Abstract
Meta-analysis is an increasingly popular and valuable tool for summarizing results across many neuroimaging studies. It can be used to establish consensus on the locations of functional regions, test hypotheses developed from patient and animal studies and develop new hypotheses on structure–function correspondence. It is particularly valuable in neuroimaging because most studies do not adequately correct for multiple comparisons; based on statistical thresholds used, we estimate that roughly 10–20% of reported activations in published studies are false positives. In this article, we briefly summarize some of the most popular meta-analytic approaches and their limitations, and we outline a revised multilevel approach with increased validity for establishing consistency across studies. We also discuss multivariate methods by which meta-analysis can be used to develop and test hypotheses about co-activity of brain regions. Finally, we argue that meta-analyses can make a uniquely valuable contribution to predicting psychological states from patterns of brain activity, and we briefly discuss some methods for making such predictions.
Keywords: PET, fMRI, meta-analysis, neuroimaging, analysis methods
INTRODUCTION
The number of human functional neuroimaging studies of psychological processes has risen dramatically over the last 15 years, from around 124 publications in 1991 to over 1000 last year.1 This growing body of knowledge is accompanied by a growing need to integrate research findings and establish consistency across labs and widely varying scanning procedures. Meta-analysis is a primary research tool for accomplishing this goal; quantitative meta-analyses can be used to localize the brain regions most consistently activated by a particular type of task. In addition, meta-analysis can help to develop new hypotheses about the neuroanatomy of cognition, emotion and social processes, and it can be used to test hypotheses derived from studies of brain-damaged patients, electrophysiology or other methods. But perhaps most importantly, it offers a unique opportunity to compare results across diverse task conditions, getting a picture of the psychological ‘forest’ as well as the trees.
This last point is crucial. Although patterns of brain activity can serve as a common language for scientists in diverse fields, without quantitative tools the problem of interpreting patterns of activated regions is a bit like reading tea leaves. There is a danger of interpreting results narrowly in the context of a limited set of studies—in the eyes of the hopeful imager, hippocampal activity may be taken as evidence for declarative memory, ventromedial prefrontal cortex (vmPFC) activity may imply self-reflection, and amygdala activity may imply threat. Unfortunately, individual neuroimaging studies typically provide direct evidence about brain activity, not mental states. The vmPFC may be activated by processes related to self-reflection; but observing vmPFC activity in a task does not necessarily imply that self-reflection has occurred. In order to make claims about psychological process, one would have to know that only self-related tasks activate vmPFC activity. Knowing this requires assessing the consistency of activation in all types of non-self-related tasks.
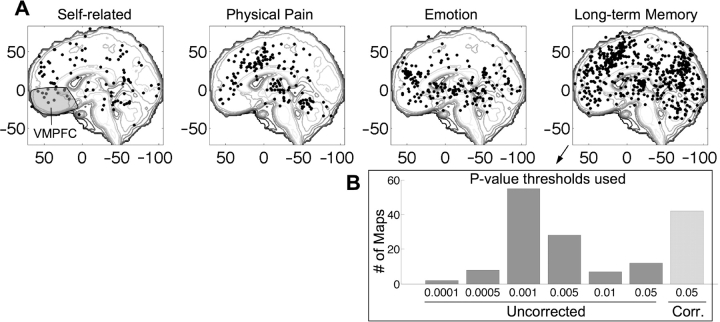
As Figure 1 shows, the vmPFC turns out to be activated by a range of different tasks, many of which do not obviously involve the self. These include viewing pleasant stimuli, perceiving tokens that symbolize reward, experiencing physical pain and retrieving items from long-term memory. Perhaps, self-related processes will turn out to be a common denominator in these tasks; however, any integrated theory about the psychological roles of this region should take all of this evidence into account. No single study to date has investigated all of these processes; across studies, however, meta-analysis can help to evaluate whether particular types of mental processes are implied by brain activation patterns.
Fig. 1.
(A) Medial activation peak coordinates within 10 mm of midline from four task domains. Coordinates from the same study comparison map within 12 mm were averaged using a recursive algorithm. Imaging studies from 1993–2003 on self-related processes (n = 14 studies), physical pain (n = 24), emotion (n = 64) and long-term memory (n = 195), all report peak activations in the vmPFC, shaded in gray in the left panel. Information from all types of studies is needed to determine how strongly vmPFC activity implies self-related processing. (B) The most common thresholds in published long-term memory literature. x-axis: P-value threshold; y-axis: number of comparison maps (whole-brain analyses of an effect of interest). Based on these thresholds and rough estimates of the number of independent comparisons per map, we estimate 663 false positives in the data set, or 17% of reported activations.
In addition to providing comparisons across diverse tasks, meta-analysis can help to separate the wheat from the chaff in imaging studies, identifying consistent activations and those that do not replicate. Because of the need to maintain statistical power with small samples and often hundreds of thousands of comparisons in each study, uncorrected or inadequately corrected statistical thresholds are the rule, and false positives are endemic in neuroimaging research. We sampled 195 studies of long-term memory (Figure 1, right panel) and noted the statistical thresholds used. The modal threshold was P < 0.001 uncorrected for multiple comparisons, with a per-study average of 1.5 separate comparison maps and on the order of 50 000–100 000 voxels tested per map. Based on smoothness estimates for studies reviewed in Nichols and Hayasaka (2003), at this threshold, we might expect roughly one false positive for every two comparison maps, or about 150 false positive activations.2 However, many thresholds were more liberal, as shown in the inset of Figure 1. Summing expected false positives across studies based on the actual thresholds used, we estimate 663 false positives, which is about 17% of the total number of reported peaks. Furthermore, because voxels are not independent [the studies reviewed by Nichols and Hayasaka have an average smoothness of about 5 mm full-width-half-max (FWHM) each direction], imposing an additional ‘cluster extent’ threshold—often cited as a guard against false positives—will not help as much as it might seem at first blush. With this smoothness, the average false-positive cluster contains around 15 contiguous 2 × 2 × 2 mm3 voxels. Additionally, many of studies that cite ‘corrected’ thresholds use inadequate methods, such as the commonly-used threshold of 0.005 with an eight-voxel extent that was reported to achieve ‘corrected’ false-positive control in simulations based on a single dataset (Forman et al., 1995). Thus, the old adage that results are best believed when replicated is particularly true in imaging. Meta-analysis can provide a basis for identifying consistency.
Meta-analyses in other research areas often combine effect sizes of a single effect to test for consistency (Rosenthal, 1991). However, in neuroimaging studies the question may fruitfully be formulated in spatial terms: ‘Where is the consistent activation?’ Individual studies sometimes use very different analyses, making it difficult to combine effect sizes in many cases, and effect sizes are only reported for a small number of ‘activated’ locations, making combined effect-size maps across the brain impossible to reconstruct from published reports. (A promising approach may be to obtain whole-brain statistic maps from the study authors). Instead, meta-analysis is typically performed on the spatial coordinates of peaks in activation reliability (‘peak coordinates’), reported in the standard coordinate systems of the Montreal Neurologic Institute (MNI) or Talairach and Tourneaux (1988).3 In spite of these difficulties, a unique positive feature of imaging meta-analyses is that many studies report peak coordinates (but not effect sizes) throughout the whole brain. Thus, if peak coordinates are used in meta-analysis, the problem of unreported activation coordinates (the ‘file drawer problem’) (Rosenthal, 1979) is greatly reduced, though not completely eliminated.
Here, we briefly summarize several popular methods for meta-analysis of neuroimaging data. We review three methods for summarizing the consistency of peak coordinates: Kernel density analysis [KDA; (Wager et al., 2003; Wager et al., 2004)], activation likelihood estimate (ALE) analysis (Turkeltaub et al., 2002; Laird et al., 2005), and a new multilevel variant of KDA designed to address several important statistical shortcomings of the other two models (Wager et al., 2007). We also briefly discuss multivariate meta-analytic methods, including spatial discriminant analysis, ‘co-activation’ maps, and classifier systems that provide information on the likelihoods that brain activity patterns are linked to particular psychological states.
Methods for summarizing consistent activations
The goal of the analyses described subsequently is to localize consistently activated regions (if any exist) in a set of studies related to the same psychological state. The methods work essentially by counting the number of activation peaks in each local area of brain tissue and comparing the observed number of peaks to a null-hypothesis distribution (usually based on a uniform distribution over the brain) to establish a criterion for significance. First, we present a side-by-side comparison of two commonly used published methods, KDA and ALE. Then, we describe a new, revised extension of KDA that eliminates some of the assumptions and shortcomings of these earlier methods.
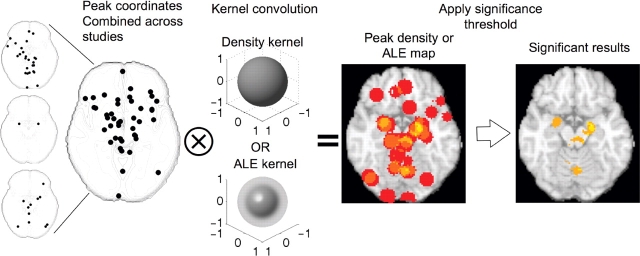
Counting the number of peaks in a local area amounts to creating a 3D histogram of peak locations and smoothing it with a kernel, as illustrated in Figure 2. Thus, the procedure is similar to kernel-based methods of analyzing distributions in many other applications. In the KDA method, the smoothing kernel is a spherical indicator function with radius r, giving the smoothed histogram an interpretation of ‘the number of peaks within r mm’. In the ALE method, the kernel is Gaussian with a width specified by the FWHM value. The kernel radius (r) or FWHM is selected by the analyst; kernels that best match the natural spatial resolution of the data are the most statistically powerful. Comparisons across KDA kernels indicate that r = 10 or 15 mm usually gives the best results (Wager et al., 2004), providing some evidence that this is the natural spread of peak locations across studies. For ALE a FWHM of 10 mm is common (Turkeltaub et al., 2002).
Fig. 2.
Example of meta-analysis using KDA or ALE analysis on three studies. The three small maps on the left show peaks reported in each study for a representative axial brain slice. Peaks are combined across studies and the combined map is smoothed with a spherical kernel (KDA) or a Gaussian kernel (ALE). The resulting peak density map (middle) or ALE map is thresholded, resulting in a map of significant results (right). In this illustration, regions with three or more peaks within 10 mm were considered ‘significant.’ In practice, the analyses use Monte Carlo resampling to determine an appropriate threshold, though the interpretation of significant results differs across KDA and ALE analyses (see text). Because peaks are combined across studies and study is thus treated as a fixed effect, some individual studies may exert undue influence on the results.
Interpretation of meta-analysis statistic values
In the KDA method, the smoothed histogram reflects the estimated density of nearby reported peaks in each brain voxel. A threshold for statistical significance is established using Monte Carlo4 procedures described subsequently. Thus, the interpretation of the KDA meta-analysis statistic with value x is ‘x peaks lie within r mm of this voxel’. Typically, this value is additionally divided by the volume of the kernel. In this case, x can be interpreted as the density (in peaks/mm3) within r mm—though dividing by this constant changes only the scale, not the statistical significance.
In the ALE method, an additional step is performed. Rather than treating the smoothed peak coordinate histograms as a measure of local activation frequency, the smoothed peaks are treated as estimates of the probability that each peak activated in that vicinity, and the union of these ‘probabilities’ is computed. This can be accomplished by assuming that the reported peaks are spatially independent (the location of each peak is independent of the others), though this assumption is questionable, because studies tend to report multiple peaks from each significantly activated region (we’ll refer to them here as ‘blobs’). (The KDA method makes this problematic assumption as well, and we return to this issue subsequently).
Though the mathematics of the ALE probabilities are straightforward given the independence assumption,5 a clear interpretation of the ALE statistic might be given through conceptual examples. By calculating the union of probabilities, the ALE statistic reflects the probability that one or more peaks truly lie in that voxel. As an example, imagine a meta-analysis of three reported peaks (though most actual meta-analyses include thousands). One peak activates in voxel v, and the others are nowhere close. If the smoothing FWHM approaches zero, then the meta-analytic ALE statistic value would be 1, which is appropriate, because we certainly know that at least one reported peak was truly in that voxel.6 Thus, the meta-analytic map is simply a restatement of the peak locations. If the smoothing FWHM is non-zero, then the ALE statistic would equal the maximum height of the smoothing kernel. This is conceptually different from the KDA statistic, which summarizes the density of reported peaks without assuming they directly reflect probabilities. In Table 1, we summarize the interpretations of the ALE and KDA statistics and other relevant points of comparison.
Table 1.
Meta-analysis method
| Property | KDA | ALE | Multilevel KDA |
|---|---|---|---|
| Kernel | Spherical | Gaussian | Spherical |
| Interpretation of statistic | Number of peaks near voxel | Probability that at least one peak lies in voxel | Number of study comparison maps activating near voxel |
| Null hypothesis | Peaks are not spatially consistent | No peaks truly activate at this voxel | Activations across studies are not spatially consistent |
| Interpretation of significant result | More peaks lie near voxel than expected by chance | One or more peaks lies at this voxel | A higher proportion of studies activate near voxel than expected by chance |
| Multiple comparisons | FWER | FDR | FWER (recommended) or FDR |
| Weighting | None, or weight peaks by z-score | None | Weight studies by sample size and fixed/random effects analysis |
| Generalize to … | New peaks from same studies | New peaks from same studies | New studies |
| Assumptions | Study is fixed effect: true between-studies differences are null | Study is fixed effect: true between-studies differences are null | Activation ‘blobs’ are spatially independent under the null hypothesis |
| Peaks are spatially independent under the null hypothesis | Peaks are spatially independent under the null hypothesis | ||
| Analyze differences across task types | Yes | Yes | Yes |
Meta-analytic significance and null hypotheses
Both KDA and ALE methods use Monte Carlo simulations to establish thresholds for statistical significance. In the KDA method, the null hypothesis is that the n peak coordinates reported in the set of studies to be analyzed are randomly and uniformly distributed throughout gray matter.7 The null hypothesis is rejected in voxels where the number of nearby peaks is greater than expected by chance. Thus, in ‘significant’ regions one can be confident that a cluster of peaks is not due to a spatially diffuse background of false positives.
In the ALE method, the null hypothesis is that the n peak coordinates are distributed uniformly over the brain, and—because the ALE statistic reflects the probability that at least one peak truly falls in a region—the null hypothesis is rejected in voxels where there are enough peaks to provide sufficient evidence that at least one of them truly falls within the voxel. In ‘significant’ regions, one can be confident that at least one peak truly lies in that voxel.
In both methods, the Monte Carlo procedure generates n peaks at random locations and performs the smoothing operation, to generate a series of statistic maps under the null hypothesis. We recommend generating at least 5000 such maps, and preferably more; as inferences are usually made on the tails of the null-hypothesis distribution, many iterations are necessary to achieve stability. Notably, the larger the included null-hypothesis region, the more spread out the peaks are under the null hypothesis, and the lower the threshold for significance becomes; thus, the gray matter mask is a more appropriate mask than a more inclusive brain mask, unless white-matter peaks are of theoretical interest.
The two methods differ in the way they correct for multiple comparisons across brain voxels. The KDA method seeks to establish a threshold that controls the chances of seeing any false positives anywhere in the brain at P < 0.05, corrected (so-called ‘familywise error rate’ control, FWER). FWER is the only type of correction that permits each significant region to be interpreted as a likely true result. In this case, the interpretation is that more peaks fall within this region than one would expect under the null hypothesis anywhere in the brain. To accomplish this, in each Monte Carlo simulation, the maximum KDA statistic over the whole brain is saved, and the critical threshold is the value that exceeds the whole-brain maximum in 95% of the Monte Carlo maps. The use of a distribution of maxima is an established method for multiple-comparisons correction using non-parametric approaches (Nichols and Holmes, 2002).
The ALE method, by contrast, seeks to identify voxels where the union of peak probabilities exceeds that expected by chance. The interpretation of a significant result is, ‘at least one reported peak truly falls in this region’. In recent implementations of the ALE approach (Laird et al., 2005), ALE statistics are subjected to false discovery rate (FDR) correction (Genovese et al., 2002), which ensures that at P < 0.05 corrected, on average, 5% of the reported voxels are false positives (i.e. no peak truly falls within that voxel). This procedure is typically more sensitive than FWER control, but it is not possible to say for sure which of the reported results are false positives—one can say with confidence that most of them are true results, but one cannot be confident about any single result.
Assumptions
Both of the methods described earlier are limited in that they make several assumptions that seriously limit the inferential power of the meta-analysis. First, the analyst assumes that peak reported coordinates are representative of the activation maps from which they come. Second, because the procedures lump peak coordinates across studies, study identity is treated as a fixed effect. Thus, the analyst assumes that true inter-study differences in number and location of peaks, smoothness, false positive rates and statistical power are zero. This assumption is critical because (i) it allows the possibility that a significant ‘meta-analytic’ result is due to only one study, and (ii) it is patently violated in every neuroimaging meta-analysis.
The pitfalls of treating random effects as fixed effects have been widely discussed in statistics (Neter et al., 1996; Shayle et al., 2006) and in psychology (Clark, 1973). Treating study as a fixed effect implies that the meta-analysis cannot be used to generalize across a population of studies—perhaps one of the most appealing potential features of a meta-analysis—and inferences are restricted to the set of peaks reported. It also means that one study alone can dominate the meta-analysis and create significant meta-analytic results, which obviates one of the purposes of doing a meta-analysis in the first place. For example, examine the peaks in Figure 2 that go into the example ‘toy’ meta-analysis. They come from three studies, shown by the three small maps at the far left. Because study is treated as a fixed effect, information about which study contributed each of the peaks is not preserved, and all the peaks are ‘lumped together’ in the analysis. Study 1 (top) contributes 26 peaks to the meta-analysis, many of them very close together, whereas Study 2 (middle) contributes only two. When the KDA map is generated (middle) and thresholded (right), three peaks within 10 mm are required to achieve significance in the meta-analysis. Study 1 has enough peaks near the amygdala to generate significant results in some regions by itself. The problematic conditions illustrated here of (i) unequal numbers of reported peaks, and (ii) clustering of reported peaks within single studies are the rule rather than the exception in meta-analysis of neuroimaging studies. In fact, the three maps shown here are from three published studies (Damasio et al., 2000; Liberzon et al., 2000; Wicker et al., 2003) included in recent meta-analyses of emotion (Wager et al., 2003; Wager et al., 2007).
These aspects of reported neuroimaging data notwithstanding, a third assumption of KDA and ALE analyses is that peaks are spatially independent within and across studies under the null hypothesis. Thus, any clustering of peaks tends to be interpreted as meaningful correspondence (even if the cluster comes from a single study). This assumption is required for the ALE probability computation to be valid and for the null hypothesis distribution in Monte Carlo simulations for both methods to be a meaningful baseline. However, inspection of the individual study maps in Figure 2 shows that studies tend to report multiple nearby peaks associated with the same activation ‘blob’ spread over the brain. Thus, this assumption is also often violated, and the implications are that single studies can have undue influence on the meta-analytic results if they report many nearby peaks. This is problematic because procedures for how many peaks to report and how close they should be has not been rigorously evaluated and standardized, and, as Turkeltaub et al. (2002) point out, often the most poorly controlled studies, the ones that use the most liberal statistical thresholds, or the ones that impose less arbitrary smoothing of the data are the ones that report many peaks.
A new and improved KDA method
Many of these problematic assumptions need not be made if study, rather than peak location, is the unit of analysis. We have developed new procedures in which the proportion of studies that activate in a region, rather than the number of peaks, is the test statistic (Wager et al., 2007). We refer to this analysis as multilevel KDA (MKDA) because it treats peaks as nested within studies. Because study is the unit of analysis and the error terms in the analysis depend on inconsistencies across studies, no one study can contribute disproportionately, and the method is appropriate for generalizing to a population of studies. In practice, the analyst may choose to include peaks from several nominally independent comparison maps within a study (e.g. a single study may include coordinates for fear—baseline activations separately for males and females). As it is often practically valuable to include data from several comparisons, one often analyzes across ‘comparison maps’, with the additional assumption that comparison maps within a study are independent.8
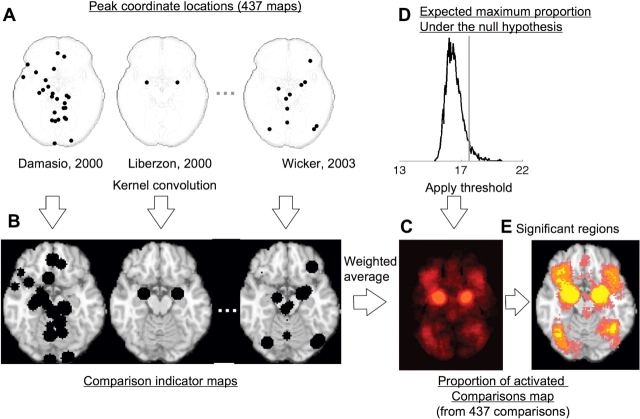
The method is diagrammed in Figure 3. The peak locations for the three sample studies discussed earlier are shown in the upper left; however, in this example, meta-analytic results shown in the lower right are based on a total of 437 comparison maps from studies of emotion (Wager et al., 2007). In this analysis, peaks from each comparison map are separately convolved with the kernel to generate comparison indicator maps (CIMs). The CIMS are limited to a maximum value of 1 (black regions in Figure 3) so that the values across brain voxels are either 1 (‘this study activated near this voxel’) or 0 (‘this study did not activate near this voxel’). The CIMs are averaged to yield the proportion of study comparison maps that activated near each voxel. The individual CIMs are weighted by the product of the square root of the sample size for that study and a discounting factor for the analysis type—neuroimaging studies treating subjects as fixed effects (mostly older studies) are given lower weight than those treating subjects as random effects, because the latter procedure is more rigorous and is statistically valid. These weights allow the larger and more rigorously performed studies to carry more weight in the meta-analysis.9 Though it is difficult to precisely determine what the discounting factor for fixed-effects studies should be, in our recent work we have used 0.75 because it down-weights these studies to a modest degree (four fixed-effects activations are required to equal three random-effects ones).
Fig. 3.
Example procedures for multilevel kernel density analysis (MKDA) of neuroimaging studies of emotion. (A) shows the peak coordinates of three of the 437 comparison maps included in this meta-analysis. Peak coordinates of each map are separately convolved with the kernel, generating comparison indicator maps (CIMs), as seen in (B). The weighted average of the CIMs (C) is thresholded based on the distribution of the maximum proportion of activated comparison maps expected under the null hypothesis (D) to produce significant results (E). Yellow voxels are FWER corrected at P < 0.05. Other colored regions are FWER corrected for spatial extent at P < 0.05 with primary alpha levels of 0.001 (orange), 0.01(pink) and 0.05 (purple).
The map of the proportion of activated comparisons is subjected to statistical thresholding via a Monte Carlo procedure similar to the one described earlier, but with several key differences. Before the Monte Carlo simulation, CIMs for each comparison are first generated and then segmented into contiguous ‘blobs’. In each Monte Carlo iteration, the locations of the centers of each blob are selected at random based on the uniform distribution over gray matter, and the CIMs and null-hypothesis proportion of activated comparisons map is created. This procedure preserves the spatial clustering of nearby peaks within each study, and thus avoids the assumption of independent peak locations within studies. It has the considerable advantage of being relatively insensitive to the conventions used for peak reporting—for example, if study authors decided to report the coordinates of every activated voxel rather than just a few peaks, the CIM for that study would be relatively unaffected, and the null-hypothesis distribution would also look much the same. This is not true of other methods.
This method also incorporates advances in statistical thresholding. FWER control is provided, as before, by calculating the 95th percentile of the maximum proportion of activated contrasts anywhere in the brain. However, it is also possible to establish an arbitrary, uncorrected threshold and then ask which voxels are part of regions that are extensive enough to be statistically significant. This is the concept behind ‘cluster extent-based’ multiple comparisons correction in SPM software (Friston et al., 1994). This can be accomplished in the Monte Carlo simulation by saving the largest cluster of contiguous voxels above a specified threshold for each iteration, and then setting a cluster extent threshold equal to the 95th percentile of these values across iterations. Significant regions are extensive enough so that one would expect a cluster this large anywhere in the brain by chance only 5% of the time. The bottom right panel of Figure 3 shows an example of applying this analysis the 437 comparison maps from emotion tasks. Yellow voxels are FWER corrected at P < 0.05, so that one voxel is enough to exceed expectations under the null hypothesis anywhere in the brain, and other colored regions have a large enough cluster extent—at primary alpha levels of 0.001 (orange), 0.01 (pink) and 0.05 (purple)—to achieve FWER corrected signifiance at P < 0.05. These regions correspond well with those identified as important for affect in animals and humans.
In summary, by treating studies (or independent comparison maps within studies) as units of analysis, a method can be devised that weights studies by the quality of their information and allows the reliability of activation to be summarized across studies without allowing studies that report more peaks to contribute disproportionately. In Table 1, we present a summary of the main features of each of the three meta-analysis types discussed earlier.
Methods for analyzing differences across task types
Any of the earlier mentioned methods can be used for testing for differences across two types of task—for example, for locating regions significantly more responsive in one type of emotion or one type of working memory than another. The logic and the same caveats about interpretation apply. During each Monte Carlo iteration, the maximum whole-brain values for the difference between the test statistic for Task A and Task B is saved (for KDA analysis) or the uncorrected difference between the union of probabilities is saved (for ALE analysis), and thresholding on these differences proceeds as described earlier. Thus, statistics are calculated over the brain for the difference in density values or ALE values between two conditions.
These methods are essentially multivariate because they provide information about the relative probabilities of activation across tasks in a region, compared with the rest of the brain. Imagine two tasks—Task A activates more across the whole brain than Task B, but in a particular region, Task A and B activate equally frequently. The KDA/MKDA or ALE ‘difference’ analyses will simulate a null hypothesis with many more activations in Task A than B, so the expectation will be that A > B in each brain region. Some areas may show greater A > B effects than expected by chance and so reach significance. With enough power, the region with equal absolute activation frequencies in Task A and Task B will show a significant B > A effect, because the frequencies in this region relative to the rest of the brain (and thus the null hypothesis) favor Task B. Thus, the density method may be sensitive in localizing the regions most strongly associated with each task, even if one task dominates the other in absolute number of activations.
However, the analyst may often wish to test the absolute difference in activation frequencies for a given region, or for each voxel over the brain. The question now is, does one task show a higher proportion of peaks (KDA), proportion of activated study comparison maps (MKDA), or likelihood that one peak lies in the voxel (ALE)? This question may be answered with the χ2 (chi-square) test, a common statistical procedure for comparing frequencies in two-way tables. In this case, the analysis is performed on each voxel or region of interest separately, so the analysis is univariate. One may code each study as either activating (‘yes’) or not (‘no’) near a voxel of interest and count frequencies of activations by task type. Maps over all voxels can be constructed as in the MKDA method described earlier; CIM maps describe the yes/no activation status in each voxel for each study, and each map is coded as belonging to one of two or more task types. The χ2 test for independence compares observed activation frequencies with the null hypothesis of equal expected frequencies across all task types. Theχ2 test can also be used to count frequencies of peaks rather than studies, although the problems associated with treating study as a fixed effect above apply.
One problem with the use of the χ2 test in neuroimaging applications, however, is that because of the low frequencies of activations, the expected activation frequencies are often low—a rule of thumb is that any cell with expected counts < 5 is problematic—and the P-values associated with the test become substantially too liberal. Fisher's exact test may be used instead for the 2-task case, although non-parametric simulation provides a general alternative for comparing two or more tasks. In this test, the task condition assignments are randomly permuted many times (∼5000 or more), and the resulting χ2 values are used to estimate the null hypothesis χ2 distribution for that voxel. The P-value for the test is the proportion of χ2 values that lie below the observed χ2 value. The test approximates exact methods for multinomial frequency analysis that are extremely computationally expensive. Study weighting may be easily incorporated into the χ2 analysis by calculating weighted frequencies (with weights normalized to be positive and with a mean of 1). However, weighting reduces the effective degrees of freedom, and without correction, the false-positive rate will be inflated. Using the non-parametric test described here this problem and is recommended.
Each of these methods—the density or ALE based ones and the non-parametric χ2 analysis—provides complementary information. As discussed, the density/ALE based methods locate regions of relative peak concentration for each task based on a null hypothesis of uniform distribution across the search volume (brain) for both tasks. The χ2 analysis, in contrast, is univariate and provides information about the absolute differences in activation frequencies across tasks.
A limitation of both voxel-based and χ2 mehods is that they do not control for other potentially confounding variables. In working memory studies, for example, many more studies of spatial working memory frequently require manipulation of information held in memory, whereas studies of object working memory very rarely do (Wager and Smith, 2003). A spatial vs object comparison in the meta-analysis thus also likely reflects a manipulation vs no-manipulation difference. Such confounding variables can be controlled for via logistic regression, treating activation in a study comparison (‘yes’ or ‘no’) as the outcome and study-level variables as predictors. Kosslyn and Thompson (2003) used logistic regression to examine the factors that influence whether studies find early visual activation in mental imagery, and Nee et al. (2007) used it to examine the stages of response conflict associated with activation in several regions during ‘conflict’ tasks. Because analyses were carried out across study comparison maps in these studies, the problems associated with treating study as a fixed effect are avoided. This approach works best when analyses are focused on regions of interest activated by a high proportion of studies; low proportions violate the assumptions and result in high false-positive rates. A non-parametric test that would avoid these issues in voxel-wise analysis has not yet been developed.
Other approaches to neuroimaging meta-analysis
The discussion earlier has by no means presented an exhaustive review of neuroimaging meta-analysis methods, but for space reasons we can only mention a few others here. Some authors have used measures of effect size directly and performed a traditional meta-analysis comparing effect sizes across studies (Davidson and Heinrichs, 2003; Zakzanis et al., 2003; Thornton et al., 2006). This approach is promising, but requires careful analysis choices about how to deal with effect sizes that may be incommensurate because of differences in analysis method—the subject-as-fixed-effect vs random-effect distinction being foremost among them. In addition, the analyst must carefully define regions of interest within which effect sizes may be treated as comparable, and the analyst must apply methods for imputing effect sizes for studies that do not report them in the region of interest.
Other approaches have used ‘data-driven’ algorithms to define regions with homogeneous activation patterns. In a creative application, Nielsen and Hansen (2004) used a matrix factorization algorithm similar to principal components analysis to decompose a matrix of both activation indicators across voxels and semantic study labels derived from publications. They identified distributed patterns of voxels across the brain that were associated with particular labels (e.g. ‘language’ or ‘spatial’). Such analyses are promising ways to identify distributed patterns for testing in new neuroimaging or lesion studies. Similar methods are now being developed to analyze ‘connectivity’ in meta-analytic databases, which will provide information about whether studies that activate one region of interest tend to also activate other regions. Identifying distributed patterns of co-activation across studies may be useful in defining multiple modes of brain function associated with different task types.
In another approach, Wager and Smith (2003) used cluster analysis to segregate reported peaks from working memory studies into spatial groupings. The clusters were defined without any knowledge of task type, and the authors subsequently examined each cluster for evidence of specialization for different types of working memory using χ2 analysis. This approach provides an alternative to specifying a priori anatomical regions of interest with arbitrary boundaries and a natural way to group peaks for subsequent analysis of differences across task types without introducing bias in these analyses.
A third multivariate application can be used to test for differences in spatial location of activation among two or more task types. Wager et al., 2004) identified parietal regions of interest using KDA analysis on attention-switching tasks. They then performed multivariate analysis of variance (MANOVA) analysis on the distribution of peak locations across task type. The x, y and z mm coordinates were dependent measures, and the procedure calculated the best linear combination for separating peaks from different types of switching, providing preliminary evidence for differential localization of different switch types in parietal cortex. This type of analysis makes the most sense when hypotheses are about spatial location—i.e. that one task activates anterior to another within the same broad region. In another approach, Murphy et al. used a 3D Kolmorogov–Smirnov test to test the global null hypothesis of equal distributions of peaks across the whole brain for multiple conditions. A rejection of the null in this test implies that two (or more) conditions differed in peak density somewhere in the brain. The limitations of treating study as a fixed effect apply in these cases as well, though this problem could be avoided in future work.
Finally, classifier analysis can be used to find distributed patterns of regions that reliably dissociate activations related to one type of task from those related to another. For example, there may be no single region of the brain reliably and uniquely associated with a particular type of emotion (e.g. fear, sadness or disgust), but distributed patterns may accurately discriminate the basic emotions. A host of methods for predicting task status given multivariate brain activity patterns may be productively applied to meta-analytic data to achieve these ends, including Fisher's linear and quadratic discriminant analysis, support-vector machines (LaConte et al., 2005; Pessoa and Padmala, 2007), and Bayesian classifiers. The goal of these analyses is to find patterns that can correctly predict the task type in a new sample, and a complex set of weights on voxels or rules is developed that gives the best predictions. Because these weights are developed using data (a ‘training set’), it is critical that the data used to test the performance is independent of the training set. It is easy to develop classifiers that predict task categories in a data set perfectly, but perform very poorly on new, independent data. Another important point is that the generalizability of a classifier analysis is only as broad as the task categories included in the analysis—a 99% correct classifiation rate in predicting Task A vs Task B does not imply good performance on classifying A vs B vs C. Meta-analysis offers a unique opportunity to assess predictive power across a wide range of different task conditions. While we are not aware of published analyses that use these techniques on meta-analytic data, they offer a rich new avenue for utilizing the wealth of data in the neuroimaging literature to make inferences on psychological processes.
Acknowledgments
Thanks to Tom Nichols for helpful discussion, and Hedy Kober, John Jonides, Susan Reading, Lisa Feldman Barrett, Seth Duncan, Eliza Bliss-Moreau, Kristen Lindquist, Josh Joseph and Jennifer Mize, for help in compiling meta-analytic databases.
Footnotes
Conflict of Interest
None declared.
A rough estimate based on an inclusive search in PsychInfo (which excludes many non-psychological medical studies) for ‘positron emission tomography’, ‘PET’, ‘functional magnetic resonance imaging’, ‘fMRI’ or ‘brain imaging’. All 124 in 1991 were Positron Emission Tomography (PET) studies, whereas about 75% of 2006 studies used fMRI.
Based on an average count of 464 RESELs (Resolution Elements), which actually substantially underestimates the number of independent comparisons made (Nichols and Hayasaka, 2003).
A standard practice is to convert coordinates into one reference space (we prefer MNI). Matthew Brett has developed useful tools for converting between Talairach and MNI space.
Sometimes referred to as ‘non-parametric tests,’ though the Monte Carlo is actually a randomization test.
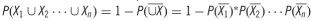 , where P(Xi) is the probability that peak Xi lies in a given voxel of interest, and
, where P(Xi) is the probability that peak Xi lies in a given voxel of interest, and 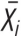 refers to the complement of Xi.
refers to the complement of Xi.
If P(X1) = 1, P(X2) = 0, P(X3) = 0, then the ALE statistic 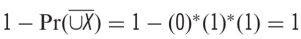 .
.
The gray-matter mask we use is smoothed to add an 8 mm border, because many studies report coordinates near the edges of gray matter.
This assumption does not strictly hold, and care must be taken to ensure that multiple highly dependent comparison maps within a single study are not entered.
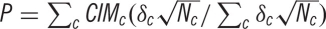 , where P is the weighted proportion of activated comparisons, c indexes comparison maps, δ is the fixed-effect discounting factor and N is the sample size that contributed to each comparison map.
, where P is the weighted proportion of activated comparisons, c indexes comparison maps, δ is the fixed-effect discounting factor and N is the sample size that contributed to each comparison map.
REFERENCES
- Clark HH. The language-as-fixed-effect fallacy: a critique of language statistics in psychological research. Journal of Verbal Learning and Verbal Behavior. 1973;12(4):335–59. [Google Scholar]
- Damasio AR, Grabowski TJ, Bechara A, et al. Subcortical and cortical brain activity during the feeling of self-generated emotions. Nature Neuroscience. 2000;3(10):1049–56. doi: 10.1038/79871. [DOI] [PubMed] [Google Scholar]
- Davidson LL, Heinrichs RW. Quantification of frontal and temporal lobe brain-imaging findings in schizophrenia: a meta-analysis. Psychiatry Research. 2003;122(2):69–87. doi: 10.1016/s0925-4927(02)00118-x. [DOI] [PubMed] [Google Scholar]
- Forman SD, Cohen JD, Fitzgerald M, Eddy WF, Mintun MA, Noll DC. Improved assessment of significant activation in functional magnetic resonance imaging (fMRI): use of a cluster-size threshold. Magnetic Resonance in Medicine. 1995;33(5):636–47. doi: 10.1002/mrm.1910330508. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Worsley KJ, Frackowiak R.SJ, Mazziotta JC, Evans AC. Assessing the significance of focal activations using their spatial extent. Human Brain Mapping. 1994;1:210–20. doi: 10.1002/hbm.460010306. [DOI] [PubMed] [Google Scholar]
- Genovese CR, Lazar NA, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage. 2002;15(4):870–8. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- Kosslyn SM, Thompson WL. When is early visual cortex activated during visual mental imagery. Psychological Bulletin. 2003;129(5):723–46. doi: 10.1037/0033-2909.129.5.723. [DOI] [PubMed] [Google Scholar]
- LaConte S, Strother S, Cherkassky V, Anderson J, Hu X. Support vector machines for temporal classification of block design fMRI data. Neuroimage. 2005;26(2):317–29. doi: 10.1016/j.neuroimage.2005.01.048. [DOI] [PubMed] [Google Scholar]
- Laird AR, Fox PM, Price CJ, et al. ALE meta-analysis: controlling the false discovery rate and performing statistical contrasts. Human Brain Mapping. 2005;25(1):155–64. doi: 10.1002/hbm.20136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon I, Taylor SF, Fig LM, Decker LR, Koeppe RA, Minoshima S. Limbic activation and psychophysiologic responses to aversive visual stimuli. Interaction with cognitive task. Neuropsychopharmacology. 2000;23(5):508–16. doi: 10.1016/S0893-133X(00)00157-3. [DOI] [PubMed] [Google Scholar]
- Murphy FC, Nimmo-Smith I, Lawrence AD. Functional neuroanatomy of emotion: a meta-analysis. Cognitive, Affective and Behavioral Neuroscience. 2003;3:207–33. doi: 10.3758/cabn.3.3.207. [DOI] [PubMed] [Google Scholar]
- Nee DE, Wager TD, Jonides J. A meta-analysis of neuroimaging studies of interference resolution. Cognitive Affective and Behavioral Neuroscience. 2007 doi: 10.3758/cabn.7.1.1. (In Press) [DOI] [PubMed] [Google Scholar]
- Neter J, Kutner MH, Wasserman W, Nachtsheim CJ. Applied Linear Statistical Models. New York: McGraw-Hill; 1996. [Google Scholar]
- Nichols T, Hayasaka S. Controlling the familywise error rate in functional neuroimaging: a comparative review. Statistical Methods in Medical Research. 2003;12(5):419–46. doi: 10.1191/0962280203sm341ra. [DOI] [PubMed] [Google Scholar]
- Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: a primer with examples. Human Brain Mapping. 2002;15(1):1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen FA, Hansen LK, Balslev D. Mining for associations between text and brain activation in a functional neuroimaging database. Neuroinformatics. 2004;2(4):369–80. doi: 10.1385/NI:2:4:369. [DOI] [PubMed] [Google Scholar]
- Pessoa L, Padmala S. Decoding near-threshold perception of fear from distributed single-trial brain activation. Cerebral Cortex. 2007;17(3):691–701. doi: 10.1093/cercor/bhk020. [DOI] [PubMed] [Google Scholar]
- Rosenthal R. The ‘file-drawer’ problem and tolerance for null results. Psychological Bulletin. 1979;86:638–41. [Google Scholar]
- Rosenthal R. Meta-Analytic Procedures for Social Research. Beverly Hills, CA: Sage; 1991. [Google Scholar]
- Shayle R, Searle GC, McCulloch CE. Variance Components. Hoboken, NJ: Wiley-Interscience; 2006. [Google Scholar]
- Talairach J, Tournoux P. Co-planar Stereotaxic Atlas of the Human Brain: 3-Dimensional Proportional System - an Approach to Cerebral Imaging. New York, NY: Thieme Medical Publishers; 1988. [Google Scholar]
- Thornton AE, Van Snellenberg JX, Sepehry AA, Honer W. The impact of atypical antipsychotic medications on long-term memory dysfunction in schizophrenia spectrum disorder: a quantitative review. Journal of Psychopharmacology. 2006;20(3):335–46. doi: 10.1177/0269881105057002. [DOI] [PubMed] [Google Scholar]
- Turkeltaub PE, Eden GF, Jones KM, Zeffiro TA. Meta-analysis of the functional neuroanatomy of single-word reading: method and validation. Neuroimage. 2002;16(3 Pt 1):765–80. doi: 10.1006/nimg.2002.1131. [DOI] [PubMed] [Google Scholar]
- Wager TD, Barrett LF, Bliss-Moreau E, et al. The neuroimaging of emotion. In: Lewis M, editor. Handbook of Emotion. 2007. [Google Scholar]
- Wager TD, Jonides J, Reading S. Neuroimaging studies of shifting attention: a meta-analysis. Neuroimage. 2004;22(4):1679–93. doi: 10.1016/j.neuroimage.2004.03.052. [DOI] [PubMed] [Google Scholar]
- Wager TD, Phan KL, Liberzon I, Taylor SF. Valence, gender, and lateralization of functional brain anatomy in emotion: a meta-analysis of findings from neuroimaging. Neuroimage. 2003;19(3):513–31. doi: 10.1016/s1053-8119(03)00078-8. [DOI] [PubMed] [Google Scholar]
- Wager TD, Smith EE. Neuroimaging studies of working memory: a meta-analysis. Cognitive Affective and Behavioral Neuroscience. 2003;3(4):255–74. doi: 10.3758/cabn.3.4.255. [DOI] [PubMed] [Google Scholar]
- Wicker B, Keysers C, Plailly J, Royet JP, Gallese V, Rizzolatti G. Both of us disgusted in my insula: the common neural basis of seeing and feeling disgust. Neuron. 2003;40(3):655–64. doi: 10.1016/s0896-6273(03)00679-2. [DOI] [PubMed] [Google Scholar]
- Zakzanis KK, Graham SJ, Campbell Z. A meta-analysis of structural and functional brain imaging in dementia of the Alzheimer's type: a neuroimaging profile. Neuropsychology Review. 2003;13(1):1–18. doi: 10.1023/a:1022318921994. [DOI] [PubMed] [Google Scholar]