Abstract
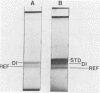
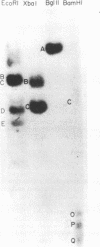
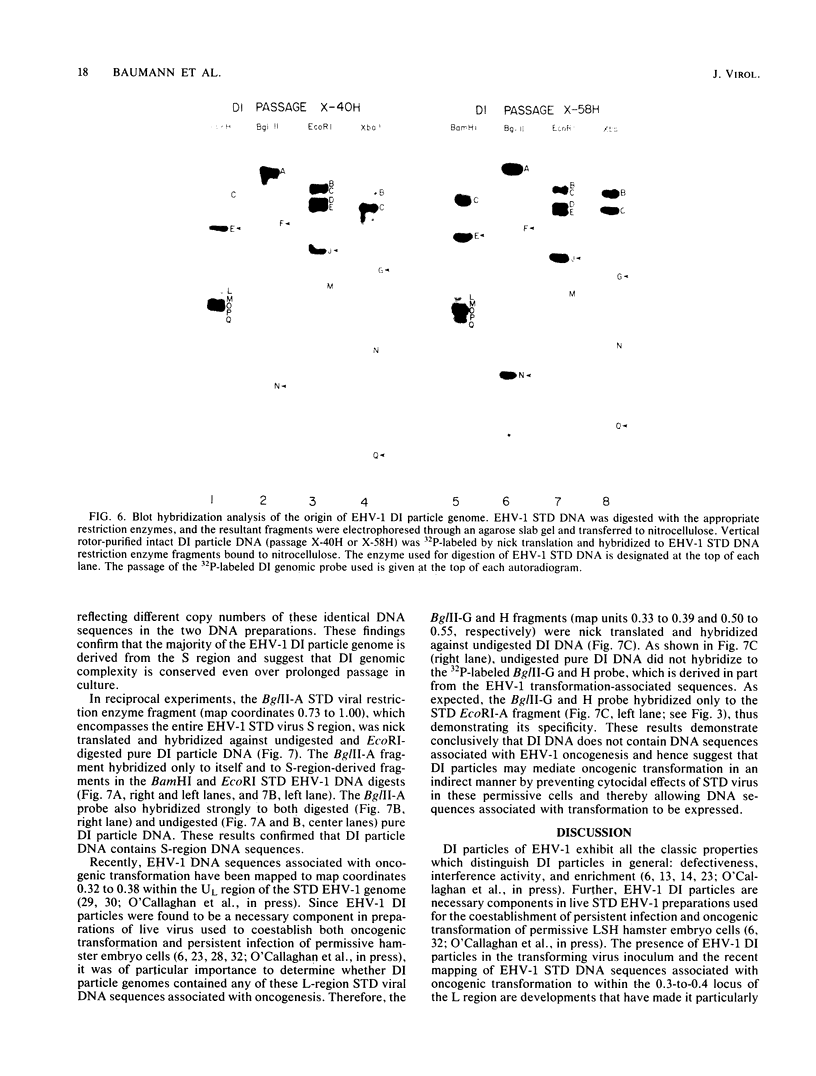
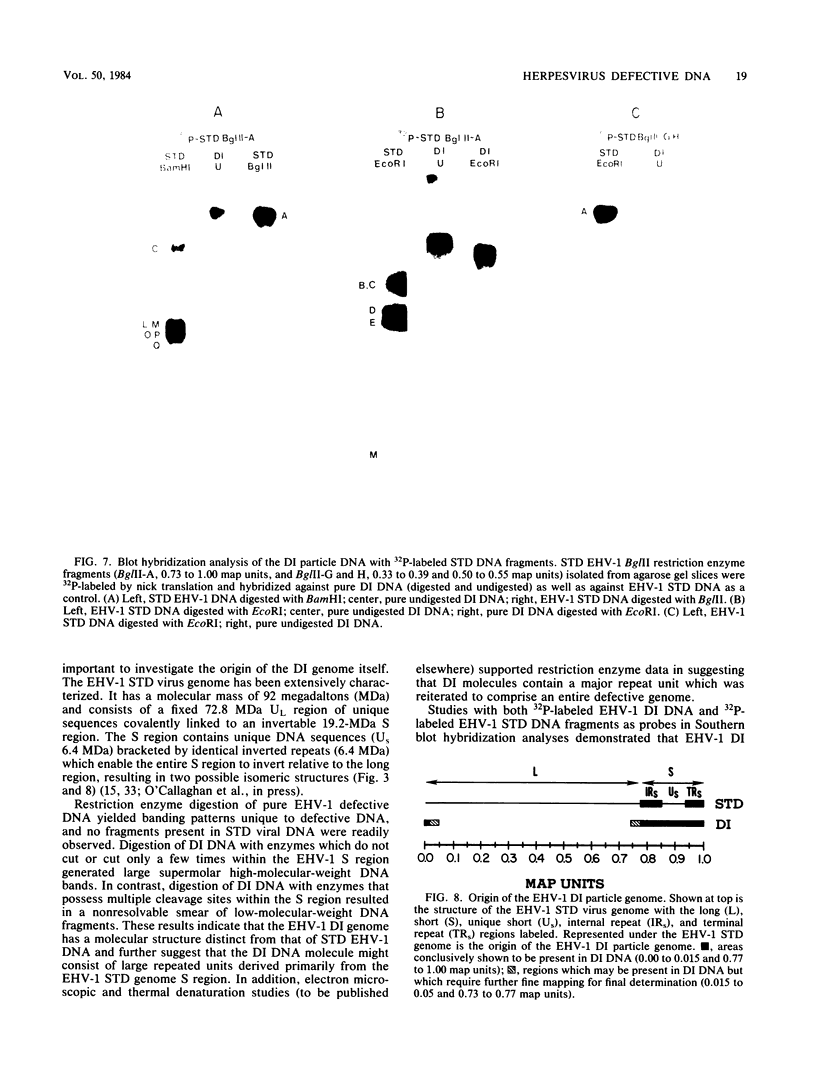
The complexity and structural organization of defective-interfering (DI) particle DNA of equine herpesvirus type 1 (EHV-1) have been elucidated by using restriction enzyme and Southern blot hybridization analyses. DI particles were generated by serial high-multiplicity passage of EHV-1 in L-M cells, and total viral DNA was extracted from virus purified from supernatants of these serial passages. EHV-1 DI particle DNA was quantitatively separated from standard (STD) DNA by several cycles of CsCl isopycnic banding in a vertical rotor. Restriction endonuclease digestion profiles of pure DI DNA were completely different from the mapped patterns observed for EHV-1 STD DNA. Digestion of pure defective DNA with restriction enzymes (Bg/II, EcoRI, and XbaI), for which there are few or no cleavage sites within the S (short) region of the EHV-1 STD genome, yielded high-molecular-weight supermolar DNA bands, suggesting that a large subgenomic repeat unit was present in defective DNA. DNA blot hybridization analysis with the Bg/II supermolar fragment of defective DNA, intact DI particle genomic DNA, and EHV-1 STD DNA restriction enzyme fragments as 32P-labeled probes indicated that the EHV-1 DI particle genome originates predominately from the STD DNA S region (0.77 to 1.00 map units) and to a lesser extent from the left terminus of the unique long (UL) region (0.00 to 0.05 map units). None of the EHV-1 DNA sequences associated to date with EHV-1 oncogenesis (0.32 to 0.38 map units; O'Callaghan et al. in B. Roizman [ed.], Herpesviruses, in press; Robinson et al., Cell 32:204-219, 1983, and Proc. Natl. Acad. Sci., U.S.A., 78:6684-6688, 1981) were detected in the DI particle DNA. The importance of these data with regard to DNA replication of DI particles and the role of DI particles in one model system of EHV-1 oncogenic transformation are discussed.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andzhaparidze O. G., Bogomolova N. N., Boriskin Y. S., Bektemirova M. S., Drynov I. D. Comparative study of rabies virus persistence in human and hamster cell lines. J Virol. 1981 Jan;37(1):1–6. doi: 10.1128/jvi.37.1.1-6.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Porat T., Rixon F. J. Replication of herpesvirus DNA. IV: analysis of concatemers. Virology. 1979 Apr 15;94(1):61–70. doi: 10.1016/0042-6822(79)90438-0. [DOI] [PubMed] [Google Scholar]
- Brockman W. W., Lee T. N., Nathans D. Characterization of cloned evolutionary variants of simian virus 40. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):119–127. doi: 10.1101/sqb.1974.039.01.017. [DOI] [PubMed] [Google Scholar]
- Campbell D. E., Kemp M. C., Perdue M. L., Randall C. C., Gentry G. A. Equine herpesvirus in vivo: cyclic production of a DNA density variant with repetitive sequences. Virology. 1976 Feb;69(2):737–750. doi: 10.1016/0042-6822(76)90502-x. [DOI] [PubMed] [Google Scholar]
- Dauenhauer S. A., Robinson R. A., O'Callaghan D. J. Chronic production of defective-interfering particles by hamster embryo cultures of herpesvirus persistently infected and oncogenically transformed cells. J Gen Virol. 1982 May;60(Pt 1):1–14. doi: 10.1099/0022-1317-60-1-1. [DOI] [PubMed] [Google Scholar]
- De B. K., Nayak D. P. Defective interfering influenza viruses and host cells: establishment and maintenance of persistent influenza virus infection in MDBK and HeLa cells. J Virol. 1980 Dec;36(3):847–859. doi: 10.1128/jvi.36.3.847-859.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckenstein B., Bornkamm G. W., Ludwig H. Repetitive sequences in complete and defective genomes of Herpesvirus saimiri. J Virol. 1975 Feb;15(2):398–406. doi: 10.1128/jvi.15.2.398-406.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckenstein B., Bornkamm G. W., Mulder C., Werner F. J., Daniel M. D., Falk L. A., Delius H. Herpesvirus ateles DNA and its homology with Herpesvirus saimiri nucleic acid. J Virol. 1978 Jan;25(1):361–373. doi: 10.1128/jvi.25.1.361-373.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkeĺ N., Locker H., Batterson W., Hayward G. S., Roizman B. Anatomy of herpes simplex virus DNA. VI. Defective DNA originates from the S component. J Virol. 1976 Nov;20(2):527–531. doi: 10.1128/jvi.20.2.527-531.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry B. E., Newcomb W. W., O'Callaghan D. J. Alterations in virus protein synthesis and capsid production in infection with DI particles of herpesvirus. J Gen Virol. 1980 Apr;47(2):343–353. doi: 10.1099/0022-1317-47-2-343. [DOI] [PubMed] [Google Scholar]
- Henry B. E., Newcomb W. W., O'Callaghan D. J. Biological and biochemical properties of defective interfering particles of equine herpesvirus type 1. Virology. 1979 Jan 30;92(2):495–506. doi: 10.1016/0042-6822(79)90152-1. [DOI] [PubMed] [Google Scholar]
- Henry B. E., Robinson R. A., Dauenhauer S. A., Atherton S. S., Hayward G. S., O'Callaghan D. J. Structure of the genome of equine herpesvirus type 1. Virology. 1981 Nov;115(1):97–114. doi: 10.1016/0042-6822(81)90092-1. [DOI] [PubMed] [Google Scholar]
- Huang A. S., Baltimore D. Defective viral particles and viral disease processes. Nature. 1970 Apr 25;226(5243):325–327. doi: 10.1038/226325a0. [DOI] [PubMed] [Google Scholar]
- Huang A. S. Defective interfering viruses. Annu Rev Microbiol. 1973;27:101–117. doi: 10.1146/annurev.mi.27.100173.000533. [DOI] [PubMed] [Google Scholar]
- Jacob R. J., Morse L. S., Roizman B. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in the generation of the four isomeric arrangements of viral DNA. J Virol. 1979 Feb;29(2):448–457. doi: 10.1128/jvi.29.2.448-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaerner H. C., Maichle I. B., Ott A., Schröder C. H. Origin of two different classes of defective HSV-1 Angelotti DNA. Nucleic Acids Res. 1979 Apr;6(4):1467–1478. doi: 10.1093/nar/6.4.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocarski E. S., Roizman B. Structure and role of the herpes simplex virus DNA termini in inversion, circularization and generation of virion DNA. Cell. 1982 Nov;31(1):89–97. doi: 10.1016/0092-8674(82)90408-1. [DOI] [PubMed] [Google Scholar]
- O'Callaghan D. J., Cheevers W. P., Gentry G. A., Randall C. C. Kinetics of cellular and viral DNA synthesis in equine abortion (herpes) virus infection of L-M cells. Virology. 1968 Sep;36(1):104–114. doi: 10.1016/0042-6822(68)90120-7. [DOI] [PubMed] [Google Scholar]
- Perdue M. L., Kemp M. C., Randall C. C., O'Callaghan D. J. Studies of the molecular anatomy of the L-M cell strain of equine herpes virus type 1: proteins of the nucleocapsid and intact virion. Virology. 1974 May;59(1):201–216. doi: 10.1016/0042-6822(74)90216-5. [DOI] [PubMed] [Google Scholar]
- Reichmann M. E., Schnitzlein W. M. Defective interfering particles of rhabdoviruses. Curr Top Microbiol Immunol. 1979;86:123–168. doi: 10.1007/978-3-642-67341-2_4. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Rixon F. J., Ben-Porat T. Structural evolution of the DNA of pseudorabies-defective viral particles. Virology. 1979 Aug;97(1):151–163. doi: 10.1016/0042-6822(79)90381-7. [DOI] [PubMed] [Google Scholar]
- Robinson R. A., Henry B. E., Duff R. G., O'Callaghan D. J. Oncogenic transformation by equine herpesviruses (EHV). I. Properties of hamster embryo cells transformed by ultraviolet-irradiated EHV-1. Virology. 1980 Mar;101(2):335–362. doi: 10.1016/0042-6822(80)90449-3. [DOI] [PubMed] [Google Scholar]
- Robinson R. A., O'Callaghan D. J. A specific viral DNA sequence is stably integrated in herpesvirus oncogenically transformed cells. Cell. 1983 Feb;32(2):569–578. doi: 10.1016/0092-8674(83)90476-2. [DOI] [PubMed] [Google Scholar]
- Robinson R. A., Tucker P. W., Dauenhauer S. A., O'Callaghan D. J. Molecular cloning of equine herpesvirus type 1 DNA: analysis of standard and defective viral genomes and viral sequences in oncogenically transformed cells. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6684–6688. doi: 10.1073/pnas.78.11.6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson R. A., Vance R. B., O'Callaghan D. J. Oncogenic transformation by by equine herpesviruses. II. Coestablishment of persistent infection and oncogenic transformation of hamster embryo cells by equine herpesvirus type 1 preparations enriched for defective interfering particles. J Virol. 1980 Oct;36(1):204–219. doi: 10.1128/jvi.36.1.204-219.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruyechan W. T., Dauenhauer S. A., O'Callaghan D. J. Electron microscopic study of equine herpesvirus type 1 DNA. J Virol. 1982 Apr;42(1):297–300. doi: 10.1128/jvi.42.1.297-300.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder C. H., Stegmann B., Lauppe H. F., Kaerner H. C. An unusual defective genotype derived from herpes simplex virus strain ANG. Intervirology. 1975;6(4-5):270–284. doi: 10.1159/000149481. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Sugden B., Sambrook J. Detection of two restriction endonuclease activities in Haemophilus parainfluenzae using analytical agarose--ethidium bromide electrophoresis. Biochemistry. 1973 Jul 31;12(16):3055–3063. doi: 10.1021/bi00740a018. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stegmann B., Zentgraf H., Ott A., Schröder C. H. Synthesis and packaging of herpes simplex virus DNA in the course of virus passages at high multiplicity. Intervirology. 1978;10(4):228–240. doi: 10.1159/000148986. [DOI] [PubMed] [Google Scholar]
- Stinski M. F., Mocarski E. S., Thomsen D. R. DNA of human cytomegalovirus: size heterogeneity and defectiveness resulting from serial undiluted passage. J Virol. 1979 Jul;31(1):231–239. doi: 10.1128/jvi.31.1.231-239.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stollar V. Defective interfering particles of togaviruses. Curr Top Microbiol Immunol. 1979;86:35–66. doi: 10.1007/978-3-642-67341-2_2. [DOI] [PubMed] [Google Scholar]
- VON MAGNUS P. Propagation of the PR8 strain of influenza A virus in chick embryos. III. Properties of the incomplete virus produced in serial passages of undiluted virus. Acta Pathol Microbiol Scand. 1951;29(2):157–181. doi: 10.1111/j.1699-0463.1951.tb00114.x. [DOI] [PubMed] [Google Scholar]
- Vlazny D. A., Frenkel N. Replication of herpes simplex virus DNA: localization of replication recognition signals within defective virus genomes. Proc Natl Acad Sci U S A. 1981 Feb;78(2):742–746. doi: 10.1073/pnas.78.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]