Abstract
In budding yeast, Tem1 is a key regulator of mitotic exit. Bfa1/Bub2 stimulates Tem1 GTPase activity as a GTPase-activating protein (GAP). Lte1 possesses a guanine-nucleotide exchange factor (GEF) domain likely for Tem1. However, recent observations showed that cells may control mitotic exit without either Lte1 or Bfa1/Bub2 GAP activity, obscuring how Tem1 is regulated. Here, we assayed BFA1 mutants with varying GAP activities for Tem1, showing for the first time that Bfa1/Bub2 GAP activity inhibits Tem1 in vivo. A decrease in GAP activity allowed cells to bypass mitotic exit defects. Interestingly, different levels of GAP activity were required to prevent mitotic exit depending on the type of perturbation. Although essential, more Bfa1/Bub2 GAP activity was needed for spindle damage than for DNA damage to fully activate the checkpoint. Conversely, Bfa1/Bub2 GAP activity was insufficient to delay mitotic exit in cells with misoriented spindles. Instead, decreased interaction of Bfa1 with Kin4 was observed in BFA1 mutant cells with a defective spindle position checkpoint. These findings demonstrate that there is a GAP-independent surveillance mechanism of Bfa1/Bub2, which, together with the GTP/GDP switch of Tem1, may be required for the genomic stability of cells with misaligned spindles.
INTRODUCTION
At the end of mitosis, cyclin-dependent kinases (CDKs) become inactivated. Mitotic exit is best characterized in the budding yeast Saccharomyces cerevisiae in which the inactivation of CDKs is regulated by the mitotic exit network (MEN). Tem1 is one of the most upstream components of the MEN, functioning as its key regulator (Lee et al., 2001; Mah et al., 2001). Activated Tem1 stimulates Cdc15 kinase, which in turn activates Dbf2 kinase and its associated factor, Mob1, leading to the release of Cdc14 phosphatase from the nucleolus to ultimately block CDK activity (reviewed in Bardin and Amon, 2001).
The idea for Tem1 regulation of mitotic exit in S. cerevisiae comes from its comparison with the septation initiation network (SIN) of Schizosaccharomyces pombe (reviewed in Bardin and Amon, 2001). Although their functions somewhat differ, homologues of most MEN components have been found in the SIN and vice versa. The SIN is regulated by the activity of spg1, a small GTPase homologue of Tem1. In vivo, spg1 is negatively regulated by byr4 and cdc16, which act as a two-component GTPase-activating protein (GAP) for spg1 in vitro (Minet et al., 1979; Song et al., 1996; Furge et al., 1998). Likewise, Tem1 is also a Ras-like GTP-binding protein whose activity can be accelerated substantially in vitro by Bfa1 and Bub2, which are highly related to byr4 and cdc16, respectively (Barbacid, 1987; Song et al., 1996; Furge et al., 1998; Geymonat et al., 2002). Indeed, Bub2 has a domain similar to GAP and requires Bfa1 to antagonize Tem1 (Neuwald, 1997; Geymonat et al., 2002). Bfa1/Bub2 GAP activity is down-regulated via Bfa1 phosphorylation by Cdc5 (Hu et al., 2001; Geymonat et al., 2003). In cdc5-2 cells that have a defect in phosphorylating Bfa1, mitotic arrest can be suppressed by BFA1 deletion (Hu et al., 2001). Accordingly, Bfa1/Bub2 GAP activity has been proposed to negatively control Tem1. Lte1 further establishes the control of Tem1 GTPase activity in mitotic exit. Δlte1 cells have a defect in mitotic exit at low temperatures, and high levels of Tem1 suppress the cold-sensitive phenotype of Δlte1 cells, indicating that Lte1 acts to positively regulate mitotic exit upstream of Tem1 (Shirayama et al., 1994a,b). In fact, Lte1 possesses an apparent guanine-nucleotide exchange factor (GEF) domain, and its overexpression promotes mitotic exit (Shirayama et al., 1994a; Bardin et al., 2000). Thus, Lte1 has been thought to function as a GEF for Tem1. It is generally believed that Tem1 is maintained in an inactive GDP-bound state by Bfa1/Bub2 GAP and that Lte1 promotes the conversion of GDP into GTP, thereby activating the MEN.
To maintain genomic integrity, mitotic exit is delayed in response to various perturbations, such as DNA damage, spindle disruption, and misaligned spindles (Wang et al., 2000). Deletion of either BFA1 or BUB2 was sufficient to trigger mitotic exit in checkpoint-arrest cells (Wang et al., 2000), suggesting that Bfa1/Bub2 is a target of multiple checkpoint pathways to inhibit mitotic exit and that the inhibition of Bfa1/Bub2 activity may be the actual trigger of mitotic exit. Indeed, Tem1 activation is likely to occur without its putative GEF, Lte1. Although Lte1 is restricted to daughter cells, Adames et al. (2001) observed that certain mutants improperly exit mitosis even though the mispositioned spindle is present within the mother cells. In addition, LTE1 deletion had little effect on the timing of mitotic exit at physiological temperature (Adames et al., 2001). These results have been explained by high intrinsic nucleotide exchange ability of Tem1 (Geymonat et al., 2002) or presence of additional ways parallel to Lte1 in facilitating mitotic exit (Hofken and Schiebel, 2002).
Nonetheless, there have been conflicting data as to whether Bfa1/Bub2 GAP activity inhibits Tem1 and mitotic exit. Bfa1 overexpression can prevent mitotic exit in Δbub2 cells, although Bfa1 surprisingly inhibits the GTP exchange activity of Tem1 in vitro in the absence of Bub2 (Geymonat et al., 2002; Ro et al., 2002). When the phosphorylation sites of Cdc5 were mutated to alanines, Bfa1-11A was expected to be constitutively active, resulting in either telophase arrest or delay (Hu et al., 2001). However, this mutant showed no significant delay in mitotic exit (Hu et al., 2001). In addition, Fraschini et al. (2006) reported that Bub2 GAP activity appears to be dispensable for Tem1 inhibition. In fact, there was no evidence that Bfa1/Bub2 GAP activity inhibited Tem1 in vivo, although it can function as a GAP for Tem1 in vitro (Geymonat et al., 2002). For these reasons, whether the GDP/GTP switch for Tem1 controls mitotic exit remains obscure.
In this study, we constructed Bfa1 mutants with various GAP activities in complex with Bub2 and showed that Bfa1/Bub2 GAP activity in vivo inhibited mitotic exit. Interestingly, different levels of GAP activity were required for preventing mitotic exit, depending on the type of perturbation. Bfa1/Bub2 GAP activity was essential to prevent mitotic exit in cells with either DNA or spindle damage, but was not necessary to activate the spindle position checkpoint. Thus, these results help us to better understand the regulatory mechanisms of Tem1 in mitotic exit and present an integrated view of current contradictory data.
MATERIALS AND METHODS
Yeast Strains, Cultures, and Cell Cycle Synchronization
Yeast cell culture and genetic techniques were carried out as described by Kim et al. (2004). All yeast strains used in this study are listed in Supplementary Table S1. The strains were constructed by PCR-based homologous recombination methods and verified by PCR and/or Western blot analysis (Longtine et al., 1998). The full-length BFA1-DR mutants (BFA1M347A, BFA1N350A, BFA1W356A, BFA1GN358AA, BFA1F366A, BFA1G411E, BFA1M413I, BFA1D416A, and BFA1W422A) were constructed by PCR-based site-directed mutagenesis using integrative pRS304-BFA1-GFP or pRS304-BFA1-TAP plasmid as templates in which BFA1-GFP and BFA1-TAP are expressed under the control of BFA1 endogenous promoter. Various genetic interactions demonstrated that green fluorescent protein (GFP) or tandem affinity purification (TAP), epitope tag fused to the C-terminus of Bfa1 had little effect on Bfa1 function as the negative regulator of mitotic exit (data not shown). For chromosomal integration of BFA1 or each BFA1-DR mutant, each integrative plasmid linearized with EcoRV was inserted into the TRP1 locus of Δbfa1 background strains, and expression was verified by fluorescence microscopy and Western blot. The copy number of integrated plasmid was also verified by Southern analysis with EcoRI-digested genomic DNA using 836-base pair HindIII-EcoRI fragment of BFA1 as a probe (Supplementary Figure S2). Cells were synchronized at G1 by the addition of either 10 μg/ml or 50 ng/ml α-factor (Sigma, St. Louis, MO) for BAR1 or bar1 cells, respectively. Overexpression of either BFA1-D8 or BFA1-D8M413I was induced by GAL promoter as described by Kim et al. (2004).
Random Mutagenesis of BFA1 and Screening for Bfa1 Mutants
Random in vitro mutagenesis was performed with hydroxylamine as described previously (Kim and Song, 2006). Transformants harboring mutagenized pRS316-PGAL-BFA1-D8-GFP or pRS316-PBFA1-BFA1-D8 were replica-plated on 2% galactose or 10 μg/ml benomyl medium, respectively. Viable colonies on galactose plates and nonviable colonies on benomyl plates were selected. From isolated cells, mutagenized plasmids were rescued, sequenced, and subcloned into intact pRS316-PGAL-GFP or pRS316-PBFA1. Subcloned plasmids were retransformed into Δbfa1 (YSK8) cells to verify viability on galactose or benomyl medium.
In Vitro GTPase Assay
Intrinsic or GAP-stimulated GTPase activity of Tem1 was monitored as described by Kim and Song (2006) with an EnzCheck Phosphate Assay Kit (E-6646; Molecular Probes, Eugene, OR). For in vitro GTPase assays, the fusion proteins glutathione S-transferase (GST)-Bub2, maltose-binding protein (MBP)-Tem1, MBP-Bfa1, -Bfa1M347A, -Bfa1G411E, -Bfa1M413A, -Bfa1D416A, and -Bfa1W422A were expressed in Escherichia coli and purified as described by Geymonat et al. (2002). The MBP part was removed by factor Xa from MBP-Tem1. To examine the Tem1 GTPase activity catalyzed by MBP-Bfa1 or each MBP-Bfa1 mutant heterodimerized with GST-Bub2, each Bfa1 protein (5 μg) was added to an 0.8-ml reaction mixture [50 mM HEPES, pH 7.6, 0.1 mM EDTA, 0.2 mM MESG (2-amino-6-mercapto-7-methylpurine riboside), 10 U purine nucleoside phosphorylase, 200 μM GTP, 4 μg Tem1 (treated with factor Xa to remove MBP), and 8 μg GST-Bub2] and the GTPase reaction was initiated by adding MgCl2 at a final concentration of 5 mM. The amount of γ-Pi released from Tem1-GTP was monitored at 30°C by measuring the absorbance at 360 nm.
Microscopy and Flow Cytometry
Fluorescence microscopy was performed as described by Kim et al. (2004). DNA content was measured by flow cytometry using a Becton Dickinson fluorescence-activated cell sorter (Mountain View, CA) as described previously (Kim et al., 2004).
Protein Analysis and Yeast Two-Hybrid Assay
Yeast lysates were prepared at 4°C as described by Kim et al. (2004). For precipitation of TAP-tagged Bfa1 or Bfa1 mutants, total cellular lysates (1 mg in 700 μl modified H-buffer containing 1% NP-40) were incubated at 4°C for 2 h with IgG Sepharose beads (Amersham Pharmacia, Piscataway, NJ). Peroxidase anti-peroxidase (PAP; Sigma), monoclonal anti-LexA (Santa Cruz Biotechnology, Santa Cruz, CA), monoclonal anti-hemagglutinin (HA; Roche, Indianapolis, IN), polyclonal anti-GFP (Santa Cruz Biotechnology), and polyclonal anti-actin (Santa Cruz Biotechnology) were used for Western blot analysis. Yeast two-hybrid assays were performed as described previously (Kim et al., 2004). The band intensity was quantitatively analyzed with a LAS-3000 image analyzer (Fujifilm, Tokyo, Japan).
RESULTS
Two Imperfect Direct Repeat Regions of Bfa1 Are Highly Conserved among Fungi
To better understand the regulatory mechanism of mitotic exit by the Bfa1/Bub2 complex, we screened BFA1 mutants with defects in preventing mitotic exit. Because the function of Bfa1 as a negative regulator of MEN critically depends on the 184 residues of its C-terminus (Kim et al., 2004; Figure 1A), BFA1-D8391-574 was randomly mutated. The pCEN-PGAL-D8-GFP and pCEN-PBFA1-D8 plasmids were respectively mutated with hydroxylamine as described in Materials and Methods and transformed into Δbfa1 (YSK8). BFA1-D8391-574 is overexpressed in pCEN-PGAL-D8-GFP under the control of GAL promoter and expressed endogenously in pCEN-PBFA1-D8 under its own promoter. Transformants harboring pCEN-PGAL-D8-GFP were replica-plated on the medium containing galactose, and the BFA1-D8M413I mutant that could grow in spite of its overexpression was subsequently isolated (Figure 1B). Cells overexpressing Bfa1-D8 arrested with 2C DNA content, whereas cells overexpressing Bfa1-D8M413I completed mitosis and entered the next cell cycle (Figure 1C). On the other hand, transformants with mutated pCEN-PBFA1-D8 were screened for defects in the mitotic checkpoint arrest in the presence of a microtubule-destabilizing drug benomyl, and BFA1-D8M413I and BFA1-D8G411E mutants were isolated (Figure 1B). As shown in Figure 1D, cells treated with the microtubule-destabilizing compound nocodazole showed extra new buds by mitotic progression in 98% of Δbfa1 (vector-transformed), 93.2% of BFA1-D8M413I, 35.7% of BFA1-D8G411E, and 24.8% of BFA1-D8 cells. These results show that the BFA1-D8M413I and BFA1-D8G411E cells have a defect in mitotic arrest compared with BFA1-D8 cells.
Figure 1.
Isolation of Bfa1 mutants that fail to induce mitotic arrest. (A) Schematic diagram of full-length BFA1 and BFA1-D8. The arrows represent two regions of imperfect direct repeats that are highly conserved among fungi: direct repeat 1 (DR1, residues 344-366) and DR2 (residues 409-422). Sequence alignment of DR1 and DR2 among fungi is shown. Identical amino acids are highlighted in black, and amino acids with high similarity are highlighted in gray. (B) Mutated Bfa1 residues in the screened mutants. Mutated bases by random mutagenesis with hydroxylamine are shaded. (C and D) Bfa1-D8M413I and Bfa1-D8G411E mutants failed to induce mitotic arrest. Δbfa1 (YSK8) cells carrying the indicated plasmids were arrested by α-factor and released into medium containing (C) galactose for overexpression or (D) 15 μg/ml nocodazole. At each time point after the release, (C) DNA content was analyzed by fluorescence-activated cell sorting (FACS; n = 50,000) and (D) cells with more than one bud were counted (n = 300). (D) The average of three independent counts is shown with SDs.
Bfa1 was first identified by its homology to byr4 in S. pombe (Song et al., 1996; Li, 1999). The region of similarity between byr4 and Bfa1 is confined to the two C-terminal imperfect direct repeats (Figure 1A). We found that these two repeat regions are also highly conserved among other fungi and contained nine identical amino acid residues (Figure 1A). The mutated residues of Met413 of Bfa1-D8M413I and Gly411 of Bfa1-D8G411E, we isolated for a defect in mitotic arrest are also located in the second repeat and highly conserved (Figure 1A). Thus, we examined whether the conserved amino acids in two repeats of Bfa1 are critical for negatively regulating mitotic exit.
To test this possibility, we constructed various full-length Bfa1 mutants by a series of site-directed mutagenesis as described in Materials and Methods, each of which contained a mutated residue in the first or the second repeat (Figure 1A). Each Bfa1 mutant generated was then tested for Bfa1 function in vivo and in vitro. Mutants in the first repeat were designated as Bfa1-direct repeat1 (Bfa1-DR1) and included Bfa1M347A, Bfa1N350A, Bfa1W356A, Bfa1GN358AA, and Bfa1F366A. Bfa1-DR2 mutants contained substitutions in the second repeat and included Bfa1G411E, Bfa1M413I, Bfa1D416A, and Bfa1W422A. In fact, Bfa1-DR1 mutants showed no difference compared with the wild-type Bfa1 in all in vitro and in vivo assays performed in this study (Supplementary Figure S1). Thus, only Bfa1-DR2 mutants were described hereafter.
Bfa1-DR2 Mutants Are Defective in Negatively Regulating Mitotic Exit
We first investigated the ability of each BFA1 mutant to function as a negative regulator of mitotic exit by analyzing its genetic interactions with mitotic exit–defective mutants, such as Δlte1, Δlte1Δste20, and cdc5-2.
Cells lacking Lte1 show arrest of the cell cycle at the end of mitosis due to its essential role in MEN activation at low temperatures (<14°C; Shirayama et al., 1994a; Figure 2, A and B). Because BFA1 deletion rescues these mitotic exit defects, we examined whether BFA1-DR mutants could prevent mitotic exit in Δlte1Δbfa1 cells. To do this, Δlte1BFA1 and Δlte1BFA1-DR mutant strains were generated by integration of pRS304-BFA1-GFP or each pRS304-BFA1-DR-GFP mutant into the TRP1 locus of Δlte1Δbfa1 cells as described in Materials and Methods. The single integration of the plasmid was confirmed by Southern blot and the expression of either BFA1 or each BFA1 mutant was verified by Western blot and fluorescence microscopy (Supplementary Figure S3). At 13°C, wild-type BFA1 blocked cell proliferation and led to anaphase arrest with a large bud and two separated nuclei, whereas the presence of BFA1-DR2 mutants had little effect on the growth of Δlte1Δbfa1 cells (Figure 2, A and B; Table 1). These results indicate that all Bfa1-DR2 mutants are defective in preventing mitotic exit.
Figure 2.
The genetic interactions of BFA1-DR2 mutants with various mitotic exit–defective cells. (A and B) The effects of BFA1-DR2 mutants on Δlte1Δbfa1 cells. (A) Δlte1BFA1 (YSK2052), Δlte1Δbfa1 (YSK2051), and Δlte1BFA1-DR2 mutants (YSK2058, 2059, 2060, and 2061) were serially diluted on YPAD and incubated at either 25°C for 2 d or 13°C for 10 d. (B) Cells (YSK2051, 2052, 2058, 2059, 2060, and 2061) were synchronized with α-factor at room temperature and released to fresh medium at 13°C. Cells were fixed and stained with DAPI. Anaphase cells were determined per indicated time point (n = 200). (C and D) The effects of BFA1-DR2 mutants on cdc5-2Δbfa1 cells. (C) Top, cdc5-2BFA1 (YSK2030), cdc5-2Δbfa1 (YSK823), and cdc5-2BFA1-DR2 mutants (YSK2036, 2037, 2038, and 2039) were serially diluted on YPAD and incubated at either 25 or 37°C. Bottom, cdc5-2BFA1-TAP cells (YSK1122) synchronized with α-factor at 25°C were released at 37°C, harvested at each time point, and their protein extracts were subjected to Western blotting with PAP. cdc15-2BFA1-TAP cells (YSK1153) incubated for 3 h at 37°C were included as a positive control for slowly migrating forms of Bfa1. Arrow indicates the slowest migrating form of Bfa1. (D) Cells (YSK823, 2030, 2036, 2037, 2038, and 2039) were synchronized with α-factor at 25°C and allowed to progress the cell cycle in YPAD at 37°C. At each time point, cells were collected to analyze DNA content by FACS (top, n = 50,000), and the cells with indicated phenotypes were counted (bottom, n = 200).
Table 1.
Summary of in vitro GAP activity and in vivo genetic interactions in each Bfa1-DR2 mutant
| Activity | Bfa1 mutants |
|||||
|---|---|---|---|---|---|---|
| Wt Bfa1 | G411E | D416A | M413I | W422A | Δbfa1 | |
| In vitro GAP activitya | 100% | 0% | ||||
| The growth rate in Δlte1Δbfa1 cellsb | ++ | − | − | − | − | − |
| The growth rate in Δlte1Δste20Δbfa1cellsc | ++ | + | − | − | − | − |
| Premature binding of Mob1 to SPBs in cdc15–2Δbfa1 cellsd | ++ | ++ | ++ | + | +/− | − |
| The growth rate in cdc5–2Δbfa1cellse | ++ | ++ | ++ | ++ | + | − |
| Spindle damage checkpoint activityf | ++ | + | − | − | − | − |
| DNA damage checkpoint activityg | ++ | ++ | + | + | − | − |
| Spindle position checkpoint activityh | ++ | ++ | − | + | ++ | − |
Kinase-dead cdc5-2 cells grown at the restrictive temperature were arrested in late anaphase by a block of Bfa1 phosphorylation, which can be suppressed by BFA1 deletion (Hu et al., 2001; Figure 2, C and D). Thus, we expected that each Bfa1-DR mutant would suppress the growth of cdc5-2Δbfa1 cells, depending on its ability to inhibit the MEN. pRS304-BFA1-GFP or each pRS304-BFA1-DR-GFP mutant was integrated into cdc5-2Δbfa1 cells as described in Materials and Methods. The single copy number of the integrated plasmid was assessed by Southern blot and the expression of either BFA1 or each BFA1 mutant was verified by Western blot and fluorescence microscopy (Supplementary Figure S4). Differently from above result, Bfa1G411E, Bfa1M413I, and Bfa1D416A suppressed the growth of cdc5-2Δbfa1 cells (Figure 2C; Table 1). Only cdc5-2BFA1W422A cells displayed increased growth rate at the restrictive temperature (Figure 2C; Table 1). To verify this result, we examined cell cycle progression of these cells, which were arrested at G1 and released at 37°C. All of cdc5-2BFA1-DR2 mutants except cdc5-2BFA1W422A were arrested as large-budded cells with 2C DNA content, similar to cdc5-2BFA1 cells, demonstrating that these Bfa1 mutants inhibited mitotic exit of cdc5-2Δbfa1 cells (Figure 2D). Conversely, in a portion of cdc5-2BFA1W422A cells, new bud began to appear, and cells with DNA content greater than 2C were accumulated similar to cdc5-2Δbfa1 cells (40% for cdc5-2Δbfa1 cells; 32% for cdc5-2BFA1W422A cells), demonstrating that Bfa1W422A failed to prevent mitotic exit.
We next investigated the growth of Δste20Δlte1BFA1-DR cells. Ste20, a cell polarity protein in the cortex, is needed for mitotic exit independent of Lte1 (Hofken and Schiebel, 2002). Thus, Δlte1Δste20 cells are not viable, likely due to a failure in MEN activation (Hofken and Schiebel, 2002; Supplementary Figure S5). Δlte1Δste20BFA1 and Δlte1Δste20BFA1-DR mutant strains were generated by integration of pRS304-BFA1-GFP or each pRS304-BFA1-DR-GFP mutant into Δlte1Δste20Δbfa1 cells with pCEN-URA3-LTE1 plasmid as described in Materials and Methods. The expression of either BFA1 or each BFA1-DR2 mutant was verified by fluorescence microscopy (Supplementary Figure S5). We found that all Bfa1-DR2 mutants did not block mitotic progression of Δlte1Δste20Δbfa1 cells similarly to the case of Δlte1Δbfa1 cells, whereas wild-type Bfa1 repressed the growth of Δlte1Δste20Δbfa1 cells (Table 1; Supplementary Figure S5). Interestingly, Δlte1Δste20BFA1G411E cells showed a slower growth rate than other Δlte1Δste20BFA1-DR2 cells (Table 1; Supplementary Figure S5).
On the basis of the results of Figure 2, A and B, we expected that Bfa1-DR2 mutants would have little effect on growth rate of Δbfa1 cells with mitotic exit defects. However, only Bfa1W422A failed to prevent mitotic exit of cdc5-2Δbfa1 cells, and Δlte1Δste20BFA1G411E cells showed slow growth rate. Therefore, these results led us to propose that Bfa1 activity in regulating mitotic exit is defective in Bfa1-DR2 mutants, but their activities are not equal and that there is a difference in the extent of Bfa1 activity for the complete inhibition of the MEN in mitotic exit–defective mutants. The function of Bfa1 as a negative regulator may be most severely impaired in Bfa1W422A among Bfa1-DR2 mutants and is less weakened in Bfa1G411E than in other Bfa1-DR2 mutants. Consistent with this notion, the extent of impaired activities also varied, as observed in the premature localization of Mob1-GFP to the spindle pole bodies (SPBs) in BFA1-DR2 and cdc15-2BFA1-DR2 cells (Table 1; Supplementary Figure S6).
GAP Activity of Each Bfa1-DR2 Mutant with Bub2 Is Impaired in Tem1 GTPase Assays In Vitro
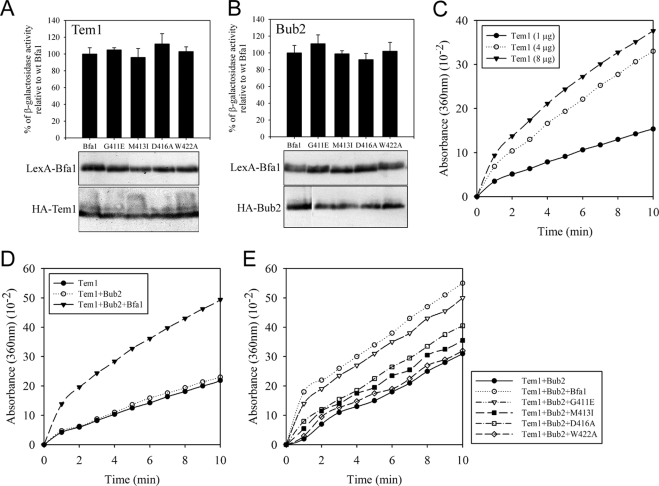
To determine whether the failure of the Bfa1-DR2 mutants in blocking mitotic exit is related to their GAP activity, we directly measured GAP activity by in vitro Tem1 GTPase assay. First, we quantitatively examined the interaction of Bfa1 mutants with Tem1 and Bub2 by yeast two-hybrid assay. The affinity of each Bfa1-DR2 mutant for either Tem1 or Bub2 was comparable to that of wild-type Bfa1 (Figure 3, A and B).
Figure 3.
The GAP activity of Bfa1 and each Bfa1-DR2 mutant complexed with Bub2 in vitro. (A and B) Yeast two-hybrid β-galactosidase assay. The interaction of wild-type Bfa1 and each Bfa1-DR2 mutant with (A) Tem1 or (B) Bub2 was measured quantitatively by yeast two-hybrid assay as described in Materials and Methods. Western blots (bottom) show that similar protein amounts were included in this assay. (C–E) In vitro Tem1 GTPase assays. The amount of Pi released by GTPase activity was monitored over time at 30°C by absorbance at 360 nm as described in Materials and Methods. (C) The intrinsic Tem1 GTPase. The indicated concentrations of Tem1 were included in each reaction mixture. (D and E) The GAP activity of wild-type Bfa1 and each Bfa1 mutant combined with Bub2. Tem1 (4 μg), Bub2 (8 μg), and wild-type Bfa1 or each Bfa1 mutant (5 μg) were included in each reaction mixture.
We then compared the Tem1 GTPase-activating capacity of the Bfa1-DR2 mutants with wild-type Bfa1 when combined with Bub2. Tem1, Bub2, Bfa1, and each Bfa1 mutant were expressed in E. coli and purified. The rate of GTP hydrolysis by Tem1 was determined by the 2-amino-6-mercapto-7-methylpurine riboside (MESG)/phosphorylase assay (Zhang and Zheng, 1998). We checked the intrinsic GTP hydrolysis activity of Tem1 (Figure 3C) by continuously monitoring absorbance at 360 nm as a reflection of γ-Pi release from Tem1-GTP by the phosphorylase-coupling reaction using MESG as a substrate. The amount of released γ-Pi increased with increasing amounts of Tem1, ensuring that the MESG/phosphorylase assay could effectively measure Tem1 GTPase activity (Figure 3C). We also analyzed the GAP activity of Bub2 for Tem1 in the absence of Bfa1. As reported by Geymonat et al. (2002), Bub2 had little effect on Tem1 GTPase activity in the absence of Bfa1 (Figure 3D). However, the addition of Bfa1 in the presence of Bub2 caused a steep increase in γ-Pi release from Tem1-GTP, demonstrating that Bfa1/Bub2 stimulated Tem1 GTPase activity (Figure 3D). We then measured the GAP reaction catalyzed by each Bfa1 mutant in the presence of Bub2. Although Tem1 GTPase activity was stimulated, the rate of γ-Pi release in each Bfa1-DR2 mutant/Bub2 complex was slower than that in the Bfa1/Bub2 complex (Figure 3E). Bfa1G411E/Bub2 GAP activity was slightly decreased. GAP activity was markedly but not completely decreased in the presence of either Bfa1M413I/Bub2 or Bfa1D416A/Bub2. Importantly, the increase in Tem1 GTPase activity was barely detectable with Bfa1W422A/Bub2, indicating that its GAP activity was almost completely eliminated. These results were consistent with their corresponding genetic interactions shown in Figure 2, which suggest that the defects of the Bfa1-DR2 mutants in negatively regulating mitotic exit results from decreases in their GAP activity for Tem1 (summarized in Table 1). Based on genetic analyses and in vitro assays, the GAP activity of the Bfa1-DR2 mutants can be listed as follows: Bfa1W422A < Bfa1M413I < Bfa1D416A < Bfa1G411E.
Mitotic Delay in Cells with Disrupted Spindles or Damaged DNA Depends on Bfa1/Bub2 GAP Activity
Bfa1-DR2 mutants with various GAP activities are currently available. Thus, we are able to investigate how GAP activity for Tem1 functions in delaying mitotic exit in vivo, particularly in response to various checkpoint-activating signals. If GAP activity for Tem1 is responsible for preventing mitotic exit, BFA1-DR2 mutants would delay mitotic exit based on their GAP activity.
To examine the delay of mitotic exit in the presence of spindle damage, pRS304-BFA1-GFP or each pRS304-BFA1-DR2-GFP plasmid was integrated into the TRP1 locus of Δbfa1 cells as described in Materials and Methods. The expression of either BFA1 or each BFA1-DR2 is verified by Western blot and fluorescence microscopy, and single integration of the plasmid was also verified by Southern blot (Figure 4A). When spindle integrity was disrupted by benomyl or nocodazole, wild-type BFA1 cells were arrested with 2C DNA content and did not exit from mitosis (Figure 4, B and C). In contrast, Δbfa1 cells completed mitosis and progressed into the next cell cycle, resulting in new bud formation and, ultimately, cell death (Figure 4, B and C). BFA1M413I, BFA1D416A, and BFA1W422A cells progressed into the next cell cycle with kinetics nearly identical to Δbfa1 cells (Figure 4, B and C). The percentage of cells with new buds also increased in BFA1G411E cells, but only slightly compared with BFA1 cells; 7 h after release, the percentage of newly budded cells was 1.5% for BFA1, 83% for Δbfa1, and 10% for BFA1G411E (Figure 4C). These results indicate that mitotic arrest in response to spindle damage relies on the GAP activity of Bfa1/Bub2 (summarized in Table 1).
Figure 4.
The ability of each BFA1-DR2 mutant to prevent mitotic exit in response to either spindle or DNA damage. (A–C) The checkpoint activity of each BFA1-DR2 mutant in response to spindle damage. Δbfa1 (YSK1077), BFA1 (YSK2083), and BFA1-DR2 mutants (YSK2089, 2090, 2091, and 2092) were used. (A) The single-copy integration of BFA1 or each BFA1-DR2 mutant in strains used were assessed by genomic Southern blots (top), and the expression of GFP-fused wild-type and each mutant Bfa1-DR2 was verified by Western blots with anti-GFP (bottom) and fluorescence microscopy (right). Actin blot was shown as a loading control (bottom). Bar, 10 μm. (B) Cells were serially diluted on either YPAD or YPAD containing benomyl and incubated at 25°C. (C) Cells were synchronized with α-factor and released into YPAD containing 15 μg/ml nocodazole at 25°C. At each time point after the release, cells were collected to analyze DNA content by FACS (top, n = 50,000) and cells with the indicated phenotypes were counted (bottom, n = 200). (D and E) The checkpoint activity of each BFA1-DR2 mutant in response to DNA damage. cdc13-1Δbfa1 (YSK1138), cdc13-1BFA1 (YSK2073), and cdc13-1BFA1-DR2 mutants (YSK 2079, 2080, 2081, and 2082) were used. (D) The single copy integration and expression of BFA1 and each BFA1-DR2 mutant were verified in indicated strains as described in A. (E) Cells were synchronized with α-factor and released into YPAD at 34°C. At each time point, the cells were collected to analyze DNA content by FACS (top, n = 50,000), and cells with the indicated phenotypes were counted (bottom, n = 200).
We then examined the in vivo function of Bfa1/Bub2 GAP activity in response to DNA damage. Each BFA1-DR2 mutant was tested for its ability to suppress mitotic exit in cdc13-1Δbfa1 cells with DNA damage induced by the cdc13-1 mutation. cdc13-1BFA1 and cdc13-1BFA1-DR2 mutant strains were generated by integration of pRS304-BFA1-GFP or each pRS304-BFA1-DR2-GFP mutant into the TRP1 locus of cdc13-1Δbfa1 cells as described in Materials and Methods. In cdc13-1BFA1 and cdc13-1BFA1-DR2 mutant cells generated, the expression of either BFA1 or each BFA1-DR2 was verified by Western blot and fluorescence microscopy, and their copy number was also verified by Southern blot (Figure 4D). As shown in Figure 4E, when shifted to 34°C after G1 arrest at 25°C, cdc13-1BFA1 cells arrested with large buds and 2C DNA content, whereas cdc13-1Δbfa1 cells exited from mitosis and rebudded, resulting in the accumulation of both newly budded cells with DNA content greater than 2C and G1 phase cells with 1C DNA content. Bfa1G411E was able to rescue the DNA checkpoint defect of cdc13-1Δbfa1 cells with kinetics similar to wild-type Bfa1; 4 h after release, the percentage of rebudded cells was 4% for cdc13-1 BFA1G411E, 3% for cdc13-1 BFA1, and 54% for cdc13-1Δbfa1 cells (Figure 4E). Conversely, in cdc13-1BFA1M413I and cdc13-1BFA1D416A cells, rebudding significantly increased; 4 h after release, the percentage of rebudding was 21% for cdc13-1BFA1M413I and 22% for cdc13-1BFA1D416A cells (Figure 4E). Of the cdc13-1BFA1-DR2 mutants, cdc13-1BFA1W422A cells exhibited the most dramatic checkpoint defect; 4 h after release, ∼51% of these cells rebudded, which is comparable to 54% rebudded cells in cdc13-1Δbfa1 (Figure 4E). These results demonstrate that when DNA is damaged, Bfa1/Bub2 GAP activity inhibits mitotic exit (summarized in Table 1).
BFA1W422A Cells, Although Lacking GAP Activity for Tem1, Prevent Mitotic Exit in Response to Spindle Misalignment
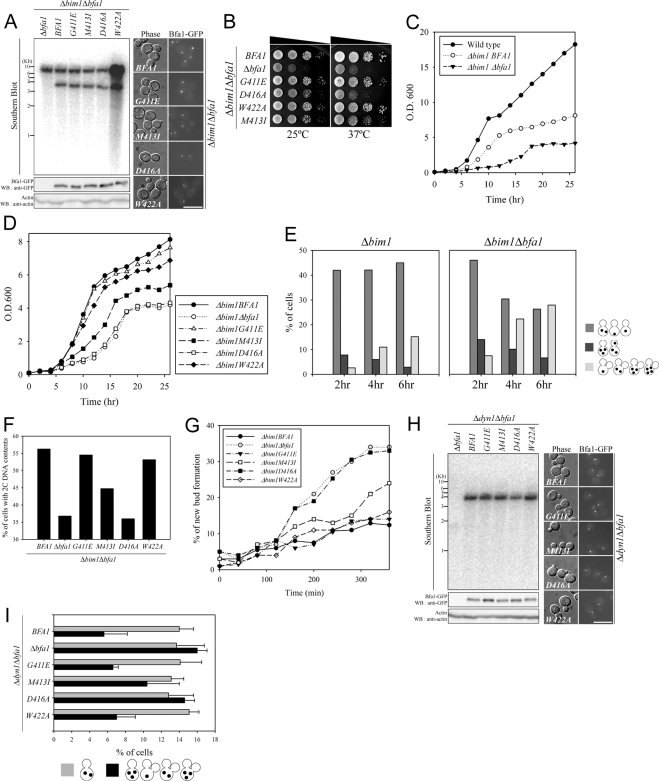
Correct positioning of the mitotic spindle relies on two separate pathways involving the microtubule motor, dynein, and Bim1, a plus-end microtubule-binding protein (Li et al., 1993; Lee et al., 2000). When the anaphase spindle is misaligned in the mother cell body as in Δbim1 or Δdyn1 cells, BFA1 deletion overrides mitotic arrest and both multinucleate and anucleate cells accumulate, ultimately leading to cell death, indicating that Bfa1 is essential for the activation of the spindle position checkpoint (Bardin et al., 2000; Bloecher et al., 2000; Pereira et al., 2000). To determine whether Bfa1/Bub2 GAP activity in vivo controls mitotic arrest in cells with mispositioned spindles, we constructed Δbim1BFA1-DR2 mutants and examined the effects of BFA1-DR2 mutants in Δbim1Δbfa1 cells. We integrated pRS304-BFA1-GFP or each pRS304-BFA1-DR2-GFP mutant into the TRP1 locus of Δbim1Δbfa1 cells as described in Materials and Methods, constructing Δbim1BFA1 and Δbim1BFA1-DR2 mutant strains. The single integration of the plasmid was assessed by Southern blot and the expression of either BFA1 or each BFA1-DR2 mutant was verified by Western blot and fluorescence microscopy (Figure 5A). Compared with BIM1 wild-type, Δbim1 cells grew slowly (Figure 5, B–D), and the number of cells in which either the nucleus was mispositioned or anaphase occurred within the mother cell increased (Figure 5E), leading to the accumulation of cells with 2C DNA content particularly at 37°C (Figure 5F). BFA1 deletion had a deleterious effect on the growth rate of Δbim1 cells (Figure 5, B–D). In addition, in the Δbim1Δbfa1 mutant, cells with either no nucleus or multiple nuclei accumulated (Figure 5E), and cells with 2C DNA content decreased compared with Δbim1 cells because of the failure of mitotic arrest (Figure 5F).
Figure 5.
The ability of each BFA1-DR2 mutant to prevent mitotic exit in response to spindle misalignment. (A–G) The ability of Bfa1-DR2 mutants to prevent mitotic exit in Δbim1Δbfa1 cells. Δbim1Δbfa1 (YSK1867), Δbim1BFA1 (YSK2093), and Δbim1BFA1-DR2 mutants (YSK2099, 2100, 2101, and 2102) were used. (A) A genomic Southern blot (top) assessed the single-copy integration of BFA1 or each BFA1-DR2 mutant in indicated strains as described in Materials and Methods. In the genomic Southern blot, the lower bands indicate specific one copy integration of the wild-type or mutant BFA1 gene. Both Western blot with anti-GFP (bottom) and fluorescence microscopy (right) verified the expression of GFP-fused wild-type and each mutant Bfa1-DR2. Actin blot was shown as loading control (bottom). Bar, 10 μm. (B–D) The growth rate of Δbim1BFA1, Δbim1Δbfa1, and Δbim1BFA1-DR2 mutants. (B) Cells were serially diluted on YPAD and incubated at either 25 or 37°C. (C and D) The growth of indicated strains (wild-type W303a) was monitored at 37°C by measuring the absorbance at 600 nm every 2 h after inoculation at the same density (OD600 = 0.1). (E–G) Cells were grown at 25°C to midlog phase, synchronized with α-factor, and released into YPAD at 37°C. (E) The number of cells arrested in mitosis or with improper mitotic exit was quantified in Δbim1BFA1 and Δbim1Δbfa1 cells. At each time point after the release, cells were fixed and stained with propidium iodide to score the indicated cell types (n = 200). (F) The number of cells with 2C DNA content was quantified. After release for 6 h, DNA content was analyzed by FACS (n = 50,000). (G) At each time point after the release, cells with an extra new bud were scored (n = 200). (H and I) The ability of Bfa1-DR2 mutants to prevent mitotic exit in Δdyn1Δbfa1 cells. Δdyn1BFA1 (YSK2103), Δdyn1Δbfa1 (YSK1129), and Δdyn1BFA1-DR2 mutants (YSK2105, 2106, 2107, and 2108) were used. (H) The single-copy integration and expression of BFA1 and each BFA1-DR2 mutant were verified in indicated cells as described in A. (I) The number of cells exhibiting improper mitotic exit was quantified. Cells were synchronized with α-factor at 30°C and released into YPAD at 16°C for 24 h. Cells with indicated phenotypes were quantified (n = 300) after staining with DAPI. The average of three independent counts was plotted with SDs.
Surprisingly, BFA1-DR2 mutants showed unexpected responses to spindle misalignment. Δbim1BFA1G411E cells displayed phenotypes similar to Δbim1BFA1 cells (Figure 5, B, D, and F). Conversely, the growth rate and proportion of cells with 2C DNA content decreased rapidly in Δbim1BFA1D416A cells as in Δbim1Δbfa1 cells and partially in Δbim1BFA1M413I cells (Figure 5, B, D, and F). Importantly, Bfa1W422A completely repressed the synthetic growth defect of Δbim1Δbfa1 cells and decrease of cells with 2C DNA content similar to wild-type Bfa1 (Figure 5, B, D, and F). Of note, Bfa1W422A/Bub2 rarely stimulated Tem1 GTPase activity, and its GAP activity was less than that of either Bfa1M413I/Bub2 or Bfa1D416A/Bub2 (Figure 3). Hence, we could not observe any correlation between Bfa1/Bub2 GAP activity and the inhibition of mitotic exit in cells with misaligned spindles (summarized in Table 1).
Newly budded Δbim1Δbfa1 cells prominently increased compared with Δbim1 cells (Figure 5, E and G). To further verify the role of GAP activity in activating the spindle position checkpoint, new bud formation was examined after release at 37°C from G1 arrest (Figure 5G). Consistent with the above results, the presence of BFA1W422A in Δbim1Δbfa1 cells decreased the number of cells with extra buds as also seen in Δbim1BFA1 cells, whereas BFA1D416A could not suppress new bud formation of Δbim1Δbfa1 cells; 6 h after release, the percentages of newly budded cells were ∼16% for Δbim1BFA1, 34% for Δbim1Δbfa1, 14% for Δbim1BFA1G411E, 24% for Δbim1BFA1M413I, 33% for Δbim1BFA1D416A, and 16% for Δbim1BFA1W422A (Figure 5G). These observations strongly suggest that Bfa1 is essential for the delay of mitotic exit when the spindle is not properly oriented, but Bfa1/Bub2 GAP activity is not necessary for the complete activation of the spindle position checkpoint.
These results were further confirmed with Δdyn1BFA1-DR2 mutants. A lack of DYN1 also induces anaphase spindle misalignment in the mother cell body and thus triggers mitotic arrest (Li et al., 1993). Because this defect is much more pronounced at 16°C, we monitored mitotic arrest in each Δdyn1BFA1-DR2 mutant after α-factor–synchronized cells were released at 16°C. To do this, Δdyn1BFA1 and Δdyn1BFA1-DR2 mutant strains were generated by integration of pRS304-BFA1-GFP or each pRS304-BFA1-DR2-GFP mutant into the TRP1 locus of Δdyn1Δbfa1 cells as described in Materials and Methods. The single integration of the plasmid was assessed by Southern blot, and the expression of either BFA1 or each BFA1-DR2 mutant was verified by Western blot and fluorescence microscopy (Figure 5H). Consistently with the previous results, BFA1G411E and BFA1W422A mutants were able to restore the improper mitotic exit of Δdyn1Δbfa1 cells similar to BFA1, whereas Δdyn1BFA1D416A cells showed a comparable number of both newly budded cells and cells with three and more nuclei in the mother cell body to Δdyn1Δbfa1 cells (Figure 5I). BFA1M413I partially suppressed these phenotypes of improper mitotic exit (Figure 5I).
Interaction of Bfa1 and Kin4 Decreases in BFA1D416A Cells with a Defective Spindle Position Checkpoint
We showed that Bfa1/Bub2 GAP activity is essential for the arrest of mitotic exit in response to either spindle or DNA damage but not to spindle misalignment. Recently, a novel kinase Kin4 was identified as a component of the spindle position checkpoint, which has been suggested to counteract Cdc5 and inhibit mitotic exit via maintenance of Bfa1/Bub2 GAP activity (D'Aquino et al., 2005; Pereira and Schiebel, 2005; Maekawa et al., 2007). Thus, we carefully studied the relationship between Kin4 and Bfa1/Bub2 GAP activity in controlling mitotic exit in the presence of misoriented spindles. As reported previously, the loss of Kin4 led to the accumulation of cells with new buds in Δbim1 and Δdyn1 cells (D'Aquino et al., 2005; Pereira and Schiebel, 2005; Figure 6, A and B). However, even in Δbim1BFA1W422A and Δdyn1BFA1W422A cells that lack GAP activity toward Tem1, Kin4 was essential for suppression of mitotic progression (Figure 6, A and B). These observations are not consistent with the proposed function of Kin4 to sustain Bfa1/Bub2 GAP in response to spindle misorientation.
Figure 6.
The function of Kin4 in regulating Bfa1 and activating the spindle position checkpoint. (A) The effect of KIN4 deletion on Δbim1BFA1-DR2 cells. Δbim1KIN4BFA1 (YSK2093), Δbim1Δkin4Δbfa1 (YSK2109), Δbim1Δkin4BFA1 (YSK2110), and Δbim1Δkin4BFA1-DR2 mutants (YSK2111, 2112, 2113, and 2114) were synchronized with α-factor at 25°C and released into YPAD at 37°C. At each indicated time point, cells with new buds were scored (n = 300). (B) The effect of KIN4 deletion on Δdyn1BFA1-DR2 cells. Δdyn1KIN4BFA1 (YSK2103), Δdyn1Δkin4Δbfa1 (YSK2115), Δdyn1Δkin4BFA1 (YSK2116), and Δdyn1Δkin4BFA1-DR2 mutants (YSK2117, 2118, 2119, and 2120) were synchronized with α-factor at 30°C and released into YPAD at 16°C for 24 h. Cells were stained with DAPI, and the indicated phenotypes were quantified (n = 300). The average of three independent counts was plotted with SDs. (C) The phosphorylation of Bfa1 in Δkin4 cells. KIN4 (YSK2121) and Δkin4 (YSK2122) cells synchronized with α-factor were released into YPAD containing 15 μg/ml nocodazole for 3 h. Cells were harvested and protein extracts were subjected to Western blotting with PAP. cdc15-2BFA1-TAP cells (YSK1153) incubated for 3 h at 37°C were included as a positive control for slowly migrating forms of Bfa1. The arrow indicates the slowly migrating hyperphosphorylated form of Bfa1. (D) The genetic interaction between KIN4 and Δlte1Δste20 cells. Δlte1Δste20Δbfa1KIN4 (YSK2062), Δlte1Δste20BFA1KIN4 (YSK2063), and Δlte1Δste20BFA1Δkin4 cells (YSK2124) containing pCEN-URA3-LTE1 plasmid were serially diluted on YPAD or YPAD containing 5-fluoroorotic acid and incubated at 25°C. (E) The ability of Bfa1-D8 (391-574) to induce cell cycle arrest in Kin4-overexpressing cells. pCEN-BFA1-HA, pCEN-BFA1-D1302-574-HA, and pCEN-BFA1-D8391-574-HA were transformed into Δbfa1 cells with PGAL-HA-KIN4 (YSK2353). Left, cells were serially diluted on YPAD or YPAG (the induction of GAL promoter). Right, expression of Kin4, full-length Bfa1, Bfa1-D1, and Bfa1-D8 was verified by Western blotting with anti-HA. (F) The localization of Bfa1-GFP in cells with misaligned spindles. Δdyn1BFA1 (YSK2103), Δdyn1BFA1D416A (YSK2107), and Δdyn1BFA1W422A (YSK2108) cells were synchronized with α-factor and released at 16°C for 24 h. Bar, 5 μm. (G) Coprecipitation of Kin4 with Bfa1. KIN4-3HAΔbfa1 (YSK2125), KIN4-3HA BFA1-TAP (YSK2126), KIN4-3HA BFA1D416A-TAP (YSK2127), and KIN4-3HA BFA1W422A-TAP (YSK2128) cells were grown at 25°C to midlog phase and harvested. Extracts were prepared, and Bfa1 was precipitated with IgG beads as described in Materials and Methods. Bfa1 and Kin4 were detected with PAP and anti-HA, respectively. The intensities of copurified Kin4 were normalized by purified Bfa1 or Bfa1 mutants. (H) The localization of Kin4-GFP in cells with mispositioned spindles. Δdyn1BFA1 (YSK2363), Δdyn1BFA1D416A (YSK2365), and Δdyn1BFA1W422A (YSK2367) cells were synchronized with α-factor and released at 16°C for 24 h. Kin4-GFP was observed by fluorescence microscopy, and the expression of Bfa1 and Bfa1 mutants was verified by Western blotting with PAP. Bar, 5 μm.
To understand the function of Kin4 in regulating Bfa1, we first examined the phosphorylation of Bfa1 in Δkin4 cells because Bfa1 phosphorylation by Cdc5 reduces its GAP activity for Tem1 in vitro (Geymonat et al., 2003). When KIN4 and Δkin4 cells synchronized with α-factor were released into medium containing nocodazole, Bfa1 was not phosphorylated in KIN4 wild-type cells and was only partially phosphorylated in Δkin4 cells compared with fully phosphorylated Bfa1 in anaphase-arrested cdc15-2 cells (Figure 6C), suggesting that KIN4 deletion may only slightly diminish Bfa1/Bub2 GAP activity.
Then, we measured Bfa1/Bub2 GAP activity in Δkin4 cells in vivo. It was technically difficult to directly quantify Bfa1/Bub2 GAP activity for Tem1 in KIN4 and Δkin4 cells. As an alternative, we investigated the genetic interactions between KIN4 and mitotic exit–defective mutants. Given that Kin4 maintains Bfa1/Bub2 GAP activity, its deletion would allow growth of mitotic exit–defective mutants. As reported previously, the loss of Kin4 suppressed the mitotic exit defect of Δlte1 cells, similar to that of BFA1 deletion (Pereira and Schiebel, 2005; Supplementary Figure S7). The growth defect of Δlte1Δste20 cells was also restored by KIN4 deletion, but the growth rate of Δlte1Δste20Δkin4 cells was not comparable to that of Δlte1Δste20Δbfa1 cells (Figure 6D). On the contrary, KIN4 deletion failed to ameliorate the mitotic exit defect of cdc5-2 (Supplementary Figure S8), and did not prematurely localize Mob1-GFP to SPBs (Supplementary Figure S9). These results are very similar to the genetic interactions of BFA1G411E with mitotic exit–defective mutants; BFA1G411E completely restored the growth defect in Δlte1 cells (Figure 2, A and B) and partially in Δlte1Δste20 cells (Supplementary Figure S5). In contrast, BFA1G411E could suppress the mitotic exit of cdc5-2Δbfa1 cells (Figure 2, C and D), and the premature localization of Mob1-GFP to SPBs in cells lacking BFA1 (Supplementary Figure S6). Thus, these similarities suggest that the GAP activity of Bfa1/Bub2 in Δkin4 cells might be comparable to that of Bfa1G411E/Bub2. The GAP activity of Bfa1G411E/Bub2 was only mildly decreased, and BFA1G411E cells were able to inhibit new bud formation in kinetics nearly similar to that of wild-type BFA1 cells in response to spindle and DNA damage (Figures 3 and 4). Indeed, Δkin4 cells also showed nearly proficient checkpoint responses to spindle and DNA damage (Supplementary Figures S10 and S11). On the other hand, we found that Bfa1-D8391-574 is essential to prevent mitotic exit in Kin4-overexpressing cells similar to that of full-length Bfa1, suggesting that this C-terminal domain interacts with Kin4 to activate the spindle position checkpoint (Figure 6E).
What then makes the difference between BFA1D416A and BFA1W422A cells in inducing Kin4-dependent mitotic arrest? The symmetric localization of either Bfa1 or Kin4 on both spindle poles has been proposed to, at least in part, be responsible for mitotic arrest when the spindle is misaligned (Pereira et al., 2001; Maekawa et al., 2007). However, spindle position checkpoint-deficient Bfa1D416A was present on both SPBs in cells with misoriented spindles as the checkpoint-proficient wild-type Bfa1 and Bfa1W422A (Figure 6F). We then examined the physical interaction between Bfa1 and Kin4. Bfa1 coprecipitated with Kin4 (Figure 6G). Interestingly, the amount of Kin4 coprecipitated with Bfa1D416A decreased compared with that of either wild-type Bfa1 or Bfa1W422A (Figure 6G). Hence, it is likely that the failure of Bfa1D416A in activating the spindle position checkpoint might be related to its decreased ability to physically interact with Kin4, probably leading to the failure of Kin4 association with SPBs in response to spindle misorientation. However, Kin4 was still on both SPBs in Δdyn1BFA1D416A cells with mispositioned spindles (Figure 6H), suggesting that Kin4 association with SPBs does not rely on its interaction with Bfa1; we rarely detected differences in the abundance of Kin4-GFP bound to SPBs in Δdyn1BFA1, Δdyn1BFA1D416A, or Δdyn1BFA1W422A cells when the spindle was not properly aligned.
DISCUSSION
Mitotic exit must be tightly coupled with nuclear position in order to ensure that daughter cells inherit each nucleus after cell division. In budding yeast, Tem1 GTPase activity and the spatial separation of MEN components provide an attractive model for how mitotic exit is temporally and spatially coordinated with spindle positioning. Lte1 is sequestered predominantly at the bud cortex, whereas Bfa1, Bub2, and Tem1 colocalize preferentially to the SPB directed toward the bud (Bardin et al., 2000; Pereira et al., 2000). Hence, mitotic exit could only become activated when the nucleus is pulled into the bud and Tem1 encounters Lte1 (Bardin et al., 2000). Nonetheless, there are confusing data as to whether Tem1 GTPase activity controls mitotic exit in vivo. We previously identified a Bfa1 mutant, Bfa1E438K, whose GAP activity with Bub2 is slightly decreased (Kim and Song, 2006). Interestingly, BFA1E438K cells were partially defective in activating the spindle assembly checkpoint, while completely blocked mitotic exit in response to DNA damage or spindle misorientation (Kim and Song, 2006). Thus, we further screened for Bfa1 mutants with defects in preventing mitotic exit in order to understand the regulatory mechanism for mitotic exit. Here, we found that the DR2 region of Bfa1 is essential for acting in concert with Bub2 to stimulate Tem1 GTPase activity. Because Bfa1-DR2 mutants normally interact with Bub2 and Tem1, the DR2 domain may be responsible for generating GAP activity for Tem1. Importantly, these mutants showed a tight correlation between their GAP activities and negative control of the MEN, providing the first evidence in vivo that Bfa1/Bub2 GAP activity inhibits Tem1 to control mitotic exit.
Surprisingly, we observed that different types of perturbations prevent mitotic exit with different levels of Bfa1/Bub2 GAP activity. The checkpoint activity was partially deficient by DNA damage and was fully lacking in response to spindle damage in BFA1M413I and BFA1D416A cells, whereas BFA1W422A cells were similar to Δbfa1. Based on their in vitro GAP activities, these observations indicate that more Bfa1/Bub2 GAP activity is needed for spindle damage than for DNA damage to activate the checkpoint. Conversely, when the spindle is misaligned, mitotic delay does not absolutely rely on Bfa1/Bub2 GAP activity.
Kin4 supported that the control of GAP activity is not the only mechanism that regulates mitotic exit against misaligned spindles. The phosphorylation status of Bfa1 in Δkin4 cells and various genetic interactions of KIN4 showed that Bfa1/Bub2 GAP activity became only slightly reduced in Δkin4 cells as in BFA1G411E mutant. Considering that BFA1G411E cells are proficient in the spindle position checkpoint, the failure of mitotic arrest in Δkin4 cells with mispositioned spindles is not absolutely due to a decrease in GAP activity. Indeed, Kin4 was required to activate the spindle position checkpoint even in GAP activity-deprived BFA1W422A cells. Additionally, we observed that Bfa1-D8391-574 was sufficient to prevent mitotic exit by Kin4, although Maekawa et al. (2007) showed that SPB-associated Kin4 phosphorylates serines-150 and 180 of Bfa1, making it resistant to inactivation by Cdc5. Therefore, there may be another mechanism by which Kin4 regulates the Bfa1/Bub2 to prevent mitotic exit in response to spindle misorientation. These results also explain why Kin4 is not essential for DNA or spindle damage.
How then can GAP activity-deprived cells delay mitotic exit in response to spindle misorientation? Along with Bfa1/Bub2 GAP activity, the SPB association of MEN components appears to be important in the regulation of mitotic exit. Bfa1 and Kin4 are present on both SPBs in cells with misaligned spindles (Pereira et al., 2001; Pereira and Schiebel, 2005). Indeed, we observed that Bfa1 localization onto both SPBs delays MEN activation (Kim and Song, unpublished data). Forced targeting of Kin4 to both SPBs also delayed mitotic exit (Maekawa et al., 2007). However, Bfa1D416A and Bfa1W422A were on both SPBs when the spindle was misoriented (Figure 6F), demonstrating that misaligned spindle–induced localization of Bfa1 on both SPBs is not under control of Bfa1/Bub2 GAP activity. Interestingly, spindle position defects promoted the physical interaction between Bfa1 and Kin4 regardless of Bfa1/Bub2 GAP activity (Figure 6G), but this did not affect the recruitment of Kin4 onto SPB: Kin4 still associated with both SPBs in Δdyn1BFA1D416A and Δdyn1BFA1W422A cells when the spindle was improperly aligned (Figure 6H). Recently, D'Aquino et al. (2005) showed that Tem1 recruitment onto SPBs appears to be regulated by spindle position and Kin4: KIN4 deletion restored the localization of Tem1 to SPBs in a majority of Δdyn1 mutants with misaligned spindles and Kin4 overexpression led to Tem1 displacement from the SPBs. Thus, it will be interesting to examine whether Kin4 competes with Tem1 to bind Bfa1 for the recruitment onto SPBs. Now, although how Bfa1-Kin4 interaction affects on mitotic exit is unclear, this may also be needed for yet unknown GAP-independent surveillance mechanism(s), because Bfa1-D8391-574, having no target residues for Kin4 phosphorylation, is essential for Kin4-dependent arrest (Figure 6E). We propose that Kin4 may prevent mitotic exit by at least two different ways: 1) maintaining Bfa1/Bub2 GAP activity via blocking inhibitory phosphorylation by Cdc5 and 2) promoting the interaction between Bfa1 and Kin4, which may contribute to the activation of the spindle position checkpoint in a Bfa1/Bub2 GAP-independent manner.
Although KIN4 deletion allowed Δdyn1 or Δbim1 cells with mispositioned spindles to exit from mitosis, the extent to which both multinucleate and anucleate cells accumulated was less than that seen with simultaneous deletion of KIN4 and BFA1 (Figure 6, A and B). These observations suggest that there may be other element(s) except Kin4 that regulate Bfa1/Bub2 against spindle misorientation. Several groups have already shown that the interaction of astral microtubules with the bud neck/cortex can control both the localization of the Bfa1/Bub2 complex and mitotic exit (Adames et al., 2001; Pereira et al., 2001; Nelson and Cooper, 2007). This mechanism may also contribute to mitotic arrest induced by spindle misorientation, although how information from bud neck/cortex is transmitted to Bfa1/Bub2 is not understood. Taken together, we suggest that the GTP/GDP exchange of Tem1, the SPB association of MEN components, and microtubule-bud neck/cortex interactions are mutually interdependent and may provide multiple mechanisms for maintaining genomic integrity against spindle misalignment. Hence, our results are consistent with the current GTP/GDP switch model of mitotic exit and, at the same time, could integrate contradictory observations regarding this model.
Although several homologues of the MEN components in budding yeast have been reported, those of Bfa1, Bub2, and Tem1 have not been identified in mammalian cells. We found that the conserved residues of DR2 domain in byr4 of S. pombe were also essential for regulating cytokinesis and septation (Supplementary Figure S12). Hence, we expect that Bfa1-DR2 domain may help to identify the mammalian ortholog(s) of Bfa1. Together with this, understanding the regulatory mechanisms of mitotic exit in budding yeast may provide insight into how cells monitor spindle position and coordinate with mitotic exit for faithful chromosomal segregation in the asymmetric division of eukaryotic cells.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Drs. S. Elledge (Harvard University) and K. S. Lee (National Institutes of Health), the Yeast Resource Center (Washington University), and EUROSCARF for yeast strains and constructs. We also appreciate Dr. A. Amon (Massachusetts Institute of Technology) for giving helpful comments on this manuscript. This work was supported by a grant from the 21C Frontier Microbial Genomics and Application Center Program, Ministry of Science and Technology of the Republic of Korea. J.K. was supported by the fellowship of Brain Korea 21 Project (2006–2007) and the Korea Research Foundation Grant (KRF-2006-8-1284) funded by the Korean Government Ministry of Education (MOEHRD).
Abbreviations used:
- Bfa1-DR
Bfa1-direct repeat
- GAP
GTPase-activating protein
- GEF
guanine-nucleotide exchange factor
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- GTPase
guanine triphosphatase
- MBP
maltose-binding protein
- MEN
mitotic exit network
- MESG
2-amino-6-mercapto-7-methylpurine riboside
- PAP
peroxidase anti-peroxidase.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-02-0149) on July 30, 2008.
REFERENCES
- Adames N. R., Oberle J. R., Cooper J. A. The surveillance mechanism of the spindle position checkpoint in yeast. J. Cell Biol. 2001;153:159–168. doi: 10.1083/jcb.153.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbacid M. Ras genes. Annu. Rev. Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- Bardin A. J., Amon A. Men and sin: what's the difference? Nat. Rev. Mol. Cell Biol. 2001;2:815–826. doi: 10.1038/35099020. [DOI] [PubMed] [Google Scholar]
- Bardin A. J., Visintin R., Amon A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 2000;102:21–31. doi: 10.1016/s0092-8674(00)00007-6. [DOI] [PubMed] [Google Scholar]
- Bloecher A., Venturi G. M., Tatchell K. Anaphase spindle position is monitored by the BUB2 checkpoint. Nat. Cell Biol. 2000;2:556–558. doi: 10.1038/35019601. [DOI] [PubMed] [Google Scholar]
- D'Aquino K. E., Monje-Casas F., Paulson J., Reiser V., Charles G. M., Lai L., Shokat K. M., Amon A. The protein kinase Kin4 inhibits exit from mitosis in response to spindle position defects. Mol. Cell. 2005;19:223–234. doi: 10.1016/j.molcel.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Fraschini R., D'Ambrosio C., Venturetti M., Lucchini G., Piatti S. Disappearance of the budding yeast Bub2-Bfa1 complex from the mother-bound spindle pole contributes to mitotic exit. J. Cell Biol. 2006;172:335–346. doi: 10.1083/jcb.200507162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furge K. A., Wong K., Armstrong J., Balasubramanian M., Albright C. F. Byr4 and Cdc16 form a two-component GTPase-activating protein for the Spg1 GTPase that controls septation in fission yeast. Curr. Biol. 1998;8:947–954. doi: 10.1016/s0960-9822(98)70394-x. [DOI] [PubMed] [Google Scholar]
- Geymonat M., Spanos A., Smith S. J., Wheatley E., Rittinger K., Johnston L. H., Sedgwick S. G. Control of mitotic exit in budding yeast. In vitro regulation of Tem1 GTPase by Bub2 and Bfa1. J. Biol. Chem. 2002;277:28439–28445. doi: 10.1074/jbc.M202540200. [DOI] [PubMed] [Google Scholar]
- Geymonat M., Spanos A., Walker P. A., Johnston L. H., Sedgwick S. G. In vitro regulation of budding yeast Bfa1/Bub2 GAP activity by Cdc5. J. Biol. Chem. 2003;278:14591–14594. doi: 10.1074/jbc.C300059200. [DOI] [PubMed] [Google Scholar]
- Hofken T., Schiebel E. A role for cell polarity proteins in mitotic exit. EMBO J. 2002;21:4851–4862. doi: 10.1093/emboj/cdf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F., Wang Y., Liu D., Li Y., Qin J., Elledge S. J. Regulation of the Bub2/Bfa1 GAP complex by Cdc5 and cell cycle checkpoints. Cell. 2001;107:655–665. doi: 10.1016/s0092-8674(01)00580-3. [DOI] [PubMed] [Google Scholar]
- Kim J., Jeong J., Song K. The C-terminus of Bfa1p in budding yeast is essential to induce mitotic arrest in response to diverse checkpoint-activating signals. Genes Cells. 2004;9:399–418. doi: 10.1111/j.1356-9597.2004.00731.x. [DOI] [PubMed] [Google Scholar]
- Kim J., Song K. The study of Bfa1p(E438K) suggests that Bfa1 control the mitotic exit network in different mechanisms depending on different checkpoint-activating signals. Mol. Cell. 2006;21:251–260. [PubMed] [Google Scholar]
- Lee L., Tirnauer J. S., Li J., Schuyler S. C., Liu J. Y., Pellman D. Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science. 2000;287:2260–2262. doi: 10.1126/science.287.5461.2260. [DOI] [PubMed] [Google Scholar]
- Lee S. E., Frenz L. M., Wells N. J., Johnson A. L., Johnston L. H. Order of function of the budding-yeast mitotic exit-network proteins Tem1, Cdc15, Mob1, Dbf2, and Cdc5. Curr. Biol. 2001;11:784–788. doi: 10.1016/s0960-9822(01)00228-7. [DOI] [PubMed] [Google Scholar]
- Li R. Bifurcation of the mitotic checkpoint pathway in budding yeast. Proc. Natl. Acad. Sci. USA. 1999;96:4989–4994. doi: 10.1073/pnas.96.9.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. Y., Yeh E., Hays T., Bloom K. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc. Natl. Acad. Sci. USA. 1993;90:10096–10100. doi: 10.1073/pnas.90.21.10096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Maekawa H., Priest C., Lechner J., Pereira G., Schiebel E. The yeast centrosome translates the positional information of the anaphase spindle into a cell cycle signal. J. Cell Biol. 2007;179:423–436. doi: 10.1083/jcb.200705197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah A. S., Jang J., Deshaies R. J. Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex. Proc. Natl. Acad. Sci. USA. 2001;98:7325–7330. doi: 10.1073/pnas.141098998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minet M., Nurse P., Thuriaux P., Mitchison J. M. Uncontrolled septation in a cell division cycle mutant of the fission yeast Schizosaccharomyces pombe. J. Bacteriol. 1979;137:440–446. doi: 10.1128/jb.137.1.440-446.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson .A, Cooper J. A. A novel pathway that coordinates mitotic exit with spindle position. Mol. Biol. Cell. 2007;18:3440–3450. doi: 10.1091/mbc.E07-03-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald A. F. A shared domain between a spindle assembly checkpoint protein and Ypt/Rab-specific GTPase-activators. Trends Biochem. Sci. 1997;22:243–244. doi: 10.1016/s0968-0004(97)01073-6. [DOI] [PubMed] [Google Scholar]
- Pereira G., Hofken T., Grindlay J., Manson C., Schiebel E. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol. Cell. 2000;6:1–10. [PubMed] [Google Scholar]
- Pereira G., Tanaka T. U., Nasmyth K., Schiebel E. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 2001;20:6359–6370. doi: 10.1093/emboj/20.22.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Schiebel E. Kin4 kinase delays mitotic exit in response to spindle alignment defects. Mol. Cell. 2005;19:209–221. doi: 10.1016/j.molcel.2005.05.030. [DOI] [PubMed] [Google Scholar]
- Ro H. S., Song S., Lee K. S. Bfa1 can regulate Tem1 function independently of Bub2 in the mitotic exit network of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2002;99:5436–5441. doi: 10.1073/pnas.062059999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M., Matsui Y., Tanaka K., Toh-e A. Isolation of a CDC25 family gene, MSI2/LTE1, as a multicopy suppressor of ira1. Yeast. 1994a;10:451–461. doi: 10.1002/yea.320100404. [DOI] [PubMed] [Google Scholar]
- Shirayama M., Matsui Y., Toh E. A. The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M phase. Mol. Cell. Biol. 1994b;14:7476–7482. doi: 10.1128/mcb.14.11.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song K., Mach K. E., Chen C. Y., Reynolds T., Albright C. F. A novel suppressor of ras1 in fission yeast, byr4, is a dosage-dependent inhibitor of cytokinesis. J. Cell Biol. 1996;133:1307–1319. doi: 10.1083/jcb.133.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Hu F., Elledge S. J. The Bfa1/Bub2 GAP complex comprises a universal checkpoint required to prevent mitotic exit. Curr. Biol. 2000;10:1379–1382. doi: 10.1016/s0960-9822(00)00779-x. [DOI] [PubMed] [Google Scholar]
- Zhang B., Zheng Y. Regulation of RhoA GTP hydrolysis by the GTPase-activating proteins p190, p50RhoGAP, Bcr, and 3BP-1. Biochemistry. 1998;37:5249–5257. doi: 10.1021/bi9718447. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.