Abstract
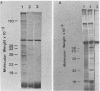
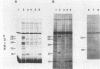
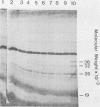
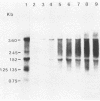
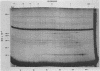
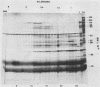
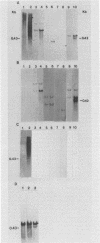
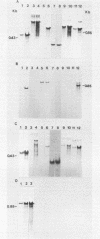
Three late transcripts have been mapped to the vaccinia virus 7.1-kilobase (kb) EcoRI F fragment, which is located approximately 85 kb from the left end of the viral genome. Hybrid selection and translation of RNA have demonstrated that these mRNAs encode three polypeptides with apparent molecular weights of 32,000 (32K), 26K, and 19K. The transcripts encoding the 32K and 19K polypeptides are synthesized in a leftward direction and overlap each other as well as the RNA encoding the early 110K polypeptide. In addition, the RNA encoding the 32K polypeptide also overlaps an early transcript of 1.35 kb (see accompanying paper). The transcript encoding the 26K polypeptide overlaps the early 2.45-, 0.6- to 0.7-, and 1.7-kb RNAs, and all four mRNAs are transcribed from left to right. The protein coding sequences for the 26K and 32K polypeptides lie outside the 7.1-kb DNA fragment. Due to their heterogeneity in size, each of the three late transcripts is undetectable as a distinct size by filter hybridization. In addition, although S1 mapping has detected the 5' terminus of the late RNA which maps entirely within this fragment, it has been unable to size and locate the 3' ends of all three transcripts, and this result indicates that the heterodisperse size of the RNAs is due to heterogeneity at the 3' ends of these transcripts. The cause of this heterogeneity in termination of transcription is not known.
Full text
PDF










Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alt F. W., Bothwell A. L., Knapp M., Siden E., Mather E., Koshland M., Baltimore D. Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3' ends. Cell. 1980 Jun;20(2):293–301. doi: 10.1016/0092-8674(80)90615-7. [DOI] [PubMed] [Google Scholar]
- Alwine J. C., Kemp D. J., Parker B. A., Reiser J., Renart J., Stark G. R., Wahl G. M. Detection of specific RNAs or specific fragments of DNA by fractionation in gels and transfer to diazobenzyloxymethyl paper. Methods Enzymol. 1979;68:220–242. doi: 10.1016/0076-6879(79)68017-5. [DOI] [PubMed] [Google Scholar]
- Bailey J. M., Davidson N. Methylmercury as a reversible denaturing agent for agarose gel electrophoresis. Anal Biochem. 1976 Jan;70(1):75–85. doi: 10.1016/s0003-2697(76)80049-8. [DOI] [PubMed] [Google Scholar]
- Baroudy B. M., Moss B. Purification and characterization of a DNA-dependent RNA polymerase from vaccinia virions. J Biol Chem. 1980 May 10;255(9):4372–4380. [PubMed] [Google Scholar]
- Boone R. F., Parr R. P., Moss B. Intermolecular duplexes formed from polyadenylylated vaccinia virus RNA. J Virol. 1979 Apr;30(1):365–374. doi: 10.1128/jvi.30.1.365-374.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Cooper J. A., Wittek R., Moss B. Extension of the transcriptional and translational map of the left end of the vaccinia virus genome to 21 kilobase pairs. J Virol. 1981 Sep;39(3):733–745. doi: 10.1128/jvi.39.3.733-745.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. A., Wittek R., Moss B. Hybridization selection and cell-free translation of mRNA's encoded within the inverted terminal repetition of the vaccinia virus genome. J Virol. 1981 Jan;37(1):284–294. doi: 10.1128/jvi.37.1.284-294.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early P., Rogers J., Davis M., Calame K., Bond M., Wall R., Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980 Jun;20(2):313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- Hruby D. E., Ball L. A. Mapping and identification of the vaccinia virus thymidine kinase gene. J Virol. 1982 Aug;43(2):403–409. doi: 10.1128/jvi.43.2.403-409.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isle H. B., Venkatesan S., Moss B. Cell-free translation of early and late mRNAs selected by hybridization to cloned DNA fragments derived from the left 14 million to 72 million daltons of the vaccinia virus genome. Virology. 1981 Jul 15;112(1):306–317. doi: 10.1016/0042-6822(81)90636-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mahr A., Roberts B. E. Organization of six early transcripts synthesized from a vaccinia virus EcoRI DNA fragment. J Virol. 1984 Feb;49(2):497–509. doi: 10.1128/jvi.49.2.497-509.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. S., Ricciardi R. P., Roberts B. E., Paterson B. M., Mathews M. B. Arrangement of messenger RNAs and protein coding sequences in the major late transcription unit of adenovirus 2. J Mol Biol. 1980 Oct 5;142(4):455–488. doi: 10.1016/0022-2836(80)90258-2. [DOI] [PubMed] [Google Scholar]
- Moss B., Salzman N. P. Sequential protein synthesis following vaccinia virus infection. J Virol. 1968 Oct;2(10):1016–1027. doi: 10.1128/jvi.2.10.1016-1027.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J. R., Joklik W. K. Isolation and properties of the vaccinia virus DNA-dependent RNA polymerase. J Biol Chem. 1977 Oct 10;252(19):6930–6938. [PubMed] [Google Scholar]
- Pelham H. R., Jackson R. J. An efficient mRNA-dependent translation system from reticulocyte lysates. Eur J Biochem. 1976 Aug 1;67(1):247–256. doi: 10.1111/j.1432-1033.1976.tb10656.x. [DOI] [PubMed] [Google Scholar]
- Reichardt L., Kaiser A. D. Control of lambda repressor synthesis. Proc Natl Acad Sci U S A. 1971 Sep;68(9):2185–2189. doi: 10.1073/pnas.68.9.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J., Early P., Carter C., Calame K., Bond M., Hood L., Wall R. Two mRNAs with different 3' ends encode membrane-bound and secreted forms of immunoglobulin mu chain. Cell. 1980 Jun;20(2):303–312. doi: 10.1016/0092-8674(80)90616-9. [DOI] [PubMed] [Google Scholar]
- Salzman N. P., Sebring E. D. Sequential formation of vaccinia virus proteins and viral deoxyribonucleic acid replication. J Virol. 1967 Feb;1(1):16–23. doi: 10.1128/jvi.1.1.16-23.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A., Berk A. J., Berget S. M. Transcription maps of adenovirus. Methods Enzymol. 1980;65(1):750–768. doi: 10.1016/s0076-6879(80)65071-x. [DOI] [PubMed] [Google Scholar]
- Spencer E., Shuman S., Hurwitz J. Purification and properties of vaccinia virus DNA-dependent RNA polymerase. J Biol Chem. 1980 Jun 10;255(11):5388–5395. [PubMed] [Google Scholar]
- Weir J. P., Bajszár G., Moss B. Mapping of the vaccinia virus thymidine kinase gene by marker rescue and by cell-free translation of selected mRNA. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1210–1214. doi: 10.1073/pnas.79.4.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittek R., Cooper J. A., Moss B. Transcriptional and translational mapping of a 6.6-kilobase-pair DNA fragment containing the junction of the terminal repetition and unique sequence at the left end of the vaccinia virus genome. J Virol. 1981 Sep;39(3):722–732. doi: 10.1128/jvi.39.3.722-732.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittek R., Menna A., Schümperli D., Stoffel S., Müller H. K., Wyler R. HindIII and Sst I restriction sites mapped on rabbit poxvirus and vaccinia virus DNA. J Virol. 1977 Sep;23(3):669–678. doi: 10.1128/jvi.23.3.669-678.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittek R., Moss B. Colinearity of RNAs with the vaccinia virus genome: anomalies with two complementary early and late RNAs result from a small deletion or rearrangement within the inverted terminal repetition. J Virol. 1982 May;42(2):447–455. doi: 10.1128/jvi.42.2.447-455.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Hagenbüchle O., Schibler U. A single mouse alpha-amylase gene specifies two different tissue-specific mRNAs. Cell. 1981 Feb;23(2):451–458. doi: 10.1016/0092-8674(81)90140-9. [DOI] [PubMed] [Google Scholar]