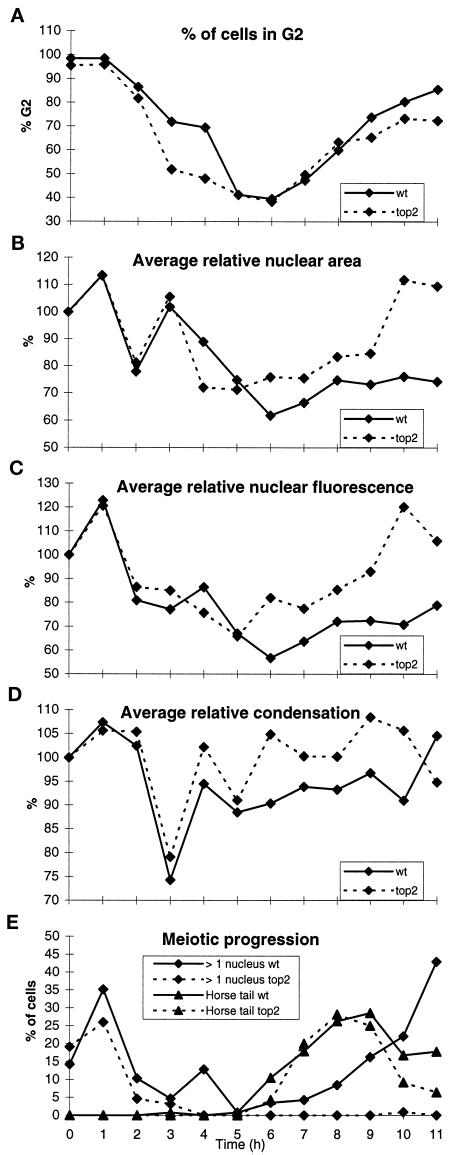
Figure 2.
In a meiotic time course, DNA content, nuclear area, nuclear fluorescence, and chromatin condensation were determined in wild type and top2 at 34°C. For detailed explanation see text and MATERIALS AND METHODS. (A) For every hour the percentage of cells that are in G2 is shown. (B and C) The average relative nuclear area (B) and the average relative nuclear fluorescence (C) are shown for every hour. (D) The nuclear area divided by the DNA content gives a measure of chromatin condensation. (E) The timing of meiotic events in this particular time course is shown. wt, wild type; >1 nucleus, percentage of cells that performed the first meiotic division; Horse tail, percentage of cells that show the typical prophase nuclear morphology after DAPI staining.
The size of a nucleus is determined both by its DNA content and its degree of chromatin condensation. To get a measure of the degree of chromatin condensation, we determined for every time point the nuclear area and total nuclear fluorescence (measure of DNA content) of at least 200 cells with only one nucleus. A repetition of this time course experiment with formaldehyde-fixed cells gave similar results (our unpublished results). For details see MATERIALS AND METHODS. This method permits the quantitative analysis of large numbers of nuclei in a meiotic time course experiment. The preparation of the cells is simple and requires only a few steps. No harsh treatment, possibly creating artifacts, is needed, which is an advantage compared with studying condensation with in situ hybridization methods.
Determination of the nuclear area is shown in Figure 2B. The average values were standardized to the average nuclear area at 0 h (100%). At 2 h, for both wild type and top2, there was a sharp dip in the average relative area because of the mitotic division, which took place at 1 h (see Figure 2E) and reduced the DNA content of the cells. Then, for both strains, there was a remarkable increase in average relative area at 3 h. This cannot be explained by an increase in DNA content (Figure 2, A and C). Then the average relative area decreased again to a level somewhat lower than at 2 h. Starting at 5 (top2) and 6 h (wild type) the average relative area started to increase slowly as a result of DNA replication (see Figure 2A). In wild type the average relative area reached a plateau at ∼8 h. This was because the effect of DNA replication on the average relative nuclear area was counteracted by meiotic chromatin condensation shortly before the first meiotic division (see below; Robinow, 1977). In top2 the average relative area increased at the end of the meiotic time course to a level somewhat higher than the mitotic average relative area at 0 h.
We also determined the total nuclear fluorescence of each of the cells, giving a measure of the DNA content (Figure 2C). Again the average values were standardized to the average nuclear fluorescence at 0 h (100%). At 2 h there was a sharp decrease in average relative fluorescence as a result of the transition of the cells from G2 to G1 because of the mitotic division (Figure 2, A and E). Starting at 5 (top2) and 6 h (wild type) the average relative fluorescence increased again, in top2 to a level similar to mitotic (G2) cells. In wild type the increase in DNA content was underestimated because of the progression of the replicated meiotic prophase cells into cells with two and four nuclei, which were left out of the analysis.
To get a measure of the degree of chromatin condensation, for each nucleus the nuclear fluorescence was divided by its nuclear area (Figure 2D). For each time point the average condensation was standardized to the average condensation at 0 h (100%). A sharp decrease in the average relative condensation was seen at 3 h. At 4 h the average relative condensation increased again to a level somewhat lower than at 0 h. At 11 h the average relative condensation in wild type increased, whereas it decreased in top2.
From the above observations we concluded that right after the mitotic division, a remarkable decondensation and recondensation takes place in both wild-type and top2 cells. This decondensation and recondensation was also seen in wild-type meiosis at 30°C (our unpublished results) and appears to be independent of topoisomerase II. In wild type, at the end of meiotic prophase, the nuclei condense again (also see Robinow, 1977). This wild-type condensation was underestimated in the time course experiment, because it was immediately followed by meiosis I (cells with two or four nuclei were left out of the analysis). In top2-arrested cells (see below) the nuclei were not condensed at the end of the meiotic time course. Thus the condensation shortly before the first meiotic division is topoisomerase II dependent. These changes in chromatin condensation are illustrated in Figure 3. For explanation see figure legend.
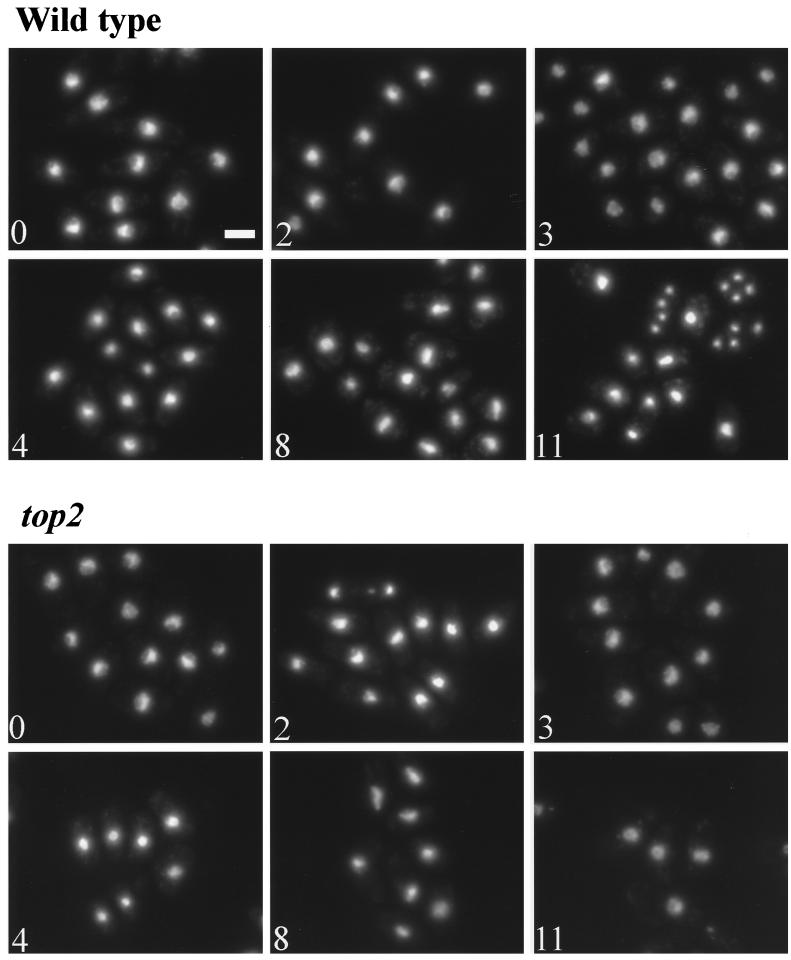
Figure 3.
Illustration of the changes in chromatin condensation during meiotic time courses in wild-type and top2 cells at 34°C. Quantitative data on these time courses are presented in Figure 2. At 0 h the majority of the still mitotic cells are in G2 (also see Figure 2A) and show a dense nuclear staining. At 2 h the cells have performed a mitotic division (see Figure 2E), are smaller, and have a lower DNA content. The nuclei are densely stained, and the level of chromatin condensation is similar to that at 0 h (see Figure 2D). At 3 h the cells still have a DNA content similar to 0 h but are larger because of chromatin decondensation (see Figure 2D) and show a more open chromatin structure. At 4 h the cells have recondensed again, showing the dense nuclear staining. At 8 h nuclei are visible, which show the typical prophase nuclear morphology (horse tail nuclei). During this time DNA replication takes place. In wild type at 11 h, shortly before the first meiotic division, the nuclei condense again, showing a dense nuclear staining. In the top2 mutant at 11 h the cells do not condense and show a more open chromatin structure. Bar, 10 μm.
Nuclear Structures in Meiotic Prophase Are Normal in top2 Cells at Restrictive Temperature
We performed three meiotic time course experiments in which we compared different characteristics between the top2 and wild-type strains. In nuclear spread preparations no obvious differences in morphology of the linear elements were visible between the two strains at 24 and 34°C (our unpublished results). Also, the dynamics of appearance and disappearance of the linear elements was similar between the two strains at 24 and 34°C (Figure 4). An interesting observation, beyond the scope of this study, is that for both the wild-type and top2 strains the percentage of linear-element containing cells was reduced at 34°C compared with 24°C. Because the sporulation efficiencies (>1 nucleus) of the wild-type strain were similar at 24 and 34°C, the decrease of linear elements might reflect the reduction in recombination frequency at 34°C compared with 25°C (Bähler et al., 1991). Also the percentage of horse tail nuclei was reduced at 34°C compared with 24°C (Figure 4). By DAPI staining no obvious differences in morphology of the prophase nuclei (horse tail nuclei; see INTRODUCTION) were visible between the two strains at 24 and 34°C (our unpublished results).
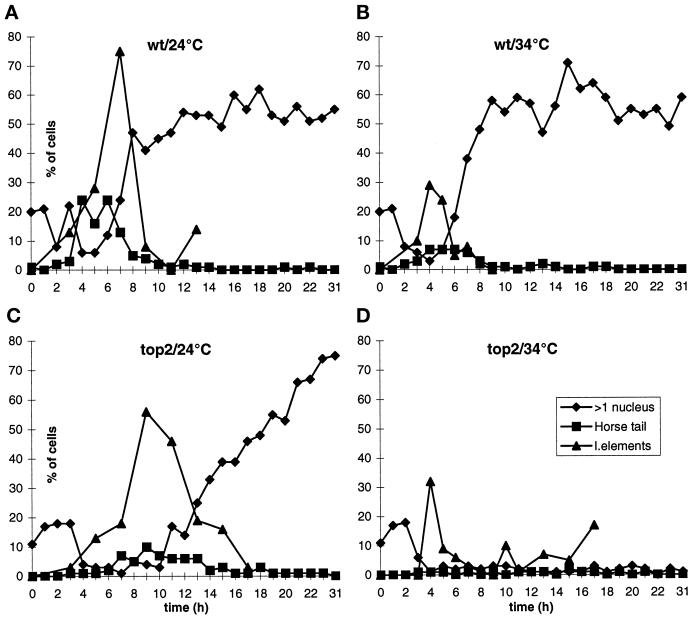
Figure 4.
Meiotic time courses with the top2 mutant and the wild type (wt) at 24°C (permissive temperature) and 34°C (restrictive temperature). For details see MATERIALS AND METHODS. The delay in linear element formation in the top2 strain at 24°C (C) was within the normal variation between the different time courses. In the wild-type cells at 24°C (A, 13 h) and in the top2 cells at 34°C (D, 17 h) the percentage of linear element containing cells seemed to increase after a while. This was due to a spreading artifact, caused by selection against cells that were in late stages of meiosis and were not properly protoplasted. This led to an overrepresentation of early cells late in the time courses. In the top2 strain at 24°C (C) the meiotic divisions were delayed, possibly because the top2 mutant is not completely wild type at permissive temperature (also see Figure 1 legend). >1 nucleus, percentage of cells that performed the first meiotic division; Horse tail, percentage of cells that show a nuclear morphology typical of prophase after DAPI staining and fluorescence microscopy; l. elements, percentage of cells that contain linear elements (electron microscopy).
The top2 Cells Arrest before the First Meiotic Division, but the Spindle Pole Body Cycle Continues
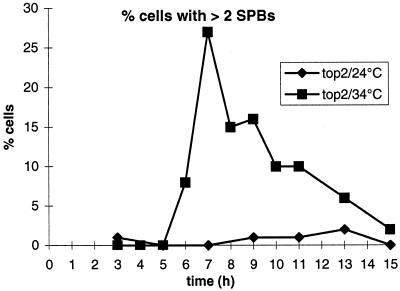
Although DNA replication was normal, and no clear phenotype was visible in meiotic prophase of the top2 strain at restrictive temperature, the cells were not able to perform the first meiotic division. As can be seen in Figure 4D, the percentage of cells that contained more than one nucleus stayed close to zero at later time points.
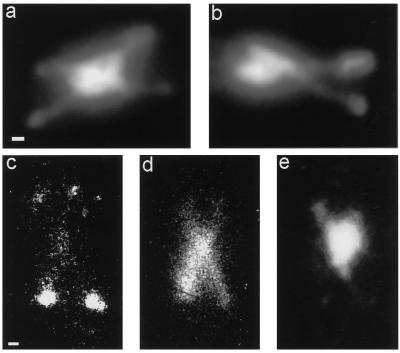
In spread preparations of the arrested top2 cells, often four spindle pole bodies were visible in one nucleus (Figure 5). As much as 27% of the nuclei showed this morphology at 7 h (Figure 6), but afterward the percentage decreased again. This was probably due to an overrepresentation of early cells late in the time course because of the poor spreading of advanced cells. At later time points at restrictive temperature, DAPI-stained top2 cells were found with chromatin fibers stretched crosswise out of the nucleus (Figure 7, a and b). Because this suggested that the cells were actively trying to separate the chromatin, we also did a double-staining immunofluorescence experiment in whole cells with antibodies that recognize the spindle and spindle pole body, respectively. At 7 h a small portion of the meiotic cells showed two spindles and four spindle pole bodies (Figure 7c–e). The cell shown has four spindle pole bodies (Figure 7c) and two spindles (Figure 7d), probably representing both spindles of the second meiotic division. Only one nucleus is visible, with chromatin fibers stretching out along the spindle (Figure 7e). This striking morphology was never observed at permissive temperature or in the wild-type strain. The final arrest stage was also observed during 2 h in several living cells with video fluorescence microscopy. During this time the spindle attempted to segregate the chromosomes, because transient protrusions from opposite sides of the nucleus were observed (our unpublished results). Thus at restrictive temperature the spindle pole body cycle continues in the top2 mutant, although the first meiotic division was not completed.
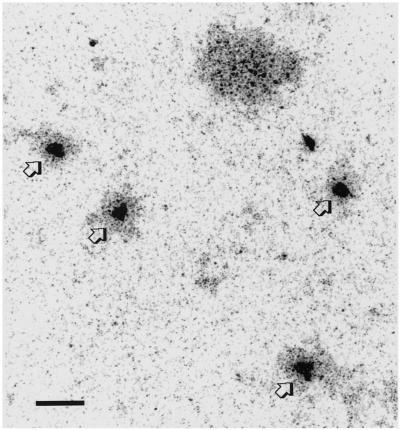
Figure 5.
Electron micrograph of a spread preparation of an arrested top2 cell. Four spindle pole bodies (arrow) are visible in one nucleus. The larger dark-staining area represents the nucleolus. Bar, 1 μm.
Figure 6.
Quantitation of spindle pole bodies. For different time points at permissive and restrictive temperatures, the percentage of top2 cells containing more than two spindle pole bodies is shown. The data are from the same meiotic time course as presented in Figure 4. For further explanation see RESULTS.
Figure 7.
(a and b) Two examples of DAPI-stained, arrested top2 cells. Chromatin fibers are stretched crosswise out of the nucleus. Bar, 1 μm. (c–e) Immunofluorescence micrographs of an arrested top2 cell at restrictive temperature. Spindle pole bodies (c) and spindles (d) were stained with specific antibodies (for details see MATERIALS AND METHODS). The nucleus is visualized by DAPI staining (e). Four spindle pole bodies (c) and two crossed meiosis II spindles (d) are visible in a cell that contains only one nucleus, of which DNA fibers are pulled out (e). Bar, 1 μm.
Arrest Is Partly Solved in a top2 rec7 Double Mutant
It has been proposed that topoisomerase II is required for the segregation of sister chromatids of recombined chromosomes during meiosis I (Rose et al., 1990). To test this hypothesis in S. pombe we studied meiosis in a top2 rec7 double mutant (see MATERIALS AND METHODS). In the rec7-102 mutant, intergenic recombination is strongly reduced (DeVeaux and Smith, 1994), whereas the morphology and dynamics of the linear elements is similar to wild type (Molnar, personal communication). The morphology and dynamics of appearance and disappearance of the linear elements in the top2 rec7 strain at permissive and restrictive temperature were also normal (our unpublished results). In three different time courses the percentage of cells able to perform the first meiotic division was determined. Data were collected for strains carrying only top2 or both top2 and rec7 at permissive and restrictive temperatures. For comparison, the sporulation efficiencies at restrictive temperature were adjusted by dividing them by the sporulation efficiencies of the parallel cultures at permissive temperature (Table 1). In the top2 rec7 strain the adjusted average value for successful meiosis I was approximately four times higher than in the top2 strain. Thus we concluded that the top2 arrest is partly solved in the top2 rec7 double mutant.
Table 1.
Partial resolution of the top2 arrest in the double mutant top2 rec7
| Exp. | Cells performing meiosis I (%)
|
|||||
|---|---|---|---|---|---|---|
|
top2
|
top2 rec7
|
|||||
| 24°C | 34°C | 34°C adjusteda | 24°C | 34°C | 34°C adjusteda | |
| 1 | 75 | 1 | 1 | 46 | 6 | 13 |
| 2 | 83 | 11 | 13 | 52 | 22 | 42 |
| 3 | 70 | 2 | 3 | 53 | 7 | 13 |
| Average | 76 | 5 | 6 | 50 | 12 | 23 |
The adjusted values were obtained by standardization to the efficiency of meiosis I at 24°C. For further explanation see RESULTS.
DISCUSSION
We studied meiosis in a heat-sensitive top2 mutant of S. pombe. With a complementary temperature shift experiment we showed that the function of topoisomerase II is not required during meiotic prophase but at the time of the meiotic divisions. DNA replication is normal. Topoisomerase II is necessary for chromatin condensation shortly before meiosis I but not for early decondensation and recondensation. Nuclear morphology and the dynamics of the linear elements during meiotic prophase are normal at restrictive temperature. The cells are blocked at the first meiotic division, but the spindle pole body cycle continues. Finally, we showed that the arrest is partly solved in a top2 rec7 double mutant.
Interlocks
During the process of meiotic chromosome pairing, chromosomes (or chromosome pairs) can get trapped between the partners of a homologous chromosome pair. This is called an interlock. Different mechanisms have been proposed to prevent the formation of interlocks during early prophase. In general, in late pachytene almost all of the interlocks are resolved (von Wettstein et al., 1984; Kleckner and Weiner, 1993). It has been proposed that topoisomerase II might have a function in the resolution by resolving the DNA component of the interlock (Rasmussen, 1986). Rose and Holm (1993) suggested that the regulatory arrest that they observed in a S. cerevisiae cs top2 mutant at restrictive temperature could be triggered by the presence of unresolved interlocks and that the synaptonemal complex could play a role in their detection. In spread preparations of the S. cerevisiae top2 mutant no interlocks or abnormalities are visible (Rose and Holm, 1993). This suggests that interlocks are not formed at a very high frequency in S. cerevisiae and thus are unlikely to be the basis for the complete meiosis I arrest. Assuming that the rad50 mutation does not prevent the formation of interlocks, the result that the S. cerevisiae top2 rad50 double mutant is able to perform the first meiotic division (Rose et al., 1990) indicates that in the double mutant interlocks are not preventing chromosome separation in S. cerevisiae.
Throughout S. pombe meiotic prophase a chromosome bouquet with clustered telomeres and centromeres is maintained (Scherthan et al., 1994), making it unlikely that interlocks are formed at high frequency. It is hardly conceivable that failure of interlock resolution explains the complete nonregulatory arrest at meiosis I in the top2 mutant, even when topoisomerase II does resolve interlocks during meiotic prophase.
Untangling of Sister Chromatids and Chromatin Condensation
In mitosis topoisomerase II is necessary for the segregation of sister chromatids after DNA replication (see INTRODUCTION). At the first meiotic division the homologous chromosomes are separated. For separation of homologous chromosomes having one or more crossovers the same task has to be fulfilled as in mitosis: separation of sister chromatids that are entangled because of meiotic DNA replication (Rose et al., 1990). So it is conceivable that the inability of a topoisomerase II mutant to separate sister chromatids leads to arrest at the first meiotic division. This hypothesis is supported by our observation that in a top2 rec7 mutant the arrest is partly solved. We estimate that the reduction in crossover frequency in the rec7 mutation is ∼40-fold (calculated from data of DeVeaux and Smith, 1994). This means that on average each cell forms approximately one crossover (45 in wild type; Munz, 1994). If we assume that the residual crossovers are distributed according to Poisson, 37% of the meiocytes gets no crossover at all. This is a rough estimate of the fraction of cells that are expected to complete the first meiotic division in the top2 rec7 double mutant. As shown in Table 1, the observed value is somewhat lower (23%).
In S. pombe topoisomerase II is necessary for mitotic chromosome condensation (Uemura et al., 1987). Holm (1994) suggested that chromosome condensation may provide the directionality for disentanglement of sister chromatids by topoisomerase II. Different studies suggest that the processes of chromatid separation and chromatin condensation are linked (reviewed by Yanagida, 1995; Koshland and Strunnikov, 1996). Untangling of sister chromatids possibly takes place in concert with chromatin condensation, and a defect in one of these processes could lead to a defect in the other. If this is true, what could be the primary meiotic defect in the S. pombe top2 mutant, chromatin condensation or sister chromatid separation? We have shown that early in a meiotic time course, before meiotic DNA replication (see Figure 2, A and D), topoisomerase II is not necessary for decondensation and recondensation. Because these events happen after the mitotic division, at a time when meiotic chromosome pairing begins (Scherthan et al., 1994), we think that these are early meiotic prophase events. The recondensation may be comparable with the leptotene chromosome condensation that marks the beginning of meiotic prophase in other organisms. Whereas topoisomerase II is not needed for early meiotic prophase chromatin condensation, it is required for condensation shortly before meiosis I. This is a remarkable observation and suggests that topoisomerase II is not primarily responsible for chromatin condensation but for the separation of sister chromatids. The defect in decatenating the sister chromatid DNA would then lead to loss of chromatin condensation shortly before meiosis I. This interpretation is supported by the observation that in the top2 mutant of S. cerevisiae no defect in early meiotic chromosome condensation is visible (Rose and Holm, 1993). In this organism chromosome condensation takes place in early prophase, after DNA replication (Padmore et al., 1991; Scherthan et al., 1992). The observation that in S. pombe vegetative cells topoisomerase II is not required for prophase condensation (Uemura et al., 1987) is consistent with our results. Thus, although chromosome condensation seems to be necessary for proper segregation (for review, see Yanagida, 1995; Koshland and Strunnikov, 1996), topoisomerase II might not be the enzyme that is directly responsible for chromatin condensation.
Andreassen et al. (1997) found that in (nonmeiotic) mammalian cells, override of a checkpoint imposed by VM-26 led to the formation of fully condensed chromosomes, whereas override of an ICRF-193 checkpoint arrest only resulted in partially condensed chromosomes. VM-26 blocks the topoisomerase II strand passing reaction at a step at which the enzyme is covalently bound to a cleaved DNA double strand, whereas ICRF-193 locks the enzyme in a closed clamp formation before DNA cleavage or strand passage. They proposed that in mammalian cells topoisomerase II not only plays an enzymatic role in decatenation but also has a nonenzymatic, structural function, which is necessary for the final steps of chromosome condensation (Andreassen et al., 1997, and references cited therein).
VM-26 causes severe fragmentation of the DNA, ranging from 20 to 800 kb (Roberge et al., 1990). The double-strand breaks caused by VM-26 possibly relieve catenation between the two sister chromatids, permitting the final steps of DNA condensation. In contrast, ICRF-193 does not cause DNA fragmentation (Andreassen et al, 1997). Thus the differences between checkpoint-overridden VM-26 and ICRF-193 arrest could alternatively be explained by the different mechanisms in which both drugs act. The partial condensation in the checkpoint-overridden ICRF-193 arrest suggests that catenation prevents complete chromosome condensation.
We suggest that the inability to disentangle sister chromatid DNA is the primary defect in the top2 mutant causing loss of chromatin condensation and failure of sister chromatid separation.
DNA Catenation and Sister Chromatid Cohesion
In mitosis a system is needed to keep the sister chromatids together until the mitotic division, to ensure proper segregation (for review, see Holm, 1994). Murray and Szostak (1985) suggested that entangling of the sister chromatid DNA is the primary force that keeps them together until mitotic anaphase. However, in vegetative cells of S. cerevisiae, catenation is resolved immediately after S phase (Koshland and Hartwell, 1987; Guacci et al., 1994; for review, see Holm, 1994). From the temperature shift experiment we concluded that in S. pombe topoisomerase II is not needed until the time of the meiotic divisions. However, it cannot be excluded that normally in wild type topoisomerase II decatenates the sister chromatid DNA earlier, but that decatenation can be postponed until shortly before the first meiotic division. Molnar et al. (1995) have shown with fluorescence in situ hybridization that in a S. pombe rec8 mutant the sister chromatid cohesion is lost during meiotic prophase. When in S. pombe catenation is resolved at the end of meiotic prophase, as suggested by the temperature shift experiment, this means that catenation is not able to prevent loss of sister chromatid cohesion in the rec8 mutant. Alternatively, catenation is resolved earlier during meiotic prophase (see above). Either way, it suggests that in S. pombe meiosis, catenation is not necessary for sister chromatid cohesion.
Nonregulatory Arrest
S. pombe top2 cells arrest before meiosis I. The formation of two spindles and the stretching of chromatin fibers out of the undivided nucleus suggest that the cell tries to perform not only the first but even the second meiotic division. The small percentage of cells containing two spindles at later time points suggests that the spindle formation is transient. Arrested cells contain four spindle pole bodies. Thus the spindle pole body cycle continues despite the blocked nuclear division, indicating that the top2 arrest is nonregulatory. Also in mitosis the top2 arrest in S. cerevisiae and S. pombe is nonregulatory (see INTRODUCTION). Downes et al (1994) showed that in mammalian cells a catenation-sensitive checkpoint exists. Rose and Holm (1993) concluded that in S. cerevisiae the meiotic top2 arrest is regulatory and proposed that the synaptonemal complex might play a role in triggering the regulatory arrest. The fact that in S. pombe, in which no synaptonemal complex is formed, the meiotic arrest is nonregulatory is consistent with this proposal.
Conclusions
It has already been known for some time that topoisomerase II has a role in sister chromatid separation as well as in chromatin condensation (see INTRODUCTION). But, to our knowledge, a link between these events has not been established. The use of a meiotic system not only enabled us to study the meiotic function of topoisomerase II, but also showed that topoisomerase II is not necessary for chromatin condensation per se, which has implications for the general function of topoisomerase II. Based on our results, we suggest that the primary defect in the top2 mutant is the inability to disentangle sister chromatid DNA after replication. This leads to failure of sister chromatid separation, a loss of chromatin condensation shortly before meiosis I, and a nonregulatory arrest.
ACKNOWLEDGMENTS
We thank I. Hagan, M. Yanagida, and K. Gull for the generous gift of antibodies and the developers of the ImageTool program for providing such an excellent image analysis program. We thank M. Molnar for communication of unpublished results and Y. Hiraoka for his generosity with respect to the analysis of living top2 cells in his laboratory by one of the authors. This project was supported by grants from the Swiss National Science Foundation and from the Roche Research Foundation.
Abbreviations used:
- cs
cold sensitive
- DAPI
4′,6-diamino-2-phenylindole
- ts
temperature sensitive
REFERENCES
- Andreassen PR, Lacroix FB, Margolis RL. Chromosomes with two intact axial cores are induced by G2 checkpoint override: evidence that DNA decatenation is not required to template the chromosome structure. J Cell Biol. 1997;136:29–43. doi: 10.1083/jcb.136.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J, Schuchert P, Grimm C, Kohli J. Synchronized meiosis and recombination in fission yeast: observations with pat1-114 diploid cells. Curr Genet. 1991;19:445–451. doi: 10.1007/BF00312735. [DOI] [PubMed] [Google Scholar]
- Bähler J, Wyler T, Loidl J, Kohli J. Unusual structures in meiotic prophase of fission yeast: a cytological analysis. J Cell Biol. 1993;121:241–256. doi: 10.1083/jcb.121.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach D, Rodgers L, Gould J. ran1+ controls the transition from mitotic division to meiosis in fission yeast. Curr Genet. 1985;10:297–311. doi: 10.1007/BF00365626. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Ding D-Q, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y. Telomere-led premeiotic chromosome movement in fission yeast. Science. 1994;264:270–273. doi: 10.1126/science.8146661. [DOI] [PubMed] [Google Scholar]
- DeVeaux LC, Smith GR. Region-specific activators of meiotic recombination in Schizosaccharomyces pombe. Genes Dev. 1994;8:203–210. doi: 10.1101/gad.8.2.203. [DOI] [PubMed] [Google Scholar]
- DiNardo S, Voelkel K, Sternglanz R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc Natl Acad Sci USA. 1984;81:2616–2620. doi: 10.1073/pnas.81.9.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes CS, Clarke DJ, Mullinger AM, Giménez-Abián JF, Creighton AM, Johnson RT. A topoisomerase II-dependent G2 cycle checkpoint in mammalian cells. Nature. 1994;372:467–470. doi: 10.1038/372467a0. [DOI] [PubMed] [Google Scholar]
- Goto T, Wang JC. Yeast DNA topoisomerase II is encoded by a single-copy, essential gene. Cell. 1984;36:1073–1080. doi: 10.1016/0092-8674(84)90057-6. [DOI] [PubMed] [Google Scholar]
- Guacci V, Hogan E, Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz H, Leslot H, Leopold U, Loprieno N. Handbook of Genetics. Vol. 1. R.C. King, New York: Plenum Publishing; 1974. Schizosaccharomyces pombe; pp. 395–446. [Google Scholar]
- Hagan I, Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan IM, Hyams JS. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Hirata A, Tanaka K. Nuclear behavior during conjugation and meiosis in the fission yeast Schizosaccharomyces pombe. J Gen Appl Microbiol. 1982;28:263–274. [Google Scholar]
- Holm C. Coming undone: how to untangle a chromosome. Cell. 1994;77:955–957. doi: 10.1016/0092-8674(94)90433-2. [DOI] [PubMed] [Google Scholar]
- Holm C, Goto T, Wang JC, Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985;41:553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- Holm C, Stearns T, Botstein D. DNA topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Mol Cell Biol. 1989;9:159–168. doi: 10.1128/mcb.9.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N, Weiner BM. Potential advantages of unstable interactions for pairing of chromosomes in meiotic, somatic, and premeiotic cells. Cold Spring Harbor Symp Quant Biol. 1993;58:553–565. doi: 10.1101/sqb.1993.058.01.062. [DOI] [PubMed] [Google Scholar]
- Koshland D, Hartwell LH. The structure of sister minichromosome DNA before anaphase in Saccharomyces cerevisiae. Science. 1987;238:1713–1716. doi: 10.1126/science.3317838. [DOI] [PubMed] [Google Scholar]
- Koshland D, Strunnikov A. Mitotic chromosome condensation. Annu Rev Cell Dev Biol. 1996;12:305–333. doi: 10.1146/annurev.cellbio.12.1.305. [DOI] [PubMed] [Google Scholar]
- Molnar M, Bähler J, Sipizki M, Kohli J. The rec8 gene of Schizosaccharomyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics. 1995;141:61–73. doi: 10.1093/genetics/141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munz P. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics. 1994;137:701–707. doi: 10.1093/genetics/137.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S, Yanagida M, Niwa O. A large circular minichromosome of Schizosaccharomyces pombe requires a high dose of type II DNA topoisomerase for its stabilization. Mol Gen Genet. 1995;246:671–679. doi: 10.1007/BF00290712. [DOI] [PubMed] [Google Scholar]
- Murray AW, Szostak JW. Chromosome segregation in mitosis and meiosis. Annu Rev Cell Biol. 1985;1:289–315. doi: 10.1146/annurev.cb.01.110185.001445. [DOI] [PubMed] [Google Scholar]
- Olson LW, Edén U, Egel-Mitani M, Egel R. Asynaptic meiosis in fission yeast? Hereditas. 1978;89:189–199. [Google Scholar]
- Padmore R, Cao L, Kleckner N. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. Cerevisiae. Cell. 1991;66:1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- Ponticelli AS, Smith GR. Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. Genetics. 1989;123:45–54. doi: 10.1093/genetics/123.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SW. Initiation of synapsis and interlocking of chromosomes during zygotene in Bombyx spermatocytes. Carlsberg Res Commun. 1986;51:401–432. [Google Scholar]
- Roberge M, Th’ng J, Hamaguchi J, Bradbury EM. The topoisomerase II inhibitor VM-26 induces marked changes in histone H1 kinase activity, histones H1 and H3 phosphorylation and chromosome condensation in G2 phase and mitotic BHK cells. J Cell Biol. 1990;111:1753–1762. doi: 10.1083/jcb.111.5.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinow CF. The number of chromosomes in Schizosaccharomyces pombe: light microscopy of stained preparations. Genetics. 1977;87:491–497. doi: 10.1093/genetics/87.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose D, Holm C. Meiosis-specific arrest revealed in DNA topoisomerase II mutants. Mol Cell Biol. 1993;13:3445–3455. doi: 10.1128/mcb.13.6.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose D, Thomas W, Holm C. Segregation of recombined chromosomes in meiosis I requires DNA topoisomerase II. Cell. 1990;60:1009–1017. doi: 10.1016/0092-8674(90)90349-j. [DOI] [PubMed] [Google Scholar]
- Scherthan H, Bähler J, Kohli J. Dynamics of chromosome organization and pairing during meiotic prophase in fission yeast. J Cell Biol. 1994;127:273–285. doi: 10.1083/jcb.127.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H, Loidl J, Schuster T, Schweitzer D. Meiotic chromosome condensation and pairing in Saccharomyces cerevisiae studied by chromosome painting. Chromosoma. 1992;101:590–595. doi: 10.1007/BF00360535. [DOI] [PubMed] [Google Scholar]
- Spell R, Holm C. Nature and distribution of chromosomal intertwinings in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:1465–1476. doi: 10.1128/mcb.14.2.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda A, Bähler J, Kohli J. Microtubule-driven nuclear movements and linear elements as meiosis-specific characteristics of the fission yeasts Schizosaccharomyces versatilis and Schizosaccharomyces pombe. Chromosoma. 1995;104:203–214. doi: 10.1007/BF00352185. [DOI] [PubMed] [Google Scholar]
- Uemura T, Ohkura H, Adachi Y, Morino K, Shiozaki K, Yanagida M. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987;50:917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- Uemura T, Yanagida M. Isolation of type I and II DNA topoisomerase mutants from fission yeast: single and double mutants show different phenotypes in cell growth and chromatin organization. EMBO J. 1984;3:1737–1744. doi: 10.1002/j.1460-2075.1984.tb02040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Yanagida M. Mitotic spindle pulls but fails to separate chromosomes in type II DNA topoisomerase mutants: uncoordinated meiosis. EMBO J. 1986;5:1003–1010. doi: 10.1002/j.1460-2075.1986.tb04315.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wettstein D, Rasmussen SW, Holm PB. The synaptonemal complex in genetic segregation. Annu Rev Genet. 1984;18:331–413. doi: 10.1146/annurev.ge.18.120184.001555. [DOI] [PubMed] [Google Scholar]
- Watt PM, Hickson ID. Structure and function of type II DNA topoisomerases. Biochem J. 1994;303:681–695. doi: 10.1042/bj3030681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A, Sherwin T, Sasse R, McRae TH, Baines AJ, Gull K. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J Cell Sci. 1989;93:491–500. doi: 10.1242/jcs.93.3.491. [DOI] [PubMed] [Google Scholar]
- Yanagida M. Frontier questions about sister chromatid separation in anaphase. Bioessays. 1995;17:579–526. doi: 10.1002/bies.950170608. [DOI] [PubMed] [Google Scholar]