INTRODUCTION
“The greatest treasure by far, however, was what Grandfather referred to as his ’stereopticon.’”
Artist Frederick Franck points in this passage to the origin of his singular eye for detail (Franck, 1997). Ironically, Franck believes it was a device for viewing space in flat paper images that awakened him as a child to the richness of the actual world. The principle of the stereopticon has also served biologists well, showing them glimpses of the microscopic world from their accustomed binocular point of view, letting them “crawl” into spaces of submicron dimension. Human visual perception has evolved with special emphasis on making sense of a colorful, three-dimensional (3D) reality that we move in and that moves around us. With the development of new imaging and labeling technologies, light microscopy has now evolved so that visual data collected from a single living specimen can go beyond the domain of the stereopticon, to engage the viewer’s full capacity for seeing in space and through time. A challenge arising out of these advances is how to display these data so that the microscopist can sense and actively explore the recording’s full spatial and temporal content, to better understand visually what cannot be seen directly through the microscope eyepiece. Confronted with this opportunity in my own work on live fluorescence imaging of Caenorhabditis elegans development, I have invested some experimental effort in representing multidimensional microscope data interactively on the computer screen. In this essay, I describe my recent experience of trying to make the most of the observer’s ability to see and comprehend microscopic specimens in more than two dimensions.
The basis for 3D visualization in light microscopy is the acquisition of optical sections, 2D images that uniquely portray features found at specific depths of focus within the specimen. A stack of optical sections through a sample defines its 3D structure, and repeated collection of such stacks over time produces a 4D recording. Simultaneous 4D acquisition of separate optical channels, in turn, yields a 5D data set. Software designed to acquire, archive, and play back 4D and 5D recordings has allowed the viewer to animate the data both over time and slice-by-slice through focus, to freely and repeatedly reenact the experience of observing the live specimen on the microscope (Hird and White, 1993; Fire, 1994; Inoue and Inoue, 1994; Thomas et al., 1996). This standard “through-focus 4D” representation is quite valuable, especially in transmitted light microscopy, where subtle image contrast prohibits use of volume-rendering techniques.
Yet in many cases a specimen’s full spatial content can be nicely represented in a single image. Fluorescence microscopy, with its high degree of contrast, often lends itself well to volume reconstruction (Minden et al., 1989; Chen et al., 1996; Amberg, 1998; Mohler and White, 1998b). Computational rendering of 3D structure can be extremely powerful (Drebin et al., 1988; Ford-Holevinski et al., 1991). It produces real views of the specimen that can never actually be seen on the microscope and allows the viewer to use stereo vision and a sense of spatial motion to discern the position and trajectory of details within the greater space; however, high-resolution fluorescence imaging has been notoriously toxic to the sample, limiting the time dimension of 4D recording. Recently, epifluorescence techniques that spare the viability of the specimen while producing optical sections have become more available. Multiphoton laser-scanning microscopy and low-light, wide-field deconvolution microscopy both minimize the noxious effects of excitation and fluorescence to a point where long 4D recordings of living fluorescent samples can now be obtained (Minden et al., 1989; Denk et al., 1990; Mohler and White, 1998a) (J. A. Waddle, personal communication). The increase in use of vital fluorescent probes and green fluorescent protein (GFP) fusion transgenes has combined with these new imaging approaches to bring live time-lapse fluorescence microscopy to new prominence. Thus with low-damage fluorescence imaging we may now fruitfully combine 4D and 5D recording with 3D volume reconstruction to watch the behavior of an entire space-filling fluorescent specimen.
A worthwhile goal then is to present the viewer with a time-animated volume rendering of a such a specimen that can be handled manually and experienced visually on a time scale suitable to the viewer’s intrinsic sensitivity to space and motion. To pursue this goal, I have focused on adapting functions of existing graphics programs to the specific task of portraying these 4D and 5D microscope data sets. Data reconstruction has been performed on either of two different levels of computational sophistication: the inexpensive personal computer and the advanced graphics workstation. In both cases, however, public domain “open source” software is the basis for creating and viewing data reconstructions. My hope is that visualization methods like these may become streamlined enough so that biologists, rather than computer technophiles, may easily use such technology to explore their subjects.
MATERIALS AND METHODS
“The antique contrivance consisted of twin lenses set in a leather-covered housing lined with red velvet.”
The details of apparatus used in this work, although less elegant than Franck’s grandfather’s, are provided here. The relative strengths and weaknesses of each reconstruction scheme are presented.
Microscopy
C. elegans embryos were imaged by multiphoton or confocal laser-scanning microscopy, as previously described. Image stacks were recorded with commercial acquisition software in Bio-Rad “.PIC” file format.
Stereo–4D Reconstruction
On the Apple Macintosh platform, I have used a customized combination of three programs to produce and view rotatable, time-animated volume (“stereo-4D”) reconstructions (Mohler and White, 1998b) (http://www.loci.wisc.edu/stereo4d). NIH Image v1.62 was developed by Wayne Rasband at the National Institutes of Health and is available on the Internet at http://rsb.info.nih.gov/nih- image/. 4D Turnaround v3.23 and 4D Viewer v4.12 are available at http://www.loci.wisc.edu/4d (Thomas et al., 1996). In essence, a stack of optical sections from each time point is converted to a stack of projections of the 3D specimen seen from incrementally rotated angles of view. The whole set of all projections from all time points is then collated into a single QuickTime movie file. Once a stereo–4D reconstruction has been made, the observer uses the keyboard to roam forward or backward in time, turning the specimen to examine it from different perspectives. The viewing experience is strictly a matter of movie playback; all calculations of projected views are performed before the reconstruction is observed. Using video compression algorithms standard within QuickTime, the final stereo–4D movie file can be relatively compact, occupying a small fraction of the disk space filled by the original raw image files. The program 4D Viewer plays movies from disk and thus demands little operating memory, an ideal situation for distribution of full reconstructions to colleagues with unassuming personal computers.
NIH Image, the program doing the lion’s share of the work during reconstruction, has many useful options for customizing the display of data. In addition, the stereo–4D macros allow for isolation (“cropping”) or removal (“coring”) of user-defined volumes of data from within the greater imaged volume; however, any such settings must be made at the start of the stereo–4D reconstruction process and cannot be changed while the reconstruction is viewed. Nonetheless, with some experience, suitable settings can be anticipated. Alternative stereo–4D renderings or standard slice-by-slice 4D movies of the same data must each be made into separate movie file. Furthermore, simultaneous display of multiple colored channels (“stereo–5D”) requires merging of separate movies within the advanced video-editing program Adobe Premiere v5.1.
Vis5D
The program Vis5D v5.0 is an advanced graphics application capable of interactively displaying and reconstructing 5D animations directly from raw data (Hibbard et al., 1994). Designed to plot weather-modeling data by the Visualization Project at the University of Wisconsin Space Science and Engineering Center, the package runs on a number of computer platforms and supports rendering by either 3D graphics hardware or the Mesa software library. My work has been performed running Vis5D within the IRIX 6.2 operating system on a Silicon Graphics Onyx Reality Engine with 512 MB of system RAM. The interface in Vis5D is almost entirely mouse-driven and gives the user a kinesthetic sense of control over the rendered object, including stepwise or running temporal animation, free rotation around any point, and zoom magnification within the display window. The viewer also can probe the intensity values of individual pixels within the volume and change display parameters such as color lookup tables or threshold values while actively examining the recording. In addition to isosurface or volumetric 3D rendering of the entire volume, Vis5D allows reslicing to create optical sections at any angle relative to the original image plane. Display of each separate channel may be controlled independently using any of these options, either within a single reconstruction encompassing all variables or in separate parallel display windows that are synchronized in time and space. Alternatively, Vis5D-formatted data can be displayed in the program Cave5D, which allows live rendering using immersive “virtual reality” presentation hardware.
Although it is able to work from disk, Vis5D performs most smoothly when all data are resident in system memory. Furthermore, the program’s running memory demand while rendering often equals up to 30 times the size of the raw data itself. Thus unlike playback of a QuickTime stereo–4D movie from disk, live viewing in Vis5D is governed by limitation of the size of the data set in proportion to the computer’s system memory capacity. In practice, I have often dramatically reduced the pixel–resolution of raw data before entry into Vis5D to allow display of the full spatiotemporal range of a recording. Fortunately, because the forte of the workstation is polished graphical rendering, this reduction in data resolution (up to 36-fold minimization in some cases) can have a surprisingly slight effect on the quality of the final image. The intensive polygonal interpolation performed by the graphics hardware does a remarkable job of restoring accurate continuity between the sparse data points.
The main issue in basic adaptation of Vis5D to microscopy has been conversion of data to “.v5d” file format. Fortunately, pixel bytes are ordered similarly both in Bio-Rad .PIC image stack files and in .v5d grid files, and a simple data-reformatting program was rather easy to devise. Still under development is a “transparent” data converter that will import image file data into Vis5D—automatically reducing pixel density, if desired— with minimal attention from the user.
RESULTS AND DISCUSSION
“Then, pressing the velvet edge to your face, you saw through the lenses an oak tree, not flat, as in picture, but all in the round: a living presence.”
In my own experience, the act of examining a reconstructed microscope recording can be no less remarkable than Franck’s view through the stereopticon. The videos presented here should demonstrate some of the potential for increased spatial comprehension that can be fulfilled by these sorts of multidimensional reconstruction methods. Unfortunately, publication limits dictate that in each case only a short representative linear sequence from a full interactive multidimensional recording is shown. Especially in Videos 3 through 5, the main purpose is to portray the important aspects of the viewing experience rather than specific features of the individual specimens.
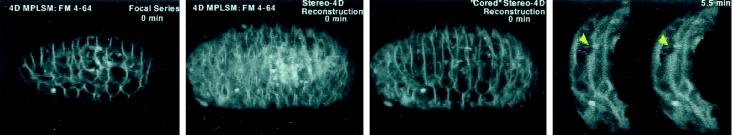
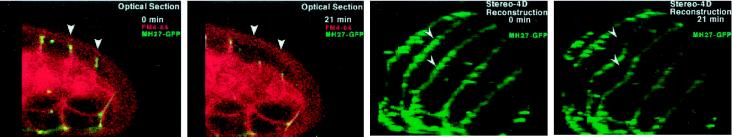
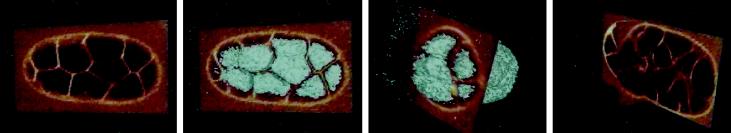
Videos 1 and 2 (reproduced with permission from Mohler et al., 1998) are examples of stereo–4D reconstruction applied to imaging of cell membrane fusions within live C. elegans embryos. In Video 1 (Figure 1) the complementary attributes of slice-by-slice standard 4D and volume-rendered stereo–4D format are apparent. Cell fusions occur within the hypodermis, an epithelial monolayer that envelops the embryo. Because these cells are curved, no single plane of section can capture the full extent of the cell boundaries; however, stereo–4D rendering—taking additional advantage of the ability to “core” out internal embryonic structures—yields a rotatable animated reconstruction of the entire living skin of the embryo, viewed in isolation. Thus, the progressive disappearance of membranes between fusing cells can be observed and compared both in cross section and in three dimensions. In Video 2 (Figure 2), the same principles are applied to the same problem, but this time 5D recording is used to observe both the membranes and a fluorescent protein (MH27::GFP) within the intercellular junctional complex. The outcome of this work is a clear view, for the first time, of the behavior of membranes in actively fusing cells, as well as two surprising but vivid conclusions. The first is that the fusion origin is situated at a point “above” the apical adherens junctions between neighboring cells, and the second is that the wave-like widening of the fusion aperture actually displaces the adherens junction to the basal surface of the cell (Mohler et al., 1998).
Figure 1.
Dynamics of cell membrane fusion seen in stereo–4D reconstructions of C. elegans embryos labeled with FM 4–64 and imaged by multiphoton microscopy (Mohler et al., 1998). This video illustrates the processes of 3D rendering and “cored” reconstruction, as well as demonstrating the widening of the cell fusion aperture both in cross section and 3D. Embryos measure ∼60 × 30 μm.
Figure 2.
Relative movement of membranes and intercellular junctions shown by dual-channel stereo–4D reconstruction (Mohler et al., 1998). FM 4–64–labeled membranes (red) and MH27::GFP-labeled adherens junctions (green) were imaged simultaneously by confocal microscopy. The signals are shown together and separately, in both optical sections and 3D reconstructions, to demonstrate localization of junctional protein to the receding edge of fusing plasma membranes.
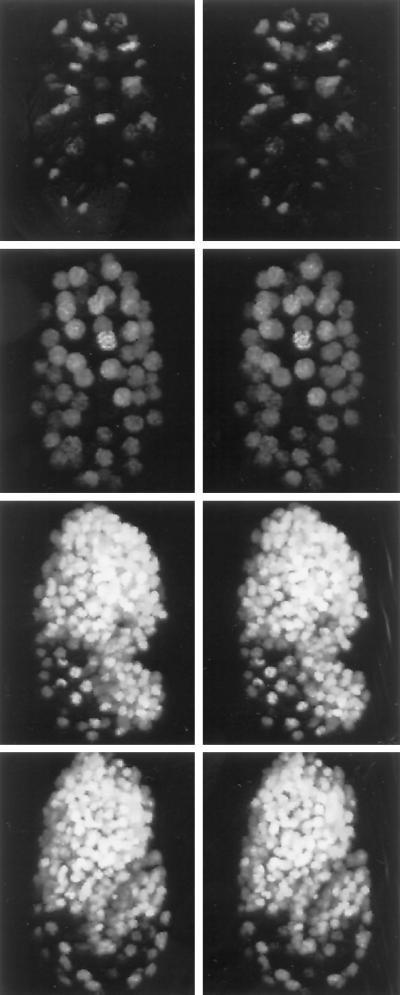
Video 3 (Figure 3) shows stereo–4D footage of cell nuclei during C. elegans embryonic cleavage, gastrulation, and early morphogenesis. Here, in a technique being developed for automated lineage analysis (J. A. Waddle, personal communication), a histone-H1::GFP chimeric protein binds to DNA, allowing 4D fluorescence imaging of nuclei and condensed chromatin during all phases of the cell cycle. Stereo–4D reconstruction provides perspective, within a single field of view, on the relative positioning and temporal synchrony of the many cell divisions and migrations captured during this recording. Each cell division within the embryo, even if directed out of the imaging plane, can be characterized as to its orientation and the timing of its various stages. Meanwhile, the entirety of these individual divisions can gradually be seen to produce and precisely arrange the ∼550 component cells, such that cell divisions largely cease before differentiation and morphogenesis begin to mold the various organs and the animal takes shape. Such stereoscopic perspective should certainly complement the slice-by-slice analysis of 4D cell lineage recordings for which this nuclear imaging strategy was developed.
Figure 3.
Nuclei of a developing C. elegans embryo labeled with histone-H1::GFP and followed during development by stereo–4D reconstruction from a multiphoton recording. Adjacent images should be viewed in stereo.
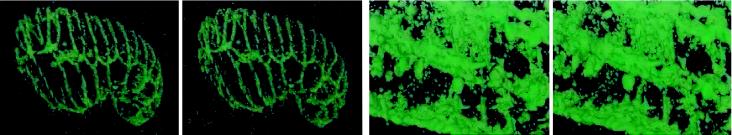
Videos 4 and 5 portray Vis5D reconstruction of microscope recordings. It is worth emphasizing here that unlike the stereo–4D sequences above, these show “real-time” unedited recordings of viewing sessions in the Vis5D environment. In Video 4 (Figure 4), an embryo expressing the junctional MH27::GFP marker is shown during a short period of C. elegans development spanning the fusion of several hypodermal cells. The zoom, free-rotation, and continuous animation features of the Vis5D interface are apparent. The 3D isosurface rendering shown here is comparable to stereo–4D projections in Video 2 of a similar specimen. Yet in Vis5D viewing, the embryo can be watched from any angle of view, and internal epithelial structures can be magnified and examined with outer layers “dissected” away. Any choice to change the viewing parameters can be exercised immediately according to the viewer’s whim.
Figure 4.
Adherens junctions in a C. elegans embryo labeled with MH27::GFP, imaged by multiphoton microscopy, and rendered in Vis5D. The free rotation, time animation, zoom, and clipping-plane features of Vis5D are illustrated. This sequence shows an embryo at the onset of elongation, and several dorsal hypodermal cells fuse during the time-course loop portrayed.
In another example, the plasma membranes of a midcleavage stage C. elegans embryo were imaged in the recording shown in Video 5 (Figure 5). In this sequence, optical sections —in the original “XY” orientation, as well as perpendicular “XZ” and arbitrarily tilted angles—are shaded with a custom pseudocolor palette. At the same time, isosurfaces defining the shapes of all cells within the embryo are rendered in three dimensions. Each of these representations may be toggled on or off, and they may all be viewed simultaneously while they are manipulated in space and animated through time. This method highlights the cell borders, complementing the imaging of DNA shown in Video 3, to yield a converse record of the cell lineage. The symmetry or asymmetry of each division and the sizes and shapes of individual cells are revealed, as are the surface areas and durations of transient, and potentially inductive, intercellular contact events.
Figure 5.
Plasma membranes of dividing cells within a cleaving C. elegans embryo. FM 4–64–labeled membranes were imaged in a multiphoton recording and displayed in Vis5D. Simultaneous time animation, free rotation, reslicing, and isosurface rendering are illustrated.
CONCLUSION
“Grandfather’s gadget … gave me the first hints that one’s everyday eye can become an awakened eye, an eye that can do infinitely more than merely look at things, recognize, classify, label them.”
Although the artist’s wont is to take from a subject its abstract and spiritual essence, to build imagination on reality, a microscopist’s passion for recognizing, classifying, and labeling the features of life can be similarly awakened by a holistic gaze upon the specimen. It is often a biologist’s goal in microscopy to move from conceptual models, abstracted from less visual modes of experimentation, to confirmation based on our instinct to see and believe. The sorts of “living” reconstructions described in this essay simplify the viewer’s ability to transition from collecting data on the microscope, to observing and reflexively recognizing an important event or detail within the fully rendered specimen, to analytically scrutinizing an isolated element in a single optical section or time point.
Microscopes, computers, and software continue to improve. As their design becomes more focused on the specific task of multidimensional imaging, and the simplicity and flexibility in making and viewing recordings increases, the ability to share and publish the full abundance of these data sets will become imperative. The software required for both methods I have described here is freely distributed to the scientific community and is being actively adapted for use across the Internet. This is essential to the goal of sharing the view with biologists at large, letting eyes with different experience and perspective explore the same rich space, so that high-tech microscopic imaging progresses from an exclusive “gee-whiz” art form to a facile form of communication. Life scientists will soon learn, as Frederick Franck advises:
“… that seeing things and beings in this way is an excellent substitute—to say the least—for lots of thinking and reading about them.”
Supplementary Material
ACKNOWLEDGMENTS
I thank John White for advice and for use of microscopes at the Integrated Microscopy Resource; David Wokosin and Victoria Centonze Frohlich for instrument support; Bill Hibbard for discussions and demonstration of Vis5D; Michael Redmond for access to the Silicon Graphics workstation at the University of Wisconsin Model Advanced Facility; Charles Thomas, Kevin Eliceiri, Joshua Leach, Andrew Gardner, and Jean-Yves Sgro for computing guidance; Melanie Dunn and Geraldine Seydoux for histone-H1::GFP transgenic worms; and James Waddle for discussion of unpublished data. This work was supported by fellowship GM18200-02 and grant RR00570 from the National Institutes of Health and by a research development grant from the Muscular Dystrophy Association.
Footnotes
REFERENCES
- Amberg D. Three-dimensional imaging of the yeast actin cytoskeleton through the budding cell cycle. Mol Biol Cell. 1998;9:3259–3262. doi: 10.1091/mbc.9.12.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Hughes DD, Chan TA, Sedat JW, Agard DA. IVE (Image Visualization Environment): a software platform for all three-dimensional microscopy applications. J Struct Biol. 1996;116:56–60. doi: 10.1006/jsbi.1996.0010. [DOI] [PubMed] [Google Scholar]
- Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- Drebin RA, Carpenter L, Hanrahan P. Volume rendering. Comput Graphics. 1988;22:65–78. [Google Scholar]
- Fire A. A four-dimensional digital image archiving system for cell lineage tracing and retrospective embryology. Comput Appl Biosci. 1994;10:443–447. doi: 10.1093/bioinformatics/10.4.443. [DOI] [PubMed] [Google Scholar]
- Ford-Holevinski TS, Castle MR, Herman JP, Watson SJ. Microcomputer-based three-dimensional reconstruction of in situ hybridization autoradiographs. J Chem Neuroanat. 1991;4:373–385. doi: 10.1016/0891-0618(91)90044-d. [DOI] [PubMed] [Google Scholar]
- Franck F. The stereopticon. Parabola. 1997;22:33–36. [Google Scholar]
- Hibbard WL, Paul BE, Santek DA, Dyer CR, Battaiola AL, Voidrot-Martinez MF. Interactive visualization of earth and space science computations. Computer. 1994;27:65–72. [Google Scholar]
- Hird SN, White JG. Cortical and cytoplasmic flow polarity in early embryonic cells of Caenorhabditis elegans. J Cell Biol. 1993;121:1343–1355. doi: 10.1083/jcb.121.6.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Inoue TD. Through-focal and time-lapse stereoscopic imaging of dividing cells and developing embryos in DIC and polarization microscopy. Biol Bull. 1994;187:232–233. doi: 10.1086/BBLv187n2p232. [DOI] [PubMed] [Google Scholar]
- Minden JS, Agard DA, Sedat JW, Alberts BM. Direct cell lineage analysis in Drosophila melanogaster by time-lapse, three-dimensional optical microscopy of living embryos. J Cell Biol. 1989;109:505–516. doi: 10.1083/jcb.109.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler W, Simske J, Williams-Masson E, Hardin J, White J. Dynamics and ultrastructure of developmental cell fusions in the Caenorhabditis elegans hypodermis. Curr Biol. 1998;8:1087–1090. doi: 10.1016/s0960-9822(98)70447-6. [DOI] [PubMed] [Google Scholar]
- Mohler WA, White JG. Multiphoton laser scanning microscopy for four-dimensional analysis of C. elegans embryonic development. Optics Express. 1998a;3:325–331. doi: 10.1364/oe.3.000325. [DOI] [PubMed] [Google Scholar]
- Mohler WA, White JG. Stereo-4-D reconstruction and animation from living fluorescent specimens. Biotechniques. 1998b;24:1006–1010. , 1012. [PubMed] [Google Scholar]
- Thomas C, DeVries P, Hardin J, White J. Four-dimensional imaging: computer visualization of 3D movements in living specimens. Science. 1996;273:603–607. doi: 10.1126/science.273.5275.603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.