Abstract
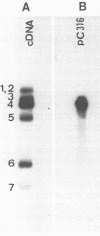
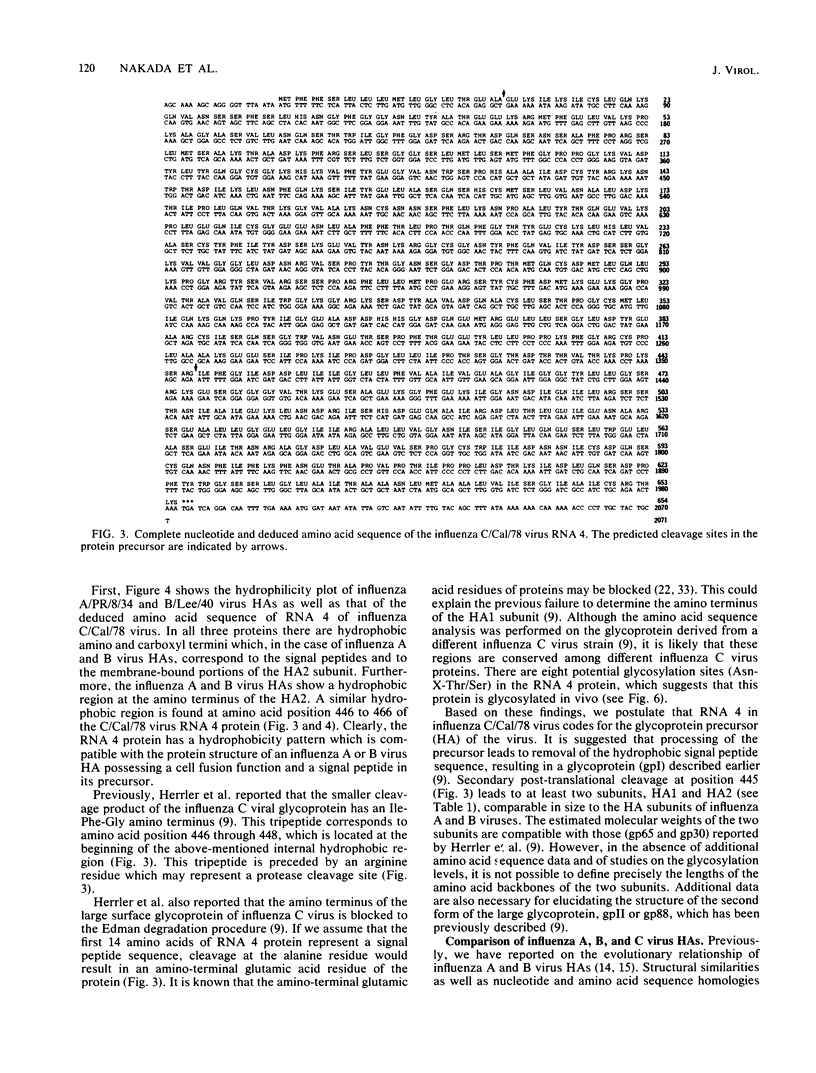
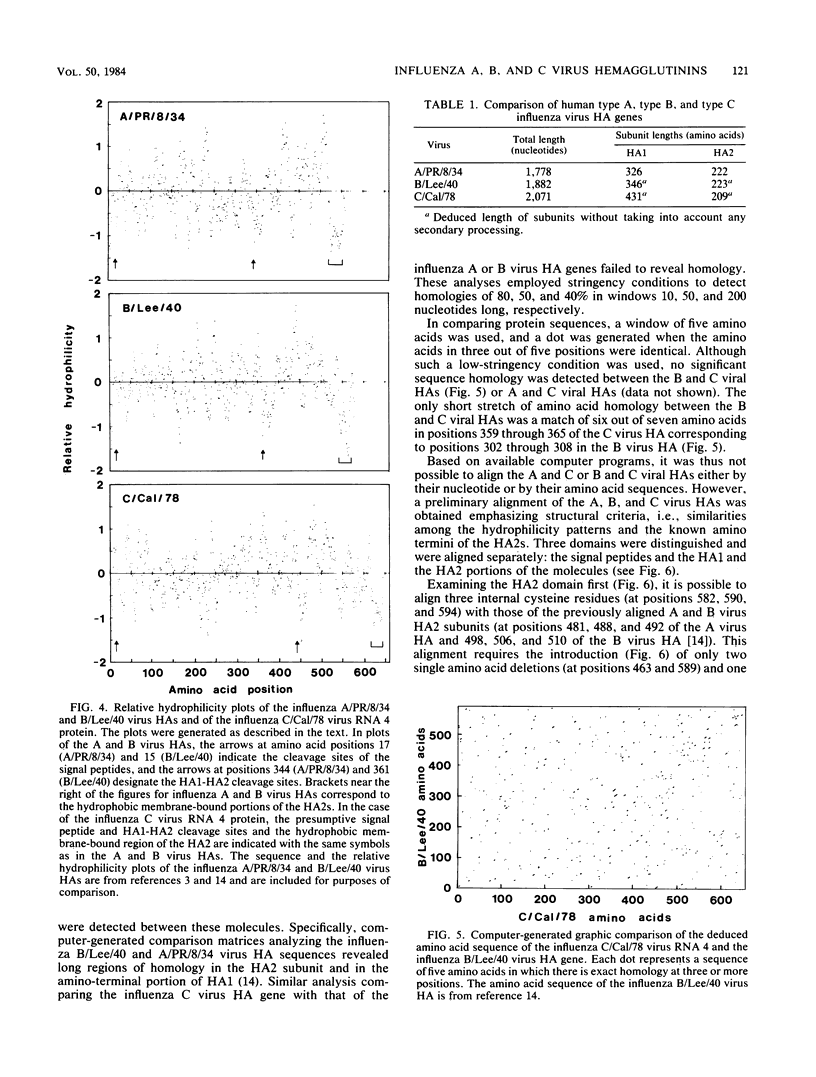
The complete nucleotide sequence of the influenza C/California/78 virus RNA 4 was obtained by using cloned cDNA derived from the RNA segment. This gene is 2,071 nucleotides long and can code for a polypeptide of 654 amino acids. Although there are no convincing sequence homologies between RNA 4 and the hemagglutinin genes of influenza A and B viruses, we suggest, on the basis of structural features, that RNA 4 of the influenza C virus codes for the hemagglutinin. The structural features which are common to the hemagglutinins of influenza A, B, and C viruses include (i) a hydrophobic signal peptide, (ii) an arginine cleavage site between the hemagglutinin 1 and 2 subunits, (iii) hydrophobic regions at the amino and carboxyl termini of the hemagglutinin 2 subunit, and (iv) several conserved cysteine residues. Additional evidence that RNA 4 of influenza C virus codes for the hemagglutinin is that the tripeptide Ile-Phe-Gly, known to be present at the amino terminus of the hemagglutinin 2 subunit of influenza C virus, is encoded by RNA 4 at a point immediately adjacent to the presumptive arginine cleavage site. The lack of primary sequence homology between the influenza C virus hemagglutinin and the influenza A or B virus hemagglutinins, which all have similar functions, might be attributed to convergent rather than divergent evolution. However, the structural similarities among the influenza A, B, and C virus hemagglutinins strongly suggest that the three hemagglutinin genes have diverged from a common precursor.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alwine J. C., Kemp D. J., Parker B. A., Reiser J., Renart J., Stark G. R., Wahl G. M. Detection of specific RNAs or specific fragments of DNA by fractionation in gels and transfer to diazobenzyloxymethyl paper. Methods Enzymol. 1979;68:220–242. doi: 10.1016/0076-6879(79)68017-5. [DOI] [PubMed] [Google Scholar]
- Baez M., Taussig R., Zazra J. J., Young J. F., Palese P., Reisfeld A., Skalka A. M. Complete nucleotide sequence of the influenza A/PR/8/34 virus NS gene and comparison with the NS genes of the A/Udorn/72 and A/FPV/Rostock/34 strains. Nucleic Acids Res. 1980 Dec 11;8(23):5845–5858. doi: 10.1093/nar/8.23.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compans R. W., Bishop D. H., Meier-Ewert H. Structural components of influenza C virions. J Virol. 1977 Feb;21(2):658–665. doi: 10.1128/jvi.21.2.658-665.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desselberger U., Palese P. Molecular weights of RNA segments of influenza A and B viruses. Virology. 1978 Jul 15;88(2):394–399. doi: 10.1016/0042-6822(78)90297-0. [DOI] [PubMed] [Google Scholar]
- Desselberger U., Racaniello V. R., Zazra J. J., Palese P. The 3' and 5'-terminal sequences of influenza A, B and C virus RNA segments are highly conserved and show partial inverted complementarity. Gene. 1980 Feb;8(3):315–328. doi: 10.1016/0378-1119(80)90007-4. [DOI] [PubMed] [Google Scholar]
- Gething M. J., Bye J., Skehel J., Waterfield M. Cloning and DNA sequence of double-stranded copies of haemagglutinin genes from H2 and H3 strains elucidates antigenic shift and drift in human influenza virus. Nature. 1980 Sep 25;287(5780):301–306. doi: 10.1038/287301a0. [DOI] [PubMed] [Google Scholar]
- Grütter M. G., Weaver L. H., Matthews B. W. Goose lysozyme structure: an evolutionary link between hen and bacteriophage lysozymes? Nature. 1983 Jun 30;303(5920):828–831. doi: 10.1038/303828a0. [DOI] [PubMed] [Google Scholar]
- Herrler G., Nagele A., Meier-Ewert H., Bhown A. S., Compans R. W. Isolation and structural analysis of influenza C virion glycoproteins. Virology. 1981 Sep;113(2):439–451. doi: 10.1016/0042-6822(81)90173-2. [DOI] [PubMed] [Google Scholar]
- Hiti A. L., Davis A. R., Nayak D. P. Complete sequence analysis shows that the hemagglutinins of the H0 and H2 subtypes of human influenza virus are closely related. Virology. 1981 May;111(1):113–124. doi: 10.1016/0042-6822(81)90658-9. [DOI] [PubMed] [Google Scholar]
- Kendal A. P. A comparison of "influenza C" with prototype myxoviruses: receptor-destroycing activity (neuraminidase) and structural polypeptides. Virology. 1975 May;65(1):87–99. doi: 10.1016/0042-6822(75)90009-4. [DOI] [PubMed] [Google Scholar]
- Kester W. R., Matthews B. W. Comparison of the structures of carboxypeptidase A and thermolysin. J Biol Chem. 1977 Nov 10;252(21):7704–7710. [PubMed] [Google Scholar]
- Kitame F., Sugawara K., Ohwada K., Homma M. Proteolytic activation of hemolysis and fusion by influenza C virus. Arch Virol. 1982;73(3-4):357–361. doi: 10.1007/BF01318090. [DOI] [PubMed] [Google Scholar]
- Krystal M., Elliott R. M., Benz E. W., Jr, Young J. F., Palese P. Evolution of influenza A and B viruses: conservation of structural features in the hemagglutinin genes. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4800–4804. doi: 10.1073/pnas.79.15.4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystal M., Young J. F., Palese P., Wilson I. A., Skehel J. J., Wiley D. C. Sequential mutations in hemagglutinins of influenza B virus isolates: definition of antigenic domains. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4527–4531. doi: 10.1073/pnas.80.14.4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B. W., Grütter M. G., Anderson W. F., Remington S. J. Common precursor of lysozymes of hen egg-white and bacteriophage T4. Nature. 1981 Mar 26;290(5804):334–335. doi: 10.1038/290334a0. [DOI] [PubMed] [Google Scholar]
- Matthews B. W., Remington S. J., Grütter M. G., Anderson W. F. Relation between hen egg white lysozyme and bacteriophage T4 lysozyme: evolutionary implications. J Mol Biol. 1981 Apr 25;147(4):545–558. doi: 10.1016/0022-2836(81)90399-5. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Nerome K., Ishida M., Nakayama M. Absence of neuraminidase from influenza C virus. Arch Virol. 1976;50(3):241–244. doi: 10.1007/BF01320578. [DOI] [PubMed] [Google Scholar]
- Ohuchi M., Ohuchi R., Mifune K. Demonstration of hemolytic and fusion activities of influenza C virus. J Virol. 1982 Jun;42(3):1076–1079. doi: 10.1128/jvi.42.3.1076-1079.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov Y. A., Abdulaev N. G., Feigina M. Y., Kiselev A. V., Lobanov N. A. The structural basis of the functioning of bacteriorhodopsin: an overview. FEBS Lett. 1979 Apr 15;100(2):219–224. doi: 10.1016/0014-5793(79)80338-5. [DOI] [PubMed] [Google Scholar]
- Palese P., Racaniello V. R., Desselberger U., Young J., Baez M. Genetic structure and genetic variation of influenza viruses. Philos Trans R Soc Lond B Biol Sci. 1980 Feb 25;288(1029):299–305. doi: 10.1098/rstb.1980.0005. [DOI] [PubMed] [Google Scholar]
- Racaniello V. R., Palese P. Isolation of influenza C virus recombinants. J Virol. 1979 Dec;32(3):1006–1014. doi: 10.1128/jvi.32.3.1006-1014.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchey M. B., Palese P., Kilbourne E. D. RNAs of influenza A, B, and C viruses. J Virol. 1976 May;18(2):738–744. doi: 10.1128/jvi.18.2.738-744.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J. S., Schubert M., Lazzarini R. A. Polyadenylation sites for influenza virus mRNA. J Virol. 1981 Apr;38(1):157–163. doi: 10.1128/jvi.38.1.157-163.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertus J. D., Alden R. A., Birktoft J. J., Kraut J., Powers J. C., Wilcox P. E. An x-ray crystallographic study of the binding of peptide chloromethyl ketone inhibitors to subtilisin BPN'. Biochemistry. 1972 Jun 20;11(13):2439–2449. doi: 10.1021/bi00763a009. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Staden R. A strategy of DNA sequencing employing computer programs. Nucleic Acids Res. 1979 Jun 11;6(7):2601–2610. doi: 10.1093/nar/6.7.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Further procedures for sequence analysis by computer. Nucleic Acids Res. 1978 Mar;5(3):1013–1016. doi: 10.1093/nar/5.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Sequence data handling by computer. Nucleic Acids Res. 1977 Nov;4(11):4037–4051. doi: 10.1093/nar/4.11.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I. A., Skehel J. J., Wiley D. C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981 Jan 29;289(5796):366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- Wong R. S., Hofmann T., Bennick A. The complete primary structure of a proline-rich phosphoprotein from human saliva. J Biol Chem. 1979 Jun 10;254(11):4800–4808. [PubMed] [Google Scholar]