Abstract
Stress conditions affecting the functions of the endoplasmic reticulum (ER) cause the accumulation of unfolded proteins. ER stress is counteracted by the unfolded-protein response (UPR). However, under prolonged stress the UPR initiates a proapoptotic response. Mounting evidence indicate that the ER chaperone calnexin is involved in apoptosis caused by ER stress. Here, we report that overexpression of calnexin in Schizosaccharomyces pombe induces cell death with apoptosis markers. Cell death was partially dependent on the Ire1p ER-stress transducer. Apoptotic death caused by calnexin overexpression required its transmembrane domain (TM), and involved sequences on either side of the ER membrane. Apoptotic death caused by tunicamycin was dramatically reduced in a strain expressing endogenous levels of calnexin lacking its TM and cytosolic tail. This demonstrates the involvement of calnexin in apoptosis triggered by ER stress. A genetic screen identified the S. pombe homologue of the human antiapoptotic protein HMGB1 as a suppressor of apoptotic death due to calnexin overexpression. Remarkably, overexpression of human calnexin in S. pombe also provoked apoptotic death. Our results argue for the conservation of the role of calnexin in apoptosis triggered by ER stress, and validate S. pombe as a model to elucidate the mechanisms of calnexin-mediated cell death.
INTRODUCTION
The endoplasmic reticulum (ER) is a specialized organelle playing essential and central roles in the biology of the cell. The ER is the site of synthesis and folding of secreted, membrane-bound and some organelle-targeted proteins (Bukau et al., 2000; Fewell et al., 2001). Protein folding in the ER is assisted by a battery of molecular chaperones and foldases (Bukau et al., 2000; Fewell et al., 2001; Trombetta and Parodi, 2003; Helenius and Aebi, 2004). In addition, the ER contains several factors required for optimum protein folding, including, ATP, Ca2+, and an oxidizing environment to allow disulphide-bond formation. Proper ER function is critical for numerous aspects of cell physiology, including vesicle trafficking and lipid and membrane biogenesis, as well as protein targeting and secretion (Lai et al., 2007).
The ER is highly sensitive to stresses perturbing the cellular energy levels and ER lipid or glycolipid imbalances or changes in the redox state or Ca2+ concentration (Breckenridge et al., 2003; Boyce and Yuan, 2006; Szegezdi et al., 2006). Such stresses reduce the protein folding capacity of the ER, which results in the accumulation and aggregation of unfolded proteins, a condition referred to as ER stress. When the capacity of the ER to fold proteins properly is compromised or overwhelmed, a highly conserved unfolded-protein response (UPR) signal-transduction pathway is activated. The ER response to stress is basically conserved from yeast to mammalian cells (Patil and Walter, 2001; Ron and Walter, 2007). To counter ER stress, the UPR halts general protein synthesis and up-regulates the transcription of genes encoding ER resident chaperones and other regulatory components of the secretory pathway, giving the cell a chance to correct the environment within the ER (Breckenridge et al., 2003; Schroder and Kaufman, 2005; Xu et al., 2005; Boyce and Yuan, 2006; Szegezdi et al., 2006; Wu and Kaufman, 2006). Moreover, in coordination with the UPR, the cell increases the capacity of the proteasome-dependent ER-associated degradation (ERAD) to remove misfolded proteins from the ER (Ahner and Brodsky, 2004; Meusser et al., 2005; Marciniak and Ron, 2006).
In Saccharomyces cerevisiae, ER stress is monitored by Ire1p, an ER transmembrane protein. According to current models (Patil and Walter, 2001; McCracken and Brodsky, 2005; Boyce and Yuan, 2006; Ron and Walter, 2007), the ER lumenal domain of a single Ire1p molecule interacts with BiP/Kar2p, an ER lumenal chaperone that assists the folding of nascent polypeptides entering the lumen. Under ER stress conditions, BiP is titrated away from Ire1p due to its binding to unfolded or misfolded proteins. The removal of BiP permits the activation of Ire1p by oligomerization leading to the efficient induction of UPR, which up-regulates genes coding for ER chaperones and factors involved in all stages of the secretory pathway. The mammalian UPR retains several key features of the yeast program like the role of BiP, but is significantly more complex and is mediated through three ER transmembrane receptors: PERK, ATF6, and IRE1. In normal conditions, the suite of responses activated by the UPR pathway succeeds in restoring ER homeostasis. However, if protein aggregation is persistent and the stress cannot be resolved, the UPR signaling switches from prosurvival to proapoptotic. On prolonged ER stress the PERK, ATF6, and IRE1 proapoptotic signals ultimately execute cell death through the activation of kinases, proteases, and transcription factors, thereby affecting the expression and activities of the Bcl-2 family of proteins and their modulators (Breckenridge et al., 2003; Xu et al., 2005; Szegezdi et al., 2006).
Apoptosis is a form of programmed cell death that is characterized by several specific morphological and biochemical changes including cell rounding and shrinkage, chromatin breakage, nuclear fragmentation, and activation of caspases (Kerr et al., 1972; Kerr, 2002). Apoptosis was first described and extensively studied in multicellular organisms, where it plays critical roles in development and the control of many diseases in adult organisms. Evidence accumulated in the last 10 y has demonstrated that apoptosis-like processes do also occur in S. cerevisiae and in Schizosaccharomyces pombe yeasts. Yeasts undergo apoptosis-like death under a variety of conditions and cell processes including DNA damage, mitotic catastrophe, replication defects, deficiency in triacylglycerols, ER stress, aging, and mating (Burhans et al., 2003; Zhang et al., 2003; Fahrenkrog et al., 2004; Wissing et al., 2004; Low et al., 2005; Cho et al., 2006; Hauptmann et al., 2006; Roux et al., 2006; Walter et al., 2006; Frohlich et al., 2007). Both S. cerevisiae and S. pombe encode several homologues of proteins characterized for their implication in apoptosis including AIF, HtrA2/Omi, and IAP. Yeasts also encode a metacaspase called Yca1/Mca1 in S. cerevisiae and Pca1 in S. pombe (Uren et al., 2000; Madeo et al., 2002; Lim et al., 2007). However, programmed-cell death pathways in yeast can be metacaspase-dependent or -independent (Madeo et al., 2002; Bettiga et al., 2004; Fannjiang et al., 2004; Herker et al., 2004; Wadskog et al., 2004; Ivanovska and Hardwick, 2005; Reiter et al., 2005; Liang et al., 2008; Mazzoni and Falcone, 2008). It has been proposed that apoptotic processes are useful in unicellular organisms for their survival under conditions of environmental stresses and are relevant in populations of cells such as colonies and biofilms (Skulachev, 2002; Buttner et al., 2006). Because of their powerful genetics, S. cerevisiae and S. pombe have become interesting models to study the core mechanisms of apoptosis (Ink et al., 1997; Ligr et al., 1998; Madeo et al., 2002; Priault et al., 2003; Hardwick and Cheng, 2004; Madeo et al., 2004; Rodriguez-Menocal and D'Urso, 2004; Burhans and Weinberger, 2007; Frohlich et al., 2007; Almeida et al., 2008; Fabrizio and Longo, 2008; Low and Yang, 2008).
The molecular chaperone calnexin plays key roles in the translocation of nascent polypeptides and in the folding and quality control of newly synthesized proteins (Bukau et al., 2000; Fewell et al., 2001; Williams, 2006). Structurally, calnexin is a type I ER transmembrane protein, with a large lumenal domain, a transmembrane domain (TM), and a short cytosolic tail (see Figure 1A). The lumenal domain of calnexin contains the highly conserved central domain (hcd), which is the portion of the protein that is the most conserved across species. This region of calnexin interacts with client proteins in a glycan-lectin manner or via protein–protein interactions (Arunachalam and Cresswell, 1995; Fernandez et al., 1996; Jannatipour et al., 1998; Beaulieu et al., 1999; Saito et al., 1999; Parodi, 2000; Marechal et al., 2004; Hebert et al., 2005; Thammavongsa et al., 2005; Williams, 2006).
Figure 1.
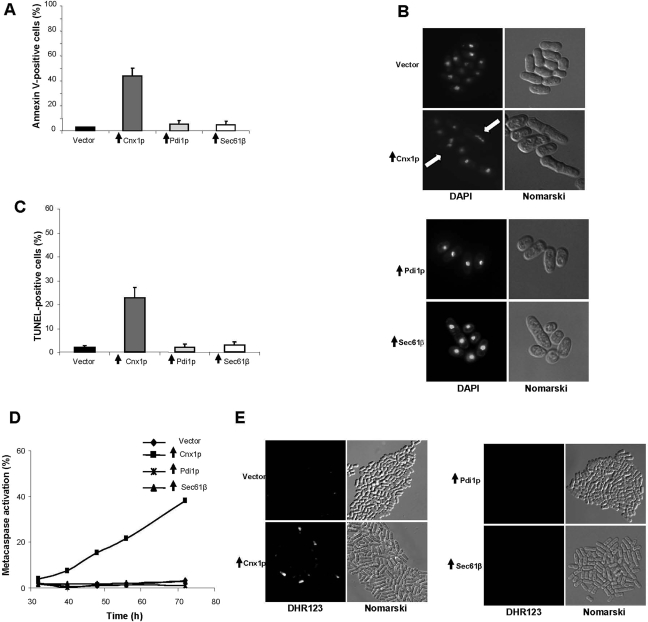
Overexpression of cnx1+ induces cell death. (A) Schematic representation of S. pombe calnexin, Cnx1p. SP corresponds to the signal peptide (22 amino acids; aa), followed by the N-terminal part (98 aa; symbolized by the forward-hatched box). The highly conserved central domain (hcd) is represented by a white box (317 aa). The C-terminal part of lumenal domain of calnexin is 52-aa long and is represented by the backward-hatched box. The transmembrane domain (TM, black box) and the cytosolic tail (punctuated box) span 23 and 48 aa, respectively. (B) Survival of cells overexpressing Cnx1p (strain SP8007R) and control strains (empty vector, SP7975R; Pdi1p, SP16058; and Sec61β, SP15302) was assayed by serial dilution on inducing plates (without thiamine) or repressing plates (with thiamine). Samples of 10 μl of four 10-fold serial dilutions (10−1–10−4) of cells at 0.5 OD595 were spotted on selective MM and incubated at 30°C for 7 d (see Materials and Methods). (C) Percentage of dead cells measured by staining with fluorescent vital dye Phloxin B. Cells overexpressing Cnx1p (strain SP8007R) and control strains (empty vector, SP7975R; Pdi1p, SP16058; and Sec61β, SP15302) were stained with Phloxin B after 48 h of induction of overexpression and fluorescent cells were quantified by FACS. Stained cells were considered as dead. A global ANOVA showed that the values obtained among the strains were significantly different (p = 0.000). In C, vertical black arrows symbolize overexpression of the indicated protein.
Pointing to its critical biological roles, the knockout of calnexin causes early-postnatal death in mouse and is lethal in the fission yeast S. pombe (Jannatipour and Rokeach, 1995; Parlati et al., 1995a; Denzel et al., 2002). Intriguingly, some S. pombe calnexin mutants devoid of chaperone activity are viable, demonstrating therefore that the essential cellular role of calnexin is not its chaperone function (Elagoz et al., 1999; Marechal et al., 2004; Hajjar et al., 2007).
Cumulating evidence point to the implication of calnexin in apoptosis induced by ER stress. First indication resulted from studies in S. pombe that led to the identification of calnexin as a lethal partner of the proapoptotic mammalian proteins Bak and Bax (Torgler et al., 1997). The authors showed that the expression of Bak is lethal in S. pombe cells expressing full-length calnexin but not in cells expressing a calnexin mutant devoid of the C-terminal tail, suggesting a role as signal transducer for calnexin by recruiting lethal partners (Torgler et al., 1997). In the case of mammalian cells, it has been shown that cells devoid of calnexin are relatively resistant to ER-stress–induced apoptosis (Zuppini et al., 2002; Groenendyk et al., 2006). It was also proposed that mammalian calnexin acts as a scaffold for the cleavage by caspase-8 of the ER transmembrane protein Bap31 under ER-stress conditions, such as in the presence of tunicamycin (Delom et al., 2006, 2007). Moreover, mammalian calnexin was shown to be cleaved by caspases under stress conditions, and it was suggested that this cleavage may have a role in the mediation of the apoptotic signal (Takizawa et al., 2004). Nevertheless, much remains to be elucidated about the mechanistic details of the role of calnexin in ER-stress–induced apoptosis. Certainly, the study of the involvement of calnexin in apoptosis caused by ER stress could greatly benefit from the use of a model organism.
Here, we demonstrate that calnexin is involved in the induction of apoptosis triggered by ER stress in S. pombe. We further show that the apoptotic effect of calnexin is counteracted by overexpression of Hmg1/2p, the S. pombe homologue of the mammalian antiapoptotic protein HMGB1 (high-mobility group box-1 protein). Interestingly, the overexpression of mammalian calnexin also induced apoptosis in S. pombe, suggesting the functional conservation of the role of calnexin in apoptosis.
MATERIALS AND METHODS
Yeast Strains, Media, and Vectors
Experiments were carried out using the S. pombe strains described in Table 1. All strains were cultured in Edinburgh minimal medium (here denoted as MM) supplemented with required supplements (Moreno et al., 1991) at 30°C. DNA transformations into S. pombe were performed by the PEG-lithium acetate procedure, as previously described (Elbe, 1992). S. pombe homozygous, haploid strains deleted for the homologue of IRE1 (geneID SPAC167.01), for the homologue of YCA1 (geneID SPCC1840.04, pca1::kanMX4; strain SP8167R), and for the gene encoding the homologue of Bap31 (geneID SPAC9E9.04, dma1::kanMX4; strain SP8083R) were purchased from Bioneer (Daejeon, Korea).
Table 1.
Yeast strains used for this study
| Strain | Genotype | Source |
|---|---|---|
| SP556 | h+ade6-M216 ura4-D18 leu 1-32 | Paul Nurse lab |
| SP7975R | SP556 + pREP3X | This lab |
| SP16058 | SP556 + pREP2pdi1+ | This lab |
| SP15302 | SP556 + pREP2sbh1+ | This lab |
| SP8007R | SP556 + pREP1cnx1+ | This lab |
| SP7965R | SP556 + pREP1mini_cnx1 | This lab |
| SP16053 | SP556 + pREP1lumenal_cnx1 | This lab |
| SP8056R | SP556 + lumenalTM_cnx1 | This lab |
| SP8125R | SP556 + pREP1C-termTM_cnx1_cmyc | This lab |
| SP248 | h−his3-D1 ade6-M216 ura4-D18 leu1-32 | Burke and Gould (1994) |
| SP8075R | SP248 Δcnx1::his3 + pREP41lumenalTM_cnx1 + pREP42cnx1+ | This study |
| SP8160R | SP248 Δcnx1::his3 + pREP41C-termTM_cnx1_cmyc + pREP42cnx1+ | This study |
| SP7951R | SP248 Δcnx1::his3 + pREP41cnx1+ + pREP42 | Elagöz et al. (1999) |
| SP8490R | SP248 Δcnx1::his3 + pREP41mini_cnx1 + pREP42 | Elagöz et al. (1999) |
| SP8488R | SP248 Δcnx1::his3 + pREP41lumenal_cnx1 + pREP42 | Elagöz et al. (1999) |
| SP8085R | SP248 Δcnx1::his3 + pREP41lumenalTM_cnx1 + pREP42 | This study |
| SP16081 | SP556 + pREP1cnx1+ + pREP2hmg1/2+ | This study |
| SP16084 | SP556 + pREP1human calnexin | This study |
| SP8083R | h+ ade6-M210 ura4-D18 leu 1-32 KanMX4::dma1 (Δdma1/bap31) | BIONEER |
| SP8167R | h+ ade6-M210 ura4-D18 leu 1-32 KanMX4::pca1 (Δpca1)) | BIONEER |
| SP8212R | h+ ade6-M210 (or M216) ura4-D18 leu 1-32 KanMX4::SPAC167.01 (Δire1) | BIONEER |
| SP8145R | SP8083R + pREP1 | This study |
| SP8136R | SP8083R + pREP1cnx1+ | This study |
| SP8178R | SP8167R + pREP3X | This study |
| SP8081R | SP8167R + pREP1cnx1+ | This study |
| SP8227R | pREP3X | This study |
| SP8231R | pREP1cnx1+ | This study |
pREP is a family of multicopy expression vectors for S. pombe. All vectors contain the S. pombe ars1 origin of replication and a version of thiamine-repressible nmt promoter (Maundrell, 1993). The nmt1 promoter in pREP1, pREP2, and pREP3X is full strength, and these vectors were used to overexpress wild-type calnexin (strains SP8007R, SP8231R, SP8136R, and SP8081R), mutants of calnexin (strains SP7965R, SP16053, SP8056R, and SP8125R), and controls (strains SP16058, SP15302, SP8227R, SP8145R, and SP8178R; Forsburg, 1993). The pREP3X vector was derived from the pREP family, has the promoter strength like pREP1 and pREP2, but has an XhoI site replacing the ATG at the 5′ end of its polylinker (Forsburg, 1993). This empty vector was used as control (strain SP7975R). The nmt promoter in the vectors pREP41 and pREP42 is medium strength and expressed calnexin at levels comparable to the endogenous genomic copy of the gene (Jannatipour et al., 1998). pREP41 differs from pREP42 in that it contains the S. pombe LEU2 marker instead of ura4+, and likewise for the pREP1 and pREP2 vectors, respectively. To test the ability to confer viability the calnexin mutants lumenalTM_cnx1 and C-termTM_cnx1_cmyc were cloned in the pREP41 and pREP42 vectors and were shuffled against wild-type calnexin (strains SP8075R and SP8160R, respectively). The wild-type calnexin and the viable mutants on pREP41 vector were used to study ER stress induced by tunicamycin (strains SP7951R, SP8490R, SP8488R, and SP8085R).
Construction of cnx1 Mutants and Plasmids
The construction of mutants mini_cnx1 and lumenal_cnx1 was previously described (Elagöz et al., 1999). The mutant lumenalTM_cnx1 was obtained by PCR amplification with the following primers: A2: 5′-AAACATATGAAGTACGGAAAGGTATCT-3′ and L: 5′-AAAGGATCCTTAAGCAAAGAAATAAAAGTAACA-3′. These two primers contained restriction sites for NdeI and BamHI to allow the cloning of the PCR product into the pREP1 and pREP41. For the mutant C-termTM_cnx1_cmyc, two independent fragments were first amplified by PCR. One fragment corresponded to the signal peptide (SP) of the wild-type calnexin and was amplified with the primers A2 and F2: 5′-AGGATCAGCAAGTGATCCCCG-3′. The second fragment corresponding to the transmembrane domain with the cytosolic tail in fusion with a c-myc tag was obtained with primers M2: CTTGCTGATCCTATTGGGATTGCAATTGTTGCCGTT-3′ and c-mycSTOPcnx1: 5′-GGATCCTTACATGGCATTCAAGTCCTCTTCAGAAATGAGCTTTTGCTCCATGTCTTCATTCTTCGCAGT-3′. These two PCR products were mixed together to perform an overlap PCR amplification with the oligonucleotides primers A2 and c-mycSTOPcnx1. These two oligonucleotide primers contain restriction sites (NdeI for A2 and BamHI for c-mycSTOPcnx1), allowing their cloning into the pREP vectors family. The final product was digested with the restriction enzymes NdeI and BamHI, as it was done for the lumenalTM_cnx1 mutant and cloned into the pREP1 and pREP41 vector. The homologue of HMGB1 in S. pombe is Hmg1/2 that is encoded by the open reading frame (ORF) SPBC28F2.11, which was amplified from genomic DNA by PCR with the following primers: HMG1/2 NdeI forward: 5′-AAACATATGGCTCAAAACTCAACCC-3′ and HMG1/2 SalI reverse: 5′-AAAGTCGACTCAATTAGCAACTTTGGC-3′, and cloned into the cloning vector pDRIVE (PCR Cloning Kit; QIAGEN, Chatsworth, CA) following the manufacturer's recommendations. The gene hmg1/2 was then cut out from pDRIVE-HMG1/2 with the restriction enzymes NdeI and SalI and cloned into pREP42 and pREP2 vectors. The cDNA encoding human calnexin was kindly provided by Dr. Michael Brenner (Brigham Women's Hospital, Boston, MA; Rajagopalan et al., 1994) The gene was amplified with the following oligonucleotide primers: cnxh1-NdeI-F: 5′-AAACATATGGAAGGGAAGTGGTTGCTG-3′ and cnxh1-BamHI-R: 5′-AAAGGATCCTCACTCTCTTCGTGGCTT-3′ and cloned into the cloning pDRIVE vector (QIAGEN PCR Cloning Kit) following the manufacturer' recommendations. The cDNA encoding human calnexin was then digested with restriction enzymes NdeI and BamHI and cloned into the pREP41 and pREP1.
Plasmid Shuffling and Viability of Calnexin Mutants
To test the viability of new calnexin mutants, S. pombe (cnx1Δ + pcnx1+) cells containing a deletion of genomic calnexin (cnx1Δ) and a plasmid bearing a wild-type copy of the calnexin gene (cnx1+) were transformed with plasmids bearing the calnexin mutants to be tested (strains SP8075R and SP8160R). S. pombe strains bearing the two plasmids were grown for 7 d at 30°C in 5 ml liquid nonselective media (MM supplemented with adenine and appropriate supplements, i.e., uracil and/or leucine). Cells were plated on the same solid nonselective media, and auxotrophy for uracil and/or leucine was analyzed after 5–7 d by toothpicking on selective plates. The viability of the calnexin mutants was determined by examining the cell's capacity to conserve only the plasmid encoding mutant calnexin without the pcnx1+ plasmid.
Membrane Extraction
Microsomal membranes were prepared as previously described (Elagoz et al., 1999). S. pombe microsomal membranes were treated for 15 min at 4°C by mixing 1 vol of either 1 M NaCl or 0.2% SDS. Membrane lysates were spun at 80,000 × g for 1 h at 4°C in a L8–70M Sorvall ultracentrifuge (Newton, CT), and then the pellet from this spin was resuspended in 0.1 ml of 3× Laemmli's sample buffer (P fraction). Proteins in the supernatant fraction were treated for 30 min at 4°C in the presence of 6% ice-cold trichloroacetic acid (TCA) and spun at 2000 × g for 45 min at 4°C. The pellet was washed twice in ice-cold 80% acetone and dissolved in 0, 1 ml 3× Laemmli's sample buffer (S fraction). Before SDS-PAGE, samples were boiled for 5 min.
Immunoprecipitations
Immunoprecipitations from cells (the equivalent of OD595 0.8–1.0), treated or not with 10 μg/ml tunicamycin for 24 h, were carried out as previously described (Jannatipour et al., 1998).
Immunoblotting
Protein extractions were carried out as previously described (Elagoz et al., 1999) Protein extracts were migrated on a 10 or 12% (wt/vol) SDS-PAGE gel. Proteins were transferred onto nitrocellulose membrane according to the manufacturer's instructions. Immunoblotting to detect Cnx1p and mutants of calnexin was carried out with the anti-Cnx1p rabbit polyclonal antibody (LAR223), dilution 1:30,000 or with the anti-cmyc mouse mAb 9E10, dilution 1:500. Immunoblotting to detect BiP was performed with the anti-BiP rabbit polyclonal antibody (LAR311), dilution 1:30,000 (Collin et al., 2004). When necessary, the intensity of the immunoblotting bands was quantified with Bio-Rad Quantity One 4.6.5 Basic program (Richmond, CA).
Genetic Screen for Suppressors of the Lethality of Overexpression of Calnexin
Briefly, cells overexpressing calnexin were transformed with the S. pombe pURSP1 genomic bank (Barbet et al., 1992) containing insertions between 2.5 and 10 Kb (the generous gift of Dr. Anthony Carr, University of Sussex, United Kingdom). About 150,000 clones were screened and 50 potential suppressors were selected. All 50 potential candidate strains were subjected to a secondary screening by Western blotting, to verify that the incoming genomic plasmid did not affect the levels of overexpression of calnexin. After verification, only one candidate was a real suppressor of cell death by overexpression, without it affecting the levels of calnexin. The ORF-identified SPBC28F2.11 encoding Hmg1/2p, the homologue of the mammalian nuclear protein HMGB1, was cloned into the pREP2 overexpression vector.
Apoptosis Induction Assays
Overexpression of Wild-Type and Calnexin Mutants.
Cells overexpressing wild-type or mutants of calnexin and control strains (see Table 1) were cultured for 43 h until saturation in MM containing the required supplements in the presence of 5 μg/ml thiamine to repress the nmt1 promoter of the pREP1 or the pREP2 vector. At saturation, the cells were diluted to OD595 = 0.3 in the same media and cultured for 19 h. Then, to induce the nmt1 promoter, an aliquot containing 5 × 107 cells was taken, and the cells were washed twice with 1 ml of media without thiamine, after which the cells were resuspended in an appropriate volume and 1 × 107 cells were inoculated in 3 ml of media without thiamine. Maximal levels of wild-type or mutants of calnexin were reached after 18 h of culture in MM without thiamine. In all experiments, the time point zero was defined as the time point where the thiamine is removed from the medium.
Tunicamycin Treatment.
Cells only expressing full-length calnexin or viable calnexin mutants at basal levels (see Table 1) were cultured for 43 h until saturation in MM with the required supplements. To obtain exponentially growing cells, the cells were diluted and cultured overnight until they reached OD595 = 0.3–0.8. A volume of 10 ml of culture was adjusted to OD595 0.3, and cells were treated with 10 μg/ml tunicamycin for 40 h at 30°C. Tests for metacaspase activation and nuclear fragmentation [DAPI (4′,6-diamidino-2-phenylindole) staining] were performed at this time.
Viability Assays
The survival of cells was measured by two different techniques: by serial 10-fold dilutions (drop tests on plates) and by cytometry with the vital fluorescent dye Phloxin B. For serial-dilution spotting experiments, an equivalent of OD595 nm = 0.8 was taken from exponentially growing cells in media containing thiamine. These cells were washed twice in media free of thiamine and adjusted to an equivalent of OD595 nm = 0.5. The cells were serially diluted (10−1–10−4), spotted on solid media with or without thiamine, and incubated for 7 d at 30°C. Viability assays with the Phloxin B fluorescent vital dye were carried out as previously described (Roux et al., 2006).
Detection of Apoptotic Markers
Metacaspase Activation.
Culture samples were taken at time points 32, 40, 48, 56, and 72 h after gene induction in the media depleted of thiamine or after 40 h of tunicamycin treatment. Aliquots containing 1 × 107 cells were washed once in 1 ml of 1× PBS, pH 7.4 (136 mM NaCl, 25 mM KCl, 12 mM NaHPO4, 18 mM KH2PO4) and resuspended in 150 μl of 1× PBS, pH 7.4, containing 10 μM FITC-VAD-FMK (valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone; CasPACE, Promega, Madison, WI). After incubation for 20 min at 30°C, cells were washed once in 1× PBS, pH 7.4, and resuspended in 100 μl 1× PBS, pH 7.4, to be analyzed by fluorescence-activated cell sorting (FACS).
TUNEL Assay.
TUNEL (Terminal uridine deoxynucleotidyl transferase dUTP nick end labeling) assay was performed with the APO-BRDU TUNEL Kit (Phoenix Flow Systems, San Diego, CA), essentially following the manufacturer's recommendations. After 96 h of gene induction by culturing cells in media without thiamine, 1.4 × 107 cells were taken and fixed with 1 ml of 3.7% formaldehyde. After fixation, the cell wall was digested by resuspending the cell pellet in 200 μl of sorbitol buffer (1.2 M sorbitol, 0.5 mM MgCl2, potassium phosphate, pH 6.8) containing 5 mg/ml lysing enzymes (Sigma, Oakville, ON, Canada), and incubating for 90 min at room temperature, followed by 30-min incubation at 37°C. The cell pellet was resuspended in 500 μl of permeabilization solution (0.1% Triton in 0.1% sodium citrate) and kept on ice for 2 min, washed twice with 400 μl of WASH solution, and incubated in 50 μl of TUNEL solution for 30 min at 30°C. After incubation, the cells were washed twice in WASH solution and incubated 30 min at room temperature in the dark with 100 μl antibodies solution (anti-BrdU antibodies). Staining of the cells was analyzed by flow cytometry (FACS).
Annexin V Staining.
Annexin V staining was performed with the Annexin V-FITC Apoptosis Detection Kit (BioVision Research Products, Mountain View, CA) essentially following the manufacturer's recommendations. After 48 h of gene induction by culturing cells in media without thiamine, 1.4 × 107 cells were centrifuged, resuspended, and incubated for 2 h at room temperature in 200 μl of sorbitol buffer (1.2 M sorbitol, 0.5 mM MgCl2, potassium phosphate, pH 6.8) containing 10 mg/ml Zymolyase 20T (Seikagaku, , Tokyo, Japan) in order to digest the cell wall. The cell pellet was washed with 400 μl of binding buffer (10 mM HEPES/NaOH, 140 mM NaCl, 2.5 mM CaCl2, and 1.2 M sorbitol) and resuspended in 35 μl of binding buffer containing 5 μl of Annexin V-FITC and incubated at room temperature in the dark for 20 min. After incubation, the cells were washed once in 400 μl of binding buffer and resuspended in 500 μl of 1× binding buffer (Annexin V-FITC Apoptosis Detection Kit, BioVision Research Products). Staining of the cells was analyzed by flow cytometry (FACS).
DAPI Staining.
For DAPI staining, samples containing 1.4 × 107 cells were taken after 96 h of induction or after 40 h of tunicamycin treatment. Cells were fixed for 10 min in a solution of 3.7% formaldehyde and washed once in 1, PBS, pH 7.4, containing 1% Nonidet P-40 and twice in 1× PBS, pH 7.4. The cells were resuspended in 100 μl 1× PBS, pH 7.4, to a final concentration of 5 × 107–1 × 108 cells/ml. Suitable quantities of cells were applied to a poly-lysine–coated coverslips, washed, and let dry. The slides were mounted with a DAPI-containing mounting media (1 μg/ml DAPI, 1 mg/ml p-phenylenediamine, 90% glycerol). Microscopic analysis was performed using a fluorescence inverted microscope Nikon TE2000U (Melville, NY). Images were acquired using a motion-picture camera CCD coolSnapFX M 12 bit (Roper Scientific, Tucson, AZ) and treated with the UIC Metamorph software (Universal Imaging, West Chester, PA).
In Vivo Detection of ROS Accumulation.
The accumulation of reactive oxygen species (ROS) was determnined essentially as previously described (Roux et al., 2006). Culture samples were taken after 24 h of induction. Samples containing 1–2 × 107 cells were incubated with 30 μM dihydrorhodamine123 (Sigma), for 30 min at 30°C. A suitable quantity of cells were applied to a poly-lysine-coated coverslip, washed with 1× PBS, pH 7.4, and let dry. Fluorescence was observed with a fluorescence microscope Nikon E800. Images were acquired using a motion-picture camera CCD coolSnapFX M 12 bit and treated with UIC Metamorph software.
Flow Cytometry Analyses.
Cells were stained with Phloxin B, FITC-VAD-FMK or with the TUNEL assay as described above. Flow cytometry analyses were performed using a FACS Calibur (BD Biosciences, Mountain View, CA) device, on 10,000 cells. Emission from the argon laser was at 488 nm; emission settings were 515–545 nm (filter FL-1) for FITC-VAD-FMK and fluorescein staining or 560–600 nm (filter FL-2) for Phloxin B staining. The percentage of positive stained cells was determined as the population of fluorescent cells with a higher fluorescent intensity than a stained negative control. Parameters of the stained negative control were adjusted with an unstained negative control. Each experiment was repeated three times.
Statistical Analyses
The significance of the variations of results among strains was determined by a global ANOVA as described in http://www.physics.cbsju.edu/stats/anova.html. In certain figures, the significance to variation with respect to controls or Cnx1p was evaluated by a Student's t test.
RESULTS
Overexpression of Wild-Type Calnexin Causes Cell Death in S. pombe
We have previously demonstrated that although calnexin is an essential chaperone playing key roles in protein folding and quality control in the ER, its vital function in S. pombe is not its chaperone activity (Elagoz et al., 1999; Marechal et al., 2004). Because the overexpression of a gene generally leads to a gain of function (Ramer et al., 1992), to obtain clues as to the essential cellular role of calnexin we overexpressed cnx1+ in the fission yeast. Interestingly, we observed that the overexpression of calnexin resulted in a severe decline in the viability of the culture. To characterize this phenotype, we assayed the viability of cells overexpressing calnexin by measuring their capacity to grow on inducing media, i.e., without thiamine (Figure 1B). When cells containing the pREP1cnx1+ plasmid were spotted on media free of thiamine, we observed a dramatic reduction in the ability to form colonies compared with the control strain containing the pREP3X vector. By contrast, no difference with the control was observed when cells were spotted on thiamine-containing media, which represses the overexpression of calnexin (Figure 1B). The same phenotype was observed when cells were stained with Phloxin B, a fluorescent vital dye that accumulates within dead cells (Roux et al., 2006). At 48 h after induction, ∼50% of cells overexpressing calnexin were stained with Phloxin B compared with nearly 0% for cells harboring the empty vector control (Figure 1C). To assess whether this death phenotype was specific to calnexin and was not due to an overloading of the ER lumen, we overexpressed Pdi1p, a soluble lumenal ER foldase involved in the formation of disulfide bonds of newly synthesized polypeptides (Maattanen et al., 2006). No cell death was observed by spotting on media free of thiamine or by Phloxin B staining when cells overexpressed Pdi1p (Figure 1, B and C). However, the strain overexpressing Pdi1p appeared to grow more slowly than the control cells containing the empty vector (Figure 1B). Because, calnexin is an ER integral-membrane protein, it remained possible that the cell-death phenotype could be the result of saturating the ER membrane and not due to a specific effect of a calnexin function. To investigate this possibility, we overexpressed the Sec61β subunit of the translocon (Sbh1p; see Table 1), which is an ER-membrane protein (Romisch, 1999). As depicted in Figures 1, B and C, the overexpression of Sec61β did not cause cell death. Finally, to ascertain that overexpression of calnexin does not disrupt the ER membrane, the ER membranes were extracted and treated with Tris buffer alone (mock) as control, with a high concentration of NaCl to disrupt ionic interactions, and with SDS to break all interactions. This experiment showed that overexpression of calnexin does not alter the integrity of the membrane because no proteins were detected in the supernatant of the mock treatment (see Supplemental Figure S1). Together, these observations demonstrate that the death phenotype of cells overexpressing calnexin is specifically inherent to this ER chaperone and not due to an artifact caused by the overloading of the capacity of the ER membrane and/or lumen.
Cells Dying from Overexpression of Calnexin Display Typical Apoptotic Features
Next we investigated whether the death of S. pombe cells overexpressing calnexin is due to an apoptotic process or whether these cells undergo unspecific necrotic death. Cells undergoing apoptosis display typical morphological and biochemical markers, which necrotic cells do not exhibit (Kerr et al., 1972; Jin and Reed, 2002; Madeo et al., 2004). Typical markers of apoptosis include phosphatidyl serine externalization, caspase activation, chromatin breakage, and nuclear fragmentation. Cells entering in the apoptotic cascade usually show phosphatidyl serine externalization as a first phenotype associated with this kind of death. About 45% of cells overexpressing Cnx1p were stained with Annexin V-FITC compared with 0% for the control cells (overexpressing Pdi1p or Sec61β, or harboring the empty vector; Figure 2A). This demonstrates that phosphatidyl serine is specifically externalized due to calnexin overexpression. Fluorescence microscopy with DAPI revealed nuclear fragmentation in cells overexpressing calnexin, whereas the nuclei of control cells (overexpressing Pdi1p or Sec61β or harboring the empty vector) remained intact for the same time point (Figure 2B). To assess if nuclear fragmentation was due to chromatin breakage, the TUNEL assay was performed and quantified by FACS analysis. Cells overexpressing calnexin exhibited a significant TUNEL-positive phenotype compared with the controls (cells overexpressing Pdi1p or Sec61β or harboring the empty vector), which is consistent with the DAPI phenotype (Figure 2C). We and others (Hauptmann et al., 2006; Roux et al., 2006) have previously shown that caspase-like (metacaspase; Uren et al., 2000) activation can be assessed in S. cerevisiae and in S. pombe by the permeable fluorescent maker FITC-VAD-FMK. FACS analysis revealed that ∼40% of cells overexpressing calnexin were metacaspase positive at 72 h after induction (Figure 2D). In contrast, no FITC-VAD-FMK fluorescent cells were observed in the control cultures of the strains overexpressing Pdi1p or Sec61β or harboring the empty vector. High levels of ROS are associated with apoptosis due to ER stress in mammalian and yeast cells (Carmody and Cotter, 2001). We measured this apoptotic marker by staining cells with dihydrorhodamine 123 (DHR123), which is oxidized to fluorescent rhodamine by ROS. As shown by fluorescent microscopy analysis, only cells overexpressing calnexin produced ROS, thus indicating that overexpression of Cnx1p may cause ER stress (Figure 2E). Collectively, these observations show that overexpression of calnexin in S. pombe leads to cell death with the typical markers associated with apoptosis.
Figure 2.
The overexpression of calnexin induces typical apoptotic phenotypes. (A) Phosphatidyl serine externalization. The Annexin V assay was carried out after 48 h of induction of overexpression for the Cnx1p (strain SP8007R) and control strains (empty vector, SP7975R; Pdi1p, SP16058; and Sec61β, SP15302). The percentage of stained cells was measured by FACS. A global ANOVA showed that the values obtained among the strains were significantly different (p = 0.000). (B) Nuclear fragmentation. Cells overexpressing Cnx1p (strain SP8007R) and control strains (empty vector, SP7975R; Pdi1p, SP16058; and Sec61β, SP15302) were stained with DAPI, after 96 h of induction of overexpression. Cells were visualized under fluorescence microscopy; Nomarski fields are shown. White arrows indicate fragmented nuclei. (C) DNA fragmentation. The TUNEL assay was carried out after 96 h of induction of overexpression for the Cnx1p (strain SP8007R) and control strains (empty vector, SP7975R; Pdi1p, SP16058; and Sec61β, SP15302). The percentage of stained cells was measured by FACS. A global ANOVA showed that the values obtained among the strains were significantly different (p = 0.000). (D) Metacaspase activation. The fluorescent probe FITC-VAD-FMK was used to measure metacaspase activation by FACS, at different time points after the overexpression of Cnx1p (strain SP8007R) and control strains (empty vector, SP7975R; Pdi1p, SP16058; and Sec61β, SP15302), as described in the Materials and Methods. The experiment was repeated three times, and this graph is a representation of a typical experiment. (E) Accumulation of ROS. ROS accumulation in the Cnx1p overexpressing strain (SP8007R) and control strains (empty vector, SP7975R; Pdi1p, SP16058; and Sec61β, SP15302) was determined by using the ROS probe DHR123 and fluorescence microscopy. In this figure, vertical black arrows symbolize overexpression of the indicated protein.
The Transmembrane Domain of Calnexin Is Required for the Induction of Apoptotic Death by Overexpression
To identify molecular determinants of calnexin involved in the triggering apoptotic death, we overexpressed cnx1 truncation mutants. We began our analysis with lumenal_Cnx1p and mini_Cnx1p, two viable mutants previously characterized (Elagoz et al., 1999). The lumenal_cnx1 mutant encodes the entire lumenal domain of calnexin (Figure 3A), which was shown to have the same level of chaperone activity as wild-type calnexin (Elagoz et al., 1999; Marechal et al., 2004). The mini_cnx1 mutant encodes a truncated calnexin protein spanning the last 52 amino acids (aa) of the lumenal domain, the TM and the cytosolic tail (Figure 3A). This mutant exhibits no detectable chaperone activity (Elagoz et al., 1999; Marechal et al., 2004).
Figure 3.
The transmembrane domain (TM) is required to induce cell death by overexpression of calnexin. (A) Structures of calnexin mutants used in this study. Four different mutants were used. The mutant lumenal_Cnx1p encodes the lumenal domain of calnexin (415 aa) and is truncated of the TM and the cytosolic tail. The mutant mini_Cnx1p encodes 123 residues corresponding to last 52 aa at the C-terminal end of the lumenal domain, the TM (23 aa), and the cytosolic tail (48 aa). The lumenalTM_Cnx1p mutant spans the lumenal domain and the TM of calnexin (437 aa). The C-termTM_Cnx1p_cmyc mutant encodes the TM and the cytosolic of calnexin (71 aa) and encodes also the c-myc tag for immunodetection. For the sake of simplicity, the SP of calnexin is not represented in this figure. The capacity of the mutants to sustain the viability of the cells in a Δcnx1 background is indicated at the right of the figure by + or − signs and was determined by a plasmid-shuffling experiment (see Materials and Methods and Elagoz et al., 1999). (B) Immunoblot detection of Cnx1p and mutants of calnexin. Samples corresponding to 10 μg of protein extracts from cells overexpressing mini_Cnx1p (strain SP7965R) and C-termTM_Cnx1p_cmyc (strain SP8125R) and 2 μg from cells overexpressing lumenal_Cnx1p (strain SP16053) and lumenalTM_Cnx1p (strain SP8056) were loaded on a 12% (wt/vol) SDS-PAGE gel. Calnexin proteins were detected by immunoblotting with the anti-Cnx1p rabbit polyclonal serum (1:30,000) for Cnx1p, mini_Cnx1p, lumenal_Cnx1p, and lumenalTM_Cnx1p mutants and with the 9E10 anti-cmyc mouse mAb (1:500) for C-termTM_Cnx1p_cmyc mutant. The positions of the molecular mass markers (in kDa) are indicated on the left. The bands corresponding to the overexpressed are identified with a asterisk (*). The band marked with a black circle (●) corresponds to the endogenous calnexin present in all the strains but that is visible only in the mini_Cnx1p well due to the amount of proteins loaded. (C) Survival of cells overexpressing Cnx1p (strain SP8007R), the mutants of calnexin (mini_Cnx1p, SP7965R; lumenal_Cnx1p, SP16053; lumenalTM_Cnx1p, SP8056R; C-termTM_Cnx1p_cmyc, SP8125R) and the control strain (empty vector, SP7975R) was assayed by serial dilution on inducing plates (without thiamine) and repressing plates (with thiamine). Samples of 10 μl of four 10-fold serial dilutions (10−1–10−4) of cells at 0.5 OD595 were spotted on selective MM and incubated at 30°C for 7 d (see Materials and Methods). (D) Percentage of dead cells measured by staining with fluorescent vital dye Phloxin B. Cells overexpressing Cnx1p (strain SP8007R), the mutants of calnexin (mini_Cnx1p, SP7965R; lumenal_Cnx1p, SP16053; lumenalTM_Cnx1p, SP8056R; C-termTM_Cnx1p_cmyc, SP8125R) and the control strain (empty vector, SP7975R) were stained with Phloxin B after 48 h of induction of overexpression, and fluorescent cells were quantified by FACS. Stained cells were considered as dead. A global ANOVA showed that the values obtained among the strains were significantly different (p = 0.000). The significance of differences in the results was evaluated by a Student's t test, pairwise calculated with respect to the control for Cnx1p and C-termTM_Cnx1p_cmyc and to Cnx1p for the calnexin mutants. **p < 0.01 and *p < 0.05.
Remarkably, overexpression of lumenal_Cnx1p did not provoke cell death, as measured by spotting on inducing media or by Phloxin B (Figure 3, C and D). In agreement with these results, cells overexpressing lumenal_Cnx1p did not exhibit apoptotic markers (nuclear fragmentation, DNA breakage, and metacaspase activation; Figure 4, A–C). In contrast, the overexpression of mini_Cnx1p resulted in significant cell death (Figure 3, C and D). Overexpression of mini_Cnx1p also provoked apoptotic phenotypes similar to those caused by overexpression of wild-type calnexin (Figure 4, A–C). Therefore, removing the TM and the cytosolic tail completely abolished the apoptotic effect triggered by overexpression of calnexin.
Figure 4.
The TM of calnexin is required for induction of apoptotic cell death due to overexpression. (A) Nuclear fragmentation. Cells overexpressing the mutants of calnexin (mini_Cnx1p, SP7965R; lumenal_Cnx1p, SP16053; lumenalTM_Cnx1p, SP8056R; C-termTM_Cnx1p_cmyc, SP8125R) and the control strain (empty vector, SP7975R) were stained with DAPI, after 96 h of induction of overexpression. Cells were visualized under fluorescence microscopy; Nomarski fields are shown. White arrows indicate fragmented nuclei. (B) DNA fragmentation. The TUNEL assay was carried out after 96 h of induction of overexpression for the Cnx1p (strain SP8007R), the mutants of calnexin (mini_Cnx1p, SP7965R; lumenal_Cnx1p, SP16053; lumenalTM_Cnx1p, SP8056R; C-termTM_Cnx1p_cmyc, SP8125R) and the control strain (empty vector, SP7975R). The percentage of stained cells was measured by FACS. A global ANOVA showed that the values obtained among the strains were significantly different (p = 0.000). The significance of differences in the results was evaluated by a Student's t test, pairwise calculated with respect to the control for Cnx1p and C-termTM_Cnx1p_cmcyc and to Cnx1p for the calnexin mutants. **p < 0.01 and *p < 0.05. (C) Metacaspase activation. The fluorescent probe FITC-VAD-FMK was used to measure metacaspase activation by FACS, at different time points after the overexpression of Cnx1p (strain SP8007R), the mutants of calnexin (mini_Cnx1p, SP7965R; lumenal_ Cnx1p, SP16053; lumenalTM_Cnx1p, SP8056R; C-termTM_Cnx1p_cmyc, SP8125R) and the control strain (empty vector, SP7975R) as described in the Materials and Methods. The experiment was repeated three times, and this graph is a representation of a typical experiment. In this figure, vertical black arrows symbolize overexpression of the indicated protein.
To further map the determinants involved in the induction of apoptosis by overexpression of calnexin, we constructed two additional mutants (Figure 3A). One of them is called lumenalTM_cnx1 and encodes the lumenal domain of calnexin and includes the TM anchor. The other mutant is C-termTM_cnx1_cmyc and encodes the TM and the cytosolic tail of calnexin with a c-myc tag to facilitate immunodetection (Figure 3B). The mutant lumenalTM_Cnx1p conferred viability, whereas C-termTM_Cnx1p_cmyc could not sustain viability in a Δcnx1 strain (Figure 3A). That the encoded mutant proteins were indeed correctly integrated into the ER membrane was verified using cell fractionation and detergent solubilization, as we have previously described for wild-type calnexin and mini_Cnx1p (Elagoz et al., 1999; see Supplemental Figure S1).
As for wild-type calnexin, the overexpression of lumenalTM_Cnx1p and C-termTM_Cnx1p_cmyc mutants resulted in apoptotic cell death (Figure 3, C and D), as judged by the typical markers nuclear fragmentation, DNA breakage, and metacaspase activation (Figure 4, A–C). However, in the case of the mutant lumenalTM_Cnx1p, the apoptotic death phenotype was stronger than with wild-type calnexin, whereas in the case of the C-termTM_Cnx1p_cmyc mutant the apoptotic death phenotype was weaker. For instance, cells overexpressing lumenalTM_Cnx1p were not able to form colonies on media without thiamine, whereas cells overexpressing C-termTM_Cnx1p_cmyc formed colonies, albeit not as efficiently as the control strains (Figure 3C). Moreover, ∼70% of the cells overexpressing the lumenalTM_Cnx1p mutant were dead as judged by Phloxin B staining, whereas only 21% of dead cells were observed for the strain overexpressing C-termTM_Cnx1p_cmyc (Figure 3D). Cells overexpressing these two mutants showed distinctive nuclear fragmentation, DNA breakage, and metacaspase activation (Figure 4, A–C). The strongest difference in apoptotic markers was observed for metacaspase activation measurements because 70% of cells overexpressing lumenalTM_Cnx1p were positive after 72 h of induction, compared with ∼27% for C-termTM_Cnx1p_cmyc (Figure 4C). Together these experiments indicate that the TM is required for the induction of apoptotic cell death by calnexin overexpression and suggest that some calnexin sequences on either or both sides of the ER membrane might be necessary for this lethal phenotype. Importantly, as the mini_Cnx1p and C-termTM_Cnx1p_cmyc mutants do not exhibit chaperone activity, it clearly appears that the role of calnexin in apoptosis is distinct from its chaperone function in the ER lumen.
Calnexin Is Involved in ER-Stress–induced Apoptosis
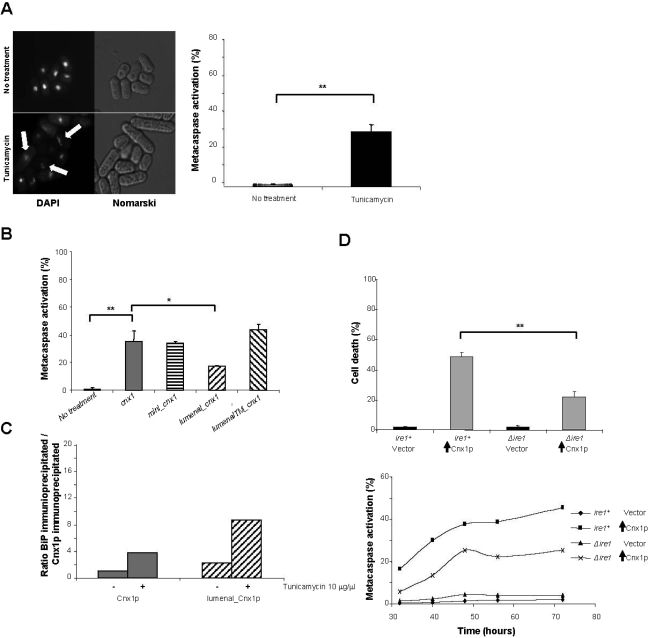
Because its overexpression induces apoptotic death, we wanted to investigate whether calnexin is involved in apoptosis provoked by ER stress. Tunicamycin is an inhibitor of N-glycosylation that strongly induces apoptosis in mammalian cells and in the budding yeast (Perez-Sala and Mollinedo, 1995; Hacki et al., 2000; Hauptmann et al., 2006; Hauptmann and Lehle, 2008). We have previously reported that tunicamycin efficiently inhibits protein N-glycosylation and induces ER stress in S. pombe (Jannatipour and Rokeach, 1995; Jannatipour et al., 1998; Beaulieu et al., 1999). As depicted in Figure 5A, S. pombe cells treated with tunicamycin displayed typical apoptotic makers, such as nuclear fragmentation and metacaspase activation. To investigate if calnexin is involved in apoptosis induced by the ER stressor tunicamycin, we constructed S. pombe strains solely expressing the mutant version of calnexin at the basal level, in a Δcnx1 genetic background. The genes coding for the calnexin mutants to be tested were cloned into the pREP41 vector, which expresses calnexin at the basal endogenous level (Jannatipour et al., 1998). It is important to note that because calnexin is essential for viability in S. pombe, only viable calnexin mutants can be tested in the Δcnx1 background. The mutants tested were mini_cnx1, lumenal_cnx1, and lumenalTM_cnx1. After treatment with tunicamycin, ∼35% of wild-type calnexin cells were positive for metacaspase activation as measured by staining with the fluorescent marker FITC-VAD-FMK (Figure 5B). The same level of metacaspase activation was observed with the strains mini_cnx1 and lumenalTM_cnx1 mutants (Figure 5B). Interestingly, only 17% of the lumenal_cnx1 mutant cells treated with tunicamycin stained positive for metacaspase activation (Figure 5B). This diminution in metacaspase activity corresponds to a reduction of 50% of the total metacaspase-positive wild-type calnexin cells.
Figure 5.
Involvement of calnexin in ER-mediated apoptosis in S. pombe. (A) Tunicamycin induces apoptotic cell death in S. pombe. S. pombe cells carrying a wild-type genomic copy of calnexin (strain SP556) were treated with 5 μg/ml tunicamycin or with the solvent (no treatment) and subsequently stained with DAPI to detect nuclear fragmentation and with FITC-VAD-FMK to determine the % of metacaspase activation. For DAPI staining, cells were examined by fluorescence microscopy. White arrows indicate fragmented nuclei. The % of metacaspase-positive cells was measured by FACS as described in Materials and Methods. (B) Truncation of the TM and the cytosolic tail of calnexin dramatically reduces the levels of metacaspase activation. Cells expressing cnx1+ (strain SP7951R) or mutants of calnexin (mini_cnx1, SP8490R; lumenal_cnx1, SP8488R; and lumenalTM_cnx1, SP8085R) at basal levels were treated with 10 μg/ml tunicamycin, and subsequently the % of metacaspase-positive cells was determined with FITC-VAD-FMK by FACS, as described in Materials and Methods. A global ANOVA showed that the values obtained among the strains were significantly different (p = 0.000). The significance of differences in the results was evaluated by a Student's t test, pairwise calculated with respect to the control for Cnx1p and to Cnx1p for the calnexin mutants. **p < 0.01 and *p < 0.05. (C) Association with BiP. Cells at OD 0.8–1.0 expressing Cnx1p (strain SP7951R) or lumenal_Cnx1p (SP8488R) were treated or not with 10 μg/ml tunicamycin. Immunoprecipitations were performed with anti-Cnx1p antibodies and the membrane was blotted with anti-Cnx1p antibodies or anti-BiP antibodies. Bands corresponding to Cnx1p, lumenal_Cnx1p or BiP were quantified with the Bio-RAD Quantity One 4.6.5 Basic program and reported as a ratio to Cnx1p untreated on a graph. The graph is representative of three different experiments. (D) Implication of Ire1p in apoptosis induced by calnexin overexpression. Cells overexpressing Cnx1p in a wild-type background (SP8007R) or in a Δire1 strain (SP8231R) and the control strain (empty vector, SP7975R and SP8227R) were stained with Phloxin B after 48 h of induction of overexpression to measure cell death. Stained cells were considered as dead. At different time points after the overexpression of Cnx1p cells were stained with the fluorescent probe FITC-VAD-FMK to measure metacaspase activation. Fluorescent cells were quantified by FACS. A global ANOVA showed that the values obtained among the strains were significantly different (p = 0.000). The significance of differences in the results was evaluated by a Student's t test, pairwise calculated with respect to the control for Cnx1p. **p < 0.01 and *p < 0.05. For metacaspase activation, the experiment was repeated three times, and this graph is a representation of a typical experiment. In this figure, vertical black arrows symbolize overexpression of the indicated protein.
BiP anchoring to the ER membrane was reported in mammalian cells subjected to ER stress (Rao et al., 2002; Reddy et al., 2003). In previous reports, we have shown that calnexin and BiP associate in a complex, (Jannatipour et al., 1998; Elagoz et al., 1999; Marechal et al., 2004). To investigate whether the BiP-Cnx1p association is affected in the presence of tunicamycin, we immunoprecipitated Cnx1p or lumenal_Cnx1p and verified the presence of BiP in the precipitate. The treatment with tunicamycin increased the interaction between the two partners for both, wild-type calnexin and the lumenal_Cnx1p mutant, with a higher ratio of BiP for the latter (Figure 5C).
Ire1p is a mediator of ER stress that is conserved from yeast to mammals (Patil and Walter, 2001; Breckenridge et al., 2003). Therefore, we wanted to investigate if apoptosis induced by calnexin overexpression involves Ire1p. We overexpressed calnexin in the Δire1 background and measured cell death and metacaspase activation. Indicating the involvement of Ire1p, we observed ∼50% diminution in the levels of cell death and metacaspase activation induced by calnexin overexpression in the Δire1 background (Figure 5D).
To sum up, apoptosis induced by the ER-stressor tunicamycin is strongly reduced in the background of lumenal_Cnx1p, the only mutant tested that does not provoke apoptosis by overexpression. This observation demonstrates the involvement of calnexin in ER-mediated apoptosis and support the notion that the TM and/or the anchoring of calnexin to the ER membrane are important for the proapoptotic function of calnexin in S. pombe. That apoptosis induced by calnexin overexpression is significantly reduced in the absence of Ire1p suggests that this mediator plays a role in the apoptosis pathway involving calnexin.
The S. pombe Metacaspase Pca1p and the Homologue of Bap31 Are Not Required for the Induction of Apoptosis by Calnexin Overexpression
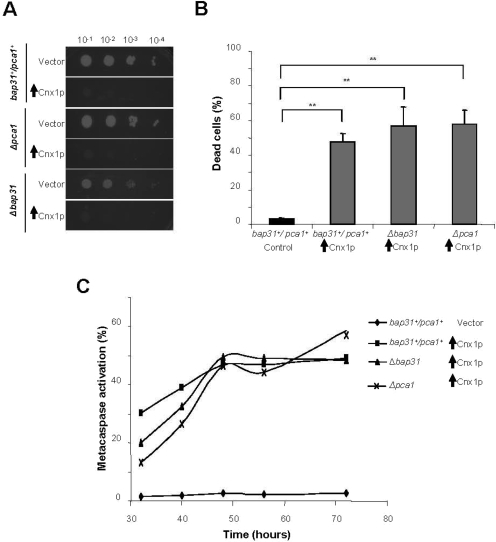
S. pombe encodes several homologues of known actors of the apoptotic pathway (Uren et al., 2000; Madeo et al., 2002; Lim et al., 2007). So far, only one caspase-like protein was identified in yeast, and the dependence of apoptosis on this metacaspase is variable, depending of the apoptotic inducer (Madeo et al., 2002; Bettiga et al., 2004; Fannjiang et al., 2004; Herker et al., 2004; Wadskog et al., 2004; Ivanovska and Hardwick, 2005; Reiter et al., 2005; Liang et al., 2008; Mazzoni and Falcone, 2008). To test whether the apoptosis induced by calnexin overexpression is dependent of the metacaspase Pca1p, death was assayed by spotting cells on inducing media and by staining cells with Phloxin B in the Δpca1 strain containing an empty vector (Vector) or overexpressing calnexin. No reduction in the death levels was observed in the Δpca1 background (Figure 6, A and B). Metacaspase activation was analyzed with the fluorescent probe FITC-VAD-FMK. We observed a slower kinetics of caspase-like activation in Δpca1 cells; however the level of final activation was similar to the pca1+ strain (Figure 6C). Therefore, apoptosis induced by calnexin overexpression is not absolutely dependent on Pca1p. Interestingly, these results demonstrate the presence of other, yet uncharacterized, caspase-like activities in S. pombe.
Figure 6.
The S. pombe metacaspase Pca1p and the homologue of Bap31 are not required for apoptosis induced by calnexin overexpression. (A) Survival of cells overexpressing Cnx1p in a Δpca1 or in a Δdma1/Bap31 strain (SP8081R and SP8136R) and the control strains (empty vector, SP7975R, SP8167R, and SP8145R), was assayed by serial dilution on inducing plates (without thiamine) and repressing plates (with thiamine). Samples of 10 μl of four 10-fold serial dilutions (10−1–10−4) of cells at 0.5 OD595 were spotted on selective MM and incubated at 30°C for 7 d (see Materials and Methods). (B) Percentage of dead cells measured by staining with fluorescent vital dye Phloxin B. Cells overexpressing Cnx1p in a wild-type background (SP8007R), in a Δpca1 strain (SP8081R) or in a ΔBap31 strain (SP8136R) and the control strain (empty vector, SP7975R) were stained with Phloxin B after 48 h of induction of overexpression and fluorescent cells were quantified by FACS. Stained cells were considered as dead. A global ANOVA showed that the values obtained among the strains were significantly different (p = 0.000). The significance of differences in the results was evaluated by a Student's t test, pairwise calculated with respect to the control for Cnx1p. **p < 0.01 and *p < 0.05. (C) Metacaspase activation. The fluorescent probe FITC-VAD-FMK was used to measure metacaspase activation by FACS, at different time points after the overexpression of Cnx1p in a wild-type background strain (SP8007R), in a Δpca1 strain (SP8081R) or in a ΔBap31 strain (SP8136R) and the control strain (empty vector, SP7975R) as described in the Materials and Methods. The experiment was repeated three times, and this graph is a representation of a typical experiment. In this figure, vertical black arrows symbolize overexpression of the indicated protein.
In mammalian cells, calnexin was demonstrated to associate with the transmembrane ER protein Bap31. Bap31 mediates apoptosis after its cleavage by caspase 8, and calnexin was proposed to act as a scaffold in this process (Delom et al., 2006, 2007). To assess if the homologue of Bap31 in S. pombe is required for apoptosis induction by calnexin overexpression, we measure death rate and metacaspase activation in cells deleted for the homologue of Bap31 (Δdam1/bap31). Overexpression of calnexin in Δdam1/bap31 strain caused death levels similar to that of the wild-type strain (Figure 6, A and B). Here again, a delay in metacaspase induction was observed in the Δdam1/bap31 strain, reaching the final levels similar that of the wild-type strain (Figure 6C). Therefore, Bap31 is not crucial in S. pombe to mediate apoptosis induced by calnexin overexpression.
The S. pombe Homologue of Human HMGB1 Prevents Apoptosis Due to Calnexin Overexpression
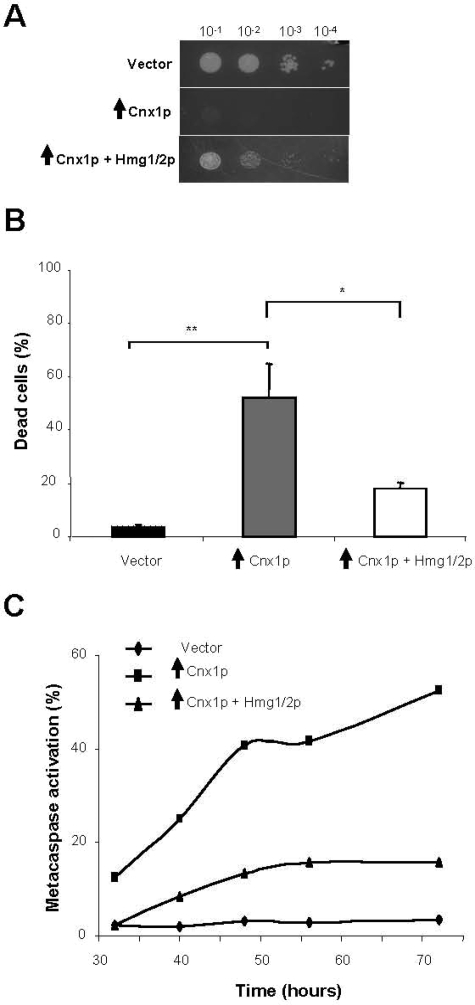
To identify cellular factors involved in cell death mediated by calnexin, we carried out a genetic screen to isolate suppressors of the lethality caused by the overproduction of calnexin. Briefly, cells overexpressing calnexin were transformed with the pURSP1 S. pombe genomic bank, and 150,000 transformants were screened for survival, i.e., the ability to form colonies. Of 50 potential candidates isolated in the primary screen, one was an authentic suppressor (Figure 7A). This suppressor clone encoded Hmg1/2p, the homologue of the mammalian nuclear protein HMGB1. This protein is part of an HMG family that interacts with DNA for replication, transcription, and DNA repair. Importantly, human HMGB1 was reported as an antiapoptotic protein in mammalian and in fission yeast cells (Brezniceanu et al., 2003).
Figure 7.
The S. pombe homologue of the human antiapoptotic HMGB1 inhibits apoptotic death induced due to calnexin overexpression. (A) Survival of cells overexpressing Cnx1p (strain SP8007R) alone and co-overexpressing Hmg1/2p, the S. pombe homologue of the human antiapoptotic HMGB1 protein, (strain SP16081) and the control strain (empty vector, SP7975R) was assayed by serial dilution on inducing plates (without thiamine) and repressing plates (with thiamine). Samples of 10 μl of four 10-fold serial dilutions (10−1–10−4) of cells at 0.5 OD595 were spotted on selective MM and incubated at 30°C for 7 d (see Materials and Methods). (B) Percentage of dead cells measured by staining with fluorescent vital dye Phloxin B. Cells overexpressing Cnx1p (strain SP8007R) alone and co-overexpressing Hmg1/2p, the S. pombe homologue of the human antiapoptotic HMGB1 protein, (strain SP16081) and the control strain (empty vector, SP7975R) were stained with Phloxin B after 48 h of induction of overexpression and fluorescent cells were quantified by FACS. Stained cells were considered as dead. A global ANOVA showed that the values obtained among the strains were significantly different (p = 0.000). The significance of differences in the results was evaluated by a Student's t test, pairwise calculated with respect to the control for Cnx1p and to Cnx1p for the Cnx1p + Hmg1/2p strain. **p < 0.01 and *p < 0.05. (C) Metacaspase activation. The fluorescent probe FITC-VAD-FMK was used to measure metacaspase activation by FACS, at different time points after the overexpression of Cnx1p (strain SP8007R) alone and co-overexpressing Hmg1/2p, the S. pombe homologue of the human antiapoptotic HMGB1 protein, (strain SP16081) and the control strain (empty vector, SP7975R) as described in the Materials and Methods. The experiment was repeated three times, and this graph is a representation of a typical experiment. In this figure, vertical black arrows symbolize overexpression of the indicated protein.
To investigate the suppressor effect of Hmg1/2p on death by calnexin overexpression, we synthesized its encoding gene (SPBC28F2.11) by PCR amplification and cloned it into an overexpression vector. Simultaneous co-overexpression of Hmg1/2p with calnexin significantly reduced the levels of cell death, as judged by spotting on inducing media and by Phloxin B staining (Figure 7, A and B). About 50% of cells stained positively with Phloxin B when calnexin was overproduced alone, compared with 20% of cells when was Hmg1/2p overproduced in concert (Figure 7B). Examination of the metacaspase activation revealed that overexpression of Hmg1/2p reduced the levels of induction of apoptosis (Figure 7C). About 45% of the cells overexpressing solely wild-type calnexin were positive for metacaspase activation, whereas only 10% of the cells co-overexpressing Hmg1/2p stained positively with the fluorescent marker FITC-VAD-FMK (Figure 7C). Western blotting showed that the suppression of cell death caused by the simultaneous overexpression of Hmg1/2p was not due to a reduction in the calnexin levels (data not shown). These observations indicate that Hmg1/2p has antiapoptotic function in S. pombe and that this protein may have a regulatory role in apoptosis involving calnexin.
Overexpression of Human Calnexin Induces Apoptosis in S. pombe
S. pombe calnexin is a close sequence homologue of its human counterpart, especially when compared with the S. cerevisiae molecule (Jannatipour and Rokeach, 1995). Recent studies in mammalian cells have demonstrated the involvement of calnexin in the apoptosis induced by ER stress in mammals (Zuppini et al., 2002; Takizawa et al., 2004; Tomassini et al., 2004; Delom et al., 2006; Groenendyk et al., 2006; Delom et al., 2007). Because several studies reported interchangeability between mammalian and fission yeast proteins in apoptotic processes (Komatsu et al., 2000), we asked whether human calnexin could also induce apoptosis when overexpressed in S. pombe.
Indeed, the overproduction of human calnexin in S. pombe provoked cell death in ∼30% of cells, as measured by Phloxin B staining (Figure 8A). Moreover, ∼30% of cells overexpressing human calnexin exhibited metacaspase activation as measured with the fluorescent probe FITC-VAD-FMK. Although these levels of death and caspase activity are lower than those observed with wild-type S. pombe calnexin, these results argue for the conservation of the mechanisms of calnexin-mediated apoptotic cell death, at least in part, between human and S. pombe cells.
Figure 8.
Overexpression of human calnexin induces apoptotic death in S. pombe. (A) Overexpression of human calnexin provokes cell death in S. pombe. Samples of 10 μl of four serial 10-fold dilutions (10−1–10−4) of 0.5 OD595 cells overexpressing human calnexin (strain SP16084), or control cells harboring the empty vector (strain SP7975R) were plated on selective minimal media and incubated at 30°C for 7 d. (B) Percentage of dead cells as measured with the vital fluorescent dye Phloxin B. Cells overexpressing human calnexin (strain SP16084) or control cells harboring the empty vector (strain SP7975R) were stained with Phloxin B after 48 h of induction of overexpression and fluorescent cells were quantified by FACS. Stained cells were considered as dead. The significance of differences in the results was evaluated by a Student's t test, pairwise calculated with respect to the control for human calnexin. **p < 0.01 and *p < 0.05. (C) Metacaspase activation. The fluorescent probe FITC-VAD-FMK was used to measure metacaspase activation by FACS, at different time points after the overexpression of human calnexin (SP16084), or control cells harboring the empty vector (strain SP7975R), as described in the Materials and Methods. The experiment was carried out three times and this graph represents a typical experiment. In this figure, vertical black arrows symbolize overexpression of the indicated protein.
DISCUSSION
Cell survival under ER-stress conditions is largely dependent on the UPR pathway (Patil and Walter, 2001; Schroder and Kaufman, 2005; Marciniak and Ron, 2006; Zhao and Ackerman, 2006; Lai et al., 2007; Ron and Walter, 2007). However, if the overload caused by ER stressors cannot be counteracted, the UPR response switches from its prosurvival function to the signaling of apoptotic death. A major current question regarding the cell's response to ER stress is how the switch from prosurvival to prodeath is mechanistically decided. According to currents models, the mammalian UPR senses intralumenal stress through the three upstream ER transducers: IRE1, PERK, and ATF6; signaling via these stress receptors is regulated by the ER molecular chaperone BiP (Lee et al., 1999; Li and Lee, 2006; Ni and Lee, 2007). Nevertheless, it is unclear whether other upstream regulators of the mechanism ER-stress apoptosis do exist, and much remains to be elucidated about the downstream factors and interactions involved in the transduction of the cell-death signal.
Recent work showed that mammalian calnexin is required for the cleavage of Bap31 and thus for the generation of the proapoptotic p20 fragment under tunicamycin stress; these authors proposed that calnexin acts as a scaffold for Bap31 processing by caspase 8 (Zuppini et al., 2002; Delom et al., 2006; Groenendyk et al., 2006; Delom et al., 2007). These observations raise the possibility that calnexin could be involved in the early steps relaying the signal toward apoptotic death initiated by overwhelming ER stress.
Here, we demonstrate that the overexpression of calnexin in S. pombe induces cell death with typical apoptotic features including early death, phosphatidyl serine exposure, metacaspase activation, ROS production, nuclear fragmentation, and DNA breakage. This effect is specific to calnexin and is not due to overloading of the ER capacity, because overproduction of the ER proteins PDI or Sec61β did not induce the apoptotic-death phenotypes and is not due to the loss of ER membrane integrity, as confirmed by microsome analysis.
It has been previously reported that the expression of mammalian Bak in the fission yeast is lethal and that Bak requires interactions with S. pombe calnexin to mediate this cell-death phenotype (Torgler et al., 1997). The authors proposed that the interaction between Bak and calnexin results in a dominant lethal effect due to the propagation of a death signal or the recruitment of additional interactors to the Bak–calnexin complex (Torgler et al., 1997). Our results show that when overexpressed, calnexin induces apoptosis without exogenous factors or ER-stress conditions. In general, overexpression causes a bypass in the regulation of the pathway studied (Ramer et al., 1992). This suggests that calnexin overexpression may disturb the control of an apoptotic pathway in which calnexin takes part. A possibility is that the overexpression of calnexin mimics conditions of ER stress in otherwise resting cells.
Truncation of the TM and cytosolic tail of calnexin abolished the apoptotic death caused by overexpression. Using various deletion mutants, we demonstrated that the anchoring of calnexin to the ER membrane is required to induce apoptosis by overexpression. Although the overexpression of the cytosolic tail anchored to the membrane induced cell death, the strongest apoptotic effect was observed by overproducing a calnexin mutant spanning the lumenal domain with the TM. These results may imply that residues on both the intralumenal portion and the cytosolic tail of calnexin could play roles in the induction of apoptosis and that the sequences on either side of the TM may participate in different prodeath interactions. Supporting the importance of the cytosolic tail and the TM, Torgler et al. (1997) reported that expression of human Bak was lethal in an S. pombe strain expressing wild-type calnexin but not in a strain expressing a mutant calnexin lacking both the TM and the cytosolic tail. Importantly, our results demonstrate that sequences within the lumenal domain of calnexin are also involved in proapoptotic interactions. The requirement of the TM to elicit apoptotic death by overexpression suggests that the anchoring of calnexin to the ER membrane may be required for its interaction with some key partner(s) for the assembly of a lethal complex.
Our observations provide first direct evidence that altering the levels of calnexin causes death with apoptotic features. That its overexpression induces programmed cell death is indicative that calnexin could play a role in apoptotic processes induced by “natural” ER stresses. An increase in the calnexin levels due to ER stress may constitute a part of a branch in the mechanism of induction of apoptotic death. Tunicamycin is a potent inhibitor of N-glycosylation that is currently used as an elicitor of ER stress in the study of the apoptosis mechanisms in mammalian cells and in the budding yeast (Perez-Sala and Mollinedo, 1995; Hacki et al., 2000; Hauptmann et al., 2006; Hauptmann and Lehle, 2008). We have previously reported that tunicamycin induces ER stress in S. pombe and increases the expression of cnx1+, the gene encoding calnexin (Jannatipour and Rokeach, 1995). In this work, we show that also in the case of S. pombe, tunicamycin provokes cell death with typical apoptosis markers.
Using S. pombe strains expressing calnexin mutants at basal levels, we demonstrated that apoptotic cell death induced by tunicamycin is significantly less efficient in cells expressing calnexin without its TM. Cells expressing only the lumenal portion of calnexin exhibited a 50% reduction in apoptotic cell death due to exposure to tunicamycin. Our observations implicate calnexin in apoptosis caused by ER stress. In agreement with our results, calnexin-deficient rodent cells are relatively resistant to apoptosis induced by ER stress (Zuppini et al., 2002; Groenendyk et al., 2006). However, it has been reported that siRNA inhibition of calnexin expression increases the sensitivity to tunicamycin of apoptosis-resistant human-breast carcinoma MCF-7 cells (Delom et al., 2006). This disparity in the results could be due to differences in the cell lines used in these studies. Nevertheless, transfection of a ΔE calnexin mutant lacking most of the lumenal domain into MCF-7 cells restored the sensitivity to tunicamycin-induced apoptosis (Delom et al., 2006). Together, these observations clearly implicate calnexin in ER-stress–induced apoptosis, in mammalian and in S. pombe.
S. pombe encodes several homologues of proteins characterized for their implication in apoptosis including the thus far only metacaspase identified Pca1p (geneID: SPCC1840.04) and a Bap31 homologue (geneID: SPAC9E9.04). Our results demonstrate that the knockout of these genes does not affect the final level of apoptosis provoked by calnexin overexpression; however, we observed a slower kinetics of induction. Likewise, induction of apoptosis by tunicamycin was not blocked in the Δpca1 and Δdma1/bap31 strains (data not shown). Importantly, our results clearly demonstrate that another, yet uncharacterized, caspase-like activity is involved in this apoptotic process. Similarly, the homologue of Pca1p in S. cerevisiae, Yca1p, is not required for apoptotic death induced by numerous conditions (Madeo et al., 2002; Bettiga et al., 2004; Fannjiang et al., 2004; Herker et al., 2004; Wadskog et al., 2004; Ivanovska and Hardwick, 2005; Reiter et al., 2005; Liang et al., 2008; Mazzoni and Falcone, 2008). More recently, it was reported that Yca1p is not required for apoptosis induced by tunicamycin in S. cerevisiae (Hauptmann and Lehle, 2008). These observations suggest the existence of uncharacterized caspase-like activities in both, S. pombe and S. cerevisiae. That cell death occurs by calnexin overexpression in the Δdma1/bap31 background does not exclude the possibility that Bap31 could be part of a complex involving calnexin, but it is indicative that the role of Bap31 is not essential for apoptosis. In this vein, it was shown in mammalian cells that an uncleavable mutant of Bap31 does not abrogate the capacity to the cells to enter FAS-mediated apoptosis but it delays some cytoplasmic apoptotic events (Nguyen et al., 2000).
The significant reduction in the levels of tunicamycin-induced apoptosis observed with the lumenal_cnx1 mutant could be due to a diminution or a loss of effective interactions with lethal partners as observed for mammalian cells (Zuppini et al., 2002; Delom et al., 2006; Groenendyk et al., 2006; Delom et al., 2007). Ire1p could be taking part of this interaction since its presence is required to attain maximum apoptosis induction by calnexin overexpression. Further experiments are required to dissect the molecular interaction of calnexin with Ire1p and other lethal partners. Such interactions probably require the anchoring of calnexin to the ER membrane to be optimal in transducing the death signal. Interestingly, the lumenalTM_cnx1 strain exhibited the highest levels of apoptosis induced by tunicamycin. These, and our results obtained by overexpression experiments implicate lumenal sequences of calnexin in apoptosis. However, it appears that the chaperone function of calnexin is distinct from its role in apoptosis since the mutant lumenal_Cnx1p is a very effective chaperone (Marechal et al., 2004) but a poor effector of tunicamycin-induced apoptosis. Further supporting this point, mini_Cnx1p does not exhibit chaperone activity (Marechal et al., 2004) but is highly efficient in inducing apoptosis by overexpression, and mini_cnx1 cells display levels of tunicamycin-induced apoptosis as the wild-type strain.
BiP anchoring to the ER membrane was reported in mammalian cells subjected to ER stress (Rao et al., 2002; Reddy et al., 2003). We have previously reported that the last 52 residues of the lumenal portion of calnexin are required for viability and do interact with ER chaperone BiP (Jannatipour et al., 1998; Beaulieu et al., 1999; Elagoz et al., 1999; Marechal et al., 2004). Because BiP regulates the ER stress receptors IRE1, PERK, and ATF6, it is therefore tempting to speculate that BiP could also play a regulatory role in the proapoptotic function of calnexin (Patil and Walter, 2001; Szegezdi et al., 2006; Ni and Lee, 2007; Ron and Walter, 2007). Our coimmunoprecipitation experiments showed that tunicamycin treatment enhances Cnx1p-BiP interaction in both the wild-type calnexin and the mutant lumenal_Cnx1p. However, apoptosis is significantly reduced in the case of the lumenal_Cnx1p mutant. These observations raise the possibility that stronger interaction between Cnx1p and BiP is required for ER-stress–induced apoptosis and that this complex needs to be anchored to the ER membrane to efficiently transduce the apoptotic signal. In such scenario, the diminution of tunicamycin-induced apoptosis in lumenal_Cnx1p cells could be due to a mislocalization of the Cnx1p-BiP complex, because this complex is not localized to the ER membrane in the case of this mutant.
In a genetic screen designed to isolate suppressors of lethality caused by calnexin overexpression we identified Hmg1/2p, the S. pombe homologue of HMGB1. The HMGB1 protein is part of the HMG family that interacts with DNA for replication, transcription, and DNA repair and is a protein overexpressed in human breast carcinoma (Brezniceanu et al., 2003; Ulloa and Messmer, 2006). Importantly, HMGB1 is an antiapoptotic protein that was identified as an inhibitor of Bak-mediated cell death in mammalian cells and in S. pombe (Brezniceanu et al., 2003). In this work we showed that like its mammalian counterpart, Hmg1/2p inhibits the ER-derived apoptotic pathway involving calnexin. Because Bak-induced apoptotic death in S. pombe is dependent on calnexin (Torgler et al., 1997) and is inhibited by HMGB1 and because Hmg1/2 represses death by overexpression of calnexin in S. pombe, we hypothesize that the apoptotic pathway involving calnexin are similar in S. pombe and in mammalian cells. Although no sequence homologue of Bak was described in the fission yeast, it is likely that some S. pombe protein plays an analogous function to that of mammalian Bak. The discovery of this lethal partner will be important for the elucidation of the apoptotic processes caused by ER stress in S. pombe and are likely to contribute to our understanding of the mechanism of apoptosis in mammals.
Human calnexin and S. pombe calnexin are very similar in their structure (Jannatipour and Rokeach, 1995). Here we report that like S. pombe calnexin, the overexpression of its human orthologue in the fission yeast induces cell death with typical apoptotic features. These results indicate that human calnexin interacts with S. pombe apoptotic machinery and overrides the mechanisms that regulate apoptosis involving endogenous calnexin. Collectively, these observations argue for the conservation between the mammalian and the S. pombe mechanisms of apoptosis involving calnexin and validate S. pombe as a model organism.
Most of our current knowledge on the apoptotic response due to ER stress derives from studies with mammalian cells (Breckenridge et al., 2003; Xu et al., 2005; Kim et al., 2006; Szegezdi et al., 2006). Evidence accumulated in the last 10 years has proven that yeasts are interesting models to investigate the mechanism of apoptosis (Ink et al., 1997; Madeo et al., 1997; Ligr et al., 1998; Madeo et al., 1999; Madeo et al., 2002; Priault et al., 2003; Hardwick and Cheng, 2004; Madeo et al., 2004; Rodriguez-Menocal and D'Urso, 2004; Burhans and Weinberger, 2007; Frohlich et al., 2007; Almeida et al., 2008). Although human and S. pombe calnexin share significant sequence similarity (Jannatipour and Rokeach, 1995), the S. cerevisiae molecule (Cne1p; Parlati et al., 1995b) is the most distant known homologue. Thus, the study of the role of calnexin in the mechanisms of ER-stress apoptosis in S. pombe should bring crucial information regarding this cellular process in mammals.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Michael Brenner for the gift of the plasmid bearing the human calnexin cDNA, and Dr. Anthony car for supplying the pURSP1 S. pombe genomic library. We also thank Dr. Gerardo Ferbeyre and the members of the Rokeach Lab for critical reading of the manuscript. This work was supported by research grants from CIHR (Grant MOP-62703) and NSERC (Grant 171325) to L.A.R. R.G. was the recipient of FES-Département de biochimie studentships.
Abbreviations used:
- aa
amino acid
- BiP
immunoglobulin binding protein
- cnx1+
S. pombe gene encoding calnexin
- Cnx1p
S. pombe calnexin
- DAPI
4′,6-diamidino-2-phenylindole
- DHR123
dihydrorhodamine123
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- FITC-VAD-FMK
valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone
- hcd
highly conserved central domain
- PDI
protein-disulfide isomerase
- ROS
reactive oxygen species
- TM
transmembrane domain
- TUNEL assay
terminal uridine deoxynucleotidyl transferase dUTP nick end labeling assay
- UPR
unfolded protein response.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-02-0188) on August 13, 2008.
REFERENCES
- Ahner A., Brodsky J. L. Checkpoints in ER-associated degradation: excuse me, which way to the proteasome? Trends Cell Biol. 2004;14:474–478. doi: 10.1016/j.tcb.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Almeida B., Silva A., Mesquita A., Sampaio-Marques B., Rodrigues F., Ludovico P. Drug-induced apoptosis in yeast. Biochim. Biophys. Acta. 2008;1783:1436–1448. doi: 10.1016/j.bbamcr.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Arunachalam B., Cresswell P. Molecular requirements for the interaction of class II major histocompatibility complex molecules and invariant chain with calnexin. J. Biol. Chem. 1995;270:2784–2790. doi: 10.1074/jbc.270.6.2784. [DOI] [PubMed] [Google Scholar]
- Barbet N., Muriel W. J., Carr A. M. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene. 1992;114:59–66. doi: 10.1016/0378-1119(92)90707-v. [DOI] [PubMed] [Google Scholar]
- Beaulieu H., Elagoz A., Crine P., Rokeach L. A. Interaction of mammalian neprilysin with binding protein and calnexin in Schizosaccharomyces pombe. Biochem. J. 1999;340(Pt 3):813–819. [PMC free article] [PubMed] [Google Scholar]
- Bettiga M., Calzari L., Orlandi I., Alberghina L., Vai M. Involvement of the yeast metacaspase Yca1 in ubp10Delta-programmed cell death. FEMS Yeast Res. 2004;5:141–147. doi: 10.1016/j.femsyr.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Boyce M., Yuan J. Cellular response to endoplasmic reticulum stress: a matter of life or death. Cell Death Differ. 2006;13:363–373. doi: 10.1038/sj.cdd.4401817. [DOI] [PubMed] [Google Scholar]
- Burke J. D., Gould K. L. Molecular cloning and characterization of the Schizosaccharomyces pombe his3 gene for use as a selectable marker. Mol. Gen. Genet. 1994;242:169–176. doi: 10.1007/BF00391010. [DOI] [PubMed] [Google Scholar]
- Breckenridge D. G., Germain M., Mathai J. P., Nguyen M., Shore G. C. Regulation of apoptosis by endoplasmic reticulum pathways. Oncogene. 2003;22:8608–8618. doi: 10.1038/sj.onc.1207108. [DOI] [PubMed] [Google Scholar]
- Brezniceanu M. L., Volp K., Bosser S., Solbach C., Lichter P., Joos S., Zornig M. HMGB1 inhibits cell death in yeast and mammalian cells and is abundantly expressed in human breast carcinoma. FASEB J. 2003;17:1295–1297. doi: 10.1096/fj.02-0621fje. [DOI] [PubMed] [Google Scholar]
- Bukau B., Deuerling E., Pfund C., Craig E. A. Getting newly synthesized proteins into shape. Cell. 2000;101:119–122. doi: 10.1016/S0092-8674(00)80806-5. [DOI] [PubMed] [Google Scholar]
- Burhans W. C., Weinberger M. Yeast endonuclease G: complex matters of death, and of life. Mol. Cell. 2007;25:323–325. doi: 10.1016/j.molcel.2007.01.030. [DOI] [PubMed] [Google Scholar]
- Burhans W. C., Weinberger M., Marchetti M. A., Ramachandran L., D'Urso G., Huberman J. A. Apoptosis-like yeast cell death in response to DNA damage and replication defects. Mutat. Res. 2003;532:227–243. doi: 10.1016/j.mrfmmm.2003.08.019. [DOI] [PubMed] [Google Scholar]
- Buttner S., Eisenberg T., Herker E., Carmona-Gutierrez D., Kroemer G., Madeo F. Why yeast cells can undergo apoptosis: death in times of peace, love, and war. J. Cell Biol. 2006;175:521–525. doi: 10.1083/jcb.200608098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody R. J., Cotter T. G. Signalling apoptosis: a radical approach. Redox. Rep. 2001;6:77–90. doi: 10.1179/135100001101536085. [DOI] [PubMed] [Google Scholar]
- Cho N. C., Kang H. J., Lim H. W., Kim B. C., Park E. H., Lim C. J. Stress-dependent regulation of Pbh1, a BIR domain-containing protein, in the fission yeast. Can. J. Microbiol. 2006;52:1261–1265. doi: 10.1139/w06-081. [DOI] [PubMed] [Google Scholar]
- Collin P., Beauregard P. B., Elagoz A., Rokeach L. A. A non-chromosomal factor allows viability of Schizosaccharomyces pombe lacking the essential chaperone calnexin. J. Cell Sci. 2004;117:907–918. doi: 10.1242/jcs.00943. [DOI] [PubMed] [Google Scholar]
- Delom F., Emadali A., Cocolakis E., Lebrun J. J., Nantel A., Chevet E. Calnexin-dependent regulation of tunicamycin-induced apoptosis in breast carcinoma MCF-7 cells. Cell Death Differ. 2007;14:586–596. doi: 10.1038/sj.cdd.4402012. [DOI] [PubMed] [Google Scholar]
- Delom F., Fessart D., Chevet E. Regulation of calnexin sub-cellular localization modulates endoplasmic reticulum stress-induced apoptosis in MCF-7 cells. Apoptosis. 2007;12:293–305. doi: 10.1007/s10495-006-0625-4. [DOI] [PubMed] [Google Scholar]
- Denzel A., Molinari M., Trigueros C., Martin J. E., Velmurgan S., Brown S., Stamp G., Owen M. J. Early postnatal death and motor disorders in mice congenitally deficient in calnexin expression. Mol. Cell. Biol. 2002;22:7398–7404. doi: 10.1128/MCB.22.21.7398-7404.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elagoz A., Callejo M., Armstrong J., Rokeach L. A. Although calnexin is essential in S. pombe, its highly conserved central domain is dispensable for viability. J. Cell Sci. 1999;112(Pt 23):4449–4460. doi: 10.1242/jcs.112.23.4449. [DOI] [PubMed] [Google Scholar]
- Elbe R. A simple and efficient procedure for transformation of yeasts. Biotechniques. 1992;13:18–19. [PubMed] [Google Scholar]
- Fabrizio P., Longo V. D. Chronological aging-induced apoptosis in yeast. Biochim. Biophys. Acta. 2008;1783:1280–1285. doi: 10.1016/j.bbamcr.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog B., Sauder U., Aebi U. The S. cerevisiae HtrA-like protein Nma111p is a nuclear serine protease that mediates yeast apoptosis. J. Cell Sci. 2004;117:115–126. doi: 10.1242/jcs.00848. [DOI] [PubMed] [Google Scholar]
- Fannjiang Y., Cheng W. C., Lee S. J., Qi B., Pevsner J., McCaffery J. M., Hill R. B., Basanez G., Hardwick J. M. Mitochondrial fission proteins regulate programmed cell death in yeast. Genes Dev. 2004;18:2785–2797. doi: 10.1101/gad.1247904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez F., Jannatipour M., Hellman U., Rokeach L. A., Parodi A. J. A new stress protein: synthesis of Schizosaccharomyces pombe UDP–Glc:glycoprotein glucosyltransferase mRNA is induced by stress conditions but the enzyme is not essential for cell viability. EMBO J. 1996;15:705–713. [PMC free article] [PubMed] [Google Scholar]
- Fewell S. W., Travers K. J., Weissman J. S., Brodsky J. L. The action of molecular chaperones in the early secretory pathway. Annual Rev. Genet. 2001;35:149–191. doi: 10.1146/annurev.genet.35.102401.090313. [DOI] [PubMed] [Google Scholar]
- Forsburg S. L. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 1993;21:2955–2956. doi: 10.1093/nar/21.12.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich K. U., Fussi H., Ruckenstuhl C. Yeast apoptosis—from genes to pathways. Semin. Cancer Biol. 2007;17:112–121. doi: 10.1016/j.semcancer.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Groenendyk J., Zuppini A., Shore G., Opas M., Bleackley R. C., Michalak M. Caspase 12 in calnexin-deficient cells. Biochemistry. 2006;45:13219–13226. doi: 10.1021/bi061428z. [DOI] [PubMed] [Google Scholar]
- Hacki J., Egger L., Monney L., Conus S., Rosse T., Fellay I., Borner C. Apoptotic crosstalk between the endoplasmic reticulum and mitochondria controlled by Bcl-2. Oncogene. 2000;19:2286–2295. doi: 10.1038/sj.onc.1203592. [DOI] [PubMed] [Google Scholar]
- Hajjar F., Beauregard P. B., Rokeach L. A. The 160 N-terminal residues of calnexin define a novel region supporting viability in Schizosaccharomyces pombe. Yeast. 2007;24:89–103. doi: 10.1002/yea.1440. [DOI] [PubMed] [Google Scholar]
- Hardwick J. M., Cheng W. C. Mitochondrial programmed cell death pathways in yeast. Dev. Cell. 2004;7:630–632. doi: 10.1016/j.devcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Hauptmann P., Lehle L. Kex1 protease is involved in yeast cell death induced by defective N-glycosylation, acetic acid and during chronological aging. J. Biol. Chem. 2008;283:19151–19163. doi: 10.1074/jbc.M801303200. [DOI] [PubMed] [Google Scholar]
- Hauptmann P., Riel C., Kunz-Schughart L. A., Frohlich K. U., Madeo F., Lehle L. Defects in N-glycosylation induce apoptosis in yeast. Mol. Microbiol. 2006;59:765–778. doi: 10.1111/j.1365-2958.2005.04981.x. [DOI] [PubMed] [Google Scholar]
- Hebert D. N., Garman S. C., Molinari M. The glycan code of the endoplasmic reticulum: asparagine-linked carbohydrates as protein maturation and quality-control tags. Trends Cell Biol. 2005;15:364–370. doi: 10.1016/j.tcb.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Helenius A., Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- Herker E., Jungwirth H., Lehmann K. A., Maldener C., Frohlich K. U., Wissing S., Buttner S., Fehr M., Sigrist S., Madeo F. Chronological aging leads to apoptosis in yeast. J. Cell Biol. 2004;164:501–507. doi: 10.1083/jcb.200310014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ink B., Zornig M., Baum B., Hajibagheri N., James C., Chittenden T., Evan G. Human Bak induces cell death in Schizosaccharomyces pombe with morphological changes similar to those with apoptosis in mammalian cells. Mol. Cell. Biol. 1997;17:2468–2474. doi: 10.1128/mcb.17.5.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska I., Hardwick J. M. Viruses activate a genetically conserved cell death pathway in a unicellular organism. J. Cell Biol. 2005;170:391–399. doi: 10.1083/jcb.200503069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannatipour M., Callejo M., Parodi A. J., Armstrong J., Rokeach L. A. Calnexin and BiP interact with acid phosphatase independently of glucose trimming and reglucosylation in Schizosaccharomyces pombe. Biochemistry. 1998;37:17253–17261. doi: 10.1021/bi981785c. [DOI] [PubMed] [Google Scholar]
- Jannatipour M., Rokeach L. A. The Schizosaccharomyces pombe homologue of the chaperone calnexin is essential for viability. J. Biol. Chem. 1995;270:4845–4853. doi: 10.1074/jbc.270.9.4845. [DOI] [PubMed] [Google Scholar]
- Jin C., Reed J. C. Yeast and apoptosis. Nat. Rev. Mol. Cell Biol. 2002;3:453–459. doi: 10.1038/nrm832. [DOI] [PubMed] [Google Scholar]
- Kerr J. F. History of the events leading to the formulation of the apoptosis concept. Toxicology. 2002;181–182:471–474. doi: 10.1016/s0300-483x(02)00457-2. [DOI] [PubMed] [Google Scholar]
- Kerr J. F., Wyllie A. H., Currie A. R. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R., Emi M., Tanabe K., Murakami S. Role of the unfolded protein response in cell death. Apoptosis. 2006;11:5–13. doi: 10.1007/s10495-005-3088-0. [DOI] [PubMed] [Google Scholar]
- Komatsu K., Hopkins K. M., Lieberman H. B., Wang H. Schizosaccharomyces pombe Rad9 contains a BH3-like region and interacts with the anti-apoptotic protein Bcl-2. FEBS Lett. 2000;481:122–126. doi: 10.1016/s0014-5793(00)01975-x. [DOI] [PubMed] [Google Scholar]
- Lai E., Teodoro T., Volchuk A. Endoplasmic reticulum stress: signaling the unfolded protein response. Physiology. 2007;22:193–201. doi: 10.1152/physiol.00050.2006. [DOI] [PubMed] [Google Scholar]
- Lee Y. K., Brewer J. W., Hellman R., Hendershot L. M. BiP and immunoglobulin light chain cooperate to control the folding of heavy chain and ensure the fidelity of immunoglobulin assembly. Mol. Biol. Cell. 1999;10:2209–2219. doi: 10.1091/mbc.10.7.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Lee A. S. Stress induction of GRP78/BiP and its role in cancer. Curr. Mol. Med. 2006;6:45–54. doi: 10.2174/156652406775574523. [DOI] [PubMed] [Google Scholar]
- Liang Q., Li W., Zhou B. Caspase-independent apoptosis in yeast. Biochim. Biophys. Acta. 2008;1783:1311–1319. doi: 10.1016/j.bbamcr.2008.02.018. [DOI] [PubMed] [Google Scholar]
- Ligr M., Madeo F., Frohlich E., Hilt W., Frohlich K. U., Wolf D. H. Mammalian Bax triggers apoptotic changes in yeast. FEBS Lett. 1998;438:61–65. doi: 10.1016/s0014-5793(98)01227-7. [DOI] [PubMed] [Google Scholar]
- Lim H. W., Kim S. J., Park E. H., Lim C. J. Overexpression of a metacaspase gene stimulates cell growth and stress response in Schizosaccharomyces pombe. Can. J. Microbiol. 2007;53:1016–1023. doi: 10.1139/W07-067. [DOI] [PubMed] [Google Scholar]
- Low C. P., Liew L. P., Pervaiz S., Yang H. Apoptosis and lipoapoptosis in the fission yeast Schizosaccharomyces pombe. FEMS Yeast Res. 2005;5:1199–1206. doi: 10.1016/j.femsyr.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Low C. P., Yang H. Programmed cell death in fission yeast Schizosaccharomyces pombe. Biochim. Biophys. Acta. 2008;1783:1355–1349. doi: 10.1016/j.bbamcr.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Maattanen P., Kozlov G., Gehring K., Thomas D. Y. ERp57 and PDI: multifunctional protein disulfide isomerases with similar domain architectures but differing substrate-partner associations. Biochem. Cell Biol. 2006;84:881–889. doi: 10.1139/o06-186. [DOI] [PubMed] [Google Scholar]
- Madeo F., Frohlich E., Frohlich K. U. A yeast mutant showing diagnostic markers of early and late apoptosis. J. Cell Biol. 1997;139:729–734. doi: 10.1083/jcb.139.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F., Frohlich E., Ligr M., Grey M., Sigrist S. J., Wolf D. H., Frohlich K. U. Oxygen stress: a regulator of apoptosis in yeast. J. Cell Biol. 1999;145:757–767. doi: 10.1083/jcb.145.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F., et al. A caspase-related protease regulates apoptosis in yeast. Mol. Cell. 2002;9:911–917. doi: 10.1016/s1097-2765(02)00501-4. [DOI] [PubMed] [Google Scholar]
- Madeo F., Herker E., Wissing S., Jungwirth H., Eisenberg T., Frohlich K. U. Apoptosis in yeast. Curr. Opin. Microbiol. 2004;7:655–660. doi: 10.1016/j.mib.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Marciniak S. J., Ron D. Endoplasmic reticulum stress signaling in disease. Physiol. Rev. 2006;86:1133–1149. doi: 10.1152/physrev.00015.2006. [DOI] [PubMed] [Google Scholar]
- Marechal A., Tanguay P. L., Callejo M., Guerin R., Boileau G., Rokeach L. A. Cell viability and secretion of active proteins in Schizosaccharomyces pombe do not require the chaperone function of calnexin. Biochem. J. 2004;380:441–448. doi: 10.1042/BJ20031546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Mazzoni C., Falcone C. Caspase-dependent apoptosis in yeast. Biochim. Biophys. Acta. 2008;1783:1320–1327. doi: 10.1016/j.bbamcr.2008.02.015. [DOI] [PubMed] [Google Scholar]
- McCracken A. A., Brodsky J. L. Recognition and delivery of ERAD substrates to the proteasome and alternative paths for cell survival. Curr. Top. Microbiol. Immunol. 2005;300:17–40. doi: 10.1007/3-540-28007-3_2. [DOI] [PubMed] [Google Scholar]
- Meusser B., Hirsch C., Jarosch E., Sommer T. ERAD: the long road to destruction. Nat. Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Meth. Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Nguyen M., Breckenridge D. G., Ducret A., Shore G. C. Caspase-resistant BAP31 inhibits fas-mediated apoptotic membrane fragmentation and release of cytochrome c from mitochondria. Mol. Cell. Biol. 2000;20:6731–6740. doi: 10.1128/mcb.20.18.6731-6740.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M., Lee A. S. ER chaperones in mammalian development and human diseases. FEBS Lett. 2007;581:3641–3651. doi: 10.1016/j.febslet.2007.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlati F., Dignard D., Bergeron J. J., Thomas D. Y. The calnexin homologue cnx1+ in Schizosaccharomyces pombe, is an essential gene which can be complemented by its soluble ER domain. EMBO J. 1995a;14:3064–3072. doi: 10.1002/j.1460-2075.1995.tb07309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlati F., Dominguez M., Bergeron J. J., Thomas D. Y. Saccharomyces cerevisiae CNE1 encodes an endoplasmic reticulum (ER) membrane protein with sequence similarity to calnexin and calreticulin and functions as a constituent of the ER quality control apparatus. J. Biol. Chem. 1995b;270:244–253. doi: 10.1074/jbc.270.1.244. [DOI] [PubMed] [Google Scholar]
- Parodi A. J. Protein glucosylation and its role in protein folding. Annu. Rev. Biochem. 2000;69:69–93. doi: 10.1146/annurev.biochem.69.1.69. [DOI] [PubMed] [Google Scholar]
- Patil C., Walter P. Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 2001;13:349–355. doi: 10.1016/s0955-0674(00)00219-2. [DOI] [PubMed] [Google Scholar]
- Perez-Sala D., Mollinedo F. Inhibition of N-linked glycosylation induces early apoptosis in human promyelocytic HL-60 cells. J. Cell. Physiol. 1995;163:523–531. doi: 10.1002/jcp.1041630312. [DOI] [PubMed] [Google Scholar]
- Priault M., Camougrand N., Kinnally K. W., Vallette F. M., Manon S. Yeast as a tool to study Bax/mitochondrial interactions in cell death. FEMS Yeast Res. 2003;4:15–27. doi: 10.1016/S1567-1356(03)00143-0. [DOI] [PubMed] [Google Scholar]
- Rajagopalan S., Xu Y., Brenner M. B. Retention of unassembled components of integral membrane proteins by calnexin. Science. 1994;263:387–390. doi: 10.1126/science.8278814. [DOI] [PubMed] [Google Scholar]
- Ramer S. W., Elledge S. J., Davis R. W. Dominant genetics using a yeast genomic library under the control of a strong inducible promoter. Proc. Natl. Acad. Sci. USA. 1992;89:11589–11593. doi: 10.1073/pnas.89.23.11589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R. V., Peel A., Logvinova A., del Rio G., Hermel E., Yokota T., Goldsmith P. C., Ellerby L. M., Ellerby H. M., Bredesen D. E. Coupling endoplasmic reticulum stress to the cell death program: role of the ER chaperone GRP78. FEBS Lett. 2002;514:122–128. doi: 10.1016/s0014-5793(02)02289-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy R. K., Mao C., Baumeister P., Austin R. C., Kaufman R. J., Lee A. S. Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: role of ATP binding site in suppression of caspase-7 activation. J. Biol. Chem. 2003;278:20915–20924. doi: 10.1074/jbc.M212328200. [DOI] [PubMed] [Google Scholar]
- Reiter J., Herker E., Madeo F., Schmitt M. J. Viral killer toxins induce caspase-mediated apoptosis in yeast. J. Cell Biol. 2005;168:353–358. doi: 10.1083/jcb.200408071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Menocal L., D'Urso G. Programmed cell death in fission yeast. FEMS Yeast Res. 2004;5:111–117. doi: 10.1016/j.femsyr.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Romisch K. Surfing the Sec61 channel: bidirectional protein translocation across the ER membrane. J. Cell Sci. 1999;112(Pt 23):4185–4191. doi: 10.1242/jcs.112.23.4185. [DOI] [PubMed] [Google Scholar]
- Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Roux A. E., Quissac A., Chartrand P., Ferbeyre G., Rokeach L. A. Regulation of chronological aging in Schizosaccharomyces pombe by the protein kinases Pka1 and Sck2. Aging Cell. 2006;5:345–357. doi: 10.1111/j.1474-9726.2006.00225.x. [DOI] [PubMed] [Google Scholar]
- Saito Y., Ihara Y., Leach M. R., Cohen-Doyle M. F., Williams D. B. Calreticulin functions in vitro as a molecular chaperone for both glycosylated and non-glycosylated proteins. EMBO J. 1999;18:6718–6729. doi: 10.1093/emboj/18.23.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder M., Kaufman R. J. The mammalian unfolded protein response. Annual Rev. Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- Skulachev V. P. Programmed death in yeast as adaptation? FEBS Lett. 2002;528:23–26. doi: 10.1016/s0014-5793(02)03319-7. [DOI] [PubMed] [Google Scholar]
- Szegezdi E., Logue S. E., Gorman A. M., Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep. 2006;7:880–885. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa T., Tatematsu C., Watanabe K., Kato K., Nakanishi Y. Cleavage of calnexin caused by apoptotic stimuli: implication for the regulation of apoptosis. J. Biochem. 2004;136:399–405. doi: 10.1093/jb/mvh133. [DOI] [PubMed] [Google Scholar]
- Thammavongsa V., Mancino L., Raghavan M. Polypeptide substrate recognition by calnexin requires specific conformations of the calnexin protein. J. Biol. Chem. 2005;280:33497–33505. doi: 10.1074/jbc.M503648200. [DOI] [PubMed] [Google Scholar]
- Tomassini B., Malisan F., Franchi L., Nicolo C., Calvo G. B., Saito T., Testi R. Calnexin suppresses GD3 synthase-induced apoptosis. FASEB J. 2004;18:1553–1555. doi: 10.1096/fj.04-1675fje. [DOI] [PubMed] [Google Scholar]
- Torgler C. N., de Tiani M., Raven T., Aubry J. P., Brown R., Meldrum E. Expression of bak in S. pombe results in a lethality mediated through interaction with the calnexin homologue Cnx1. Cell Death Differ. 1997;4:263–271. doi: 10.1038/sj.cdd.4400239. [DOI] [PubMed] [Google Scholar]
- Trombetta E. S., Parodi A. J. Quality control and protein folding in the secretory pathway. Annu. Rev. Cell Dev. Biol. 2003;19:649–676. doi: 10.1146/annurev.cellbio.19.110701.153949. [DOI] [PubMed] [Google Scholar]
- Ulloa L., Messmer D. High-mobility group box 1 (HMGB1) protein: friend and foe. Cytokine Growth Factor Rev. 2006;17:189–201. doi: 10.1016/j.cytogfr.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Uren A. G., O'Rourke K., Aravind L. A., Pisabarro M. T., Seshagiri S., Koonin E. V., Dixit V. M. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell. 2000;6:961–967. doi: 10.1016/s1097-2765(00)00094-0. [DOI] [PubMed] [Google Scholar]
- Wadskog I., Maldener C., Proksch A., Madeo F., Adler L. Yeast lacking the SRO7/SOP1-encoded tumor suppressor homologue show increased susceptibility to apoptosis-like cell death on exposure to NaCl stress. Mol. Biol. Cell. 2004;15:1436–1444. doi: 10.1091/mbc.E03-02-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter D., Wissing S., Madeo F., Fahrenkrog B. The inhibitor-of-apoptosis protein Bir1p protects against apoptosis in S. cerevisiae and is a substrate for the yeast homologue of Omi/HtrA2. J. Cell Sci. 2006;119:1843–1851. doi: 10.1242/jcs.02902. [DOI] [PubMed] [Google Scholar]
- Williams D. B. Beyond lectins: the calnexin/calreticulin chaperone system of the endoplasmic reticulum. J. Cell Sci. 2006;119:615–623. doi: 10.1242/jcs.02856. [DOI] [PubMed] [Google Scholar]
- Wissing S., et al. An AIF orthologue regulates apoptosis in yeast. J. Cell Biol. 2004;166:969–974. doi: 10.1083/jcb.200404138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Kaufman R. J. From acute ER stress to physiological roles of the unfolded protein response. Cell Death Differ. 2006;13:374–384. doi: 10.1038/sj.cdd.4401840. [DOI] [PubMed] [Google Scholar]
- Xu C., Bailly-Maitre B., Reed J. C. Endoplasmic reticulum stress: cell life and death decisions. J. Clin. Invest. 2005;115:2656–2664. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Chieu H.K., Low C. P., Zhang S., Heng C. K., Yang H. Schizosaccharomyces pombe cells deficient in triacylglycerols synthesis undergo apoptosis upon entry into the stationary phase. J. Biol. Chem. 2003;278:47145–47155. doi: 10.1074/jbc.M306998200. [DOI] [PubMed] [Google Scholar]
- Zhao L., Ackerman S. L. Endoplasmic reticulum stress in health and disease. Current Opin. Cell Biol. 2006;18:444–452. doi: 10.1016/j.ceb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Zuppini A., Groenendyk J., Cormack L. A., Shore G., Opas M., Bleackley R. C., Michalak M. Calnexin deficiency and endoplasmic reticulum stress-induced apoptosis. Biochemistry. 2002;41:2850–2858. doi: 10.1021/bi015967+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.