Abstract
Silencing is a universal form of transcriptional regulation in which regions of the genome are reversibly inactivated by changes in chromatin structure. Sir2 (Silent Information Regulator) protein is unique among the silencing factors in Saccharomyces cerevisiae because it silences the rDNA as well as the silent mating-type loci and telomeres. Discovery of a gene family of Homologues of Sir Two (HSTs) in organisms from bacteria to humans suggests that SIR2’s silencing mechanism might be conserved. The Sir2 and Hst proteins share a core domain, which includes two diagnostic sequence motifs of unknown function as well as four cysteines of a putative zinc finger. We demonstrate by mutational analyses that the conserved core and each of its motifs are essential for Sir2p silencing. Chimeras between Sir2p and a human Sir2 homologue (hSir2Ap) indicate that this human protein’s core can substitute for that of Sir2p, implicating the core as a silencing domain. Immunofluorescence studies reveal partially disrupted localization, accounting for the yeast–human chimeras’ ability to function at only a subset of Sir2p’s target loci. Together, these results support a model for the involvement of distinct Sir2p-containing complexes in HM/telomeric and rDNA silencing and that HST family members, including the widely expressed hSir2A, may perform evolutionarily conserved functions.
INTRODUCTION
Epigenetic forms of transcriptional control regulate biological processes as diverse as growth and differentiation, dosage compensation, sex determination, and host defense (reviewed by Russo et al., 1996; Henikoff and Matzke, 1997, and references therein). Some epigenetic changes are effected by nonnuclear means, for example, plant cosuppression and prions. More common are nuclear mechanisms, such as mammalian imprinting, Drosophila position effect variegation, and plant paramutation. Nuclear gene silencing involves heritable but reversible changes in gene expression associated with structural alterations in chromatin. Gene silencing is thus a global mechanism of transcriptional control in which large regions of the genome are regulated in a position-dependent yet gene-independent manner.
Sir2p is one of several factors critical for silencing at least three loci in yeast (reviewed by Loo and Rine, 1995; Sherman and Pillus, 1997). Among the four SIR (Silent Information Regulator) genes, SIR2 is unique because it is required for silencing and suppression of recombination within the rDNA, as well as silent mating-type (HM) and telomeric silencing (Shore et al., 1984; Ivy et al., 1986; Rine and Herskowitz, 1987; Gottlieb and Esposito, 1989; Aparicio et al., 1991; Bryk et al., 1997; Fritze and Esposito, 1997; Smith and Boeke, 1997; Smith et al., 1998). A sir2Δ mutant strain exhibits complete derepression at these loci. Derepression has been correlated with increased accessibility to DNA-modifying enzymes and psoralen, indicating that these loci have a more relaxed chromatin structure in the absence of Sir2p (Nasmyth, 1982; Gottschling, 1992; Singh and Klar, 1992; Loo and Rine, 1994; Fritze and Esposito, 1997; Smith and Boeke, 1997; Smith et al., 1998).
Consistent with the direct involvement of Sir2p in maintaining silencing-competent chromatin structure, Sir2p localizes to the HM loci, to telomeres and to the rDNA within the nucleolus (Hecht et al., 1996; Gotta et al., 1997; Strahl-Bolsinger et al., 1997). Sir2p interacts with itself as well as with Sir3p and Sir4p (Moazed and Johnson, 1996; Holmes et al., 1997; Moazed et al., 1997; Strahl-Bolsinger et al., 1997), which in turn interact with the histones H3 and H4 at the HM loci and telomeres (Hecht et al., 1995, 1996). In addition, Sir2p also interacts with ubiquitination factors and/or complexes (Moazed and Johnson, 1996), and mutants in SIR2 and UBC2 (Ubiquitin-Conjugating factor 2) have similar rDNA silencing phenotypes (Bryk et al., 1997). A potential mechanism for the function of Sir2p in silencing has been suggested by the observation that SIR2 overexpression is correlated with hypoacetylation of a subset of histones (Braunstein et al., 1993). Hypoacetylated histones are often found at silenced or inactive loci in yeast and other organisms (Lin et al., 1989; Turner et al., 1992; Braunstein et al., 1993, 1996; Jeppesen and Turner, 1993; O’Neill and Turner, 1995). However, SIR2’s potential role in modulating histone deacetylation may be indirect as histone deacetylase activity has not been detected for Sir2p in vitro (Braunstein et al., 1993). These interactions thus define a role for SIR2 in silencing complexes that may be subject to and/or participate in multiple forms of regulation.
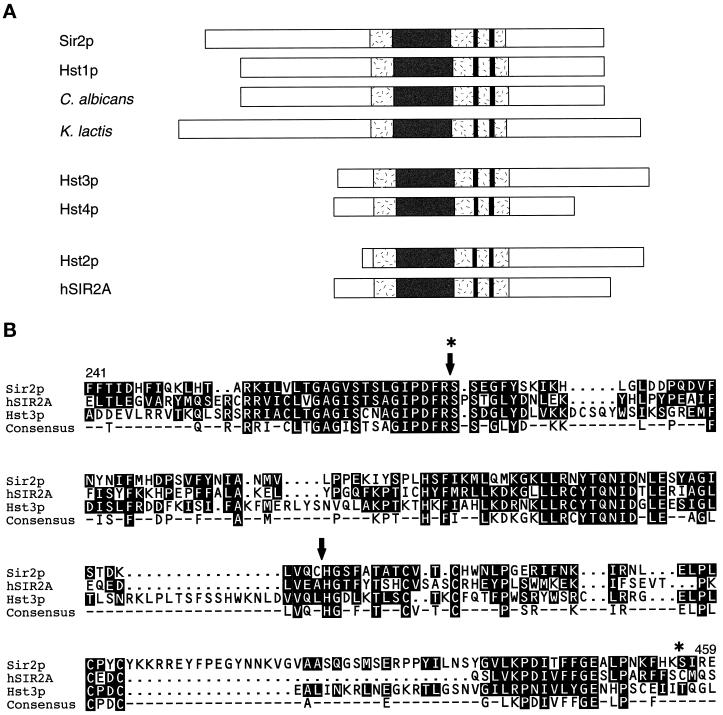
We and others discovered and characterized an evolutionarily conserved family of Homologue of Sir Two (HST) genes (Chen and Clark-Walker, 1994; Brachmann et al., 1995; Derbyshire et al., 1996; Yahiaoui et al., 1996; Tsang and Escalante-Semerena, 1998). The proteins encoded by the HSTs (Hst proteins) are ∼30–65% identical to Sir2p overall (Figure 1A). The Hst family is characterized by a conserved core domain, which is up to 84% identical to Sir2p and contains three motifs (Figure 1B). Two sequence motifs of unknown function are termed here the GAG and NID motifs to mark the beginning and end, respectively, of more extended sequences described in Figure 1. The third motif consists of four absolutely conserved cysteines that form a putative zinc finger, which may specify either protein– protein or protein–nucleic acid interactions. Alternatively, the cysteines may participate in disulfide bond formation and thus protein folding or catalysis.
Figure 1.
Alignment of yeast Sir2p and the Hst proteins. (A) The Hst proteins from bacteria to humans are ∼30–65% identical to Sir2p overall. Sir2p and the Hst proteins all contain a characteristic core domain (shaded) that includes four cysteines of a putative zinc finger (paired lines). In the most highly conserved region of this domain (dark), bounded by the GAG and NID motifs, which are of unknown function, identity reaches 84%. The Hst proteins can be grouped into three subfamilies based on the length and sequence of their relatively distinct N and C termini. Members of the first two of these subfamilies have previously been implicated in silencing (Brachmann et al., 1995). (B) Core domain sequences of members of each of the three subfamilies (Sir2p, hSir2Ap, and Hst3p) are aligned. The consensus sequence highlights the four conserved cysteines of the putative zinc finger (at positions 372, 374, 396, and 398 in Sir2p), as well as the two conserved motifs of unknown function, which generally consist of the sequences GAG(I/V)Sxxx G(I/V)PDFRS, and (Y/I)TQNID. Some additional variation at the positions in brackets is observed for published sequences and at other positions for incomplete sequences in the databases. These motifs, although somewhat degenerate, are diagnostic of members of this gene family. The arrows and stars indicate the boundaries of the region swapped in the Sir2-hSir2A/Hst3(NID) chimeras and the Sir2-hSir2A(NID+CYS) chimera, respectively. See Brachmann et al. (1995) for additional flanking sequences.
Sir2p and its homologues can be divided into three subfamilies based on the length and sequence of their N and C termini (Figure 1A) (Brachmann et al., 1995). Sir2p is grouped with its closest relative, Hst1p from Saccharomyces cerevisiae, as well as with homologues from another budding yeast Kluyveromyces lactis (Chen and Clark-Walker, 1994) and from the pathogenic filamentous yeast Candida albicans (Perez-Martin et al., 1999). In the second subfamily, there are two members from S. cerevisiae, Hst3p and Hst4p. The third subfamily is the largest and includes many of the Hst proteins from other organisms, along with a single S. cerevisiae protein, Hst2p. Homologues of SIR2 have been identified in bacteria, including Archaebacteria, protozoa, nematodes, plants, and mammals. Evolutionary conservation of the Hst and Sir2 proteins between all of the biological kingdoms suggests that they share an important function, possibly in chromatin organization. Like SIR2, members from two of three HST subfamilies have been implicated in silencing in S. cerevisiae. Overexpression of HST1 partially suppresses sir2Δ mating and HM silencing defects, but not those in telomeric silencing, rDNA silencing, or rDNA recombination (Brachmann et al., 1995; Derbyshire et al., 1996; Sherman, unpublished results). This indicates that HST1 encodes a protein capable of silencing, but which may function primarily at a different locus. An hst3Δ hst4Δ double mutant exhibits telomeric silencing defects, as well as temperature-sensitive growth, cell cycle arrest, and chromosomal instability (Brachmann et al., 1995). The involvement of several of the HSTs in silencing has led to the hypothesis that they silence as yet unidentified loci in yeast and other organisms (Brachmann et al., 1995; Sherman and Pillus, 1997).
To determine whether the conserved core is a silencing domain and whether the Hst proteins from other organisms are likely to be silencing factors, we performed a structure–function analysis of Sir2p and expression studies with a human Hst protein in yeast. We demonstrate by deletion and point mutational analyses that the conserved core domain and its motifs are essential for Sir2p silencing. This is in contrast to the nonconserved N-terminal 79 amino acids of Sir2p, which we observe are dispensable for function. Complementation and dominance studies of chimeras between SIR2 and a human homologue of SIR2 (hSIR2A) suggest that the core is an evolutionarily conserved silencing domain and that SIR-like silencing mechanisms may function in human gene regulation.
MATERIALS AND METHODS
Yeast Strains, Media, and Transformations
Genotypes of yeast strains are listed in Table 1 and are derived from the W303 (Thomas and Rothstein, 1989), YPH499 to 501 (Sikorski and Hieter, 1989), and FY2 (Winston et al., 1995) backgrounds. The sir2/sir2 homozygous diploid strain LPY3380 was constructed to facilitate immunofluorescence experiments, because the diploid nucleus is larger than the haploid nucleus. This strain maintains a haploid-specific program of gene expression, because it is also homozygous for MATa and hmlΔ::TRP1, to avoid any potential changes in Sir2p localization from haploids to diploids. The sir2Δ::HIS3 deletion (kindly provided by J. Rine, University of California, Berkeley, CA) was made by PCR-directed mutagenesis. Yeast were grown at 30°C under standard conditions (Rose et al., 1989).
Table 1.
Yeast strains
| Strain (alias) | Genotype |
|---|---|
| LPY0078 (αhis4) | MATα his4 |
| LPY0143 (ahis4) | MATa his4 |
| LPY0253 (RS927) | MATa ade2 his3 leu2 trp1 ura3 hml∷TRP1 |
| LPY0254 (RS928) | MATα ade2 his3 leu2 trp1 ura3 hml∷TRP1 |
| LPY1403 | MATα ade2 his3 leu2 trp1 ura3 hml∷TRP1 sir2Δ∷HIS3 |
| LPY1683 | MATa ade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ63 ura3-52 ADH4∷URA3 (VIIL) DIA5-1∷ADE2 (VR) |
| LPY1953 (YCB652) | MATa his3Δ200 leu2Δ1 lys2Δ202 trp1Δ63 ura3-52 sir2Δ2∷TRP1 ADH4∷URA3-TEL |
| LPY2446 (JS128) | MATα his3Δ200 leu2Δ1 ura3-52 RDN1∷Ty1-mURA3 (S6) |
| LPY2447 (JS163) | MATα his3Δ200 leu2Δ1 ura3-52 sir2Δ2∷HIS3 RDN1∷Ty1-mURA3 (S6) |
| LPY3380 | MATahmlΔ∷TRP1HMRa ade2bar1can1his3leu2lys2trp1ura3sir2Δ2∷HIS3 |
| MATa hmlΔ∷TRP1 HMRa ade2 bar1 can1 his3 leu2 lys2 trp1 ura3 sir2Δ2∷HIS3 | |
| LPY3923 | MATa ade2-1 can1-100 his3-11,15 hmrΔ∷TRP1 leu2-3,112 trp1-1 ura3-1 sir2Δ∷HIS3 |
Plasmids and SIR2/HST Mutagenesis
The sequences of the oligonucleotide primers (5′–3′) used in these studies are listed. Small letters indicate that the nucleotide is different from that of wild-type, and three slashes (///) denote deleted sequence or the junction between yeast and human sequence. Restriction sites used for cloning and/or introduced by the primer are underlined: SIR2-ATG(Jxn), ATCGCTTCGGTAGACAC; SIR2-StuI(Jxn), TGATAT-CTGGTTTGAGAACG; SIR2-ATG, CATCCAGCTTTAATGTGCCG; sir2ΔBN-NcoI, GACTTCAGATCT///CATGGCTCTTTTGCTACTGCC; sir2-CA5′, GCTACTGCCACCgcCGTTACCgcCCATTGGAACCTAC-CCGGgGAGAGGATATTTAAT; sir2-CA3′, CTCTTCTTTTTTTGTAAgcGTACGGGgcTAGTGGAAGcTCGAGGTTTCTA; sir2-HIII-SEI, G-AAGGAACCAaGcTTACGATTTC; sir2-NotI, GCGTATTTCATATGgc-GGccGcTCATCCAGCT; hsir2A-BglII, AGGAATTCCCGACTTTaGaT-CTCCATCCAC; hsir2A-NcoI, GTAGAAGGTGCCaTGgGCCTCCAC-CA; hsir2A-NotI, GGAGAAGCAGACATGGACTgCggcCGcAACT-TATTCTC; and hsir2A-NruI, GATATCTTCGCGAA///TACAGGAGA-AGAAACG.
Plasmids were constructed using standard techniques (Sambrook et al., 1989) and sequenced as appropriate. A number of SIR2+-containing plasmids were constructed by blunting a 2.7-kb BstNI fragment containing wild-type SIR2 and subcloning it into the SmaI site of YEp351 and YEp352 (Hill et al., 1986) (pLP349–350 and pLP318, respectively) and pKS+ (Stratagene, La Jolla, CA) (pLP319). To construct the sir2-ΔCORE clone (pLP387) in which aa 245–427 are deleted (FFTI244P428PYIL), pLP349 DNA was digested with BclI and StuI, blunted, and recircularized. The sir2-ΔGAG clone in which aa 245–273 are deleted and an F274D mutation is introduced (FFTI244D274RSSE) and the sir2-ΔCYS clone in which aa 364–427 are deleted (LVQC363P428PYIL) were similarly constructed. pLP349 DNA was digested with either BclI and BglII or NcoI and StuI, blunted, and religated to produce pLP385 (sir2-ΔGAG) and pLP386 (sir2-ΔCYS), respectively. Construction of the sir2-ΔNID and cysteine point mutant clones required PCR mutagenesis using either pLP349 or pLP319. To delete aa 277–363 (DFRS276H364FAT) and construct pLP656, primer sir2ΔBN-NcoI was used in combination with the sir2-HIII-SEI primer to amplify an ∼390-bp PCR fragment. This fragment was digested with BglII and StuI, and the resulting ∼200-bp fragment was cloned into pLP349 from which the wild-type ∼460-bp BglII–StuI fragment had been deleted. The cysteine to alanine point mutant clones sir2-A372A374 . . C396C398 (pLP531), sir2-C372C374 . . A396A398 (pLP555), and sir2-A372A374 . . A396A398 (pLP570) were made by multiple rounds of a three-step PCR method. In PCR I, the SIR2-ATG(Jxn) primer was used with the sir2-CA3′ primer, which mutates the second pair of cysteines, to amplify an ∼1.3-kb fragment of SIR2. In PCR II, the M13 (reverse) primer was used with sir2-CA5′, which mutates the first pair of cysteines. In PCR III, the products of PCRs I and II were used as templates for amplification with the SIR2-ATG(Jxn) primer and the internal SIR2-StuI(Jxn). The ∼1.4-kb product was digested with NcoI and StuI, and the resulting ∼190-bp fragment was cloned into pLP349 from which this piece of DNA had been removed to make pLP531 and pLP555. These clones were then used as templates for repetitions of PCRs I and II, respectively, followed by an additional PCR III step with the PCR I and II products as templates. Once again, the NcoI–StuI PCR mutagenized fragment replaced the wild-type SIR2 sequence in pLP349 to make the quadruple mutant clone sir2-A372A374 . . A396A398 (pLP570). To make the SIR2-hSIR2A(NID) chimera, the hSIR2A expressed sequence tag T66100 (pCAR258) was PCR mutagenized using the hsir2A-BglII and hsir2A-NcoI primers. The BglII and NcoI sites created were used to clone the ∼270-bp core-containing hSIR2A fragment into the SIR2 backbone. The sequence of the resulting chimera on YEp351 (pLP999) was Sir2p - - PDFR275(S61PST - - - LVEA149)H364GSF - - Sir2p. Construction of the SIR2-HST3(NID) chimera (pLP745) [Sir2p - - PDFR275(S74SDG - - - VVQL186)H364GSF - - Sir2p in which an F186L change was also made] was similar (our unpublished results). Likewise, PCR mutagenesis of the hSIR2A gene on pCAR258 with the hsir2A-BglII and hsir2A-NruI primers was used to make the necessary restriction enzyme sites to create the SIR2-hSIR2A(NID + CYS) chimera (Sir2p - - PDFR275(S61PST - - - FFSC209)I457RED - - Sir2p) on YEp351 (pLP905). An ∼350-bp BglII fragment and an ∼110-bp BglII–NruI fragment were cloned into the BglII–NruI SIR2 backbone. The CEN-SIR2-hSIR2A(NID) and CEN-SIR2-hSIR2A(NID+CYS) clones were made by digesting pLP999 and pLP905 with EcoRI and SalI, blunting, and cloning this ∼2.7-kb fragment into the SmaI site of pRS315 (Sikorski and Hieter, 1989) to create pLP1022 and pLP888, respectively. The CEN-SIR2+ clone was constructed starting with a CEN-sir2-ΔCORE construct (pLP416). pLP416 was made by isolating the PvuII fragment (∼2.5-kb) containing sir2-ΔCORE as well as the polylinker sites from pLP387 (described above) and subcloning it into the PvuII sites of pRS315. The ∼740-bp NdeI fragment from pLP416 was then replaced with the ∼1.3-kb SIR2+ NdeI fragment to create pLP907. To create sir2-ΔN (pLP411) in which aa 2–79 are deleted (M1E80LK), the fragment of SIR2 from the promoter through the ATG was generated by PCR amplifying an ∼270-bp fragment of SIR2 from pLP319 using the T7 and SIR2-ATG primers. The PCR product was digested with EcoRI to leave an EcoRI site at the 5′ end and a blunt ATG at the 3′ end of the coding strand. The fragment encoding aa 80–458 of Sir2p was obtained by isolating an ∼1.1-kb ClaI–NruI fragment of SIR2 and blunting the ClaI site. The EcoRI-ATG PCR product and blunt-ended ClaI–NruI fragment were then cloned into the polylinker EcoRI site and SIR2 NruI site in pLP318 replacing the ∼1.6-kb wild-type fragment to create pLP383. The ∼2.7-kb PvuII fragment from pLP383 was subsequently subcloned onto the high-copy LEU2 vector YEp351 to make pLP411. To construct hSIR2A cloned behind the SIR2 promoter, SIR2 was first modified to create a NotI site just downstream of the initiator ATG by a three-step PCR mutagenesis protocol. SIR2 was amplified from pLP319 using the T7 and sir2-NotI (PCR I) and T3 and SIR2-ATG(Jxn) primers (PCR II). The products of PCRs I and II (∼0.2 and 2.5 kb, respectively) were used as templates for amplification with the T7 and T3 primers in PCR III. The ∼2.7-kb product of PCR III was subsequently digested with PstI and BglII, and the resulting ∼1.1-kb fragment was cloned into these sites in pLP349 to create the SIR2-NotI clone, pLP1015. Creation of the NotI site in SIR2 led to a change in the N-terminal sequence of Sir2p from M1TIPH to M1SGAH. Genes, like hSIR2A, inserted in frame via the NotI site behind the SIR2 promoter contain the M1S2 sequence from modified Sir2p. An additional step was required before cloning hSIR2A behind the SIR2 promoter using the NotI site. A NotI site at the 3′ end of the hSIR2A gene had to be eliminated by isolating an ∼1.8-kb HindIII–NotI fragment containing hSIR2A from pCAR258, blunting, and cloning it into the SmaI site of YEp351. Both orientations of this clone were obtained (pLP659 and pLP660). The ∼630-bp PstI–BglII fragment from pLP660 (in which the 5′ end of the gene is on the side of the HindIII site) was replaced with the ∼1.1-kb PstI–BglII fragment from pLP1015 containing the SIR2 promoter and NotI site as well as additional SIR2 sequence (pLP997). pLP659 was used as the template for PCR mutagenesis with the hsir2A-NotI and M13 (forward) primers to create an ∼1.7-kb product with a NotI site in the N-terminal coding region of hSIR2A. The creation of this NotI site changed the sequence of the N terminus of the hSir2Ap from M1DFLR to M1DCGR. The PCR product was digested with NotI and BglII, and this ∼510-bp fragment was used to replace the ∼810-bp of SIR2 sequence in pLP997 to construct the SIR2 (promoter)-NotI-hSIR2A clone (pLP1024). The resulting N-terminal sequence of the hSir2A protein expressed from the SIR2 promoter is M1SGR instead of M1DFLR. To monitor expression of hSir2Ap, triple hemagglutinin (HA) and protein A tags were inserted in frame as NotI fragments. To test the effects of hSIR2A overexpression, a GAL-hSIR2A construct was made. An ∼700-bp BamHI–EagI fragment containing the GAL10 promoter followed by codons for an initiator methionine and an asparagine at position 2 and then a NotI site was subcloned from pLP748 (Freeman-Cook et al., 1999) into pRS315 to make pLP1036. The ∼1.8-kb EagI fragment containing the modified hSIR2A gene from pLP1024 was subsequently cloned into the EagI site in pLP1036 to create the GAL-hSIR2A construct pLP1062. The N terminus of the hSir2A protein in this case is M1NGR. Expression of the subcloned genes was confirmed by immunoblot analysis of cell extracts prepared from transformants of sir2Δ strains and probed with appropriate antisera (see below).
Silencing Assays
For mating and HM silencing assays, cultures of LPY1953, 1403, 3923, and 253 transformants were grown overnight in selective medium, diluted to a starting A600 of 1.0 (∼107 cells) for spotting assays (see Figure 3) and 3.0 (see Figures 2B and 4, B and C) or 4.0 (see Figure 6A) for dilution assays. It should be noted that a fundamental difference between the mating and HM silencing assays is that mating complementation requires repression be maintained only long enough for a single mating event to occur, whereas repression of the reporters must be continuous to restore a Trp− phenotype. For telomeric and rDNA silencing assays, the protocol was derived from that of Gottschling et al. (1990). Transformants were grown for 3–4 d and diluted to a starting A600 of 1.0 for spotting assays (Figure 3) and 2.5 (see Figures 2C and 4D), 4.0 (see Figure 6B), or 5.0 (see Figures 2D, 4E, and 6C) for dilution assays. Different starting densities were used to enhance detection of differences in the individual assays. In the dilution assays, the cultures were then serially diluted four- or fivefold depending on the dynamic range required for the experiment, and the dilutions were stamped using a pin replicator onto the appropriate tester plates (see figure legends). For analysis of hSIR2A cloned under control of the SIR2 and GAL10 promoters, overnight cultures were grown first in glucose leu− and then in raffinose leu− and finally diluted into galactose leu− for either an overnight (mating and HM silencing) or 3- to 4-d growth period (telomeric and rDNA silencing) as described above.
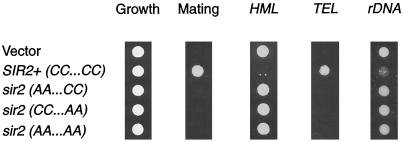
Figure 3.
Mutation of the conserved cysteines in Sir2p causes a complete loss of silencing function. sir2Δ strains were transformed with vector alone (YEp351) or containing wild-type SIR2 (pLP349) or the cysteine point mutants (pLP531 - - AA . . CC, pLP555 - -CC . . AA, and pLP570 - - AA . . AA). Equal cell densities (1 A600) for each transformant were stamped onto leu− plates (growth control) and onto selective plates to assay silencing function. Shown is a representative growth control panel and LPY1953 (MATa sir2Δ TEL::URA3) transformants stamped on a lawn of α his4 (LPY78) mating tester on minimal plates (only those that form diploids grow under these conditions) and 5-FOA to assay telomeric silencing. Also shown are LPY1403 transformants on leu−trp− plates to assay HM silencing and LPY2447 transformants on leu−ura− plates to assay rDNA silencing. Quantitation indicated that the point mutants are completely nonfunctional in silencing (our unpublished results).
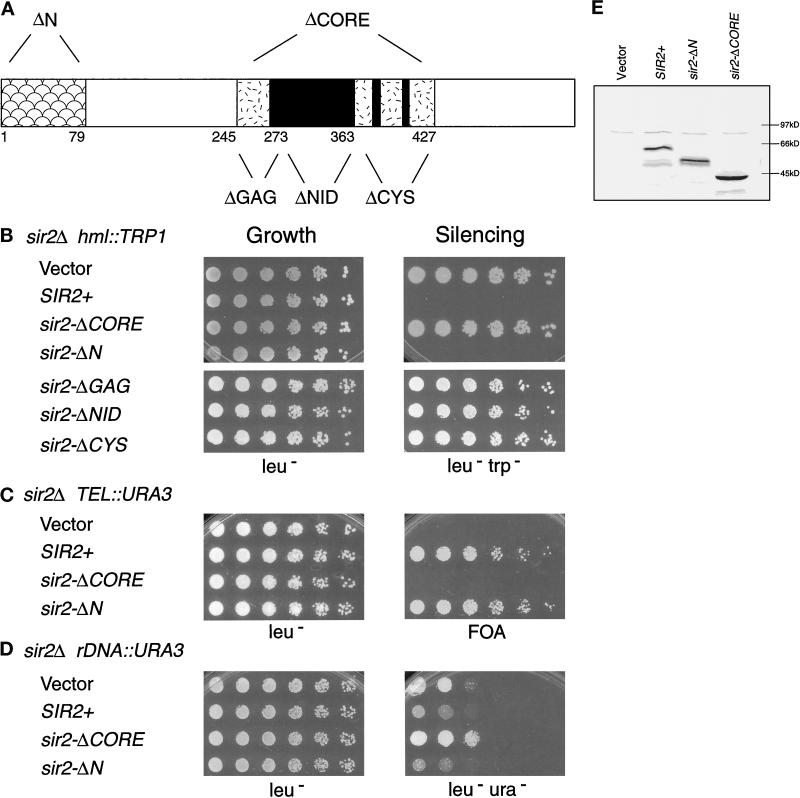
Figure 2.
The conserved core and its motifs are essential for Sir2p silencing. (A) The N- and C-terminal end points of the sir2 mutants are indicated diagramatically for the sir2-ΔN (ΔN, Δ2–79) and sir2-ΔCORE (ΔCORE, Δ245–427) constructs, as well as the smaller motif deletions [sir2-ΔGAG (ΔGAG, Δ245–273), sir2-ΔNID (ΔNID, Δ277–363), and sir2-ΔCYS (ΔCYS, Δ364–427)]. (B–D) Complementation of the sir2Δ silencing defects by the core and N-terminal deletion mutants. LPY1403 (MATα sir2Δ hml:: TRP1) (B), LPY1953 (MATa sir2Δ TEL:: URA3) (C), and LPY2447 (MATα sir2Δ rDNA:: URA3) (D) were transformed with vector alone (YEp351) or containing SIR2+ (pLP349), sir2-ΔCORE (pLP387), or sir2-ΔN (pLP411) constructs. LPY1403 was also transformed with plasmids carrying the motif deletion constructs [sir2-ΔGAG (pLP385), sir2-ΔNID (pLP656), and sir2-ΔCYS (pLP386)]. The transformants were assayed for growth by plating serial dilutions on leu− plates (left panel) and for function by plating on leu−trp− (B), 5-FOA (C), and leu−ura− (D) plates (right panel). The rDNA silencing assays were performed on leu−ura− plates to distinguish between silencing and recombination resulting in loss of the rDNA reporter. In LPY2447, such recombination occurs at high frequency on FOA-containing medium because of selection against URA3 expression and the enhanced rDNA recombination caused by a sir2Δ mutation. (E) Immunoblot analysis was performed using an antiserum directed against a C-terminal peptide derived from Sir2p (Smith et al., 1998) on whole-cell lysates of LPY1953 transformants.
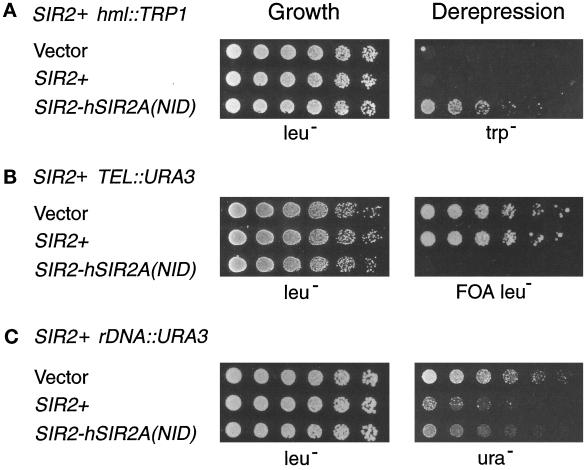
Figure 6.
The SIR2-hSIR2A(NID) chimera exhibits dominant derepression. Dilution assays were performed on LPY253 (MATa SIR2+ hml::TRP1), LPY1683 (MATa SIR2+ TEL::URA3 TEL::ADE2), and LPY2446 (MATα SIR2+ rDNA::URA3) transformed with high-copy vector alone (YEp351) or containing SIR2+ (pLP349) or the SIR2-hSIR2A(NID) chimera (pLP999). Similar results were obtained with low-copy versions of these constructs as well as with the SIR2-hSIR2A(NID+CYS) chimera on high- and low-copy plasmids (our unpublished results). Serial dilutions were plated on leu− plates as a control for growth (left panel) and onto trp−, 5-FOA leu−, and ura− to assay dominance (right panel).
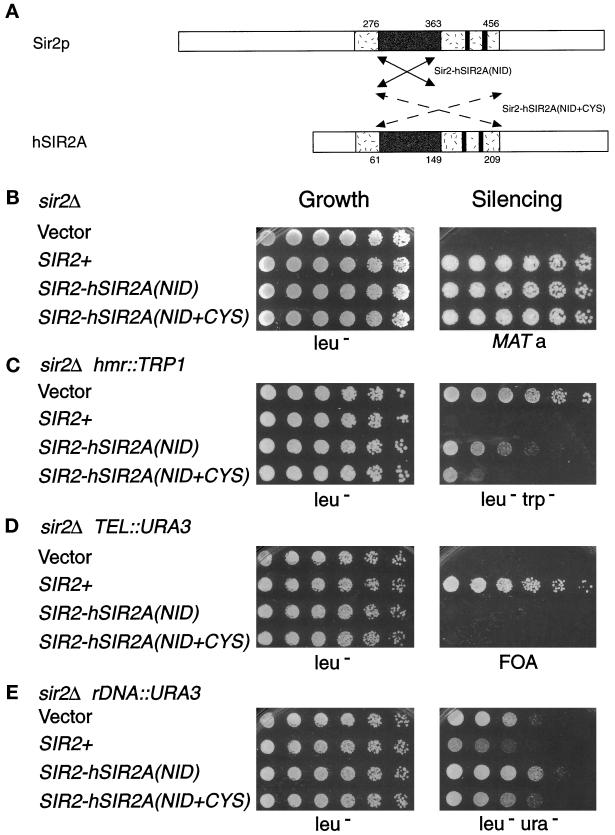
Figure 4.
The Sir2-hSir2A chimeras function in transcriptional silencing. (A) The arrows denote the region of the human Sir2A core exchanged for that of the Sir2p core in the Sir2-hSir2A(NID) chimera (solid lines indicate aa 276–363 of Sir2p were replaced with aa 61–149 of hSir2Ap) and in the Sir2-hSir2A(NID+CYS) chimera (dashed lines indicate aa 276–456 of Sir2p were replaced with aa 61–209 of hSir2Ap). (See Figure 1B for the sequences exchanged in the chimeras.) (B–E) Results of dilution assays to test the SIR2-hSIR2A(NID) (pLP999) and SIR2-hSIR2A(NID+CYS) (pLP905) chimeras’ complementation of the sir2Δ mating defect in LPY1403 (MATα sir2Δ hml:: TRP1) (B), hmr::TRP1 silencing defect in LPY3923 (MATa sir2Δ hmrΔ::TRP1) (C), telomeric silencing defect in LPY1953 (MATa sir2Δ TEL::URA3) (D), and rDNA silencing defect in LPY2447 (MATα sir2Δ rDNA::URA3) (E). The left panel depicts growth of serial dilutions on leu− plates as a control, and the right panel depicts growth on a his4 (LPY143) mating tester on minimal plates (B), leu−trp− (C), 5-FOA (D), and leu−ura− (E).
Immunoblot Analysis
Protein extracts were prepared from yeast cells using glass bead disruption according to the protocol of Rose et al. (1989). The equivalent of 1 A600 unit of each protein extract was boiled and separated on a 7.5–10% SDS-polyacrylamide gel. The proteins were then transferred in Towbin buffer containing 15% methanol to 0.2 μm nitrocellulose and processed using standard procedures (Harlow and Lane, 1988). The primary antibodies included a 1:5000 dilution of an antiserum directed against a C-terminal peptide of Sir2p (Figure 2E and our unpublished results) (Smith et al., 1998), a 1:5000 dilution of an anti-HA12CA5-E antiserum (Babco, Richmond, CA), and a 1:104 dilution of rabbit immunoglobulin G (Sigma, St. Louis, MO).
Viability Assays
Overnight cultures of LPY1683 transformed with vector alone (YEp351) or containing SIR2+ (pLP349) or with the GAL-hSIR2A construct (pLP1062) were grown in glucose leu− medium, then raffinose leu−, and finally galactose leu- medium before diluting them into galactose leu−. Growth rate was monitored by measuring A600. The expected cell number (based on the A600 reading) was compared with the actual cell number analyzed by hemocytometer counting, as well as to colony-forming units. Glucose leu− plates were chosen to increase growth rate and colony size. The colony-forming units reported thus represent a theoretical maximum for the GAL-hSIR2A transformants, because using glucose as a carbon source represses further hSIR2A expression.
Northern Analysis
Multiple-tissue Northern blot membranes were obtained from Clontech Laboratories (Palo Alto, CA) and hybridized at 68°C following the ExpressHyb hybridization protocol provided. The entire insert from expressed sequence tag T66100 (pCAR258) was used as a probe. An actin probe was used to confirm that the same relative amounts of mRNA were loaded in each lane.
Immunofluorescence Microscopy
Immunofluorescence experiments were performed as described (Gotta et al., 1997), with modifications (Ersfeld and Stone, 1999). Low-copy plasmids containing wild-type SIR2 (pLP907) or SIR2-hSIR2A(NID + CYS) (pLP888) were used to avoid the mislocalization previously observed when Sir proteins are overexpressed (for example, see Maillet et al., 1996) in which Sir3p and Sir4p overexpression is observed to result in dispersed nuclear staining of the Sir proteins). The phenotype of the Sir2-hSir2A(NID + CYS) chimera expressed from low-copy plasmid is similar to that observed for high-copy plasmid (our unpublished results). The SIR2+ and SIR2-hSIR2A(NID + CYS) plasmids were introduced into the sir2/sir2 homozygous diploid strain LPY3380, and localization was examined. Antisera directed against Sir2p (Smith et al., 1998) and Nop1p (kindly provided by J. Aris, University of Florida, Gainesville, FL; Aris and Blobel, 1988) have been described. Secondary antibodies used were fluorescein-conjugated goat anti-rabbit and Texas Red goat anti-mouse immunoglobulin G (Jackson ImmunoResearch, West Grove, PA). DNA was stained with DAPI at 1 μg/ml. Microscopy was performed with a Leica (Nussloch, Germany) DMRXA microscope with a Cooke SensiCam charge-coupled device camera, and images were captured and manipulated using the SlideBook software package (Intelligent Imaging Innovations, Denver, CO).
RESULTS
The Core of Sir2p Is Essential for Silencing
To determine whether the conserved core is important for SIR2 function, we deleted sequences encoding aa 245–427 to generate sir2-ΔCORE, which was then tested for its ability to complement sir2Δ mating, HM, telomeric, and rDNA silencing defects. In these experiments, MATa and MATα sir2Δ mutant yeast strains marked with reporter genes at the HM loci (LPY1403), telomeres (LPY1953), and within the rDNA (LPY2447) were transformed with various plasmids. The plasmids contained no SIR2 gene (vector), wild-type SIR2 (SIR2+), or sir2-ΔCORE. Modestly increased expression from high-copy plasmids under control of the endogenous SIR2 promoter was chosen to increase the likelihood of detecting silencing function for the mutant protein (Brachmann et al., 1995; Smith et al., 1998). Dilution assays (see MATERIALS AND METHODS) were used to assess growth and complementation of the sir2Δ silencing defects.
Although all of the sir2-ΔCORE-containing strains grew equivalently to the vector and SIR2 wild-type controls, the core deletion mutant was unable to complement any of the sir2Δ mutant silencing phenotypes (Figure 2, B–D). The sir2-ΔCORE mutant failed to restore silencing at HML in a sir2Δ strain containing a TRP1 reporter (Figure 2B). The sir2-ΔCORE mutant transformant remained derepressed and thus able to grow on selective medium lacking tryptophan, analogous to the vector transformant. This is in contrast to the SIR2+-transformed strain, which was unable to grow because the TRP1 reporter is silenced. Moreover, both MATa and MATα sir2Δ mutant strains (LPY1953 and 1403) transformed with the sir2-ΔCORE construct did not mate. Thus, the core sequences are essential for Sir2p silencing function at the HM loci.
The inability of the sir2-ΔCORE mutant to function in silencing was not limited to the HM loci. It was also unable to silence a telomeric URA3 reporter in a sir2Δ mutant background (Figure 2C). This strain carrying the sir2-ΔCORE mutant construct (or the vector control) was unable to grow on medium containing 5-fluoro-orotic acid (5-FOA), a suicide substrate for the URA3 gene product, because URA3 is expressed. Likewise, the sir2-ΔCORE mutant did not silence a URA3 reporter integrated in the rDNA array in a sir2Δ mutant background (Figure 2D). The sir2-ΔCORE mutant transformants grew on medium lacking uracil, whereas the wild-type SIR2 transformants grew poorly because of restored rDNA silencing. In fact, the sir2-ΔCORE mutant transformants grew slightly better and thus were slightly more URA+ than the vector transformants, suggesting that the core deletion mutant exacerbates the sir2Δ rDNA silencing defect. The failure of the sir2-ΔCORE mutant to function in silencing is not due to protein instability, because immunoblot analysis indicated that the sir2-ΔCORE mutant protein was expressed at least as well as wild-type Sir2p (Figure 2E). Thus, the conserved core of Sir2p is absolutely essential for its function in HM, telomeric, and rDNA silencing, suggesting that it is an important functional domain.
The main difference between Sir2p, which silences all three loci, and Hst1p, which can only partially silence HMR (Brachmann et al., 1995), is the length and sequence of their N termini. Sir2p has an N-terminal extension of ∼35 amino acids and is <15% identical to Hst1p over the next 75 amino acids. This led us to ask whether the N-terminus of Sir2p is important for its ability to silence genes at the HM loci, telomeres, and rDNA. In contrast to the complete loss of function observed when the core domain was deleted, deletion of 78 amino acids from the nonconserved N terminus of Sir2p did not affect its silencing function (Figure 2, B–D). The N-terminal deletion mutant (sir2-ΔN) was able to complement the sir2Δ mating, HM, telomeric, and rDNA silencing defects as well as wild-type SIR2 on both high and low copy plasmids (Figure 2B-D and our unpublished results). Thus, this nonconserved N-terminal region encompassing the first 79 amino acids of Sir2p is dispensable for mating-type silencing and does not promote the telomeric and rDNA silencing functions of SIR2 that are absent in HST1 (reviewed by Sherman and Pillus, 1997).
Sir2p Silencing Requires Conserved Sequence Motifs
We demonstrated that the core domain is required for the transcriptional silencing functions of SIR2. To test whether the three motifs found in the core are individually necessary for silencing, sequences encoding smaller regions containing each of these motifs were deleted (Figure 2A). Amino acids 244–272, which include the GAG motif, were deleted to construct the mutant denoted hereafter sir2-ΔGAG. Similarly, the NID motif was removed with a deletion of aa 275–364 to create sir2-ΔNID, and the cysteines were deleted along with aa 364–428 to yield the mutant designated sir2-ΔCYS (Figures 1B and 2A). The ability of these mutants on high-copy plasmids to complement the sir2Δ mating, HM, telomeric, and rDNA silencing defects was tested by dilution assays. Deletion of regions containing the conserved motifs did not affect growth but did lead to loss of silencing function. At HML, the sir2-ΔGAG, sir2-ΔNID, and sir2-ΔCYS mutant constructs were unable to complement the sir2Δ silencing defect of the TRP1 reporter (Figure 2B), despite being expressed at levels comparable with wild-type Sir2p (our unpublished results). Interestingly, the full core and smaller deletions were equally nonfunctional, resulting in a fully Trp+ phenotype and indicating that deletion of any one motif leads to a complete loss of function at the HM loci. The deletion mutants were similarly nonfunctional in telomeric and rDNA silencing. Thus, the regions containing these conserved motifs are individually essential for SIR2 function.
Because the sir2-ΔCYS mutant, which lacks the region of Sir2p containing the putative zinc finger, is nonfunctional, we asked whether it is the region or the cysteines themselves that are necessary for SIR2 silencing. Therefore, the four cysteines at positions 372, 374, 396, and 398 were changed to alanines (Figure 1) pairwise (cysteine 1 and 2 and cysteine 3 and 4) and together (cysteines 1–4) and assayed as described (Figure 3). Like the deletion mutants, none of the point mutant combinations complemented the sir2Δ HM, telomeric, or rDNA silencing defects (Figure 3), and each exhibited complete loss of function in dilution assays. These mutants are, however, expressed to wild-type levels. Thus, the four cysteines and the intermolecular interactions they may specify are required for SIR2 silencing function.
Sir2-hSIR2A Chimeras Promote HM Silencing
Our mutational analysis demonstrated that the core and its motifs were clearly essential for silencing by Sir2p. To learn more about the corresponding regions in other Hst proteins, we asked whether the core has a similar role in the other HST subfamilies (Figure 1) and whether this function is evolutionarily conserved. A series of chimeras between SIR2 and yeast HST3 or human SIR2A were constructed. Sequences encoding the most conserved region of the core from within the GAG motif to just C-terminal of the NID motif from HST3 or hSIR2A were exchanged for the equivalent segment of SIR2. These chimeras were then assayed for complementation of sir2Δ silencing defects.
The SIR2-HST3 chimera was made by replacing the fragment of SIR2 encoding aa 276–373 with that of HST3 encoding aa 74–186 (Figure 1B). This SIR2-HST3 chimera, despite wild-type levels of expression, did not complement any of the sir2Δ silencing defects (our unpublished results). This may be due to an additional 25 amino acids found in this region of the Hst3p core (Figure 1B). Interestingly, this chimera also failed to supply HST3 function. Although stably expressed, it did not complement the hst3Δ hst4Δ temperature-sensitive phenotype or telomeric silencing defects (our unpublished results). Thus, regions outside the core domain of Hst3p appear important for its function.
The SIR2-hSIR2A chimeras, unlike the SIR2-HST3 chimera, were able to function in yeast silencing. SIR2-hSIR2A(NID) was constructed by again replacing aa 276–373 of Sir2p, this time with aa 61–149 of hSir2Ap (Figures 1B and 4A). Despite being only 36% identical to Sir2p in this region, the SIR2-hSIR2A(NID) chimera rescued the sir2Δ mating defect in both MATα (Figure 4B) and MATa strains (our unpublished results). In fact, it silenced the mating-type information at the HM loci to wild-type levels in mating assays. Additionally, it partially silenced the TRP1 reporter genes at HMR (Figure 4C) and HML (our unpublished results). Although the SIR2-hSIR2A(NID) chimera functioned well at the HM loci, it could not silence a telomeric URA3 reporter (Figure 4D). At the rDNA, SIR2-hSIR2A(NID) had, at most, a modest repressive effect on the URA3 reporter; ∼25% of the transformants tested exhibited weak silencing, whereas others, like the isolate shown, exhibited slightly decreased silencing compared with the vector control.
Similar results were obtained with a larger SIR2-hSIR2A chimera [SIR2-hSIR2A(NID+CYS)] that extends from within the GAG motif to just C-terminal of the four cysteines (Figures 1B and 4A). This chimera was constructed by exchanging the region of hSIR2A encoding aa 61–209 for that of SIR2 encoding aa 276–456. Almost 30% of the sequence of this chimera is therefore derived from the human Sir2A protein, and the cysteines, although conserved, are spaced differently. Nevertheless, the SIR2-hSIR2A(NID+CYS) chimera restored HM silencing to near wild-type levels in sir2Δ strains (Figure 4, B and C). And, like the smaller human chimera, this chimeric protein was unable to function in telomeric or rDNA silencing (Figure 4, D and E). Thus, both Sir2-hSir2A chimeras function well in silencing at HM loci but poorly or not at all at telomeres and in the rDNA, thereby exhibiting distinct locus specificity.
hSIR2A Cannot Substitute for SIR2 Silencing Function
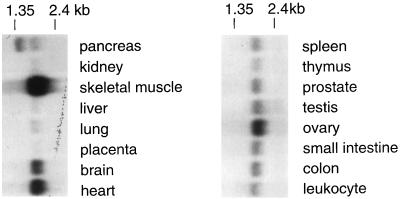
The silencing function of the human Sir2A core led us to explore the expression pattern of hSIR2A. We hybridized human tissue Northern blots with a hSIR2A probe. An ∼2.0-kb transcript, consistent with the predicted size of the hSIR2A transcript, was readily detected in all tissue types examined (Figure 5). However, the abundance varied between tissues, with the highest amounts of this transcript found in skeletal muscle, heart, and brain and the lowest in liver, lung, and kidney. Interestingly, two transcripts were detected in pancreatic tissue. The significance of this observation has yet to be determined.
Figure 5.
hSIR2A mRNA is widely distributed. A multiple-tissue RNA blot was probed with an hSIR2A cDNA probe. The blot was hybridized separately with an actin cDNA probe and shown to be evenly loaded except that the skeletal muscle sample was overloaded by aproximately twofold.
Thus, hSIR2A is widely expressed in human tissues, and the core of hSir2Ap provides silencing function in the context of the N and C termini of Sir2p (Figure 4, B and C). But, can the presumably full-length hSir2A protein function in yeast silencing on its own? To test this, we designed two constructs. In the first, hSIR2A was cloned behind the endogenous SIR2 promoter on a high-copy plasmid and tagged with HA or protein A to monitor expression. In the second construct, hSIR2A was cloned behind the GAL10 promoter to examine the effect of high levels of overexpression in yeast. The ability of hSIR2A to rescue the sir2Δ mating, telomeric, and/or rDNA silencing defects was assayed as above, except that for the GAL-hSIR2A construct, galactose-induced cells were compared on selective plates with galactose (to maintain high expression levels) or glucose (to repress further expression) as the carbon source.
Unlike the SIR2-hSIR2A chimeras, neither the untagged nor tagged versions of hSIR2A cloned behind the SIR2 promoter rescued any of the sir2Δ silencing defects (Table 2). This lack of function was not due to protein instability, because both the HA- and protein A-tagged versions of hSIR2A gave strong signals on immunoblots (our unpublished results). Nor is failure to function simply a matter of dosage, because hSIR2A expressed from the GAL10 promoter is likewise nonfunctional (Table 2). The hSir2A protein is likely expressed to even higher levels from the GAL promoter, because the GAL-hSIR2A but not the SIR2 promoter-hSIR2A is toxic to yeast. We observed that strains containing the GAL-hSIR2A construct exhibited reduced growth compared with control cultures upon diluting and plating equivalent amounts of saturated GAL-induced cultures. Analysis of growth rates under inducing conditions indicated that overexpression of hSir2Ap results in an approximately twofold increase in doubling time and a greater than threefold loss in viability when equivalent numbers of cells were plated on selective plates (Table 3). The loss of viability observed with GAL-SIR2 (Holmes et al., 1997), is similar in magnitude to the effect we measured for GAL-hSIR2A overexpression. This suggests that the properties of SIR2, which, upon overexpression, lead to increased chromosome loss and thus cell death (Holmes et al., 1997), may be shared by hSIR2A.
Table 2.
hSIR2A does not function in transcriptional silencing
| Silencing | Mating | hml∷TRP1 | TEL∷URA3 | rDNA∷URA3 |
|---|---|---|---|---|
| Vector | − | − | − | − |
| SIR2+ | + | + | + | + |
| SIR2-hSIR2A(NID) | + | + | − | − |
| SIR2-hSIR2A(NID+CYS) | + | + | − | − |
| SIR2 promoter-hSIR2A | − | − | − | − |
| GAL-hSIR2A | − | − | − | − |
Table 3.
Overexpression of hSIR2A is toxic to yeast
| Plasmid | Doubling time | Relative viability |
|---|---|---|
| Vector | 4 h 14 min | 1.00 |
| GAL-hSIR2A | 8 h 9 min | 0.32 |
Sir2-hSIR2A Chimeras Dominantly Interfere with Silencing
The ability of the SIR2-hSIR2A chimeras to complement sir2Δ HM silencing defects suggests that the function of the hSir2A core domain is at least partially conserved. Additional evidence for conserved function of the core and, thus, of hSIR2A comes from dominance analyses of the chimeras. High- and low-copy plasmids were transformed into strains analogous to those used above (Figure 4), except that they were wild type for SIR2. The effects of these constructs on silencing in the presence of SIR2+ were quantitated by dilution assays to monitor growth and silencing of reporters in the presence of 5-FOA or in the absence of tryptophan or uracil (see above).
In contrast to similarly expressed wild-type SIR2 or the SIR2-HST3 chimera, both of the SIR2-hSIR2A chimeras caused a paradoxical dominant derepression phenotype. However, they did so only at the HM loci and telomeres. The SIR2-hSIR2A chimeras caused partial derepression of HM and complete derepression of telomeric reporters (Figure 6, A and B, and our unpublished results). That is, an intermediate Trp+ phenotype was observed in SIR2-hSIR2A(NID) transformants of the SIR2+ hml::TRP1 strain. (See for comparison the complete derepression of this reporter in a sir2Δ background in Figure 2B.) However, the derepression of the telomeric URA3 marker in the SIR2+ TEL::URA3 strain was total; these transformants were fully Ura+ and did not grow on medium containing 5-FOA. This effect was not dosage dependent, because the SIR2-hSIR2A chimeras on low-copy plasmids also dominantly derepressed these loci (our unpublished results) and was not due to simple loss of function, because the sir2-ΔNID mutant did not interfere with silencing (our unpublished results). Furthermore, the chimeras derepressed a telomeric ADE2 reporter (our unpublished results) in addition to TEL::URA3 (Figure 6B), indicating that the observed telomeric dominance is locus and not gene or promoter specific. This interpretation is supported by the observation that the SIR2-hSIR2A chimeras did not dominantly derepress the URA3 gene located in the rDNA (Figure 6C). Instead, in the presence of wild-type SIR2, the SIR2-hSIR2A(NID) chimera slightly improved rDNA silencing, a dosage effect similar to that observed for increased expression of SIR2+ (Figure 6C; Fritze and Esposito, 1997; Smith et al., 1998). Together these observations are consistent with the participation of Sir2p in distinct HM/telomeric and rDNA silencing complexes.
Sir2-hSIR2A Chimeras Fail to Localize Properly Within the Nucleus
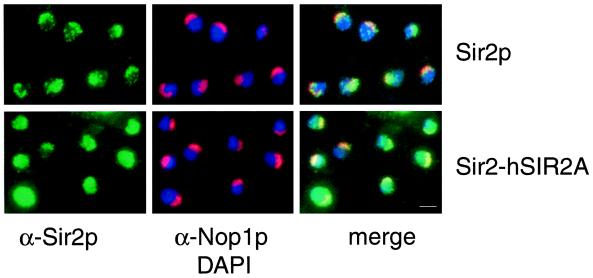
Sir2p has been localized to telomeres and the nucleolus by indirect immunofluorescence microscopy (Gotta et al., 1997). To determine whether the Sir2-hSir2A(NID+CYS) chimera localizes in the same manner as wild-type Sir2p or whether it localizes with a pattern that may reflect differences in its ability to function at these loci, we compared localization of Sir2p with that of the Sir2-hSir2A chimera. Wild-type Sir2p localized normally (Gotta et al., 1997); telomeric foci were present within the main body of DAPI-staining chromatin, and Sir2p nucleolar staining was coincident with the Nop1p nucleolar antigen (Figure 7, top row). Staining of wild-type Sir2p also revealed foci indicative of localization to subdomains within the nucleolus (Stone and Pillus, unpublished results). For wild-type Sir2p, 97% of the cells exhibited the normal telomeric and nucleolar staining pattern (n = 306). In contrast, the Sir2-hSir2A(NID + CYS) chimera localized in a diffuse and sometimes intense pattern in a majority of the nuclei examined (Figure 7, bottom row). The intense fluorescence signal does not correlate with higher expression levels of the chimeric protein as determined by immunoblotting (our unpublished observations; see DISCUSSION). A representative field is shown for cells expressing the chimera, in which mislocalization is apparent in nuclei that are either filled completely with Sir2p signal (49%) or are partially filled with occasional foci simultaneously observed (23%). Thus, a total of 72% of Sir2-hSir2A(NID + CYS) cells (n = 352) were abnormal, whereas a minority of cells (28%) retained an apparently wild-type pattern. DAPI and Nop1p staining were comparable in wild-type versus the Sir2-hSir2A(NID + CYS) chimera, indicating that the fundamental organization of chromatin and the nucleolus was not disrupted. Moreover, Rap1p staining of telomeric foci was normal for the chimera compared with wild-type Sir2p (our unpublished results). The smaller chimera Sir2-hSir2A(NID) was also examined, and a comparable pattern of mislocalization was observed (our unpublished results). Therefore, the inability of the Sir2-hSir2A proteins to function at telomeres and the rDNA is correlated with their disrupted localization.
Figure 7.
The Sir2-hSir2A-(NID+CYS) chimera fails to localize properly within the nucleus. SIR2+ and SIR2-hSIR2A-(NID+CYS) plasmids (pLP907, top row, and pLP888, bottom row, respectively) were introduced into the homozygous sir2/sir2 diploid strain LPY3380. Immunofluorescence was performed by staining cells with anti-Sir2p antibodies (to localize Sir2p and the chimeras, in green), anti-Nop1p antibodies (to identify the nucleolus, in red), and DAPI (to identify DNA, in blue). At right are merged images of the left and center panels. Bar, 2 μm.
DISCUSSION
Transcriptional activation is broadly conserved at levels of both mechanistic and molecular detail. This point is well illustrated by RNA polymerase itself, as well as regulatory complexes such as SWI–SNF and RSC (reviewed by Struhl, 1996; Kadonaga, 1998; Workman and Kingston, 1998). By contrast, although chromatin-mediated repression is a prevalent form of transcriptional regulation, evidence for molecular equivalence between divergent organisms is scarce. Arguably, silencing is best understood in molecular detail in S. cerevisiae, so the recent discovery of widespread homologues of the classically defined yeast SIR2 gene fueled the prediction that silencing, like transcriptional activation, might have conserved molecular mechanisms. SIR2’s silencing-related activities include repression of transcription at the silent mating-type loci, telomeres, and within the rDNA arrays (Shore et al., 1984; Ivy et al., 1986; Aparicio et al., 1991; Bryk et al., 1997; Fritze and Esposito, 1997; Smith and Boeke, 1997), suppression of rDNA recombination (Gottlieb and Esposito, 1989), and modulation of histone (de)acetylation (Braunstein et al., 1993). We previously showed that three of four yeast homologues can function in silencing (Brachmann et al., 1995). Here, we provide evidence from structure–function analysis of conserved domains involved in Sir2p’s functions.
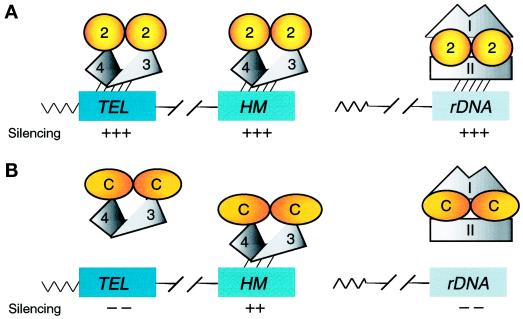
We demonstrate that a region essential for Sir2p function and comprising approximately one-third of Sir2p can be replaced with human Sir2A sequences to form a chimera that functions positively in silencing (see model in Figure 8). Interestingly, this chimera also has the capacity to dominantly interfere with Sir2p function at some loci, underscoring the concept that silencing proteins act in distinct chromatin complexes. That interacting factors may be species specific is suggested by the fact that expression of hSIR2A fails to complement. Our analysis of yeast–human chimeras and hSIR2A itself implicates the HST gene family in chromatin organization and function in organisms as diverse as yeast and humans. Furthermore, because even organisms as deeply rooted evolutionarily as the Archaebacteria have homologues of SIR2, it is possible that these molecules are among the most ancient proteins with chromatin function.
Figure 8.
Models for Sir2p silencing function and the involvement of distinct Sir2p-containing complexes in HM/telomeric and rDNA silencing. (A) Wild-type Sir2p function. Sir2p interacts with itself and Sir3p/Sir4p in a complex (Moazed and Johnson, 1996; Holmes et al., 1997; Moazed et al., 1997; Strahl-Bolsinger et al., 1997) and localizes to and silences the HM loci and telomeres (Hecht et al., 1996; Gotta et al., 1997; Strahl-Bolsinger et al., 1997). Because Sir3p and Sir4p are not required for rDNA silencing (Smith and Boeke, 1997), a second Sir2p-containing complex involving one or more unidentified factors (I and II) localizes to and silences the rDNA. Single silencing complexes are shown for simplicity, although it is believed that numerous complexes, perhaps in multimeric forms, localize to and silence each of the loci. (B) Sir2-hSir2A chimera function. The chimeras continue to dimerize or oligomerize and interact with other silencing factors. They localize to the HM loci to silence them but fail to localize properly to the telomeres and rDNA, leading to a loss of silencing. In the presence of wild-type Sir2p, Sir2p-chimera heteromers form. These heteromeric forms continue to interact with HM and telomeric silencing factor(s), e.g., Sir3p and Sir4p, and titrate them, leading to a loss of silencing at these loci. On the other hand, the Sir2p-chimera heteromers interact correctly with rDNA silencing factors (I and II) and localize subnucleolarly, leading to rDNA silencing function. Although Sir2p and the Sir2p-hSir2A chimeras are diagrammed as dimers based on wild-type Sir2p’s demonstrated ability to interact with itself (Moazed et al., 1997), the functional form of Sir2p and its variants has yet to be determined. Thus, they may function as higher-order multimers, dimers, or even monomers. Furthermore, the functional form of the Sir2-hSir2A chimeras may differ from that of wild-type Sir2p.
Motifs Conserved in Sir2p and the Hst Proteins Are Essential for Silencing
Members of the Sir2p family are defined by a core domain including three diagnostic motifs (Figure 1) (Brachmann et al., 1995; Derbyshire et al., 1996). To test the significance of the core and its motifs for SIR2 silencing function, we analyzed a series of deletion and cysteine to alanine point mutants. The results demonstrated that the core domain and smaller regions containing the conserved motifs, as well as the cysteines themselves, are absolutely essential for SIR2 silencing at the HM loci, telomeres, and within the rDNA (Figures 2, B–D, and 3). Removal of any one of the motifs leads to loss of function at all loci (Figure 2B and our unpublished results). The regions containing these motifs may specify a catalytic activity, interaction with other silencing proteins, and/or subcellular localization important for Sir2p’s function in silencing.
The four essential cysteines may mediate protein–nucleic acid or protein–protein interactions required for SIR2 activity. Alternatively, the cysteines may be important for folding or activity. Because the stable expression that we observe for the cysteine to alanine mutant proteins is consistent with proper folding (Parsell and Sauer, 1989), the cysteines are more likely to specify intermolecular interactions that contribute to silencing function. These intermolecular contacts are probably protein–protein interactions rather than protein–DNA interactions (Moran and Matthews, 1987; Coleman, 1992). Whereas Sir2p has not been shown to bind to DNA in vitro (Shore and Nasmyth, 1987; Buchman et al., 1988) or to localize to the HM loci or telomeres in the absence of other silencing factors (Hecht et al., 1996; Gotta et al., 1997; Strahl-Bolsinger et al., 1997), it has been shown by affinity chromatography and coimmunoprecipitation experiments to interact with itself as well as with two other components of the HM/telomeric silencing complex, Sir3p, and Sir4p (Hecht et al., 1996; Moazed and Johnson, 1996; Gotta et al., 1997; Holmes et al., 1997; Moazed et al., 1997; Strahl- Bolsinger et al., 1997). We predict that the cysteines are more likely to mediate Sir2p multimerization or interactions with yet unidentified proteins, rather than interactions with Sir3p and/or Sir4p. This prediction is based on the observation that deletion or mutation of the cysteines results in loss of rDNA silencing, a function requiring Sir2p, but not Sir3p or Sir4p (Smith and Boeke, 1997).
In contrast to the loss of function observed in the cysteine to alanine point mutants or with small deletions of the conserved core domain, deletion of the nonconserved N-terminal 79 amino acids of Sir2p does not affect its silencing function or locus specificity (Figure 2, B–D). This is somewhat surprising, because the length and sequence of the N terminus appears to be the major sequence difference between Sir2p and Hst1p, yet high-copy expression of Hst1p rescues only the sir2Δ HM, but not telomeric or rDNA silencing or recombination defects (Brachmann et al., 1995; Derbyshire et al., 1996; and Sherman and Pillus, unpublished results). However, other nonconserved regions outside the core (defined here as amino acids 275–427) must be important for silencing, because expression of the SIR2 core alone is not sufficient to silence the HM loci (Garcia and Pillus, unpublished results).
The conserved core of Sir2p is essential for its function in silencing. But is the function of the core evolutionarily conserved, and, thus, are the other Hst proteins likely to have a chromatin-related function? The complementation of the sir2Δ mating and HM silencing defects by the SIR2-hSIR2A(NID) chimera (Figure 4, B and C) demonstrates that the human Sir2A core can function in silencing. A larger SIR2-hSIR2A(NID + CYS) chimera is similarly functional (Figure 4, B and C). This is particularly remarkable because the human sequence in this chimera represents >40% of hSir2Ap and constitutes nearly 30% of the chimera.
Furthermore, the Sir2-hSir2A chimeras dominantly derepress the HM loci and telomeres, but not the rDNA (Figure 6). This derepression is locus and not gene or promoter specific, because they derepress both pol II- and pol III-transcribed reporters at the HM loci and radically interfere with silencing a URA3 reporter at the telomeres but not within the rDNA (Figure 6 and our unpublished results). We interpret this dominance in the context of Sir2p forming distinct complexes that act at HM and telomeric targets on one hand and at rDNA targets on the other (Figure 8; Smith and Boeke, 1997; Smith et al., 1998). Specifically, we propose that the chimeric Sir2 proteins interfere with the assembly or function of the macromolecular complexes that act in HM and telomeric silencing (Figure 6, A and B) but have no such effect on the macromolecular complexes involved in rDNA silencing (Figure 6C). In fact, expression of the Sir2-hSir2A chimeras in the presence of wild-type Sir2p promotes modestly increased silencing in the rDNA (Figure 6C), providing further evidence that distinct Sir2p-containing complexes participate in silencing specific genomic loci (Figure 8). However, the Sir2-hSir2A(NID) chimera, as well as the sir2-ΔCORE mutant protein, can interfere with the residual rDNA silencing that occurs even in the absence of Sir2p (Figures 2D and 4E). It is possible, for example, that these sir2 variants interfere with a secondary silencing mechanism that might involve Hst1p (Gotta et al., 1997).
The Core Influences Locus-specific Silencing Function
Interestingly, the SIR2-hSIR2A chimeras exhibit locus specificity; that is, they retain the ability to silence the HM loci, but not the telomeres or the rDNA in the context of a sir2Δ strain (Figures 8 and 4 B–E). Thus, there may be specificity determinants, or separable elements required for silencing specific loci, embedded in the conserved core of Sir2p. This situation is evocative of yeast and human TFIID or TATA-binding proteins in which functional differences reside in a conserved domain, rather than in the divergent N termini (Cormack et al., 1991; Gill and Tjian, 1991; Reddy and Hahn, 1991). Thus, the coincidence of conserved elements and specificity determinants may be a general feature of transcriptional regulators. In this model, one or more determinants required for HM locus function must be conserved in the human Sir2A core, because the chimeras are able to partially silence these loci. This region of the yeast Sir2p core must additionally contain sequences specifying telomeric and rDNA function, which are not found in the human Sir2A core. These determinants may influence correct Sir2p localization and/or function, because loss of telomeric and rDNA silencing by the Sir2-hSir2A chimeras correlates with loss of precise localization to telomeric foci and subnucleolar regions (Figure 7). Because the Sir2-hSir2A chimeras function at the HM loci, we assume they localize correctly there. However, their overall immunofluorescence signal is diffuse and often intense. The intensity of the signal observed in many nuclei is not likely to be due to differences in expression, because a similar level of protein to that seen for wild-type Sir2p is detected by immunoblotting (our unpublished results). Rather, the intensity may be due to a more open chromatin configuration. Or it may be due to gross mislocalization of the chimera to a soluble pool in the nucleoplasm, distinct from the normally chromatin-bound Sir2p. In either case, the Sir2-hSir2A chimera may be more accessible to the antibody probes. And either possibility would be consistent with the observed telomeric and rDNA derepression in chimeric strains.
Another possible explanation for the ability of the SIR2-hSIR2A chimeras to function only in HM silencing is that the chimeras retain enough residual function to silence the HM loci but not the telomeres or rDNA. However, we favor the interpretation that sequence differences in the human core affect locus specificity because the Sir2-hSir2A chimeras both function and dominantly interfere with wild-type Sir2p function at distinct subsets of loci, including the purportedly more stable HM loci. This suggests that the human core affects interactions with various silencing factors differently, both in the presence and absence of Sir2p. Additional evidence for this comes from the chimeras’ failure to localize properly and repress the rDNA reporter gene in sir2 mutants and their apparent ability to function in rDNA silencing in SIR2+ strains.
Alternatively, there may be competition between the HM loci and telomeres for a limiting pool of interacting factors (e.g., Sir2p or its derivatives), as has been observed between these loci for the transcriptional regulatory protein Rap1p (Buck and Shore, 1995). However, we consider competition for the Sir2-hSir2A chimeras to be an unlikely explanation for their silencing specificity for two reasons. First, telomeric silencing is independent of SIR2 dosage (Renauld et al., 1993), and the chimeric proteins are expressed at levels comparable with wild-type Sir2p. Second, deletion of the HML locus in a MATa sir2Δ TEL::URA3-marked strain does not lead to enhanced telomeric repression by the chimeras (our unpublished results), as would be predicted if competition were significant. Thus, the core domain of Sir2p not only carries out a silencing function but also influences telomeric and rDNA localization and thereby is important for determining normal locus specificity.
hSIR2A is expressed at significant levels in all tissues examined (Figure 5) and therefore is available to carry out a silencing-like function in humans, as suggested by Sir2-hSir2A chimeras’ activity in yeast. However, hSIR2A does not suppress any of the yeast sir2Δ silencing defects, thereby functionally distinguishing it from the Sir2-hSir2A chimeras (Table 2). Thus, although there are some critical silencing determinants within the core, there must also be sequences required for HM silencing residing outside the Sir2p core. These residues, which influence HM function, may not be found in the hSir2A protein, or hSir2Ap function may require species-specific factors for proper function and/or localization.
A Case for SIR-like Silencing in Humans?
That SIR-mediated silencing might occur through a complex of interacting silencing factors was originally proposed on the basis of genetic arguments (Rine and Herskowitz, 1987). Since then, it has been demonstrated that wild-type Sir2p interacts with itself as well as with Sir3p and Sir4p (Moazed and Johnson, 1996; Holmes et al., 1997; Moazed et al., 1997; Strahl-Bolsinger et al., 1997), presumably in a complex along with other silencing factors that physically associates with the HM loci and telomeres (Hecht et al., 1996; Gotta et al., 1997; Strahl-Bolsinger et al., 1997). The results presented here extend and expand this view of Sir2p’s functions in silencing. We have shown that the core of Sir2p and its motifs are essential for silencing and that the human Sir2A core can substitute for the core of Sir2p to silence. We also provide evidence that silencing of multiple genomic loci is mediated by distinct silencing complexes of which Sir2p is a common component (Figure 8). These results suggest that the HSTs from yeast and other organisms may themselves be involved in silencing and/or chromatin organization.
Like SIR2 and the HSTs, many aspects of silencing are conserved. These include two in which Sir2p has been implicated. First, overexpression of Sir2p leads to reduced acetylation of three of the four core histones (Braunstein et al., 1993). In organisms from yeast to mammals, hypoacetylated histones are associated with silenced regions of the genome (Lin et al., 1989; Turner et al., 1992; Braunstein et al., 1993, 1996; Jeppesen and Turner, 1993; O’Neill and Turner, 1995). Thus, the Hst proteins, including those from yeast and humans, may modulate histone (de)acetylation to control transcription. Second, repeat-induced silencing is also evolutionarily conserved and is involved in many basic biological processes, such as dosage compensation and host defense (reviewed in Henikoff and Matzke, 1997). This combined with the known involvement of Sir2p within the repetitive telomeric and rDNA arrays (Gottlieb and Esposito, 1989; Aparicio et al., 1991; Bryk et al., 1997; Fritze and Esposito, 1997; Smith and Boeke, 1997) suggests that the Hst proteins may also function in silencing of repeated DNAs. Although the homologues have distinct “architectural” features such as C-terminal extensions, which may be required for their individual silencing functions, we provide evidence that the core with its consensus motifs is a silencing domain. This model, in which conserved functional domains of a silencing protein specify its activity, is likely to be universal. However, whether the Hst proteins function specifically in silencing by directly interacting with chromatin or have a more general enzymatic function that contributes to silencing remains to be determined. Such catalytic activities are readily understood in the case of histone acetylases and deacetylases that are components of chromatin complexes. Other enzymatic activities may function in less obvious ways, such as that recently proposed for an inorganic pyrophosphatase, a component of the Drosophila NURF chromatin remodeling complex (Gdula et al., 1998). Although the precise roles of the Hst proteins in yeast and other organisms are not yet known, it is plausible that they function to refine gene regulation. The observation that the human Sir2A core domain can participate in yeast silencing increases the significance of this broadly conserved gene family.
ACKNOWLEDGMENTS
Strains, plasmids, and other reagents were generously provided by J. Aris, D. Gottschling, J. Rine, J. Smith, and R. Sternglanz. M. Blower and S. Garcia provided helpful comments on the manuscript. Microscopy was made possible in part by a gift from Virginia and Mel Clark. This work was supported by an American Cancer Society postdoctoral fellowship to J.M.S., a Howard Hughes Medical Institute predoctoral fellowship to L.F.C., and the National Institutes of Health.
REFERENCES
- Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- Aris JP, Blobel G. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J Cell Biol. 1988;107:17–31. doi: 10.1083/jcb.107.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Sherman JM, Devine SE, Cameron EE, Pillus L, Boeke JD. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression and chromosome stability. Genes & Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes & Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- Braunstein M, Sobel R, Allis CD, Turner BM, Broach JR. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol Cell Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk M, Banarjee M, Murphy M, Knudsen KE, Garfinkel DJ, Curcio MJ. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes & Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- Buchman AR, Kimmerly WJ, Rine J, Kornberg RD. Two DNA-binding factors recognize specific sequences at silencers, upstream activating sequences, autonomously replicating sequences, and telomeres in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:210–225. doi: 10.1128/mcb.8.1.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck SW, Shore D. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes & Dev. 1995;9:370–384. doi: 10.1101/gad.9.3.370. [DOI] [PubMed] [Google Scholar]
- Chen X-J, Clark-Walker GD. sir2 mutants of Kluyveromyces lactis are hypersensitive to DNA-targeting drugs. Mol Cell Biol. 1994;14:4501–4508. doi: 10.1128/mcb.14.7.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JE. Zinc proteins: enzymes, storage proteins, transcription factors and replication proteins. Annu Rev Biochem. 1992;61:897–946. doi: 10.1146/annurev.bi.61.070192.004341. [DOI] [PubMed] [Google Scholar]
- Cormack BP, Strubin M, Ponticelli AS, Struhl K. Functional differences between yeast and human TFIID are localized to the highly conserved region. Cell. 1991;65:341–348. doi: 10.1016/0092-8674(91)90167-w. [DOI] [PubMed] [Google Scholar]
- Derbyshire MK, Weinstock KG, Strathern JN. HST1, a new member of the SIR2 family of genes. Yeast. 1996;12:631–640. doi: 10.1002/(SICI)1097-0061(19960615)12:7%3C631::AID-YEA960%3E3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Ersfeld K, Stone EM. Simultaneous in situ detection of DNA and proteins. In: Allan V, editor. The Practical Approach Series. New York: Oxford University Press; 1999. (in press). [Google Scholar]
- Freeman-Cook, L.L., Sherman, J.M., Brachman, C.B., Allshire, R.C., Boeke, J.D., and Pillus, L. (1999). The Schizosaccharomyces pombe hst4+ gene is a SIR2 homologue with silencing and centromeric functions. Mol. Biol. Cell (in press). [DOI] [PMC free article] [PubMed]
- Fritze C, Esposito E. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 1997;16:6495–6509. doi: 10.1093/emboj/16.21.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gdula DA, Sandaltzopoulos R, Tsukiyama T, Ossipow V, Wu C. Inorganic pyrophosphatase is a component of the Drosophila nucleosome remodeling factor complex. Genes & Dev. 1998;12:3206–3216. doi: 10.1101/gad.12.20.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G, Tjian R. A highly conserved domain of TFIID displays species specificity in vivo. Cell. 1991;65:333–340. doi: 10.1016/0092-8674(91)90166-v. [DOI] [PubMed] [Google Scholar]
- Gotta M, Strahl-Bolsinger S, Renauld H, Laroche T, Kennedy BK, Grunstein M, Gasser SM. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J. 1997;16:3243–3255. doi: 10.1093/emboj/16.11.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb S, Esposito RE. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- Gottschling DE. Telomere-proximal DNA in Saccharomyces cerevisiae is refractory to methyltransferase activity in vivo. Proc Natl Acad Sci USA. 1992;89:4062–4065. doi: 10.1073/pnas.89.9.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies, a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Matzke MA. Exploring and explaining epigenetic effects. Trends Genet. 1997;13:293–295. doi: 10.1016/s0168-9525(97)01219-5. [DOI] [PubMed] [Google Scholar]
- Hill JE, Myers AM, Koerner TJ, Tzagaloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Holmes SG, Rose AB, Steuerle K, Saez E, Sayegh S, Lee YM, Broach JR. Hyperactivation of the silencing proteins, Sir2p and Sir3p, causes chromosome loss. Genetics. 1997;145:605–614. doi: 10.1093/genetics/145.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivy JM, Klar AJS, Hicks JB. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:688–702. doi: 10.1128/mcb.6.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen P, Turner BM. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993;74:282–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- Kadonaga JT. Eukaryotic transcription: an interlaced network of transcription factors and chromatin modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- Lin R, Leone JW, Cook RJ, Allis CD. Antibodies specific to acetylated histones document the existence of deposition- and transcription-related histone acetylation in Tetrahymena. J Cell Biol. 1989;108:1577–1588. doi: 10.1083/jcb.108.5.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo S, Rine J. Silencers and domains of generalized repression. Science. 1994;264:1768–1771. doi: 10.1126/science.8209257. [DOI] [PubMed] [Google Scholar]
- Loo S, Rine J. Silencing and heritable domains of gene expression. Annu Rev Cell Dev Biol. 1995;11:519–548. doi: 10.1146/annurev.cb.11.110195.002511. [DOI] [PubMed] [Google Scholar]
- Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, Gasser SM. Evidence for silencing compartments within the yeast nucleus: a role for telomere proximity and Sir protein concentration in silencer-mediated repression. Genes & Dev. 1996;10:1796–1811. doi: 10.1101/gad.10.14.1796. [DOI] [PubMed] [Google Scholar]
- Moazed D, Johnson AD. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell. 1996;86:667–677. doi: 10.1016/s0092-8674(00)80139-7. [DOI] [PubMed] [Google Scholar]
- Moazed D, Kistler A, Axelrod A, Rine J, Johnson AD. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc Natl Acad Sci USA. 1997;94:2186–2191. doi: 10.1073/pnas.94.6.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran E, Matthews MB. Multiple functional domains in the adenovirus E1A gene. Cell. 1987;48:177–178. doi: 10.1016/0092-8674(87)90418-1. [DOI] [PubMed] [Google Scholar]
- Nasmyth KA. The regulation of yeast mating-type chromatin structure by SIR: an action at a distance affecting both transcription and transposition. Cell. 1982;30:567–578. doi: 10.1016/0092-8674(82)90253-7. [DOI] [PubMed] [Google Scholar]
- O’Neill LP, Turner BM. Histone H4 acetylation distinguishes coding regions of the human genome from heterochromatin in a differentiation-dependent, but transcription-independent manner. EMBO J. 1995;14:3936–3939. doi: 10.1002/j.1460-2075.1995.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell DA, Sauer RT. The structural stability of a protein is an important determinant of its proteolytic susceptibility in E. coli. J Biol Chem. 1989;264:7590–7595. [PubMed] [Google Scholar]
- Perez-Martin J, Uria JA, Johnson AD. Phenotypic switching in Candida albicans is controlled by a SIR2 gene. EMBO J. 1999;18:2580–2592. doi: 10.1093/emboj/18.9.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy P, Hahn S. Dominant negative mutations in yeast TFIID define a bipartite DNA-binding region. Cell. 1991;65:349–357. doi: 10.1016/0092-8674(91)90168-x. [DOI] [PubMed] [Google Scholar]
- Renauld H, Aparicio OM, Zierath PD, Billington BL, Chhablani SK, Gottschling DE. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and SIR3 dosage. Genes & Dev. 1993;7:1133–1145. doi: 10.1101/gad.7.7a.1133. [DOI] [PubMed] [Google Scholar]
- Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Russo VEA, Martienssen RA, Riggs AD, editors. Epigenetic Mechanisms of Gene Regulation. New York: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1989. [Google Scholar]
- Sherman JM, Pillus L. An uncertain silence. Trends Genet. 1997;13:308–313. doi: 10.1016/s0168-9525(97)01198-0. [DOI] [PubMed] [Google Scholar]
- Shore D, Nasmyth K. Purification and cloning of a DNA binding protein from yeast that binds to both silencer and activator elements. Cell. 1987;51:721–732. doi: 10.1016/0092-8674(87)90095-x. [DOI] [PubMed] [Google Scholar]
- Shore D, Squire M, Nasmyth KA. Characterization of two genes required for the position-effect control of yeast mating-type genes. EMBO J. 1984;3:2817–2823. doi: 10.1002/j.1460-2075.1984.tb02214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Klar AJS. Active genes in budding yeast display enhanced in vivo accessibility to foreign DNA methylases: a novel in vivo probe for chromatin structure of yeast. Genes & Dev. 1992;6:186–196. doi: 10.1101/gad.6.2.186. [DOI] [PubMed] [Google Scholar]
- Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes & Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Pillus L, Boeke JD. Distribution of a limited Sir2 protein pool regulates the strength of yeast rDNA silencing and is modulated by Sir4p. Genetics. 1998;149:1205–1219. doi: 10.1093/genetics/149.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. Sir2 and Sir4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes & Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- Struhl K. Chromatin structure and the RNA polymerase II connection: implications for transcription. Cell. 1996;84:179–182. doi: 10.1016/s0092-8674(00)80970-8. [DOI] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Tsang AW, Escalante-Semerena JG. CobB, a new member of the SIR2 family of eukaryotic regulatory proteins, is required to compensate for the lack of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase activity in cobT mutants during cobalamin biosynthesis in Salmonella typhimurium LT2. J Biol Chem. 1998;273:31788–31794. doi: 10.1074/jbc.273.48.31788. [DOI] [PubMed] [Google Scholar]
- Turner BM, Birley AJ, Jayne L. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell. 1992;69:375–384. doi: 10.1016/0092-8674(92)90417-b. [DOI] [PubMed] [Google Scholar]
- Winston F, Dollard C, Ricupero-Hovasse SL. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- Yahiaoui B, Taibi A, Ouaissi A. A Leishmania major protein with extensive homology to silent information regulator 2 of Saccharomyces cerevisiae. Gene. 1996;169:115–118. doi: 10.1016/0378-1119(95)00785-7. [DOI] [PubMed] [Google Scholar]