Abstract
Small GTPases of the Ypt/Rab family are involved in the regulation of vesicular transport. Cycling between the GDP- and GTP-bound forms and the accessory proteins that regulate this cycling are thought to be crucial for Ypt/Rab function. Guanine nucleotide exchange factors (GEFs) stimulate both GDP loss and GTP uptake, and GTPase-activating proteins (GAPs) stimulate GTP hydrolysis. Little is known about GEFs and GAPs for Ypt/Rab proteins. In this article we report the identification and initial characterization of two factors that regulate nucleotide cycling by Ypt1p, which is essential for the first two steps of the yeast secretory pathway. The Ypt1p-GEF stimulates GDP release and GTP uptake at least 10-fold and is specific for Ypt1p. Partially purified Ypt1p-GEF can rescue the inhibition caused by the dominant-negative Ypt1p-D124N mutant of in vitro endoplasmic reticulum-to-Golgi transport. This mutant probably blocks transport by inhibiting the GEF, suggesting that we have identified the physiological GEF for Ypt1p. The Ypt1p-GAP stimulates GTP hydrolysis by Ypt1p up to 54-fold, has a higher affinity for the GTP-bound form of Ypt1p than for the GDP-bound form, and is specific to a subgroup of exocytic Ypt proteins. The Ypt1p-GAP activity is not affected by deletion of two genes that encode known Ypt GAPs, GYP7 and GYP1, nor is it influenced by mutations in SEC18, SEC17, or SEC22, genes whose products are involved in vesicle fusion. The GEF and GAP activities for Ypt1p localize to particulate cellular fractions. However, contrary to the predictions of current models, the GEF activity localizes to the fraction that functions as the acceptor in an endoplasmic reticulum-to-Golgi transport assay, whereas the GAP activity cofractionates with markers for the donor. On the basis of our current and previous results, we propose a new model for the role of Ypt/Rab nucleotide cycling and the factors that regulate this process.
INTRODUCTION
Transport of proteins through the secretory pathway involves their orderly progression through a series of membranous compartments. Movement between successive compartments appears to be mediated by vesicles that bud from one compartment and fuse with the next (Jamieson and Palade, 1967; Palade, 1975). Progress has been made in the last few years in understanding the machinery and mechanisms contributing to the directionality and specificity of vesicle targeting and fusion. The Ypt/Rab family of small GTPases has been shown to play a key role in vesicular trafficking in yeast and mammalian cells. These proteins are implicated in the regulation of protein transport through the exocytic, endocytic, and transcytotic pathways (Ferro-Novick and Novick, 1993; Zerial and Stenmark, 1993). In yeast, four members of the Ypt/Rab family have been shown to be essential for regulating the exocytic pathway. Ypt1p is essential for the first two steps of the pathway, endoplasmic reticulum (ER)-to-Golgi and cis-to-medial Golgi transport, mediating the targeting and/or fusion of transport vesicles in the first step (Segev et al., 1988; Rexach and Schekman, 1991; Segev, 1991; Jedd et al., 1995). We recently showed that Ypt31p and Ypt32p are essential for exit from the trans-Golgi compartment (Jedd et al., 1997). Sec4p functions at the final step of the pathway (Novick et al., 1981). It has been suggested that Ypt/Rab proteins act at the different steps of the secretory pathway to ensure the fidelity of vesicular targeting (Bourne, 1988; Novick and Brennwald, 1993; Pfeffer, 1994). However, the specific mechanism by which Ypt/Rab GTPases regulate protein transport and the means by which Ypt/Rab proteins themselves are regulated are still unknown.
Ypt/Rab proteins cycle between GTP- and GDP-bound forms by nucleotide exchange and GTP hydrolysis, reactions that are thought to switch the conformation of these proteins and to determine the partner proteins with which these proteins interact (Balch, 1990; Novick and Brennwald, 1993). The function of this nucleotide cycling by Ypt/Rab proteins has been studied by the use of mutants that are restricted to one or the other of the nucleotide-bound forms. The importance of nucleotide exchange is suggested by studies of Ypt/Rab mutants that are restricted to either the GDP form or the nucleotide-free form. These two types of mutations were shown to be dominant inhibitors of protein transport both in vivo and in vitro (Wagner et al., 1987; Walworth et al., 1989; Gorvel et al., 1991; Bucci et al., 1992; Tisdale et al., 1992; Brondyk et al., 1993; Li and Stahl, 1993; Nuoffer et al., 1994; Riederer et al., 1994). In the case of Ypt1p, the nucleotide-free mutations were shown to act via inhibition of the Ypt1p nucleotide exchanger (Jones et al., 1995). The importance of GTP hydrolysis was studied by the use of Ypt/Rab mutants that are defective in this process. Conflicting evidence suggests either that these mutations have very little effect on the functioning of the Ypt/Rab protein or that they have a dominant inhibitory or stimulatory effect. For example, we have shown that Ypt1p-mediated ER-to-Golgi transport is not affected by a mutation that severely impairs GTP hydrolysis (Richardson et al., 1998), whereas Rab5-mediated endosome fusion was shown to be stimulated by such a mutation (Gorvel et al., 1991; Bucci et al., 1992). We have suggested that GTP hydrolysis is not important for general heterotypic vesicle fusion, which is the basis for vectorial transport (e.g., Ypt1p-mediated transport), but is important for down-regulation of homotypic membrane fusion (e.g., Rab5-mediated fusion) (Richardson et al., 1998).
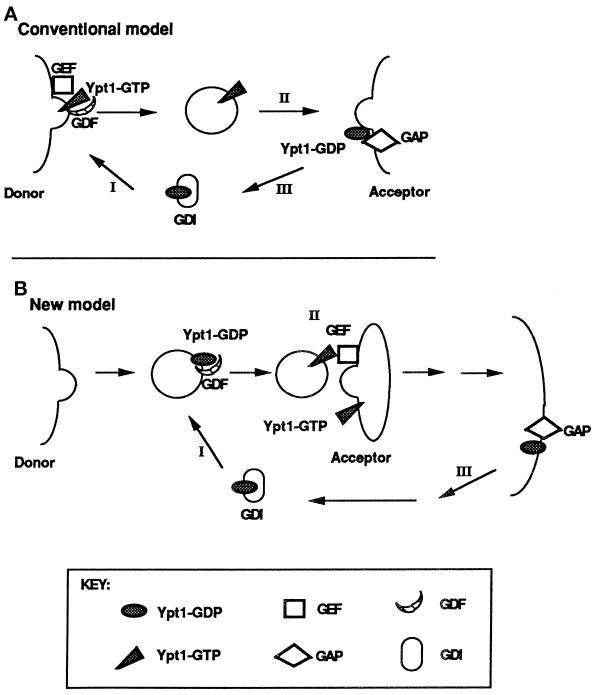
Most small GTPases have slow intrinsic rates of nucleotide exchange and GTP hydrolysis and thus require accessory factors to stimulate these reactions. GDP dissociation and GTP binding are accelerated by guanine nucleotide exchange factors (GEFs), whereas GTP hydrolysis is stimulated by GTPase-activating proteins (GAPs). Current models propose that GEF-stimulated nucleotide exchange occurs at the donor compartment and is coupled to the localization of Ypt/Rab proteins to specific membranes, whereas GAP-stimulated GTP hydrolysis occurs at the acceptor compartment and is essential for vesicle fusion or its timing (Rybin et al., 1996; Novick and Zerial, 1997) (see Figure 9A). However, there is very little evidence to support such models, and in particular, little is known regarding the localization of the regulatory factors.
Figure 9.
Two models for the role of nucleotide cycling and factors that regulate it in Ypt/Rab-mediated vesicular transport, using Ypt1p as an example. (A) Conventional model (Goud and McCaffrey, 1991; Novick and Brennwald, 1993). In addition to GEFs and GAPs, two other factors that influence nucleotide cycling of Ypt/Rab proteins are GDI and GDI-dissociation factor (GDF). GDI is implicated in the recycling of Ypt/Rab proteins between membranes (Araki et al. 1990; Soldati et al., 1993), and GDFs are thought to function as receptors or chaperones for Ypt/Rab proteins (Dirac-Svejstrup et al., 1997). Step I, recruitment of Ypt1p-GDP to the donor membrane by GDF and nucleotide exchange by GEF to yield Ypt1p-GTP are shown; Ypt1p-GTP is present on forming secretory vesicles. Step II, GTP hydrolysis is required or is coupled with fusion of secretory vesicles with the acceptor compartment. Step III, GDI recycles Ypt1p-GDP back to the donor membrane. (B) New model, based on this article and our previous work (Jones et al., 1995; Richardson et al., 1998). Step I, Ypt1p-GDP is recruited to the vesicle (or the donor membrane) by GDF. Step II, nucleotide exchange by GEF is coupled to vesicle fusion with the acceptor compartment. Step III, GTP hydrolysis occurs late in the pathway to generate Ypt1p-GDP, and GDI recycles Ypt1p-GDP for the next cycle. The important features that distinguish this model from the conventional model are the major role suggested for nucleotide exchange and the factor that mediates it (GEF), the minor role of GTP hydrolysis and GAP not in vesicle fusion but in Ypt1p recycling, and the suggested localization of these regulators. If GAP localizes to the plasma membrane, as shown here, it might have a role in GDI-mediated Ypt/Rab protein recycling (which is not required for Ypt1p function). In ypt1-Q67L mutant cells, when GTP hydrolysis is defective, Ypt1p-GTP might be recycled via a GDI-independent mechanism. If GAP localizes to the ER, it might be there to stimulate GTP hydrolysis by Ypt1p to allow better interaction with the GEF in the next cycle (see DISCUSSION).
Although GEFs and GAPs that act on members of the Ras and Rho families of small GTPases have been extensively characterized (McCormick, 1990; Boguski and McCormick, 1993), less is known about GEFs and GAPs for Ypt/Rab GTPases. To date, three genes encoding GEFs for Ypt/Rab proteins have been identified, one in yeast for Sec4p and two in mammalian cells for Rab3 and Rab5 (Horiuchi et al., 1997; Wada et al., 1997; Walch-Solimena et al., 1997). We have shown that Ypt1p mutant proteins inhibit Ypt1p-GEF activity and are also potent inhibitors of ER-to-Golgi transport in vivo and in vitro, implying that nucleotide exchange stimulated by this GEF is essential for Ypt1p function (Jones et al., 1995). In agreement with this result, the Sec4p-GEF Sec2p is essential for viability (Nair et al., 1990). Recently, a number of Ypt/Rab GAP genes have been cloned; these are the mammalian tuberin and Rab3 GAP and the yeast GYP6, GYP7, and GYP1 (Strom et al., 1993; Vollmer and Gallwitz, 1995; Fukui et al., 1997; Xiao et al., 1997; Du et al., 1998). There are several open questions regarding the mechanism of action, the localization, and the specificity of GEFs and GAPs for Ypt/Rab proteins. Identifying and characterizing these regulatory proteins should help resolve these issues. Because there is very little homology shared between the different known GEFs or GAPs for Ypt/Rab proteins, we decided to take a biochemical approach to identify factors that regulate the nucleotide cycling of Ypt1p.
In this article we describe both a GEF and a GAP for Ypt1p in yeast. The novel GEF activity stimulates both GDP release and GTP uptake by Ypt1p but not by other exocytic Ypt proteins. Contrary to the predictions of current models, which assign GEF function to the donor compartment, the Ypt1p-GEF activity is highly enriched in the P100 (100,000 × g pellet) fraction, which functions as the acceptor in a Ypt1p-mediated ER-to-Golgi cell-free transport assay. The GAP activity for Ypt1p interacts with both Ypt1p and Sec4p but not with Ypt31p or Ypt32p, indicating substrate specificity. The novel GAP activity characterized here is highly enriched in the P12 fraction, which functions as the donor in the ER-to -Golgi transport assay, even though current models would predict that Ypt1p-GAP activity should be enriched in the acceptor compartment. On the basis of the localization of these two Ypt1p regulators and our previous results (Jones et al., 1995; Richardson et al., 1998), we propose a new model for the role of nucleotide cycling and associated regulatory factors in Ypt1-mediated vesicular transport.
MATERIALS AND METHODS
Yeast Strains and Materials
The yeast strains used in this study are listed in Table 1. GYP1 and GYP7 were deleted sequentially in NSY116 using PCR products amplified from the following templates: kanMX4 (Wach et al., 1994), for disruption of the entire open reading frame of GYP1 with the kanr gene of Escherichia coli using the upstream primer 5′-ACCAA TACCG ACCAC TTAAT AAAAG TAACC ATATA CAGCT GAAGC TTCGT ACGCT-3′ and the downstream primer 5′-TACAT ACTAT ACAGT AAGTA AAATG AATAG GTCCG GCATA GGCCA CTAGT GGATCTG-3′, and pRS303 (Sikorski and Hieter, 1989), for disruption of the entire open reading frame of GYP7 with the HIS3 gene using the upstream primer 5′-AAAGT TCTAC AAGAG TCATT CATAC ATCCC CTGCT CTTGG CCTCC TCTAG-3′ and the downstream primer 5′-TATTC AATAT GTAAA GTTCC GTTTC TATTT ACCTC GTTCA GAATG ACACG-3′. Yeast strains were grown in rich medium (YEPD; 1% yeast extract, 2% bactopeptone, 2% dextrose) (Rose et al., 1988). All chemical reagents were purchased from Sigma Chemical (St. Louis, MO), unless otherwise noted. All DNA restriction endonucleases were from New England BioLabs (Beverly, MA) or Boehringer Mannheim (Indianapolis, IN). Taq DNA polymerase was from Life Technologies–Bethesda Research Laboratories (Gaithersburg, MD).
Table 1.
Yeast strains
| Strain | Genotype | Source |
|---|---|---|
| GPY60 | MATa, ura3-52, trp1, leu2, his4, pep4∷URA3 | G. Payne, UCLA |
| NSY116 | MATa, ade2, his3-Δ200, leu2-3,112, ura3-52 | D. Botstein, Stanford |
| NSY125 | MATa, his4-539am, lys2-801am, ura3-52 | D. Botstein, Stanford |
| NY929 | dss4∷URA3 | P. Novick, Yale |
| NY931 | DSS4 (wt) | P. Novick, Yale |
| NY431 | MATa, ura3-52, sec18-1 | P. Novick, Yale |
| NY418 | MATa, ura3-52, sec17-1 | P. Novick, Yale |
| NY426 | MATa, ura3-52, sec22-3 | P. Novick, Yale |
| NY13 | MATa, ura3-52 | P. Novick, Yale |
| NSY418 | NSY116 gyp1∷kanr | This study |
| NSY420 | NSY116 gyp1∷kanr gyp7∷HIS3 | This study |
Expression and Purification of Ypt Proteins
Construction of plasmids for the expression of GST-Ypt1p (pNS361), GST-Ypt1-D124N (pNS363), GST-Ypt31p (pNS210), GST-Ypt32p (pNS211), and GST-Sec4p (pNS212) is described elsewhere (Jones et al., 1995; Jedd et al., 1997). To construct the GST-Ypt1-T40K fusion protein pNS240, we used a pGEM3Zf(−) plasmid that contains the ypt1-T40K allele (Jedd et al., 1995) as a template for PCR with the upstream primer 5′-CCT GGG GAT CCA TGA ATA GCG AGT ACG ATT ACC TGT TCA AAC TGC TGT TGA TCG GG-3′ (which creates a BamHI site just upstream of the initiator methionine) and the downstream primer 5′-GGG CCC GGA TCC GAT AAG GAA GAA TG-3′. The PCR fragment was cut with BamHI and cloned into the BamHI site of pGEX-KG (Guan and Dixon, 1991) to create pNS214. GST fusion proteins were purified as described elsewhere (Jones et al., 1995) and were dialyzed against Buffer 88 [250 mM sorbitol, 20 mM HEPES, pH 6.8, 150 mM KOAc, 5 mM Mg(OAc)2] (Baker et al., 1988). Thrombin cleavage of these GST fusion proteins yields proteins with a two amino acid extension (Gly-Ser) at the amino termini.
GST-Ypt1p was used as a substrate for the geranylgeranyl transferase reaction. Geranylgeranyl transferase activity was reconstituted in vitro as described (Jiang et al., 1995). The prenylated and unprenylated forms of GST-Ypt1p were then separated by Triton X-114 phase partitioning (Bordier, 1981). The two phases were then mixed with a 50% slurry of glutathione agarose beads, and Ypt1p was purified after thrombin cleavage as described above.
Yeast Cell Extract Preparation
Crude extracts were prepared from spheroplasts and fractionated as described (Baker et al., 1990; Wuestehube and Schekman, 1992). For comparison of GEF activities in the various fractions, supernatant fractions (both S100 [100,000 × g supernatant] and S12) were dialyzed in Buffer 88 overnight. To extract endogenous Ypt1p from the particulate fraction, we treated P100 (∼10 mg of protein per milliliter) with 1% Triton X-100 or 1% n-octylglucoside for 1 h on ice. The mixture was centrifuged at 100,000 × g for 1 h, and the pellet was resuspended to the starting volume in Buffer 88 and recentrifuged. The pellet, which contained >80% of the GEF activity, was resuspended to the initial volume in Buffer 88 to generate a fraction referred to as detergent-extracted P100 (Det-P100). This fraction was further extracted with 0.5 M NaCl for 1 h on ice. A final centrifugation at 100,000 × g for 1 h yielded a supernatant, termed solubilized GEF, that contained ∼75% of the total exchange activity in the original P100 fraction.
Guanine Nucleotide Exchange Assays
[3H]GDP Release Assays.
Ten picomoles of Ypt1p were preloaded by incubating with 20 pmol of [5′,8′-3H]GDP (31.7 Ci/mmol; New England Nuclear, Boston, MA) in preload buffer (20 mM HEPES, pH 7.2, 20 mM KOAc, 1 mM DTT, 5 mM EDTA) for 10 min at 30°C. At the end of the incubation, samples were placed on ice, and MgCl2 was added to 10 mM. [3H]GDP remained stably bound to the Ypt1p for at least 1 h. The cell fractions containing the GEF activity for Ypt1p, or bovine serum albumin (BSA) as a control, were diluted into reaction buffer [20 mM HEPES, pH 7.2, 5 mM Mg(OAc)2, 0.75 mM GTP, 0.75 mM GDP, 1 mM DTT], and exchange reactions were initiated by the addition of Ypt1p-[3H]GDP. The reaction volume was 50 μl. Incubations were performed at 30°C for varying periods of time, as noted. To monitor the release of [3H]GDP from Ypt1p by filtration, we applied 5 μl samples to wet nitrocellulose filters (BA85; Schleicher & Schuell, Keene, NH) by pipetting samples into 3 ml of ice-cold wash buffer (20 mM Tris-HCl, pH 7.5, 20 mM NaCl, 5 mM MgCl2, 1 mM DTT). Filters were washed twice more with 3 ml of the same buffer and were counted with Ready Protein+ (Beckman, Fullerton, CA) or Filtron-X (National Diagnostics, Atlanta, GA) scintillation fluid. In all experiments, initial values were ∼8–10 × 103 dpm per 5 μl sample.
Ras2 protein, expressed and purified from E. coli (a gift from K. York and J. Broach), was preloaded with [3H]GDP exactly as described for Ypt1p above. His6-Sec4 protein (a gift from P. Novick) was preloaded with [3H]GDP as described (Kabcenell et al., 1990).
[32P]GTP Uptake Assays.
Bacterially expressed Ypt1p was preloaded as described above but with nonradioactive GDP. The preloaded Ypt1p was added together with cell fractions containing the GEF activity for Ypt1p or ovalbumin (as a control) to reaction buffer lacking guanine nucleotides. ATP was added to 1 mM to prevent nonspecific hydrolysis of [32P]GTP. [α-32P]GTP (3000 Ci/mmol; Amersham, Arlington Heights, IL) was diluted to a specific activity of 75 Ci/mmol. Exchange reactions were initiated by the addition of 100 pmol of GTP to a 50 μl reaction mixture containing 10 pmol of Ypt1p and various amounts of exchange-containing cellular fractions. Samples of 5 μl were removed at intervals, and the amount of [α-32P]GTP bound to Ypt1p was determined by quantitative immunoprecipitation with 2.5 μg of anti-Ypt1p immunoglobulin G purified from polyclonal serum (Segev et al., 1988) and with 20 μl of protein A sepharose (Zymed, San Francisco, CA). Immunoprecipitation was necessary to distinguish between Ypt1p and other GTP-binding proteins present in the extract. Immunoprecipitation was performed for 2 h at 4°C in reaction buffer (see above) plus 150 mM KOAc, 1% Triton X-100, and 150 μM GDP and GTP. At the end of the incubation, immune complexes were washed five times with 1 ml of wash buffer (see [3H]GDP Release Assay) plus 1% Triton X-100. Washed immune complexes were boiled for 5 min in the presence of 2% SDS, and the amount of bound radioactivity was determined by liquid scintillation counting. Immunoprecipitation under these conditions was quantitative as demonstrated by comparison of immunoprecipitation with filtration of Ypt1p-[3H]GDP from reaction mixtures identical to those above. Specificity of immunoprecipitation was demonstrated with preimmune and non-Ypt1p-specific antisera. Values for GTP uptake by trace amounts of endogenous Ypt1p in the extracts were determined by performing the reactions in the absence of bacterially produced Ypt1p. These values (equal to <5% of those for GTP uptake by the bacterially produced substrate) were subtracted from the measurements of stimulated exchange. Further background measurements were determined by sampling at the initiation of the reaction (time = 0). These values (also <5% of those for GTP uptake by the bacterially produced substrate) were subtracted from the measurements of stimulated exchange as well.
Partial Purification of GEF Activity
Gel Filtration.
Twenty-one milliliters of solubilized GEF (n-octylglucoside extracted) was prepared from 20,000 OD600 units of cells (strain GPY60) as described above. The solubilized GEF was applied to a 2.5 × 92.5 cm column of Sephacryl S-300 HR equilibrated in Buffer 88 in three separate runs using 7 ml (at 1.8 mg/ml) per run. The column was eluted at 1.4 ml/min, and 2.5 ml fractions were collected. GEF activity was measured by performing the [3H]GDP release assay on alternating fractions. Peak fractions were collected into two pools: a high-specific activity pool A and a low-specific activity pool B. The specific activities of the pools were assayed using both the [3H]GDP release assay and the [32P]GTP uptake assay. Each pool of fractions was concentrated ∼30-fold in a Centriprep-30 unit.
Hydroxyapatite Chromatography.
Concentrated material (2.3–2.9 ml) from the Sephacryl S-300 HR column was applied to a 1 × 5 cm ceramic hydroxyapatite (HAP) (Bio-Rad, Richmond, CA) column in Buffer 88 with 0.5 M NaCl and 10 mM KPO4, pH 6.8 (HAP wash buffer). The column was run at 30 cm/h (∼23.6 ml/h), and 1 ml fractions were collected. The column was washed with 1–3 column volumes of HAP wash buffer until the absorbance returned to baseline. Bound protein was then eluted with a linear gradient from 10 to 200 mM KPO4 in Buffer 88 followed by one to three column volumes of 400 mM KPO4, pH 6.8. Samples were dialyzed against Buffer 88 in a microdialysis unit (Life Technologies–Bethesda Research Laboratories) for 2 h before assaying for guanine nucleotide exchange activity by the GDP loss assay. Fractions with the greatest GDP release activity were pooled and concentrated in a Centriprep-30 unit.
ER-to-Golgi Transport Assay
An in vitro ER-to-Golgi transport system using permeabilized yeast cells as the donor compartment was used for the experiments that demonstrate rescue by purified Ypt1p-GEF of Ypt1p-D124N inhibition of ER-to-Golgi transport (Ruohola et al., 1988).
GTP Hydrolysis Assays
Three methods were used to measure GTP hydrolysis: charcoal binding, TLC, or filtration through nitrocellulose. For the charcoal-binding assay (Brandt et al., 1983; Higashijima et al., 1987), Ypt1p (10 μM) was preloaded with 5 μl of [γ-32P]GTP (2000 Ci/mmol; Amersham) in preload buffer (20 mM HEPES, pH 7.2, 20 mM KOAc, 5 mM EDTA, 0.5 mg/ml BSA, 1 mM DTT) for 15 min at 30°C in a 10 μl volume. Preload reactions were diluted with 50 μl of reaction buffer [20 mM HEPES, pH 7.2, 5 mM Mg(OAc)2, 300 mM sorbitol, 1 mM DTT] plus 0.5 mg/ml BSA, and unbound nucleotide was removed at 4°C with two successive acrylamide spin columns (BioSpin6; Bio-Rad) equilibrated with reaction buffer plus 0.5 mg/ml BSA. The volume of the flow-through was adjusted to 250 μl with reaction buffer plus BSA to give a final Ypt1p concentration of 40 nM. GAP-stimulated hydrolysis was measured by incubating 2 nM preloaded Ypt1p with the indicated quantities of the indicated subcellular fractions in reaction buffer plus 1 mM each GTP, GDP, and ATP at 30°C. Intrinsic GTP hydrolysis was measured by substituting BSA (a nonspecific protein) for the subcellular fraction. Aliquots were removed at the indicated time points and processed as described (Richardson et al., 1998). When TLC was used to measure GTP hydrolysis, Ypt1p was preloaded as described above but with [32P]GTP labeled at the α position (3000 Ci/mmol; Amersham), and unbound nucleotide was removed as described above. GTP hydrolysis reactions were performed as described above. Aliquots were removed at the indicated times, and reactions were stopped by addition of an equal volume of stop buffer (50 mM Tris-HCl, pH 7.5, 40 mM EDTA, 2% SDS). Samples were heated at 65°C for 5 min. Aliquots (5 μl) were spotted on polyethyleneimine-cellulose TLC plates, and nucleotides were resolved by developing plates in water followed by 1 M LiCl (Tanaka et al., 1991). Detection and quantitation of radioactivity were performed with a radioanalytic imager (QuantProbe 3.0; Ambis Systems, San Diego, CA). When the filtration method was used to assay GTP hydrolysis, Ypt1p was preloaded with [γ-32P]GTP as described above, except that spin columns were not used to separate unbound nucleotide. GTP hydrolysis reactions were performed as described above for the charcoal assay, except that aliquots removed at the indicated time points were processed as for the GDP release assay (see above).
Extraction of GAP Activity
Yeast extracts were prepared and fractionated as described (Baker et al., 1990; Wuestehube and Schekman, 1992). The P12 fraction was used as the source of GAP activity. For trypsin extractions, digestion was performed on P12 at 10 mg/ml with trypsin at 0.1 mg/ml for 1 h on ice and then stopped by the addition of soybean trypsin inhibitor at 0.2 mg/ml. As a control, trypsin inhibitor was added before trypsin, and the mixture was incubated for 1 h on ice. An aliquot was removed and saved as “total.” The remainder was centrifuged at 12,000 × g. The supernatant was removed and saved. The pellet was resuspended in Buffer 88 to the original volume before centrifugation. Equivalent volumes of total, supernatant, and pellet fractions of extracted material were incubated with 2 nM Ypt1p preloaded with [32P]GTP as described above. GTP hydrolysis was determined by the charcoal-binding assay as described above. For detergent inhibition curves, the P12 or the trypsin-extracted activity was used as a source of GAP activity. These fractions were incubated with the indicated amounts of detergent and 2 nM Ypt1p preloaded with [32P]GTP as described above. Aliquots were removed at the indicated time periods and processed for quantification by the charcoal-binding assay or by the filtration assay as indicated. To test geranylgeranylated Ypt1p as a substrate, we preloaded 1.2 pmol of the aqueous or detergent phase of Triton X-114–partitioned Ypt1p (see below) with [γ-32P]GTP (2000 Ci/mmol) as described above; 1.2 nM preloaded Ypt1 proteins were tested for responsiveness to trypsin-extracted GAP in the absence or presence of detergent. GTP hydrolysis was monitored by the filtration assay as described above.
Competition of Ypt1p-GAP Activity
For competition experiments, Ypt1p was preloaded with [γ-32P]GTP as described above. Competitor protein (90 μM) was preloaded for 15 min at 30°C with 180 μM unlabeled nucleotide, either GDP, GTP, or GppNHp, in a 20 μl reaction that contained 20 mM HEPES, pH 7.2, 20 mM KOAc, 5 mM EDTA, 0.5 mg/ml BSA, and 1 mM DTT. After preloading, 0.1 M Mg(OAc)2 was added to a final concentration of 5 mM. For the no-competitor control, a mock preload reaction was performed that contained nucleotide but no competitor protein. Labeled Ypt1p (2 nM) was mixed with the indicated quantities of cold competitors (or the mock preload control) before the addition of the P12 GAP activity. Reactions were initiated by the addition of 0.75 mg/ml of P12, and incubation was at 30°C. Aliquots were removed at 0, 10, 20, and 30 min and were processed for quantification by the charcoal-binding assay as described above.
RESULTS
Identification of Ypt1p-GEF
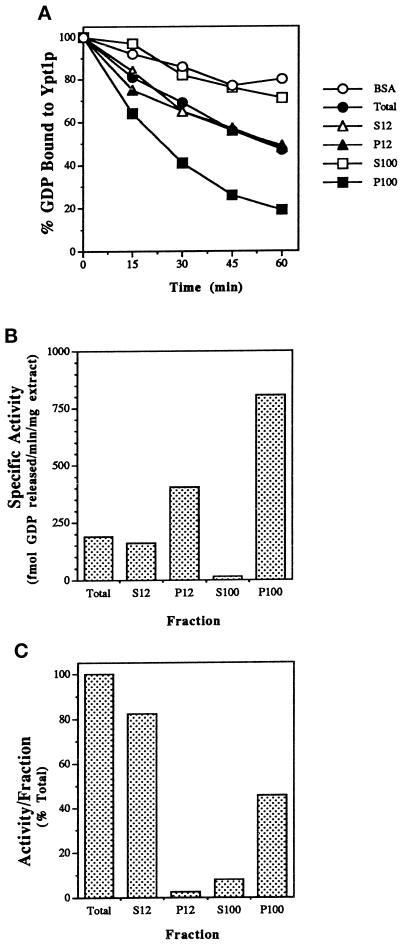
To identify a GEF activity for Ypt1p in yeast cells, we first assayed the ability of cell lysates to stimulate release of GDP from Ypt1p. Crude extracts of Saccharomyces cerevisiae strain GPY60 and fractions obtained by differential centrifugation were incubated with recombinant Ypt1p preloaded with [3H]GDP. Yeast cell extracts stimulated GDP release from Ypt1p above the intrinsic rate in a time- (Figure 1A) and concentration-dependent manner. Because we later show that this activity also promotes GTP uptake specifically by Ypt1p (see below), we refer to this activity as Ypt1p-GEF. Comparing equal amounts of protein from different cell fractions revealed that the P100 fraction was enriched in GEF activity, whereas there was no detectable or little activity in the S100, S12, and the P12 fractions, respectively (Figure 1A). The highest specific activity of Ypt1p-GEF was found in the P100 fraction (∼three- to fivefold enrichment relative to the crude lysate; Figure 1B). Approximately one-half of the Ypt1-GEF activity present in the total extract was recovered in the P100 fraction, whereas very little (∼2.5%) was found in the P12 fraction (Figure 1C), suggesting that the Ypt1p-GEF associates with a light particulate cellular compartment.
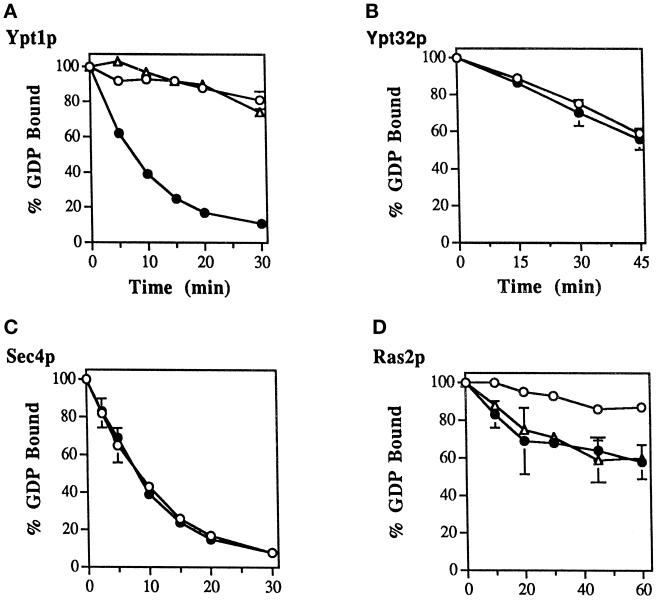
Figure 1.
Identification of a GDP release-stimulating activity for Ypt1p in the P100 fraction of yeast cells. (A) Time course of GDP release from Ypt1p in the presence of fractionated yeast extracts is shown. Ypt1p-[3H]GDP was incubated in the presence of cell extract or BSA at 1 mg/ml. Values represent the percent of label retained at the indicated time, relative to that at the beginning of the incubation. Fractions are the total extract (closed circles), S12 (open triangles), P12 (closed triangles), S100 (open squares), P100 (closed squares), and no extract (BSA; open circles). (B) Ypt1p-GEF is enriched in the P100 fraction. To determine the specific activity of GDP release in the different fractions, we varied the fraction concentration from 0.2 to 1 mg/ml in the GDP release reaction and withdrew samples at 3 min intervals over 15 min. Conditions were selected under which protein and time were limiting, and rates of GDP release were determined from the linear portion of the curve. (C) The P100 fraction contains approximately one-half of the Ypt1p-GEF activity in the total extract. The portion of activity per fraction was calculated as the product of the specific activity (Figure 1B) and amount (milligrams) of protein in the fractions obtained from ∼5000 OD600 units of cells. Results shown in this figure are representative of at least three experiments, and values from similar experiments agreed to within 5%.
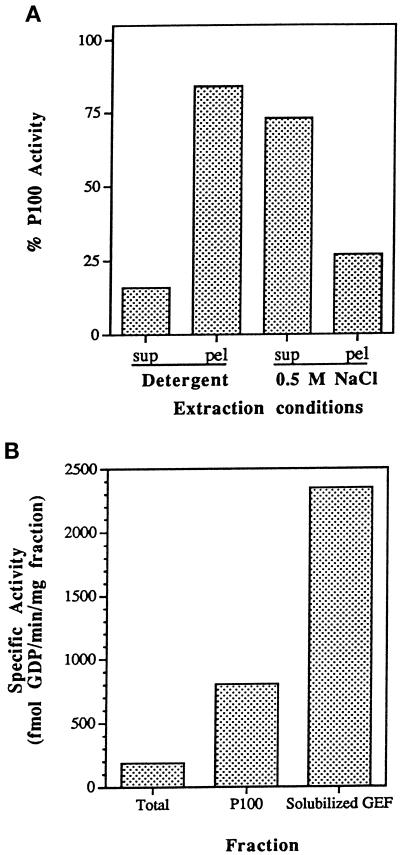
To characterize the association of the Ypt1p-GEF activity with this particulate compartment, we extracted the P100 fraction with detergents and NaCl. Although Ypt1p can be extracted from the P100 fraction by nonionic detergent treatment, the Ypt1p-GEF activity was resistant to detergent treatment. Instead, ∼75% of the GEF activity was liberated by 0.5 M NaCl (Figure 2A). Increasing the NaCl concentration beyond 0.5 M did not significantly increase the recovery of Ypt1p-GEF activity. The ability of the Ypt1p-GEF to be extracted by salt but not by detergent is consistent with an association with membranes or cytoskeletal elements by electrostatic interactions.
Figure 2.
Association of the Ypt1p-GEF with the P100 fraction and preparation of the solubilized GEF fraction. (A) Extraction of Ypt1p-GEF from the P100 fraction by salt but not by detergents is shown. P100 was treated with 1% Triton X-100, 30 mM n-octylglucoside, or 0.5 M NaCl. Equal volumes of the P100 fraction and the treated pellets and supernatants were diluted at least fivefold into the reaction mixture (NaCl concentrations were at or below 100 mM, a concentration that does not affect intrinsic or stimulated exchange) and were assayed for their ability to stimulate release of [3H]GDP from Ypt1p as described. Addition of 1% Triton X-100, 30 mM n-octylglucoside, or 0.5 M NaCl to the unfractionated P100 had no effect on its activity. Extracts were prepared at least twice and were assayed at least twice per extraction with equivalent results. Extraction with 1% Triton X-100 and 30 mM n-octylglucoside gave identical results and are presented as Detergent. (B) Sequential extraction of the P100 fraction with Triton X-100 and NaCl yields a soluble Ypt1p-GEF activity and an additional fourfold increase in specific activity. The solubilized GEF fraction is the S100 fraction from sequential extraction of the P100 fraction with 1% Triton X-100, followed by extraction of the resulting pellet with 0.5 M NaCl. The total enrichment of the Ypt1p-GEF activity in the solubilized fraction relative to the crude cell extract is 16- to 20-fold. Similar results were obtained with four independent preparations.
Salt extraction of Det-P100 yielded solubilized Ypt1p-GEF activity free of Ypt1p. The solubilized GEF activity is enriched 4-fold relative to the P100 fraction and ∼16- to 20-fold relative to the total cell extract (Figure 2B). The solubilized GEF was proteinaceous because it was sensitive to protease or heat but not to RNase or DNase. The Det-P100 fraction was used for experiments in which high concentrations of the exchange activity were needed, whereas the salt extract was used for further purification of the Ypt1p-GEF (see below; the solubilized GEF fraction could not be used in high concentrations because salt interferes with the exchange reaction and salt removal caused protein precipitation).
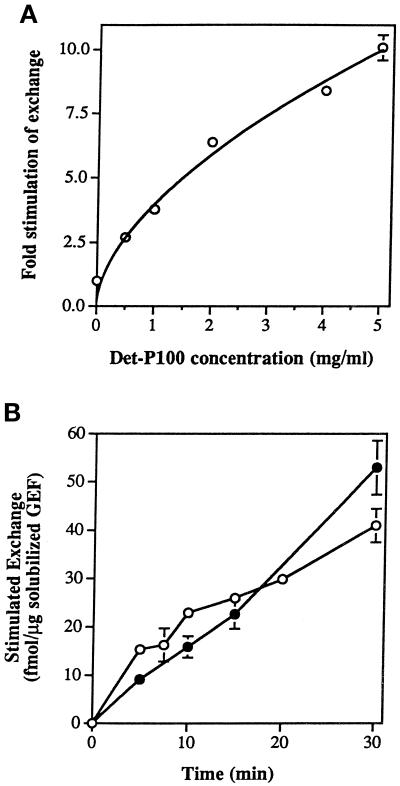
To confirm that this Ypt1p-GEF activity corresponds to a genuine exchange activity, we used the partially purified exchange factor to assay stimulation of GTP binding by recombinant Ypt1p. Recombinant Ypt1p was preloaded with nonradioactive GDP; then the binding of [α-32P]GTP was measured in the presence of either the Det-P100 fraction or the solubilized GEF. The intrinsic rates of GDP release and GTP uptake of Ypt1p were very similar (0.76 ± 0.13 and 0.87 ± 0.12 fmol/min per picomole of Ypt1p, respectively). Adding increasing amounts of the Det-P100 fraction accelerated GTP uptake by Ypt1p to a maximum 10-fold stimulation at 5 mg/ml (Figure 3A). When the solubilized GEF was used as a source of exchange activity, both GTP uptake and GDP release rates were also linear with respect to concentration and time (Figure 3B) and were essentially identical. Thus, the activity that we identified on the yeast cell membranes and that was solubilized and partially purified stimulates the exchange of Ypt1p-bound GDP for GTP at least 10-fold.
Figure 3.
Stimulation of GTP uptake by Ypt1p-GEF. (A) Increasing concentrations of the Det-P100 (Triton X-100–extracted) fraction result in increasing stimulation of GTP uptake by Ypt1p. Ten picomoles of Ypt1p preloaded with nonradioactive GDP were incubated with varying amounts of the Det-P100 fraction in reaction mixtures containing [α-32P]GTP. Reactions were performed for 15 min, and the amount of isotope bound to Ypt1p was determined by immunoprecipitation of samples removed at 2–5 min intervals. The results represent the averages of two independent experiments. Error bars represent SEM. (B) Partially purified solubilized Ypt1p-GEF stimulates GTP uptake as well as GDP release. Ten picomoles of Ypt1p preloaded with either nonradioactive or [3H]GDP were incubated with varying amounts of the solubilized GEF fraction in reaction mixtures containing [α-32P]GTP or nonradioactive GTP, respectively. Samples were taken at intervals, and the amount of isotope bound to Ypt1p was determined by immunoprecipitation ([32P]GTP uptake assay; closed circles) or nitrocellulose filtration ([3H]GDP release assay; open circles). Intrinsic rates for GDP release and GTP uptake, measured in the presence of BSA or ovalbumin, respectively, were subtracted. The results represent the averages of two to nine determinations per point. Error bars represent SEM.
A candidate for Ypt1p-GEF activity is Dss4p, a suggested GDP-release factor for the closely related Sec4p. Previous work demonstrated that purified recombinant Dss4p was capable of stimulating release of GDP from Ypt1p by 2.5-fold above the intrinsic rate (Moya et al., 1993). We compared the stimulation of GDP release for Ypt1p by yeast cell extracts prepared from a wild-type strain and a strain in which the DSS4 gene was deleted (Moya et al., 1993). P100 fractions prepared from both yeast strains yielded equivalent results and stimulated GDP release from Ypt1p by 4- to 5-fold (Table 2A). Therefore, the Ypt1p-GEF activity that we have identified is not Dss4p.
Table 2.
Activity of Ypt1p regulators in extracts of mutant strains
| A | Yeast strain | % GDP release |
|---|---|---|
| DSS4 wild-type (NY 931) | 40 | |
| Δdss4-(NY929) | 42 | |
| Wild-type (GPY60) | 39 | |
| B | Yeast strain | % GAP activity in the P12 |
| Wild type (NSY116) | 100 | |
| ΔGYP1 ΔGYP7 (NSY420) | 93.3 | |
| C | Yeast strain | GAP activity in the P12 |
| Wild type (NY13) | + | |
| sec22-3 (NY426) | + | |
| sec18-1 (NY431) | + | |
| sec17-1 (NY418) | + |
(A) The Ypt1p-GEF is not Dss4p. [3H]GDP release assays were performed in the presence of 1 mg/ml P100 fraction prepared from strains carrying either a dss4 deletion (NY929) or the wild-type DSS4 allele (NY931). Both strains yielded equivalent exchange activities, similar to the activity observed with the wild-type strain (GPY60) used for this study. Results show percent stimulated GDP release after 15 min of incubation and are representative of two experiments with results agreeing to within 5%. For comparison, <5% of the bound GDP was released after a 15-min incubation in the absence of cell extract. (B) Deletion of GYP1 and GYP7 does not affect the amount of GAP activity in the P12. Two nanomolar Ypt1p preloaded with [γ-32P]GTP was incubated at 30°C for 30 min with 0.5 mg/ml P12 generated from fractionation of yeast strains containing either the wild-type alleles of GYP1 and GYP7 or deletions of these genes. BSA (1 mg/ml) was included in the measurement of intrinsic hydrolysis. GTP hydrolysis was measured by the charcoal assay, and results are expressed as percent of the GAP activity in wild-type P12. Data shown are an average of two independent experiments, with results agreeing to within 5%. (C) The Ypt1p-GAP activity is not influenced by mutations in genes whose products are involved in vesicle fusion: SEC18, SEC17, or SEC22. GAP assays were performed as above, using the TLC method. Both the P12 and the S12 fractions were tested. Mutant cells showed the same distributions and levels of Ypt1p-GAP as wild-type cells.
Substrate Specificity of Ypt1p-GEF
To determine the specificity of the Ypt1p-GEF activity, we assayed the ability of the Det-P100 fraction to stimulate the exchange of guanine nucleotides bound to Ras2p, Ypt32p, or Sec4p. Under conditions in which the Det-P100 fraction stimulated GDP release by ∼12-fold from Ypt1p, there was an ∼4- to 6-fold stimulation of GDP release from Ras2p but no effect on the other two proteins (Figure 4). To determine whether the exchange factors for Ypt1p and Ras2p are distinct, we used a mutant Ypt1p, D124N, that inhibits the Ypt1p-GEF (Jones et al., 1995). Stimulation of nucleotide exchange by the Ypt1p was abolished by the mutant protein, while the stimulation of GDP release from Ras2p was unaffected (Figure 4, compare panels A and D), indicating that the factors that stimulate release of GDP from Ypt1p and Ras2p are distinct. Thus, the exchange activity present in the Det-P100 cellular fraction seems to be specific for Ypt1p and does not act on the other exocytic Ypt proteins. Partially purified Ypt1p-GEF (see HAP peaks A and B below) also failed to stimulate nucleotide exchange for Ypt31p and Ypt32p (our unpublished observations).
Figure 4.
The Ypt1p-GEF activity present in the Det-P100 (Triton X-100– or n-octylglucoside–extracted) fraction does not act on Ypt32p or Sec4p and is different from the exchange factor for Ras2p. Ten picomoles of Ypt1p-[3H]GDP (A), Ypt32p-[3H]GDP (B), Sec4p-[3H]GDP (C), or Ras2p-[3H]GDP (D) were incubated in the presence of the Det-P100 fraction (closed circles; 5 mg/ml, except 2 mg/ml Ypt32p) or BSA (open circles), and the stimulated and intrinsic rates of [3H]GDP release were determined by sampling at the times indicated. Including the Det-P100 fraction at 1 mg/ml resulted in ∼6-fold stimulation of GDP release from Ypt1p (our unpublished observations), but no stimulation was observed for Ypt32p at concentrations of Det-P100 up to 2 mg/ml. Including 5 mg/ml of Det-P100 fraction resulted in ∼12-fold stimulation of GDP-release from Ypt1p, no stimulation for Sec4, and approximately ∼4- to 6-fold stimulation for Ras2p. The stimulated GDP release by Ypt1p can be completely inhibited by Ypt1-D124N dominant-mutant protein (1.5 μM). However, the GEF activity for Ras2p present in this fraction is not affected by this Ypt1 mutant protein (A and D; open triangles). The results represent the averages of two experiments with duplicates for each time point. Error bars represent SEM.
At least one Rab GEF was reported to act preferentially on the prenylated form of Rab relative to the unprenylated form (Miyazaki et al., 1994). However, prenylation of the recombinant Ypt1p had no effect on the ability of the Ypt1p-GEF (P100 fraction) to stimulate nucleotide exchange (our unpublished observations).
Partial Purification of Ypt1p-GEF
To purify Ypt1p-GEF further, the P100 fraction was extracted with 1% n-octylglucoside (because it is more readily removed by dialysis than is Triton X-100), and the residual membranes were extracted with 0.5 M NaCl to generate a solubilized GEF fraction that lacks Ypt1p. Sephacryl S-300 HR gel filtration partially resolved two peaks of activity with apparent molecular sizes of ∼400–450 and ∼200 kDa (Figure 5A). The two peaks were collected as individual pools, termed pool A and pool B. Approximately 17–31% of the starting Ypt1p-GEF activity was recovered in pool A with a purification of ∼3- to 5-fold. Approximately 14–17% of the starting Ypt1p-GEF activity was recovered in pool B with a purification of ∼1.5- to 1.7-fold. The apparent molecular sizes of the Ypt1p-GEF peaks were the same regardless of whether n-octylglucoside or Triton X-100 was used in the extract preparation and did not change if the solubilized GEF fraction was dialyzed to remove residual n-octylglucoside before chromatography. Thus the apparent molecular sizes derived from gel filtration are not attributable to protein inclusion in detergent micelles. Each S-300 pool was loaded separately onto a ceramic HAP column and eluted with a 10–200 mM potassium phosphate gradient. Pool A exhibited a peak of activity at ∼105 mM phosphate, whereas pool B showed a single peak of activity at ∼80 mM phosphate (Figure 5B). We verified that these partially purified activities, which stimulate GDP release from Ypt1p, are GEFs by assaying both pools for stimulation of GTP uptake. The specific activities measured by GDP release and GTP uptake were similar (within a factor of 2) during each step of the purification procedure. The purification factor after HAP chromatography was ∼120 for peak A and ∼37 for peak B. It is not clear whether the two peaks represent two different Ypt1p-GEFs or the same GEF in two different protein complexes.
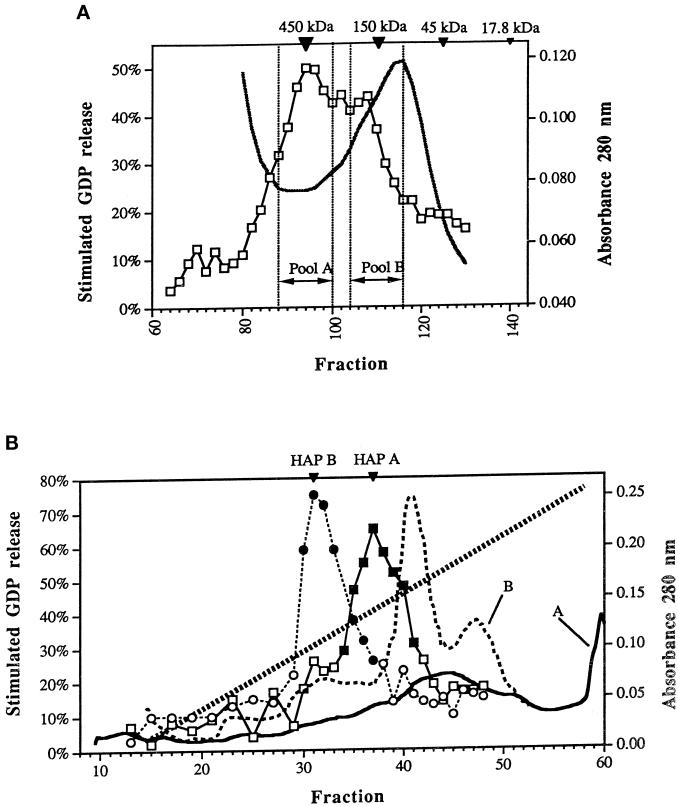
Figure 5.
Partial purification of the Ypt1p-GEF. (A) Gel filtration column. Two partially overlapping Ypt1p-GEF peaks are collected into distinct pools. The solubilized GEF fraction (Det-P100 extracted by salt) was separated on a Sephacryl S-300 HR column. Stimulated GDP loss at 30 min is graphed versus fraction number (squares). Protein concentration in the fractions as determined by absorbance at 280 nm is plotted (solid black line). The inverted triangles at the top show the positions of ferritin (450 kDa), alcohol dehydrogenase (150 kDa), ovalbumin (45 kDa), and myoglobin (17.8 kDa). Pool A and Pool B were collected separately for further analysis and purified and concentrated as described in MATERIALS AND METHODS. The chromatogram represents the average of three independent experiments performed under identical conditions. (B) HAP column. The S-300 A and S-300 B pools generate single, distinct peaks of Ypt1p-GEF activity on the hydroxyapatite column. The S-300 pool A and the S-300 pool B were loaded onto separate ceramic hydroxyapatite columns and eluted with a phosphate gradient. The results shown are the stimulated GDP loss at 30 min in fractions from the HAP column loaded with the S-300 A pool (squares), the absorbance at 280 nm of the HAP column loaded with the S-300 A pool (solid line), the stimulated GDP loss at 30 min in fractions from the HAP column loaded with the S-300 B pool (circles), and the absorbance at 280 nm of the HAP column loaded with the S-300 B pool (black dotted line). The phosphate gradient (from 10 to 200 mM) is indicated by the diagonal dotted line. Fractions with significant guanine nucleotide exchange activity (filled symbols) were combined into HAP A or HAP B pools (indicated by the inverted triangles at the top). The results represent two independent experiments.
Rescue of ER-to-Golgi Transport by Partially Purified Ypt1p-GEF
We showed previously that the Ypt1p-GEF activity present in the P100 or Det-P100 fraction is completely inhibited by the Ypt1p-D124N mutant protein (Figure 4A; Jones et al., 1995). The two peaks of Ypt1p-GEF generated by sequential purification on the S-300 and HAP columns are also inhibited to equal extents by Ypt1p-D124N (our unpublished observations). We have also shown that this dominant-mutant Ypt1p is a potent inhibitor of an ER-to-Golgi in vitro transport assay, probably because of inhibition of the Ypt1p-GEF (Jones et al., 1995). To lend support to the assertion that the partially purified Ypt1p-GEF described here is an authentic Ypt1p-GEF, we tested the most purified exchange factor, peak A from the HAP column, for its ability to restore transport function to an in vitro ER-to-Golgi transport reaction inhibited by Ypt1p-D124N. The HAP peak A restored ∼50% of the inhibited transport reaction in a concentration-dependent manner (Figure 6). These results suggest that the partially purified exchange activity described here is a physiological Ypt1p-GEF.
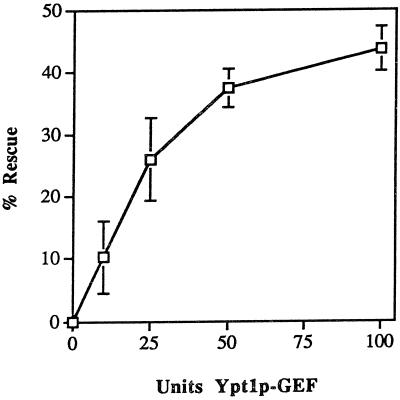
Figure 6.
Inhibition by Ypt1p-D124N of the in vitro transport reaction is relieved by Ypt1p-GEF, HAP peak A. The indicated amounts of exchange activity from HAP Pool A (1 unit is defined as 1 fmol of GDP released from 0.1 μM Ypt1p per minute per milligram of GEF) were mixed with 1.1 μg of Ypt1p-D124N protein (2 μl) in a total volume of 38 μl of Buffer 88 and were incubated on ice for 20 min. This amount of Ypt1p-D124N protein inhibits the ER-to-Golgi transport reaction 50% under these conditions. The S1 cell fraction (2 μl, ∼100 μg) was added to each reaction, and then incubations were continued on ice for 20 more minutes before the reactions were initiated by the addition of 10 μl of permeabilized yeast cells preloaded with 35S-α-factor. The reactions were incubated at 20°C for 90 min. The percent rescue of inhibition of transport was calculated by setting 0% inhibition equal to the amount of transport in the uninhibited control reaction and by setting 100% inhibition equal to the amount of ER-to-Golgi transport in the fully inhibited reaction (without HAP Pool A). This figure represents the average of two independent experiments with each sample measured as duplicates, and the error bars represent the SD of all four measurements for each point.
Identification and Characterization of a GAP Activity for Ypt1p
GAP activity was measured as the stimulation of GTP hydrolysis by recombinant Ypt1p preloaded with [32P]GTP, using the charcoal-binding method. Wild-type Ypt1p hydrolyzed GTP at a low intrinsic rate of 0.002 mol of GTP per mole of Ypt1p per minute at 30°C (Richardson et al., 1998), similar to previously reported values (Wagner et al., 1987). A GAP activity that stimulated GTP hydrolysis by Ypt1p was found to be highly enriched in the P12 fraction (see below). Treatment of this fraction at 95°C for 5 min or incubation with trypsin (1 mg/ml) ablated GAP activity, suggesting that the active factor is proteinaceous. GAP activity in the P12 fraction was linear with respect to protein concentration (0.5–1 mg/ml; Figure 7A) and time (0–30 min). Stimulation of hydrolysis approached a maximum at 5 mg/ml P12, in which the rate of GTP hydrolysis was increased 54-fold (to 0.108 mol per mole of Ypt1p per minute). Phosphate release from GTP hydrolysis in the charcoal-binding assay has been confirmed by TLC analysis that shows that the GTP is being converted to GDP. Two observations argue against the possibility that the GAP activity in the P12 fraction is a protease. 1) Ypt1p mutants that are resistant to the GAP activity have been identified (see below), and 2) the total amount of nucleotide bound to Ypt1p during the course of incubation with the P12 fraction remains constant.
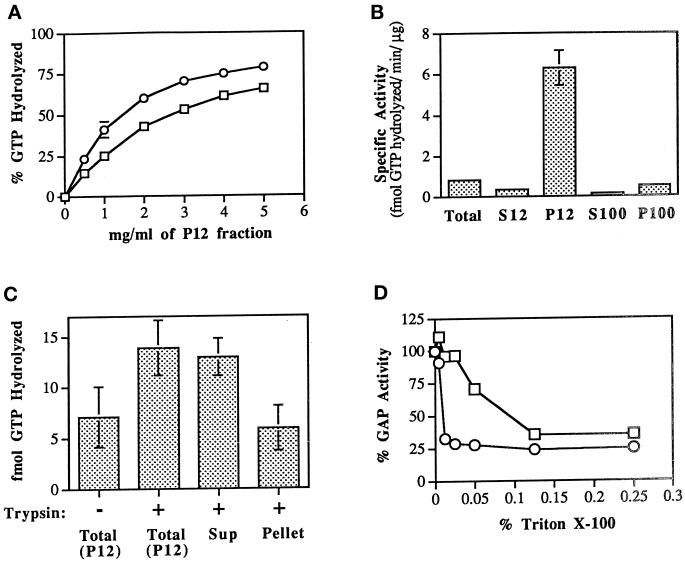
Figure 7.
Identification of a Ypt1p-GAP activity in the P12 cellular fraction and its solubilization. (A) GAP activity is linear with P12 concentration. Ypt1p (2 nM) preloaded with [γ-32P]GTP was incubated with the indicated quantities of P12 fraction at 30°C. GTP hydrolysis was measured by the charcoal-binding assay. Squares represent a 15 min incubation; circles represent a 30 min incubation. Data are expressed as the percent of the total 32P-labeled pool of GTP bound to Ypt1p that was hydrolyzed, and the intrinsic rate of GTP hydrolysis by Ypt1p was subtracted. Results are the average of two independent experiments. Error bars represent the range divided by 2. (B) Localization of GAP activity to P12 (specific activity) is shown. Crude lysates (Total) were generated from GPY60 cells. Lysates were centrifuged at 12,000 × g to generate S12 and P12. The S12 fraction was further centrifuged at 100,000 × g to generate S100 and P100. Ypt1p (2 nM; 200 fmol) preloaded with [γ-32P]GTP was incubated with the indicated cell fractions at 0.5 mg/ml for 15 min at 30°C. GTP hydrolysis was measured as described above. Specific activity is a measure of the femtomoles of GTP hydrolyzed by Ypt1p per minute per microgram of added cell fraction. Results are the average of three independent measurements performed with cell fractions from two independent fractionations. Error bars represent the SEM. (C) GAP activity is stimulated and extracted by limited trypsin digestion. Seven hundred micrograms of a P12 fraction were left untreated (Untreated Total) or treated with trypsin at 0.1 mg/ml on ice for 1 h after which time trypsin inhibitor at 0.2 mg/ml was added (Treated Total). The trypsin-treated sample was then centrifuged at 12,000 × g for 10 min to generate supernatant (Treated Sup) and pellet (Treated Pellet) fractions. The pellet was resuspended to the original volume in Buffer 88 before centrifugation. An equal volume of each sample was assayed for GAP activity as described above. Including trypsin inhibitor at the beginning of the incubation completely prevents extraction and stimulation of the GAP (our unpublished observations). Results are the average of three independent measurements. Error bars represent the SEM. (D) GAP activity is inhibited by Triton X-100. Inhibition of GAP activity in the P12 (0.5 mg/ml; squares) or in the trypsin-solubilized GAP (extracted as described in C from 0.05 mg of P12; circles) by Triton X-100 was tested. GAP assays were performed as described above in the presence of the indicated final concentrations of Triton X-100 (vol/vol) for 15 min at 30°C. Data expressed as the percent of GAP activity from the uninhibited reaction are typical of two independent experiments.
To test models of Ypt/Rab function that assign the site of action of their GAPs to the acceptor compartment, we wanted to determine the compartment in which Ypt1p-GAP resides. Lysates were fractionated by differential centrifugation into 12,000 × g supernatant (S12) and pellet (P12) fractions. The S12 was then subjected to centrifugation at 100,000 × g to generate supernatant (S100) and pellet (P100) fractions. Although detectable Ypt1p-GAP activity exists in crude lysates, 73% of the activity fractionated into the P12 fraction where it is approximately eightfold enriched over that in the cell lysate (Figure 7B). Eighteen percent of the activity was found in the P100 fraction, where its specific activity is lower than is that in cell lysates. Therefore, the activity in the P100 probably represents contamination by P12 membranes. Only 1% or less of Ypt1p-GAP activity was found in the S100 fraction. The finding that the Ypt1p-GAP is enriched in the P12 fraction, with the majority of its activity found in this fraction, suggests that it is associated with large particulate cellular structures.
Extraction of Ypt1p-GAP from Membranes
The association of Ypt1p-GAP with P12 membranes was examined using different procedures, including incubation with trypsin, high salt, high pH, and detergent. Only limited trypsin digestion (0.1 mg/ml, 1 h, 0°C) resulted in solubilization accompanied by a two- to threefold stimulation of GAP activity (Figure 7C). This indicates that Ypt1p-GAP is tightly associated with P12 membranes. The trypsin extraction may lead to elevated GAP activity by increasing substrate accessibility.
One interesting feature of the Ypt1p-GAP is that it is potently inhibited by both ionic (CHAPS) and nonionic (n-octylglucoside, Triton X-100) detergents. Triton X-100 causes a dose-dependent inhibition of GAP activity, and the trypsin-solubilized GAP is ∼10-fold more sensitive than is the insoluble GAP (Figure 7D). Ypt1p itself is still active for GTP binding under these conditions (Richardson et al., 1998). The lower sensitivity of the insoluble activity may be attributable to titration of Triton by membrane lipids and proteins. The detergent sensitivity of GAP does not appear to be attributable to interaction of GAP with the prenyl group of Ypt1p, because geranylgeranylated Ypt1p is as sensitive to detergent as unprenylated Ypt1p. Therefore, our data favor a model in which detergents exert their inhibitory effects either by unfolding GAP or by preventing interaction of GAP with Ypt1p.
Substrate Specificity of Ypt1p-GAP
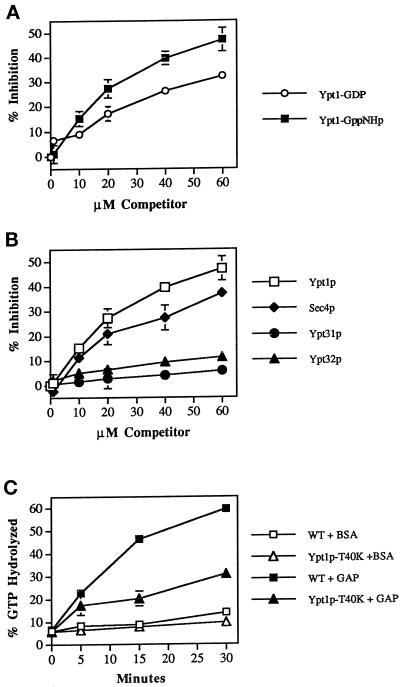
The substrate specificity of GAP was examined using competition assays. To determine whether GAP has a higher affinity for the GTP- or GDP-bound form of Ypt1p, we incubated the P12 fraction with Ypt1p preloaded with [32P]GTP, and increasing concentrations of Ypt1p preloaded with cold nucleotide were added as a competitor. Ypt1p preloaded with either GppNHp (a poorly hydrolyzable analog of GTP; Figure 8A) or GTP were both effective competitors, but high concentrations (60 μM) were needed for 50% inhibition of GAP activity. This may reflect a relatively low-affinity interaction between GAP and recombinant Ypt1p. When Ypt1p preloaded with GDP is used as a competitor, ∼60 μM gives only 32% inhibition of GAP activity. These data indicate that the GAP has a higher affinity for the GTP-bound form of Ypt1p than for the GDP-bound form.
Figure 8.
Substrate specificity of Ypt1p-GAP. (A) Ypt1 GAP has a higher affinity for the GTP form than for the GDP form of Ypt1p. Ypt1p (2 nM) preloaded with [γ-32P]GTP was mixed with the indicated concentrations of competitor Ypt1p preloaded with cold GppNHp (squares) or GDP (circles). P12 was added to a concentration of 0.75 mg/ml as a source for GAP activity. Reactions contained 1 mM each ATP, GTP, and GDP. Incubations were performed for 30 min at 30°C. The decrease in the rate of GTP hydrolysis observed is expressed as the percent inhibition of the rate determined for the uninhibited control (0 μM competitor). Data are the average of two experiments. Error bars represent SEM. (B) Competition by other exocytic Ypt proteins for Ypt1p-GAP is shown. To determine which proteins Ypt1p-GAP can interact with, we assayed various Ypt proteins for their ability to compete with Ypt1p for the Ypt1p-GAP. Reactions were performed as described in A in the presence of the indicated concentrations of Ypt1p (squares), Sec4p (diamonds), Ypt32p (triangles), or Ypt31p (circles) preloaded with GppNHp. Data are the average of two experiments. Error bars represent SEM. (C) Ypt1p-T40K is defective in GAP-stimulated GTP hydrolysis. Wild-type (squares) and Ypt1p-T40K (triangles) proteins were preloaded with [α-32P]GTP for 15 min at 30°C. GTP hydrolysis assays were performed by incubating 2 nM preloaded Ypt1p with 1.5 mg/ml P12 (GAP-stimulated hydrolysis; filled symbols) or without the P12 fraction (intrinsic hydrolysis; open symbols) at 30°C. Aliquots were removed at the indicated time points, GTP hydrolysis was determined by TLC, and radioactivity was detected with a radioanalytic imager. Hydrolysis is expressed as the percent of GDP detected divided by the total nucleotide detected (GDP + GTP). Results are the average of two independent measurements. Error bars represent the range divided by 2.
Because the P12 fraction may contain other GAP activities, we used competition assays to determine whether other Ypt proteins can compete with Ypt1p for the GAP activity. In their GppNHp forms, Ypt31p or Ypt32p, which share 42% identity with Ypt1p, were poor competitors for the GAP activity. On the other hand, Sec4p, which shares 48% identity with Ypt1p, showed 35% inhibition of GAP activity at 60 μM, as compared with 50% inhibition by Ypt1p itself (Figure 8B). Therefore, the Ypt1p-GAP is specific to a subgroup of exocytic Ypt proteins.
To assess the effect of mutations predicted to affect interactions of GTPases with their GAPs, we examined whether mutants of Ypt1p would exhibit reduced responsiveness to the activity. Specifically, we examined the behavior of a Ypt1p variant containing a mutation, T40K, in the putative effector domain, a region that is important for interaction of Ras with Ras-GAP (Boguski and McCormick, 1993; Polakis and McCormick, 1993). To determine whether Ypt1p-T40K is resistant to GAP, we assayed stimulation of GTP hydrolysis by wild-type or mutant Ypt1p preloaded with [32P]GTP in the presence of the P12 fraction. As seen in Figure 8C, the intrinsic rate of GTP hydrolysis by Ypt1p-T40K is nearly identical to that of wild type. However, the mutant protein has an ∼60% lower rate of GAP-stimulated hydrolysis. Therefore, the effector-domain mutation T40K impairs the ability of Ypt1p to respond to its GAP. GTP hydrolysis in the presence of GAP was found to be even more severely impaired in another mutant, Ypt1p-Q67L (Richardson et al., 1998). These data support the suggestion that the activity we have identified does not cause the nonspecific dissociation and breakdown of GTP from Ypt1p but is an authentic Ypt1p-GAP.
Is Ypt1p-GAP the Product of a Previously Characterized Gene?
Several yeast proteins are known to act as GAPs for Ypt proteins (Strom et al., 1993; Vollmer and Gallwitz, 1995). Although Gyp6p was reported not to have GAP activity on Ypt1p (Strom et al., 1993), possible activity of Gyp7p on Ypt1p was not reported. In addition, we found a closely related gene in the S. cerevisiae genome database (YOR070C), whose product was recently shown to have a GAP activity for Sec4p and Ypt1p and was termed GYP1 (Du et al., 1998). To test whether Gyp7p or Gyp1p are responsible for the Ypt1p-GAP activity that we have identified, we deleted GYP7 and GYP1 individually or together. No reduction in GAP activity was observed in the P12 (or S12) fraction from either the single (our unpublished observations) or double deletion strains (Table 2B). Hence, the GAP activity we have characterized is the product of a gene or genes distinct from the known GYP genes.
Among the proteins implicated in the ER-to-Golgi transport step in yeast are components and regulators of the soluble N-ethyl-maleimide sensitive factor attachment protein (SNAP) receptor (SNARE) complex, a set of proteins thought to help determine the specificity of vesicle targeting. If Ypt1p regulates the assembly of the SNARE complex in a GTP-dependent manner, as suggested in the literature (Lian et al., 1994; Sogaard et al., 1994; Lupashin et al., 1996), the assembled SNARE complex could turn off Ypt1p function by acting as a GAP or by signaling to a GAP. To test this, we determined whether preventing or promoting SNARE complex assembly would affect Ypt1p-GAP activity. To prevent SNARE complex assembly, we used extracts from sec22 mutant cells. SEC22 encodes a component of the SNARE complex, and sec22 mutants fail to form stable SNARE complexes (Lian et al., 1994; Sogaard et al., 1994). Conversely, to promote SNARE complex assembly, we generated extracts from sec17–1 and sec18–1 cells, in which the SNARE complex is stabilized (Sollner et al., 1993; Sogaard et al., 1994). The cells were shifted to the nonpermissive temperature of 37°C for 1 h before cell lysis, and all cellular fractions were tested for GAP activity, in case inactivating one of these gene products caused redistribution of the GAP activity. No differences in the localization or specific activity of GAP were observed in fractions prepared from sec18–1, sec17–1, or sec22–3 cells (Table 2C); thus, it is unlikely that the Ypt1p-GAP is either the product of these genes or influenced by the activity of these gene products.
DISCUSSION
A Novel Ypt1p-GEF
In this study we identify and present an initial characterization of both GEF and GAP activities for Ypt1p. The Ypt1p-GEF identified in the present study represents a novel enzyme and is the first GEF in yeast that has been shown to act on only one exocytic Ypt protein. We propose that our partially purified activity is the physiological Ypt1p-GEF because it can rescue the inhibition of protein transport caused by a dominant Ypt1p mutant. Stimulation of GDP release by this factor is accompanied by uptake of GTP. In contrast, a previously described GDP dissociation stimulator for the Ypt1/Sec4 GTPases in yeast, Dss4p, does not stimulate GTP uptake (Collins et al., 1997; Nuoffer et al., 1997). The Ypt1p-GEF is distinct from Dss4p because it is present in cells in which the DSS4 gene is deleted. Sec2p was recently identified as having GEF activity on Sec4p, stimulating both GDP release and GTP uptake (Walch-Solimena et al., 1997). Two pieces of data argue that the activity identified in the present study is distinct from Sec2p. 1) As shown above, Ypt1p-GEF is not active on Sec4p. 2) Purified Sec2p does not promote exchange on Ypt1p (our unpublished observations).
The high apparent molecular weight of the Ypt1p-GEF is shared with other GEFs for Ypt/Rab proteins, e.g., Rab3A-GRF (Burstein and Macara, 1992), Sec2p (Nair et al., 1990), and Rabex-5 (Horiuchi et al., 1997). With Rabex-5 and Sec2p, the high molecular mass of these exchange factors reflects the fact that they are part of larger protein complexes that are required for membrane fusion or for specification of secretory vesicle localization. Although the precise role of GEFs for Ypt/Rab proteins in vesicular transport is still unclear, our evidence suggests that the Ypt1p exchange factor described in this article is required for ER-to-Golgi transport. This conclusion is based on analysis using YPT1 dominant mutations that inhibit this exchange factor (Jones et al., 1995). Similarly, Sec2p is essential for yeast protein transport (Nair et al., 1990).
GEFs for Ypt/Rab GTPases may be important for more than simply catalyzing the guanine nucleotide exchange reaction. For example, an alternative explanation for the fractionation of Ypt1p-GEF to the P100 is that it is associated with cytoskeletal elements, which reside in this fraction. Consistent with this possibility, the exchange factor could be extracted from the particulate fraction with salt, but not with detergent. A potential association of the Ypt1p exchange factor with the cytoskeleton is interesting because an association between the secretory pathway and the cytoskeleton has been proposed (Novick and Botstein, 1985; Vale, 1987; Murphy et al., 1996). Such a connection is also implied for Sec2p, a GEF for Sec4p, by the discovery that Sec2p has a role in localization of secretory vesicles to the growing bud, a process thought to be actin dependent (Novick and Botstein, 1985; Walch-Solimena et al., 1997). In addition, we have recently found that Ypt1p-D124N, a potent inhibitor of Ypt1p-GEF, inhibits protein transport even in the presence of the GTPase-defective Ypt1p-Q67L (Richardson et al., 1998). Because the Ypt1p-Q67L mutant protein is constitutively GTP-bound and does not require GEF for achieving this state, the Ypt1p-D124N protein may inhibit the ability of Ypt1p-GEF to perform another essential function.
A Novel Ypt1p-GAP
The Ypt1p-GAP activity was found to be highly enriched in the P12 membrane fraction. A cytosolic Ypt1p-GAP activity was previously found in yeast extracts (Tan et al., 1991). However we found that the cytosolic GAP activity represented <1% of the total activity in fractionated yeast lysates; therefore this is clearly not the major Ypt1p-GAP. There are several precedents for Ypt/Rab GAP activities fractionating with membranes, e.g., a GAP activity from mammalian membranes that can work on Ypt1p (Jena et al., 1992), a Rab3A GAP activity (Burstein et al., 1991), a Sec4 GAP activity (Walworth et al., 1992), and tuberin, a Rab5 GAP (Xiao et al., 1997). Such a localization for Ypt/Rab GAPs is consistent with evidence that the active GTP-form of Ypt/Rab proteins is membrane associated, and after hydrolysis, the inactive GDP-form is removed from membranes by a GDP dissociation inhibitor (GDI) (Novick and Zerial, 1997).
We have found that Ypt1p-GAP is sensitive to detergents. A sensitivity of Ras-GAP to lipids has also been reported (Tsai et al., 1989; Serth et al., 1991). The crystal structure of Ras-GAP in complex with Ras suggests that hydrophobic interactions occur on the surfaces of contact between Ras and GAP (Scheffzek et al., 1997), and this hydrophobic interface may be disrupted by the intercalation of lipids. Ypt1p contains identical or conservatively substituted amino acids at the analogous positions and may participate in similar hydrophobic contacts with its GAP. Therefore, the detergent inhibition we have observed may be caused by the disruption of hydrophobic interactions between Ypt1p and its GAP.
The Ypt1p-GAP described here is probably an authentic GAP because the stimulation of GTP hydrolysis is significantly reduced by a mutation in the Ypt1p effector domain. Competition assays showed that Ypt1p-GAP can interact with Ypt1p, and to a lesser extent with Sec4p, but not with Ypt31p or Ypt32p. This is similar to results reported for Gyp1p (Du et al., 1998). Other GAP activities reported in the literature show a high degree of substrate specificity, e.g., Gyp6p (Strom et al., 1993), tuberin (Xiao et al., 1997), and Rab3 GAP (Fukui et al., 1997). Partially purified Ypt1p-GAP has a slightly higher affinity for the GTP-bound form of Ypt1p than for the GDP-bound form. Purified Ras-GAP has a much higher affinity for the GTP-bound form of RAS than for the GDP-bound form of Ras (Vogel et al., 1988). A comparison of the crystal structures of Ras in the GDP versus GTP forms reveals major structural rearrangements between the two forms, especially in the region of contact between Ras and GAP, termed the switch 1 region (Milburn et al., 1990). Hence, the Ypt1p-GAP, like the Ras-GAP, is likely to participate in a cycle of binding to Ypt1p-GTP, stimulation of hydrolysis, and release of Ypt1p-GDP.
A New Model for Ypt/Rab Regulation
The unexpected finding in this work is the localization of the Ypt1p-GEF and -GAP activities. Conventional models designate the donor compartment as the site of Ypt/Rab recruitment to membranes and of nucleotide exchange and the acceptor compartment as the site of GTP hydrolysis (Goud and McCaffrey, 1991; Novick and Brennwald, 1993; Novick and Zerial, 1997) (Figure 9A). In Ypt1p-mediated ER-to-Golgi vesicular transport, the donor is the ER and the acceptor is the Golgi, and these two compartments reside in the P12 and P100 cellular fractions, respectively (Baker et al., 1988, 1990). However, in subcellular localization experiments, we find that Ypt1p-GEF cofractionates in the P100 with Golgi markers, suggesting that this regulator localizes to the acceptor compartment for Ypt1p-mediated protein transport. In contrast, Ypt1p-GAP is enriched in the P12 fraction, which contains larger membranous structures, including the ER and the plasma membrane (PM).
On the basis of the intriguing localization of the Ypt1p-GEF and -GAP activities and of our previous studies of YPT1 mutations that affect nucleotide cycling, we propose a new model for the mechanism of action of the Ypt/Rab GTPases. Our combined information, from studies of a GTPase-defective mutant Ypt1p (Richardson et al., 1998) and of GAP localization, suggests that GTP hydrolysis is dispensable for vesicle fusion and that Ypt1p-GAP does not function at the acceptor compartment. We have shown previously that nucleotide exchange is essential for Ypt1p-mediated vesicular transport (Jones et al., 1995), and here we show that Ypt1p-GEF localizes to the acceptor compartment. Therefore, in our model the event that is crucial for vesicle targeting and/or fusion is the shift from the GDP- to the GTP-bound form, whereas GTP hydrolysis is needed for the recycling of Ypt/Rab proteins. We propose the following hypothesis. 1) Nucleotide exchange by GEF to generate the GTP-bound form is essential for vesicle targeting or fusion, and GEF functions at the acceptor compartment. 2) GTP hydrolysis by GAP to generate the GDP-bound form does not have a direct role in vesicle fusion but rather in a process that occurs after vesicle fusion (Figure 9B). If Ypt1p-GAP is localized to the PM, as indicated in Figure 9, GTP hydrolysis occurs at the end of the exocytic pathway and has a role in GDI-mediated recycling of Ypt/Rab proteins between membranes, a process that is not essential for Ypt1p function. If Ypt1p-GAP resides in the ER, GTP hydrolysis might occur after Ypt1p recycles back to the ER, probably by retrograde vesicular transport. The role of GTP hydrolysis in this case would be to shift the Ypt1p to its GDP-bound form, which is the preferred form for interaction with GEF. Thus, although current models predict that GEF for Ypt1p localizes to the ER and GAP for Ypt1p localizes to the Golgi, our model reflects the findings that GEF localizes to the Golgi and GAP localizes to the PM or ER.
A number of previous observations support our model. First, we propose that GEF activity is directly involved in vesicle targeting and/or fusion and is not important for the correct membrane localization of Ypt/Rab proteins. This suggestion is in agreement with the finding of a lag between the in vitro binding of Rab5 and Rab9 to membranes and the uptake of GTP by these proteins (Soldati et al., 1994; Ullrich et al., 1994), although in these experiments the function of the membranes as donor or acceptor compartments is not clear. Second, in agreement with our suggestion that GTP hydrolysis is not essential for Ypt/Rab function in vesicular transport, it has been shown that nucleotide hydrolysis is not essential for Rab5-mediated endosome fusion. In that study it was suggested that GTP hydrolysis by Rab5, although not essential for membrane fusion, is required for its timing, because inhibition of hydrolysis actually resulted in the stimulation of endosome fusion (Rybin et al., 1996). We postulate that the difference in the requirement for GTP hydrolysis between Ypt1p and Rab5 is caused by their roles in heterotypic and homotypic membrane fusion, respectively. Hence, in heterotypic fusion, GTP hydrolysis is not required to turn off Ypt1p-mediated vesicle fusion, because after the fusion of the vesicle with the acceptor compartment, the Ypt/Rab protein is neither in the right place nor in the right context to stimulate such an event (Richardson et al., 1998). Third, according to our model, Ypt1p travels to late compartments in the secretory pathway. Support for this argument comes from the finding that Ypt1p is present on late secretory vesicles that accumulate in sec mutants defective in fusion of these vesicles with the plasma membrane (Mulholland et al., 1997). And fourth, our model proposes that GTP hydrolysis has a role in GDI-mediated recycling of Ypt/Rab proteins. However, because GTP hydrolysis is not essential for Ypt1p function, we suggest that GDI-mediated recycling between membranes is also not essential for Ypt1p function. This suggestion is consistent with the observation that Ypt1p is still functional when permanently tethered to membranes via a membrane anchor (Ossig et al., 1995).
Several questions still remain about GEFs and GAPs for Ypt/Rab proteins regarding their specificity, localization, and mechanism of action. Answers to these questions will await the isolation and study of the genes encoding these factors. The identification and partial purification of Ypt1p-GEF and -GAP activities set the stage for identification of the genes encoding them.
ACKNOWLEDGMENTS
We are especially grateful to C. Palfrey and past and present members of the Segev lab for many useful discussions. We thank B. Glick, C. Palfrey, A. Turkewitz, and T. Steck for critical reading of the manuscript. We are grateful to P. Novick and Ruth Collins for the dss4 deletion strain and for a generous gift of Sec4p and Sec2p and to J. Broach and K. York for a generous gift of Ras2p. We thank Francesca Bartolini for excellent technical help in the generation of the DGYP1 and DGYP7 strains and D. Steiner for the use of protein purification equipment. This research was supported by training grants 5T32 GM-07151–20 (C.J.R.), 5T32 HL-07381 (S.J.), and 5T32 HD-07009 (R.J.L.) and by National Institutes of Health grant GM-45444 (N.S.).
REFERENCES
- Araki S, Kikuchi A, Hata Y, Isomura M, Takai Y. Regulation of reversible binding of smg p25A, a ras p21-like GTP-binding protein, to synaptic plasma membranes and vesicles by its specific regulatory protein, GDP dissociation inhibitor. J Biol Chem. 1990;256:13007–13015. [PubMed] [Google Scholar]
- Baker D, Hicke L, Rexach M, Schleyer M, Schekman R. Reconstitution of SEC gene product-dependent intercompartmental protein transport. Cell. 1988;54:335–344. doi: 10.1016/0092-8674(88)90196-1. [DOI] [PubMed] [Google Scholar]
- Baker D, Wuestehube L, Schekman R, Botstein D, Segev N. GTP-binding Ypt1 protein and Ca2+ function independently in a cell-free transport reaction. Proc Natl Acad Sci USA. 1990;87:355–359. doi: 10.1073/pnas.87.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE. Small GTP-binding proteins in vesicular transport. Trends Biochem Sci. 1990;15:473–477. doi: 10.1016/0968-0004(90)90301-q. [DOI] [PubMed] [Google Scholar]
- Boguski MS, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- Bourne HR. Do GTPases direct membrane traffic in secretion? Cell. 1988;53:669–671. doi: 10.1016/0092-8674(88)90081-5. [DOI] [PubMed] [Google Scholar]
- Brandt D, Asano T, Pedersen S, Ross E. Reconstitution of catecholamine-stimulated guanosine triphosphatase activity. Biochemistry. 1983;22:4357–4362. doi: 10.1021/bi00288a002. [DOI] [PubMed] [Google Scholar]
- Brondyk WH, McKiernan CJ, Burstein ES, Macara IG. Mutants of Rab3A analogous to oncogenic Ras mutants. J Biol Chem. 1993;268:9410–9415. [PubMed] [Google Scholar]
- Bucci C, Parton RG, Mather IA, Stunnenberg H, Simons K, Hoflack B, Zerial M. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70:715–728. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- Burstein E, Linko-Stentz K, Lu Z, Macara I. Regulation of the GTPase activity of the ras-like protein p25rab3A. J Biol Chem. 1991;266:2689–2692. [PubMed] [Google Scholar]
- Burstein ES, Macara IG. Characterization of a guanine nucleotide-releasing factor and a GTPase-activating protein that are specific for the ras-related protein p25rab3A. Proc Natl Acad Sci USA. 1992;89:1154–1158. doi: 10.1073/pnas.89.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins R, Brennwald P, Garrett M, Lauring A, Novick P. Interactions of nucleotide release factor Dss4p with Sec4p in the post-Golgi secretory pathway of yeast. J Biol Chem. 1997;272:18281–18289. doi: 10.1074/jbc.272.29.18281. [DOI] [PubMed] [Google Scholar]
- Dirac-Svejstrup A, Sumizawa T, Pfeffer S. Identification of a GDI displacement factor that releases endosomal Rab GTPases from Rab-GDI. EMBO J. 1997;16:465–472. doi: 10.1093/emboj/16.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du LL, Collins RN, Novick PJ. Identification of a Sec4p GTPase-activating protein (GAP) as a novel member of a Rab GAP family. J Biol Chem. 1998;273:3253–3256. doi: 10.1074/jbc.273.6.3253. [DOI] [PubMed] [Google Scholar]
- Ferro-Novick S, Novick P. The role of GTP-binding proteins in transport along the exocytic pathway. Annu Rev Cell Biol. 1993;9:575–599. doi: 10.1146/annurev.cb.09.110193.003043. [DOI] [PubMed] [Google Scholar]
- Fukui K, Sasaki T, Imazumi K, Matsuura Y, Nakanishi H, Takai Y. Isolation and characterization of a GTPase activating protein specific for the Rab3 subfamily of small G proteins. J Biol Chem. 1997;272:4655–4658. doi: 10.1074/jbc.272.8.4655. [DOI] [PubMed] [Google Scholar]
- Gorvel J-P, Chavrier P, Zerial M, Gruenberg J. rab5 controls early endosome fusion in vitro. Cell. 1991;64:915–925. doi: 10.1016/0092-8674(91)90316-q. [DOI] [PubMed] [Google Scholar]
- Goud B, McCaffrey M. Small GTP-binding proteins and their role in transport. Curr Opin Cell Biol. 1991;3:626–633. doi: 10.1016/0955-0674(91)90033-u. [DOI] [PubMed] [Google Scholar]
- Guan KL, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Higashijima T, Ferguson K, Smigel M, Gilman A. The effect of GTP and Mg++ on the GTPase activity and the fluorescent properties of Go. J Biol Chem. 1987;262:757–761. [PubMed] [Google Scholar]
- Horiuchi H, et al. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 1997;90:1149–1159. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- Jamieson JD, Palade GE. Intracellular transport of secretory proteins in the pancreatic exocrine cell. I. Role of peripheral elements of the Golgi complex. J Cell Biol. 1967;34:577–596. doi: 10.1083/jcb.34.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedd G, Mulholland J, Segev N. Two new Ypt GTPases are required for exit from the yeast trans-Golgi compartment. J Cell Biol. 1997;137:563–580. doi: 10.1083/jcb.137.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedd G, Richardson CJ, Litt RJ, Segev N. The Ypt1 GTPase is essential for the first two steps of the yeast secretory pathway. J Cell Biol. 1995;131:583–590. doi: 10.1083/jcb.131.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jena BP, Brennwald P, Garrett MD, Novick P, Jamieson JD. Distinct and specific GAP activities in rat pancreas act on the yeast GTP-binding proteins Ypt1 and Sec4. FEBS Lett. 1992;309:5–9. doi: 10.1016/0014-5793(92)80727-x. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Rossi G, Ferro-Novick S. Characterization of yeast type-II geranylgeranyltransferase. Methods Enzymol. 1995;257:21–41. doi: 10.1016/s0076-6879(95)57006-3. [DOI] [PubMed] [Google Scholar]
- Jones S, Litt RJ, Richardson CJ, Segev N. Requirement of nucleotide exchange for Ypt1 GTPase mediated protein transport. J Cell Biol. 1995;130:1051–1061. doi: 10.1083/jcb.130.5.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabcenell AK, Goud B, Northup JK, Novick PJ. Binding and hydrolysis of guanine nucleotides by Sec4p, a yeast protein involved in the regulation of vesicular traffic. J Biol Chem. 1990;265:9366–9372. [PubMed] [Google Scholar]
- Li G, Stahl PD. Structure-function relationship of the small GTPase rab5. J Biol Chem. 1993;268:24475–24480. [PubMed] [Google Scholar]
- Lian JP, Stone S, Jiang Y, Lyons P, Ferro-Novick S. Ypt1p implicated in v-SNARE activation. Nature. 1994;372:698–701. doi: 10.1038/372698a0. [DOI] [PubMed] [Google Scholar]
- Lupashin VV, Hamamoto S, Schekman RW. Biochemical requirements for the targeting and fusion of ER-derived transport vesicles with purified yeast Golgi membranes. J Cell Biol. 1996;132:277–289. doi: 10.1083/jcb.132.3.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick F. The world according to GAP. Oncogene. 1990;5:1281–1283. [PubMed] [Google Scholar]
- Milburn M, Tong L, DeVos A, Brünger A, Yamaizumi A, Nishimura S, Kim S. Molecular switch for signal transduction: structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247:939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- Miyazaki A, Sasaki T, Araki K, Ueno N, Imazumi K, Nagano F, Takahashi K, Takai Y. Comparison of kinetic properties between MSS4 and Rab3A GRF GDP/GTP exchange proteins. FEBS Lett. 1994;350:333–336. doi: 10.1016/0014-5793(94)00804-3. [DOI] [PubMed] [Google Scholar]
- Moya M, Roberts D, Novick P. DSS4–1 is a dominant suppressor of sec4–8 that encodes a nucleotide exchange protein that aids Sec4p function. Nature. 1993;361:460–463. doi: 10.1038/361460a0. [DOI] [PubMed] [Google Scholar]
- Mulholland J, Wesp A, Riezman H, Botstein D. Differential accumulation of GTPase-containing late secretory vesicles distinguish actin cytoskeleton mutants from late secretory mutants in Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:1481–1499. doi: 10.1091/mbc.8.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C, Saffrich R, Grummt M, Gournier H, Rybin V, Rubino M, Auvinen P, Lutcke A, Parton RG, Zerial M. Endosome dynamics regulated by a Rho protein. Nature. 1996;384:427–432. doi: 10.1038/384427a0. [DOI] [PubMed] [Google Scholar]
- Nair J, Müller H, Peterson M, Novick P. Sec2 protein contains a coiled-coil domain essential for vesicular transport and a dispensable carboxy terminal domain. J Cell Biol. 1990;110:1897–1909. doi: 10.1083/jcb.110.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Botstein D. Phenotypic analysis of temperature-sensitive yeast actin mutants. Cell. 1985;40:405–416. doi: 10.1016/0092-8674(85)90154-0. [DOI] [PubMed] [Google Scholar]
- Novick P, Brennwald P. Friends and family: the role of the Rab GTPases in vesicular traffic. Cell. 1993;75:597–601. doi: 10.1016/0092-8674(93)90478-9. [DOI] [PubMed] [Google Scholar]
- Novick P, Ferro S, Schekman R. Order of events in the yeast secretory pathway. Cell. 1981;25:461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- Novick P, Zerial M. The diversity of Rab proteins in vesicle transport. Curr Opin Cell Biol. 1997;9:496–504. doi: 10.1016/s0955-0674(97)80025-7. [DOI] [PubMed] [Google Scholar]
- Nuoffer C, Davidson HW, Matteson J, Meinkoth J, Balch WE. A GDP-bound form of Rab1 inhibits protein export from the endoplasmic reticulum and transport between Golgi compartments. J Cell Biol. 1994;125:225–237. doi: 10.1083/jcb.125.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuoffer C, Wu SK, Dascher C, Balch WE. Mss4 does not function as an exchange factor for Rab in endoplasmic reticulum to Golgi transport. Mol Biol Cell. 1997;8:1305–1316. doi: 10.1091/mbc.8.7.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossig R, Laufer W, Schmitt HD, Gallwitz D. Functionality and specific membrane localization of transport GTPases carrying C-terminal membrane anchors of synaptobrevin-like proteins. EMBO J. 1995;14:3645–3653. doi: 10.1002/j.1460-2075.1995.tb00034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palade G. Intracellular aspects of the process of protein synthesis. Science. 1975;189:347–358. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- Pfeffer S. Rab GTPases: master regulators of membrane trafficking. Curr Opin Cell Biol. 1994;6:522–526. doi: 10.1016/0955-0674(94)90071-x. [DOI] [PubMed] [Google Scholar]
- Polakis P, McCormick F. Structural requirements for the interaction of p21ras with GAP, exchange factors, and its biological effector target. J Biol Chem. 1993;268:9157–9160. [PubMed] [Google Scholar]
- Rexach MF, Schekman RW. Distinct biochemical requirements for the budding, targeting, and fusion of ER-derived transport vesicles. J Cell Biol. 1991;114:219–229. doi: 10.1083/jcb.114.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CJ, Jones S, Litt R, Segev N. GTP hydrolysis is not important for Ypt1 GTPase function in vesicular transport. Mol Cell Biol. 1998;18:827–838. doi: 10.1128/mcb.18.2.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riederer MA, Soldati T, Shapiro AD, Lin J, Pfeffer SR. Lysosome biogenesis requires Rab9 function and receptor recycling from endosomes to the trans-Golgi network. J Cell Biol. 1994;125:573–582. doi: 10.1083/jcb.125.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M, Winston F, Heiter P. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Ruohola H, Kabcenell AK, Ferro-Novick S. Reconstitution of protein transport from the endoplasmic reticulum to the Golgi complex in yeast: the acceptor Golgi compartment is defective in the sec23 mutant. J Cell Biol. 1988;107:1465–1476. doi: 10.1083/jcb.107.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybin V, Ullrich O, Rubino M, Alexandrov K, Simon I, Seabra M, Goody R, Zerial M. GTPase activity of Rab5 acts as a timer for endocytic membrane fusion. Nature. 1996;383:266–269. doi: 10.1038/383266a0. [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Kabsch W, Wiesmüller L, Lautwein A, Schmitz F, Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- Segev N. Mediation of the attachment or fusion step in vesicular transport by the GTP-binding Ypt1 protein. Science. 1991;252:1553–1556. doi: 10.1126/science.1904626. [DOI] [PubMed] [Google Scholar]
- Segev N, Mulholland J, Botstein D. The yeast GTP-binding YPT1 protein and a mammalian counterpart are associated with the secretion machinery. Cell. 1988;52:915–924. doi: 10.1016/0092-8674(88)90433-3. [DOI] [PubMed] [Google Scholar]
- Serth J, Lautwein A, Frech M, Wittinghofer A, Pingoud A. The inhibition of the GTPase activating protein–Ha-ras interaction by acidic lipids is due to physical association of the C-terminal domain of the GTPase activating protein with micellar structures. EMBO J. 1991;10:1325–1330. doi: 10.1002/j.1460-2075.1991.tb07651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;19:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogaard M, Tani K, Ye RR, Geromanos S, Tempst P, Kirchhausen T, Rothman JE, Sollner T. A rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell. 1994;78:937–948. doi: 10.1016/0092-8674(94)90270-4. [DOI] [PubMed] [Google Scholar]
- Soldati T, Reiderer MA, Pfeffer SR. Rab GDI: a solubilizing and recycling factor for rab9 protein. Mol Biol Cell. 1993;4:425–434. doi: 10.1091/mbc.4.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati T, Shapiro AD, Svejstrup ABD, Pfeffer SR. Membrane targetting of the small GTPase Rab9 is accompanied by nucleotide exchange. Nature. 1994;369:76–78. doi: 10.1038/369076a0. [DOI] [PubMed] [Google Scholar]
- Sollner T, Bennett M, Whiteheart S, Scheller R, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- Strom M, Vollmer P, Tan TJ, Gallwitz D. A yeast GTPase-activating protein that interacts specifically with a member of the Ypt/Rab family. Nature. 1993;361:736–739. doi: 10.1038/361736a0. [DOI] [PubMed] [Google Scholar]
- Tan TJ, Vollmer P, Gallwitz D. Identification and partial purification of GTPase-activating proteins from yeast and mammalian cells that preferentially act on Ypt1/Rab1 proteins. FEBS Lett. 1991;291:322–326. doi: 10.1016/0014-5793(91)81312-v. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Lin BK, Wood DR, Tamanoi F. IRA2, an upstream negative regulator of RAS in yeast, is a RAS GTPase-activating protein. Proc Natl Acad Sci USA. 1991;88:468–472. doi: 10.1073/pnas.88.2.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdale EJ, Bourne JR, Khosravi-Far R, Der CJ, Balch WE. GTP-binding mutants of Rab1 and Rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J Cell Biol. 1992;119:749–761. doi: 10.1083/jcb.119.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M, Yu C, Wei F, Stacey D. The effect of GTPase activating protein upon Ras is inhibited by mitogenically responsive lipids. Science. 1989;243:522–526. doi: 10.1126/science.2536192. [DOI] [PubMed] [Google Scholar]
- Ullrich O, Horiuchi H, Bucci C, Zerial M. Membrane association of Rab5 mediated by GDP-dissociation inhibitor and accompanied by GDP/GTP exchange. Nature. 1994;368:157–160. doi: 10.1038/368157a0. [DOI] [PubMed] [Google Scholar]
- Vale RD. Intracellular transport using microtubule-based motors. Annu Rev Cell Biol. 1987;3:347–378. doi: 10.1146/annurev.cb.03.110187.002023. [DOI] [PubMed] [Google Scholar]
- Vogel U, Dixon R, Schaber M, Diehl R, Marshall M, Scolnick E, Sigal I, Gibbs J. Cloning of bovine GAP and its interaction with oncogenic ras p21. Nature. 1988;335:90–93. doi: 10.1038/335090a0. [DOI] [PubMed] [Google Scholar]
- Vollmer P, Gallwitz D. High expression cloning, purification, and assay of Ypt-GTPase-activating proteins. Methods Enzymol. 1995;257:118–128. doi: 10.1016/s0076-6879(95)57017-9. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wada M, Nakanishi H, Satoh A, Hirano H, Obaishi H, Matsuura Y, Takai Y. Isolation and characterization of a GDP/GTP exchange protein specific for the Rab3 subfamily small G proteins. J Biol Chem. 1997;272:3875–3878. doi: 10.1074/jbc.272.7.3875. [DOI] [PubMed] [Google Scholar]
- Wagner P, Molenaar CMT, Rauh AJG, Brokel R, Schmitt HD, Gallwitz D. Biochemical properties of the ras-related YPT protein in yeast: a mutational analysis. EMBO J. 1987;6:2373–2379. doi: 10.1002/j.1460-2075.1987.tb02514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Solimena C, Collins R, Novick P. Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J Cell Biol. 1997;137:1495–1509. doi: 10.1083/jcb.137.7.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth NC, Brennwald P, Kabcenell AK, Garrett M, Novick PJ. Hydrolysis of GTP by Sec4 protein plays an important role in vesicular transport and is stimulated by a GTPase-activating protein in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2017–2028. doi: 10.1128/mcb.12.5.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walworth NC, Goud B, Kabcenell AK, Novick PJ. Mutational analysis of SEC4 suggests a cyclical mechanism for the regulation of vesicular traffic. EMBO J. 1989;8:1685–1693. doi: 10.1002/j.1460-2075.1989.tb03560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuestehube LJ, Schekman RW. Reconstitution of transport from endoplasmic reticulum to Golgi complex using endoplasmic-reticulum-enriched membrane fraction from yeast. Methods Enzymol. 1992;219:124–136. doi: 10.1016/0076-6879(92)19015-x. [DOI] [PubMed] [Google Scholar]
- Xiao G, Shoarinejad F, Jin F, Golemis E, Yeung R. The tuberin sclerosis 2 gene product, tuberin, functions as a Rab5 GTPase activating protein (GAP) in modulating endocytosis. J Biol Chem. 1997;272:6097–6100. doi: 10.1074/jbc.272.10.6097. [DOI] [PubMed] [Google Scholar]
- Zerial M, Stenmark H. Rab GTPases in vesicular transport. Curr Opin Cell Biol. 1993;5:613–620. doi: 10.1016/0955-0674(93)90130-i. [DOI] [PubMed] [Google Scholar]