Abstract
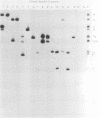
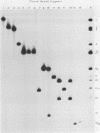
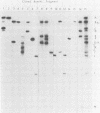
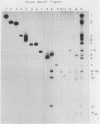
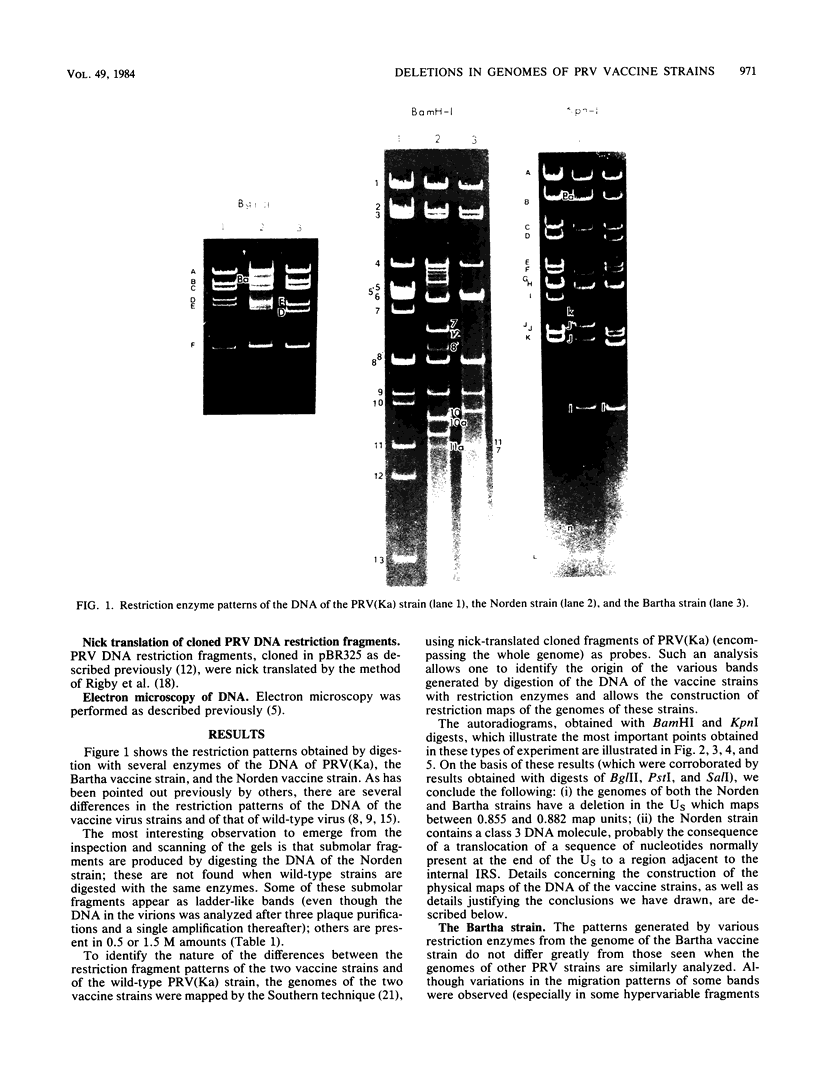
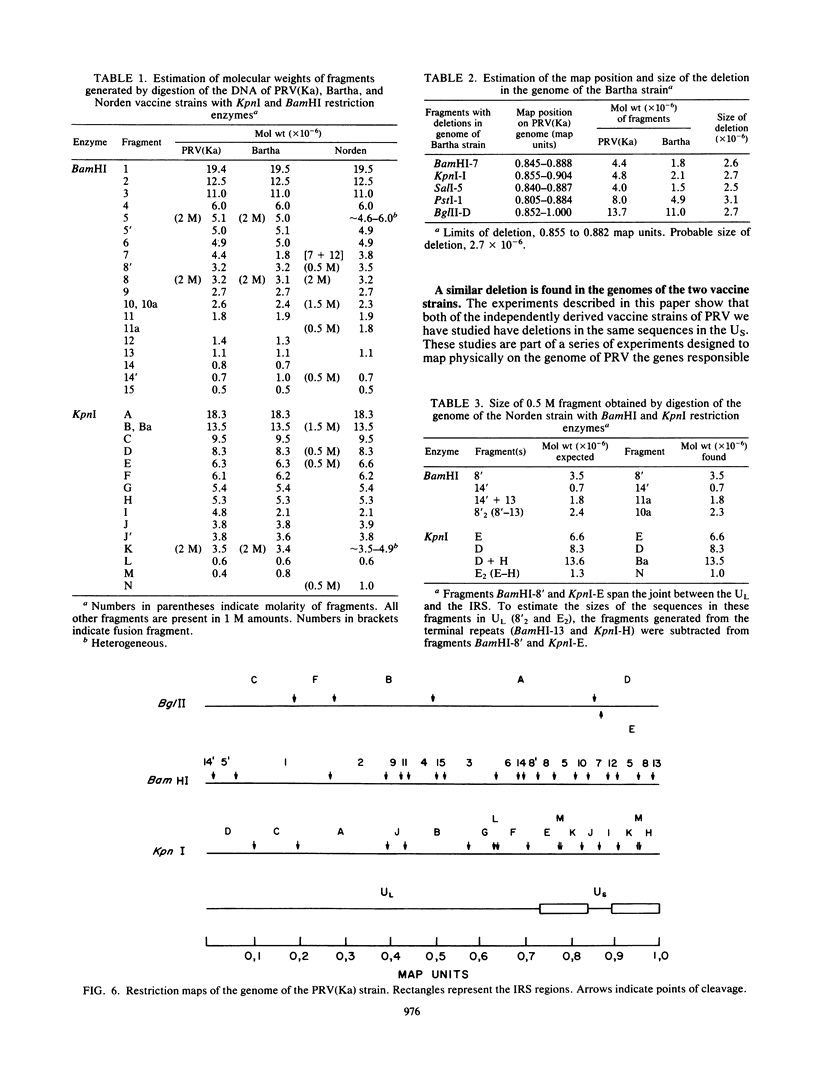
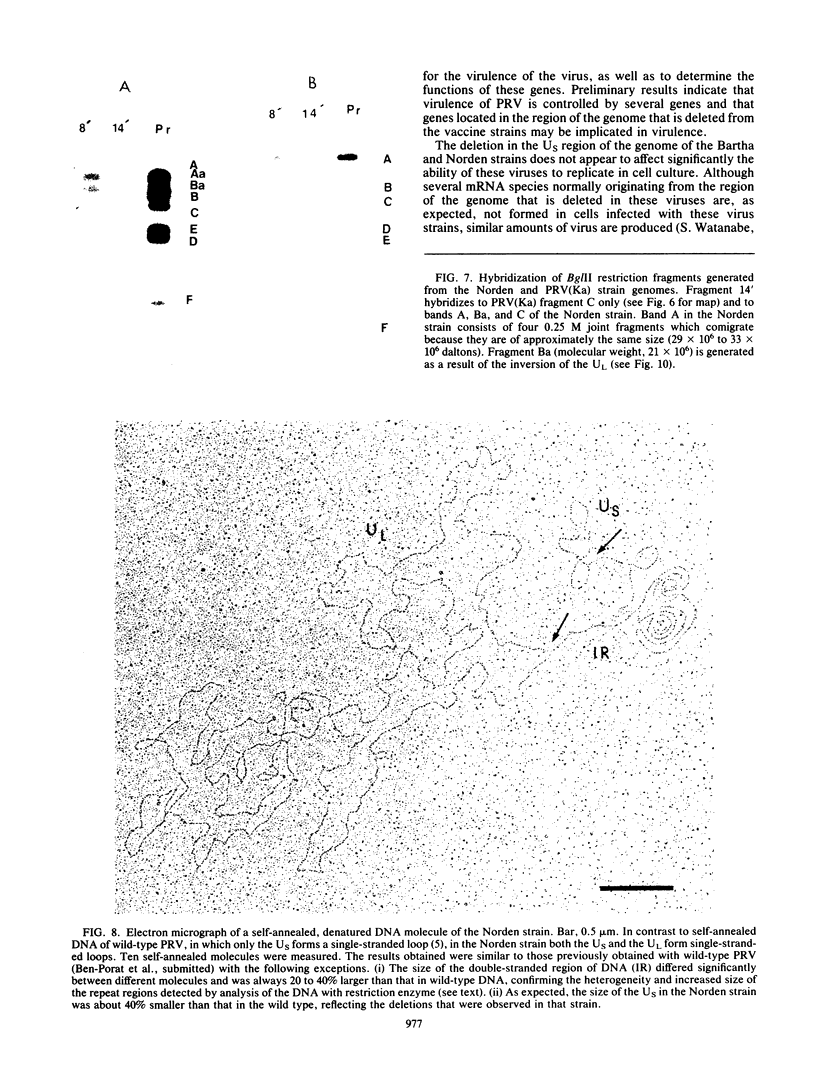
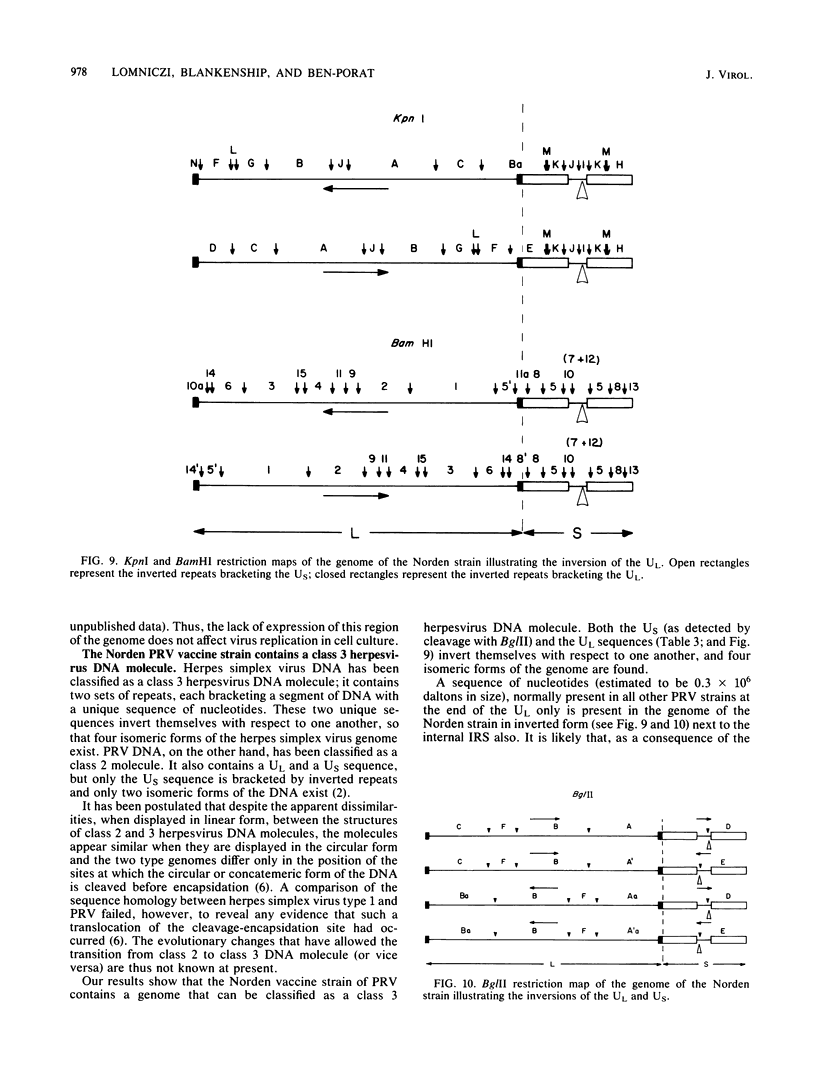
As part of a study designed to identify the genes responsible for the virulence of pseudorabies virus, we have mapped the genomes of two independently derived vaccine strains (Bartha and Norden) by restriction enzyme analysis. The structures of these genomes have been compared with that of the genome of a laboratory strain previously mapped, of restriction fragments which had been cloned. The genome of the Bartha strain was found to be very similar to that of other pseudorabies virus strains, except that a deletion of approximately 2.7 X 10(6) daltons was found in the unique short (US) region. This deletion was also observed in the genome of the Norden vaccine strain but was not observed in the genomes of any other pseudorabies virus strains that have been studied (more than 20). The genome of the Norden strain differs from that of other pseudorabies virus strains in several other respects as well. The most important difference is that in contrast to all other pseudorabies virus strains analyzed to date, which contain a type 2 herpesvirus DNA molecule (in which the US region only inverts itself relative to the unique long [UL] region), the genome of the Norden strain is a type 3 molecule in which both the US and the UL regions of the genome invert themselves, giving rise to four isomeric forms of the genome. The ability of the UL region to invert itself is probably related to the fact that a sequence normally present in all other pseudorabies virus strains at the end of the UL only is found also in inverted form at the junction of the UL and the internal inverted repeat in the Norden strain.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ben-Porat T., Demarchi J. M., Kaplan A. S. Characterization of defective interfering viral particles present in a population of pseudorabies virions. Virology. 1974 Sep;61(1):29–37. doi: 10.1016/0042-6822(74)90239-6. [DOI] [PubMed] [Google Scholar]
- Ben-Porat T., Rixon F. J., Blankenship M. L. Analysis of the structure of the genome of pseudorabies virus. Virology. 1979 Jun;95(2):285–294. doi: 10.1016/0042-6822(79)90484-7. [DOI] [PubMed] [Google Scholar]
- Ben-Porat T., Veach R. A., Ihara S. Localization of the regions of homology between the genomes of herpes simplex virus, type 1, and pseudorabies virus. Virology. 1983 May;127(1):194–204. doi: 10.1016/0042-6822(83)90383-5. [DOI] [PubMed] [Google Scholar]
- EAGLE H. Amino acid metabolism in mammalian cell cultures. Science. 1959 Aug 21;130(3373):432–437. doi: 10.1126/science.130.3373.432. [DOI] [PubMed] [Google Scholar]
- Honess R. W., Watson D. H. Unity and diversity in the herpesviruses. J Gen Virol. 1977 Oct;37(1):15–37. doi: 10.1099/0022-1317-37-1-15. [DOI] [PubMed] [Google Scholar]
- KAPLAN A. S., VATTER A. E. A comparison of herpes simplex and pseudorabies viruses. Virology. 1959 Apr;7(4):394–407. doi: 10.1016/0042-6822(59)90068-6. [DOI] [PubMed] [Google Scholar]
- Ladin B. F., Ihara S., Hampl H., Ben-Porat T. Pathway of assembly of herpesvirus capsids: an analysis using DNA+ temperature-sensitive mutants of pseudorabies virus. Virology. 1982 Jan 30;116(2):544–561. doi: 10.1016/0042-6822(82)90147-7. [DOI] [PubMed] [Google Scholar]
- Lomniczi B. Biological properties of Aujeszky's disease (pseudorabies) virus strains with special regard to interferon production and interferon sensitivity. Arch Gesamte Virusforsch. 1974;44(3):205–214. doi: 10.1007/BF01240608. [DOI] [PubMed] [Google Scholar]
- Paul P. S., Mengeling W. L., Pirtle E. C. Differentiation of pseudorabies (Aujeszky's disease) virus strains by restriction endonuclease analysis. Arch Virol. 1982;73(2):193–198. doi: 10.1007/BF01314727. [DOI] [PubMed] [Google Scholar]
- Platt K. B., Maré C. J., Hinz P. N. Differentiation of vaccine strains and field isolates of pseudorabies (Aujeszky's disease) virus: thermal sensitivity and rabbit virulence markers. Arch Virol. 1979;60(1):13–23. doi: 10.1007/BF01318093. [DOI] [PubMed] [Google Scholar]
- Platt K. B., Maré C. J., Hinz P. N. Differentiation of vaccine strains and field isolates of pseudorabies (Aujeszky's disease) virus: trypsin sensitivity and mouse virulence markers. Arch Virol. 1980;63(2):107–114. doi: 10.1007/BF01320767. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rixon F. J., Ben-Porat T. Structural evolution of the DNA of pseudorabies-defective viral particles. Virology. 1979 Aug;97(1):151–163. doi: 10.1016/0042-6822(79)90381-7. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]