Abstract
YY1 is a transcription factor that can repress or activate the transcription of a variety of genes. Here, we show that the function of YY1 as a repressor in cardiac myocytes is tightly dependent on its ability to interact with histone deacetylase 5 (HDAC5). YY1 interacts with HDAC5, and overexpression of YY1 prevents HDAC5 nuclear export in response to hypertrophic stimuli and the increase in cell size and re-expression of fetal genes that accompany pathological cardiac hypertrophy. Knockdown of YY1 results in up-regulation of all genes present during fetal development and increases the cell size of neonatal cardiac myocytes. Moreover, overexpression of a YY1 deletion construct that does not interact with HDAC5 results in transcription activation, suggesting that HDAC5 is necessary for YY1 function as a transcription repressor. In support of this relationship, we show that knockdown of HDAC5 results in transcription activation by YY1. Finally, we show that YY1 interaction with HDAC5 is dependent on the HDAC5 phosphorylation domain and that overexpression of YY1 reduces HDAC5 phosphorylation in response to hypertrophic stimuli. Our results strongly suggest that YY1 functions as an antihypertrophic factor by preventing HDAC5 nuclear export and that up-regulation of YY1 in human heart failure may be a protective mechanism against pathological hypertrophy.
INTRODUCTION
YY1 is a ubiquitously expressed transcription factor that is highly conserved across species and is involved in a variety of cellular processes, including the regulation of cardiac disease (Sucharov et al., 2003). YY1 differentially regulates a multitude of gene promoters by acting as either a repressor, an activator, and/or an initiator of transcription (reviewed in Thomas and Seto, 1999). Previous work suggests that YY1 functions primarily as a repressor, often by displacing transcriptional activators (Lee et al., 1992; Thomas and Seto, 1999) and that its function as an activator is a result of its interaction with other proteins that block its repression domain (Lee et al., 1995a; Lee et al., 1995b; Bushmeyer and Atchison, 1998). However, these examples do not fully demonstrate the multiple functions of YY1, which result from the ability of YY1 to interact with a variety of other regulatory factors. Recently, the repressor activity of YY1 was shown to be regulated by a complex mechanism that requires acetylation of its central region followed by deacetylation by HDACs and increased HDAC activity in its C-terminal region (Yao et al., 2001). YY1 was shown to repress transcription of several genes through interaction with and targeting of HDAC1 and HDAC2 to the promoter region of these genes (Luke et al., 2006; Liu et al., 2007). In muscle cells, class II HDACs play an important role in regulating gene expression, and they are phosphorylated in response to various hypertrophic stimuli, resulting in their translocation to the cytoplasm and in transcription derepression (Han et al., 2006).
At the cellular level, myocardial failure and myocyte hypertrophy are characterized by an increase in cell size and by changes in the gene expression of many components of the heart, including the contractile apparatus. These molecular changes have been described as a recapitulation of a “fetal” gene program (FGP) because many embryonically expressed genes that are down-regulated postnatally are reactivated, whereas several “adult” genes are repressed (Lompre et al., 1979; Abraham et al., 2002). Of the changes that are observed in failing hearts, increases in β myosin heavy chain (β-MyHC), skeletal α-actin, and atrial natriuretic peptide (ANP), with coordinate decreases in α myosin heavy chain (αMyHC) and sarcoplasmatic reticulum ATPase 2a (SERCA), are perhaps the most widely recognized.
In the work presented here, we show the class II HDAC HDAC5 interacts with YY1 in muscle cells and that this interaction is necessary for YY1 to function as a transcription repressor of cardiac-specific promoters. Moreover, we show that overexpression of YY1 in cardiac myocytes prevents HDAC5 nuclear export in response to hypertrophic stimuli and prevents induction of the fetal gene program. In addition, YY1 overexpression blocks increases in cell size that result from α-adrenergic receptor (α-AR)-mediated hypertrophy through a mechanism that involves interaction with and retention of HDAC5 in the nucleus. Together, these data suggest that YY1 functions in concert with HDAC5 to maintain gene-specific transcriptional repression in cardiac cells.
MATERIALS AND METHODS
Antibodies
YY1 (SC-7341X), Myc (SC-40), HDAC5 (SC-5250), Gal4 (SC-510), PKD (sc-639) and Sp1 (SC-59) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). FLAG (F3165), calnexin (C4731) and α-actinin (A 7811) antibody were purchased from Sigma-Aldrich (St. Louis, MO). The horseradish peroxidase (115-035-146) anti-mouse and anti-rabbit were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA). Alexa Fluor 594 (A11032) anti-mouse was purchased from Invitrogen (Carlsbad, CA).
DNA and Adenovirus Constructs
The YY1-green fluorescent protein (GFP) adenovirus construct was a gift from Dr. Aristidis Moustakas (Ludwig Institute of Cancer Research, Uppsala, Sweden) and the YY1-FLAG 174–200 deletion construct was a gift of Dr. Edward Seto (University of South Florida, Tampa, FL). YY1-GAL4 wild-type control and deletion constructs were a gift of Dr. Michael Atchison (University of Pennsylvania, Philadelphia, PA) and are described in detail in Bushmeyer et al. (1995) and Bushmeyer and Atchison (1998).
Cell Culture and Adenoviral Infection
Neonatal rat cardiac myocytes (NRVMs) were prepared according to the method described in Waspe et al. (1990). Cells were infected with an adenovirus expressing YY1-GFP and/or HDAC5-FLAG or with a control adenovirus at a multiplicity of infection of 7 plaque-forming units/cell.
Real-Time Polymerase Chain Reaction (PCR)
Total RNA was extracted by TRIzol (Invitrogen). 0.5 μg of RNA was reverse transcribed into cDNA using the SuperScript III first-strand cDNA synthesis kit (Invitrogen). Typically, 0.1 ng of cDNA, 12.5 nM of each primer, and Power SYBER Green PCR Master Mix (Applied Biosystems, Foster City, CA) were used in the reverse transcription (RT)-PCR reactions. Reactions were performed using the ABI7300 system. The primers used are presented on Table 1.
Table 1.
Sequence of the primers used for the RT-PCR reaction
| Primer | Sequence |
|---|---|
| αMyHC F | CCTGTCCAGCAGAAAGAGC |
| αMyHC R | CAGGCAAAGTCAAGCATTCATATTTATTGTG |
| 18S F | GCCGCTAGAGGTGAAATTCTTG |
| 18S R | CTTTCGCTCTGGTCCGTCTT |
| BNP F | GGTGCTGCCCCAGATGATT |
| BNP R | CTGGAGACTGGCTAGGACTTC |
| SERCA F | GGCCAGATCGCGCTACA |
| SERCA R | GGGCCAATTAGAGAGCAGGTTT |
| Sk α-actin F | CCACCTACAACAGCATCATGAAGT |
| Sk α-actin R | GACATGACGTTGTTGGCGTACA |
| βMyHC F | CGCTCAGTCATGGCGGAT |
| βMyHC R | GCCCCAAATGCAGCCAT |
| ANF F | GCGAAGGTCAAGCTGCTT |
| ANF R | CTGGGCTCCAATCCTGTCAAT |
All primers are presented in a 5′-3′ orientation.
Incorporation of [14C]Leucine and Cell Size Measurements
Protein synthesis was measured by the incorporation of [14C]leucine as described previously (Maass et al., 1995). Cell size was measured using ImageJ software (National Institutes of Health) as described previously (Emter et al., 2005).
Nuclear and Cytoplasmic Fractionation
Nuclear and cytoplasmic fractionation were performed using the NE-PER kit (Pierce Chemical, Rockford, IL) according to manufacturer's recommendations.
Western Blots
Western blots were performed essentially as described previously (Sucharov et al., 2003). Antibodies were diluted 1:1000 in 1× Tris-buffered saline (20 mM Tris and 500 mM NaCl, pH 7.5) containing 3% bovine serum albumin (BSA) and 0.1% Tween and incubated with the blot overnight at 4°C.
Immunoprecipitation/Immunoblotting
Immunoprecipitation experiments were done using antibodies described in the text. Experiments were done according to Santa Cruz Biotechnology recommendations, with minor modifications. After four washes with 1× radioimmunoprecipitation assay buffer (Calalb et al., 1995), the sample was incubated with 2–3× packed volume with 2× sample buffer (Bio-Rad, Hercules, CA) at room temperature for 30 min.
Immunofluorescence
Immunofluorescence was performed according to Harrison et al. (2004). Cells were washed with Tris-buffered saline/Tween 20 (TBST) and fixed with 10% formaldehyde for 20 min. Cells were again washed with TBST and incubated with 0.1% Triton X for 30 min. Cells were then blocked with 1% BSA in TBST for 1 h followed by 1-h incubation with 1:500 dilution of the FLAG antibody. Cells incubated with 1:1000 dilution of Alexa 594 anti-mouse antibody and 2 μg/ml Hoechst staining for 1 h. Images were captured at a 40× magnification with a fluorescence microscope (Nikon E800) equipped with a digital camera (AxioCam) and Axiovision, version 3.0.6.36 imaging software (Carl Zeiss, Thornwood, NY).
COS Cell Transfection
COS cells were transfected with Lipofectamine 2000 (Invitrogen). Briefly, 8.4 μg of total DNA was combined with 25 μl of Lipofectamine according to manufacturer's recommendations.
Cardiac Myocyte Transfection
Cardiac myocyte transfections were done using the nucleofaction protocol (Amaxa Biosystems, Gaithersburg, MD). This methodology results in ∼50% transfection efficiency. Briefly, 2 × 106 cells were transfected with 4 μg of plasmid DNA according to the manufacturer's recommendations.
YY1 and HDAC5 Small Interfering RNA (siRNA) Transfection
YY1 siRNA was purchased from Ambion (Austin, TX; catalog no. 16704), and HDAC5 siRNA was purchased from Thermo Fisher Scientific (Waltham, MA). siRNAs were transfected using the nucleofaction protocol (HDAC5 and YY1) or oligofectamine methodology (YY1) (Invitrogen). In both cases, 20 μM siRNA oligonucleotide was used. All results were compared with transfections containing a negative control siRNA (Ambion; catalog no. 4611).
Chromatin Immunoprecipitation (ChIP)
ChiPs were performed using the ChIP assay kit (Millipore, Billerica, MA). Cells (1 × 106) were used for each condition. Cells were sonicated four times with 10-min pulses at 40% of the power. The resulting DNA–protein complex was immunoprecipitated with YY1 agarose-conjugated antibody (Santa Cruz Biotechnology; sc-281), immunoglobulin G (IgG), or RNA polymerase antibody kit (Millipore). Cross-link was reversed and protein was digested with proteinase K. DNA was analyzed by PCR. Primers used in the PCR reaction are described in Table 2.
Table 2.
Sequence of the primers used for the PCR reaction
| Gene | Sequence | Position |
|---|---|---|
| αMyHC F | GGTAAGGGCCATGTGGGTA | −322/−304 |
| αMyHC R | CCCACGCTAAACTCCTTCTTACTTGGGAT | +36/+8 |
| BNP F | GGCAGGAAACAAGGACCTGTT | −254/−234 |
| BNP R | CCAGGCAGCTGCGATGGTGT | +67/+48 |
| GAPDH F | GCGCACACACACACGCACATAT | −763/−742 |
| GAPDH R | GGGCTGTTTGCTCCCAGCAT | −568/−587 |
| Amylase F | GCATTGAACAACTCATGTCATAGCACA | −236/−210 |
| Amylase R | CCGGTAATCTCTGTAATGCATCATGT | −99/−124 |
All primers are presented in a 5′-3′ orientation, and they are all within the promoter regions of the respective genes.
Statistical Analysis
All analyses were performed using analysis of variance with Fisher post hoc test, with a p < 0.05 in a two-tailed distribution considered to be statistically significant.
RESULTS
YY1 Prevents Phenylephrine (PE)-mediated Induction of the Fetal Gene Program and Cellular Hypertrophy
YY1 has been shown to function mainly as a repressor of muscle-specific promoters (MacLellan et al., 1993; Patten et al., 2000; Sucharov et al., 2003; Mariner et al., 2005), suggesting that its function in muscle cells is primarily that of a repressor. To determine the role of YY1 in regulating the expression of the components of the fetal gene program, NRVMs were infected with an YY1-expressing adenovirus, which resulted in 20- to 30-fold overexpression of the protein (data not shown). As determined by RT-PCR, overexpression of YY1 in NRVMs repressed the expression of all the fetal components of the fetal gene program (βMyHC, ANF, brain natriuretic peptide [BNP], and skeletal α-actin), but it up-regulated the expression of the adult isoforms αMyHC and SERCA2A (Figure 1).
Figure 1.
YY1 represses fetal gene expression and activates adult gene expression in cardiac cells. Cells were infected with YY1-GFP and harvested 48 h after infection. Total RNA was analyzed by RT-PCR. Results were compared with cells infected with a control CMV-GFP adenovirus, defined as 100% (line at 100%). n = 8.
To test whether YY1 can prevent up-regulation of the fetal gene program under conditions that induce pathological cardiac hypertrophy, NRVMs were infected with YY1 adenovirus construct and treated with the α-adrenergic receptor agonist PE 24 h after infection. Activation of the α-adrenergic receptor has been shown to increase myocyte cell size and to activate the fetal gene program, with repression of αMyHC and SERCA2A, and up-regulation of ANP, BNP, βMyHC, and skeletal α-actin (Patten et al., 1996). mRNA was extracted 48 h after treatment and analyzed by RT-PCR. As shown in Figure 2A, PE induced gene expression changes that accompany pathological hypertrophy, and YY1 overexpression either prevented or blunted PE-mediated up-regulation of the fetal components. Furthermore, YY1 overexpression blocked the increase in protein synthesis that accompanies cellular hypertrophy. As shown in Figure 2B, protein content, measured by incorporation of radiolabeled leucine, was increased in PE-treated cells infected with control adenovirus. However, cells overexpressing YY1 did not show significant changes in protein content in response to PE, suggesting that hypertrophy was blocked in these cells. This effect on cell size can also be observed visually. As shown in Figure 2C, cell size was increased in response to PE treatment, but this increase was blunted in cells overexpressing YY1. Figure 2D is representative of cardiac myocytes immunostained with α-actinin antibody. As shown in Figure 2 sarcomeric organization was present in PE-treated cells and in cells infected with YY1 adenovirus, suggesting that YY1 induces sarcomeric organization in the absence of hypertrophic stimuli.
Figure 2.
YY1 prevents up-regulation of the fetal isoforms and PE-induced cellular hypertrophy in response to PE. (A) NRVMs were treated with 10 μM PE for 48 h; white bars, no treatment; black bars, PE treatment. Total RNA was analyzed by RT-PCR. Results were compared with vehicle-treated cells infected with CMV-GFP, defined as 100%. n = 8. (B) Increase in protein synthesis was measured by leucine incorporation in untreated and PE-treated cells infected with a CMV control or YY1 adenovirus construct. (C) Cell size was measured using the ImageJ software. Fifty cells from four different experiments were measured. (D) Immunofluorescence with anti-actinin antibody of cells infected with a control or YY1-GFP virus in untreated and PE-treated cells.
Down-Regulation of YY1 Induces the Fetal Gene Program and Increases in Cell Size
If YY1 acts to repress pathological gene expression in cardiac cells, decreasing YY1 levels in these cells should derepress the fetal gene program and induce a hypertrophic response. To test this hypothesis, NRVMs were transfected with a siRNA oligonucleotide against YY1 to reduce YY1 levels. As shown in Figure 3, siRNA transfection resulted in a dramatic down-regulation of YY1 protein levels (Figure 3A), and it was accompanied by an induction of the fetal gene program (Figure 3B). Compared with cells transfected with a control siRNA oligonucleotide, the cells transfected with the YY1 siRNA showed marked increases in several fetal gene markers, no changes in SERCA expression, and a down-regulation of αMyHC mRNA. In addition, cells transfected with the YY1 siRNA showed an increase in protein content (Figure 3C) and in cell size (Figure 3, D and E), consistent with a hypertrophic response. Together, these data suggest that YY1 is required to maintain a normal myocyte phenotype by repressing the fetal gene program and preventing myocyte hypertrophy.
Figure 3.
Down-regulation of YY1 expression results in up-regulation of the fetal isoforms and in increase in cell size and protein synthesis. (A) YY1 expression is repressed by transfection with an YY1 siRNA oligonucleotide (lane 3) but not by a control or an unrelated siRNA oligonucleotide (lanes 2 and 4). Lane 1, no transfection. Total extract was used, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. (B) Cells were transfected with YY1 siRNA nucleotide and treated with vehicle (white bars) or PE (black bars) for 48 h. RNA expression was analyzed by RT-PCR and normalized to 18S. Results were compared with vehicle-treated cells transfected with a control siRNA, defined as 100%. n = 4. (C) Radiolabeled protein experiments show an increase in radiolabeled leucine incorporation in cells transfected with YY1 siRNA oligonucleotide. (D) Cell size measurements show an increase in cell size in cells that do not express YY1. Fifty-five cells from three different experiments were measured. (E) Immunofluorescence with anti-actinin antibody of cells transfected with a control or YY1 siRNA oligonucleotide.
YY1 Binds to the Endogenous αMyHC and BNP Promoters
Because YY1 has been shown to interact with αMyHC and BNP promoter constructs in electrophoretic mobility shift assays (Bhalla et al., 2001, Sucharov et al., 2003; Mariner et al., 2005), we wanted to know whether YY1 interacts with the promoter region of the endogenous genes. To test this, ChIP experiments were performed using an YY1-agarose–conjugated antibody and primers designed to amplify the promoter regions of the αMyHC and BNP genes. As shown in Figure 4, these promoters are bound by YY1 in these cells. ChIP experiments were performed six different times. No consistent difference was observed in the level of amplified product. This suggests that YY1 function in regulating gene expression likely results from a mechanism that is independent of YY1 occupancy of the promoter.
Figure 4.
YY1 binds to αMyHC and BNP promoters. ChIPs were done in noninfected cells, in cells overexpressing YY1 or in cells transfected with YY1 siRNA as described in the methods section. ChIPs from cells infected with YY1 or transfected with YY1 siRNA are a result of six independent experiments. YY1 or RNA polymerase antibodies, or IgG were used in the immunoprecipitation assay.
ChIP experiments were performed in cells transfected with YY1 siRNA. As shown in Figure 4, in ChIP experiments performed in the absence of YY1, a PCR product was not detected for αMyHC and BNP primers, showing that PCR amplification is in fact due to YY1 interaction with these promoters. Protein A (data not shown) or IgG was used as control for the immunoprecipitation experiments. RNA polymerase antibody was used as a positive control for the experiments, whereas α-amylase primer was used as a negative control due to lack of interaction of YY1 with the amylase promoter (Caretti et al., 2004).
YY1 Interacts with HDAC5 in NRVMs and in Rat Hearts
Because the activity of YY1 is known to depend on its interaction with other transcriptional regulators, we were interested to determine what cofactors are involved in the YY1-mediated regulation of the fetal gene program in cardiac cells. Previous work with HeLa cells demonstrated that YY1 can interact directly with class I HDACs (Yao et al., 2001). Furthermore, the class II HDAC HDAC5 has been shown to be regulated during hypertrophy (Zhang et al., 2002), and phosphorylation of HDAC5 in response to PE stimulation results in its nuclear export and transcription derepression (Zhang et al., 2002). To test whether YY1 interacts with HDAC5, NRVMs were infected with YY1-GFP adenovirus construct. Cells were harvested and immunoprecipitation experiments were performed with an anti-YY1 antibody. As shown in Figure 5A, YY1 and HDAC5 coimmunoprecipitated in untreated but not in PE-treated cells, suggesting that PE treatment prevents their interaction. To demonstrate that this interaction is also observed in vivo, protein extracts were prepared from rat heart tissue, and immunoprecipitation experiments were performed with anti-YY1 and anti-HDAC5 antibodies. As seen in NRVMs, endogenous YY1 and HDAC5 also coimmunoprecipitated, suggesting that they interact in vivo (Figure 5B). This interaction of YY1 with HDACs seems to be specific to HDAC5 in these cells because no interaction of YY1 with HDAC 4 or HDAC9 was observed in these experiments (Figure 5B).
Figure 5.
YY1 interacts with HDAC5 in NRVMs and in rat heart tissue. (A) Cells were infected with YY1-GFP adenovirus construct. Nuclear extracts were immunoprecipitated with anti-YY1 antibody and HDAC5 was detected by Western blot. (B) Rat heart nuclear fraction was immunoprecipitated with anti-YY1 antibody or IgG and interaction with HDAC5, HDAC4, or HDAC9 was detected by Western blot.
YY1 Prevents Nuclear Export of HDAC5 in PE-stimulated Cells
Because YY1 and HDAC5 interact in cardiac cells and HDAC5 localization seems to be important to hypertrophic responses, we hypothesized that YY1 prevents export of HDAC5 to the cytoplasm. By retaining HDAC5 in the nucleus, YY1 effectively increases HDAC5 activity in the cell and prevents the up-regulation of the fetal gene program in hypertrophy-induced cells. To test this hypothesis, NRVMs were infected with HDAC5-FLAG and YY1-GFP adenovirus constructs, or HDAC5-FLAG and cytomegalovirus (CMV)-GFP adenovirus constructs as a control. After 24 h, the cells were treated with PE for 2 h followed by harvesting and fractionation of nuclear and cytoplasmic extracts. As shown in Figure 6, A and E, HDAC5 is exported to the cytoplasm in PE-treated cells in the absence of YY1 (GFP control infection). However, in cells overexpressing YY1, HDAC5 is found in the nuclear fraction, suggesting that it is retained in the nucleus even when the cells are stimulated with PE. These results are depicted graphically in Figure 6B to show data pooled from three independent experiments. To test whether YY1 can regulate endogenous HDAC5 localization cells were infected with YY1 adenovirus, treated with PE and cytoplasmic and nuclear fractions were extracted. As shown in Figure 6C, YY1 prevented translocation of endogenous HDAC5 to the cytoplasm in PE-treated cells.
Figure 6.
YY1 prevents nuclear export of HDAC5 in PE-treated cells, but down-regulation of YY1 does not result in cytoplasmic localization of HDAC5. (A) Western blot of nuclear and cytoplasmic fractions of cells infected with HDAC5-FLAG and GFP or HDAC5-FLAG and YY1 adenovirus constructs. Cells were treated with PE for 2 h before harvesting. GAPDH and Sp1 were used as loading control for cytoplasmic fraction and nuclear fraction respectively and to show that there was no cross-contamination between fractions. Calnexin was used as a control for both cytoplasmic and nuclear fractions. (B) The graph is representative of three individual Western Blot experiments shown in A. (C) YY1 prevents export of endogenous HDAC5 to the cytoplasm in response to PE treatment. Experiments were performed as described in A, but cells were infected only with YY1 or CMV adenovirus. Endogenous HDAC5 was detected using HDAC5 antibody. (D) Western blot of nuclear and cytoplasmic fractions of cells transfected with a control siRNA or YY1 siRNA and infected with HDAC5-FLAG. Calnexin was used as a loading control. (E) Immunostaining of NRVMs infected with HDAC5-FLAG–untreated cells (1), HDAC5-FLAG–PE-treated (2), HDAC5-FLAG and YY1–PE-treated (3), HDAC5-FLAG and transfected with YY1 siRNA (4). The panels show FLAG staining for HDAC5 localization, Hoechst staining for nuclear localization and merging of HDAC5 and nuclear staining.
Down-Regulation of YY1 in NRVMs Does Not Change the Cellular Localization of HDAC5
Given the observation that YY1 overexpression prevents the nuclear export of HDAC5 in response to PE, we asked whether YY1 is required for nuclear localization under unstimulated conditions. If so, the cytoplasmic localization of HDAC5 would be expected to increase if YY1 levels were experimentally reduced. This does not seem to be the case, however. As shown in Figure 6, D and E, cellular localization of HDAC5 does not change in cells transfected with the YY1 siRNA oligonucleotide compared with cells that received the control oligonucleotide. These results suggest that HDAC5 does not require the presence of YY1 to be localized to the nucleus. Instead, HDAC5 is likely just a binding partner with YY1, and overexpression of YY1 acts to prevent HDAC5 from active nuclear export. In this scenario, YY1 knockdown would have no effect on the nuclear versus cytoplasmic localization of HDAC5 but likely plays an important role in the subnuclear localization of HDAC5. That is, decreasing YY1 levels may not cause HDAC5 to be exported to the cytoplasm, but it certainly affects which gene promoters recruit HDAC5 and are regulated by its repressor activity.
YY1 Reduces PE-mediated HDAC5 Phosphorylation
Because YY1 overexpression prevents the activated nuclear export of HDAC5 in cardiac cells, we hypothesized that the binding interaction of YY1 with HDAC5 prevents HDAC5 phosphorylation at the amino acids that are specifically phosphorylated in response to hypertrophic stimuli. To test this hypothesis, we first examined the level of HDAC5 phosphorylation in cells overexpressing YY1. As shown in Figure 7, A and B, overexpression of YY1 results in a dramatic reduction in the phosphorylation levels of HDAC5-FLAG or endogenous HDAC5. We then tested whether YY1 interacts with a region of HDAC5 that includes the serine residues (Ser259 and Ser496) that are phosphorylated in response to PE. By transfecting an HDAC5 260–615 deletion construct and the full-length YY1 into COS cells, we show that YY1 does not bind the HDAC5 deletion construct in coimmunoprecipitation assays (Figure 7, C and D), suggesting that this region of HDAC5 is necessary for YY1 interaction. Together, these data suggest that YY1 prevents HDAC5 nuclear export by directly binding its activation domain and preventing its phosphorylation.
Figure 7.
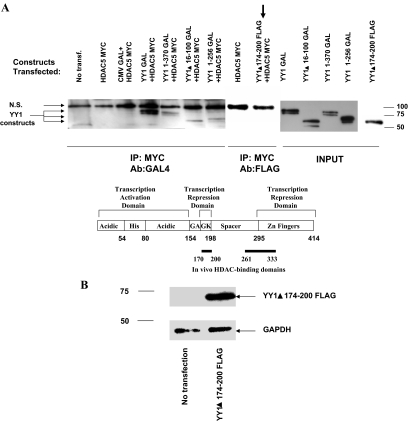
The phosphorylation domain of HDAC5 is required for its interaction with YY1, and YY1 blunts HDAC5 phosphorylation. (A and B) Overexpression of YY1 blunts HDAC5 phosphorylation in response to PE. (A) NRVMs were infected with CMV or YY1-GFP. Cells were treated with PE for 1 h and total protein was extracted. Phosphorylated Ser 259 HDAC5 and total HDAC5 were detected by Western blot, and calnexin antibody was used as a loading control. (B) Quantification of Western in A. (C and D) COS cells were transfected with YY1-GAL and wild-type HDAC5-MYC (C) or YY1-GAL and HDAC5 del 260-615-FLAG (D), and total protein was extracted. Immunoprecipitation was done with a GFP antibody (control) or a GAL4 antibody, and HDAC5 was detected by Western blot using either MYC or FLAG antibodies. N.S., nonspecific.
YY1 Construct That Lacks the HDAC Binding Domain Up-Regulates the Fetal Gene Program
To delineate which regions of YY1 were responsible for interaction with HDAC5, we next tested a series of YY1 deletion constructs for their ability to bind HDAC5 in coimmunoprecipitation assays. As shown in Figure 8A, only the YY1 174–200 deletion construct was unable to interact with HDAC5 in COS cells. Previous work with these YY1 mutants showed that this deletion construct had the unique ability to activate promoters in cells where YY1 is known to be a repressor of transcription (Sucharov et al., 2006). Using Amaxa's nucleofection technology, the YY1 174–200 deletion construct was transfected into NRVMs, resulting in the up-regulation of all fetal mRNAs tested. PE treatment resulted in a higher mRNA expression of the fetal isoforms when compared with PE-control transfection (Figure 8C). These results suggest that an interaction with HDAC5 is required for YY1 to function as a transcription repressor of the fetal gene program.
Figure 8.
YY1 is a transcription activator in the absence of or lack of interaction with HDAC5. (A) COS7 cells were transfected with various YY1 deletion constructs tagged with GAL4 or FLAG, and HDAC5-MYC. Total extract was immunoprecipitated using the MYC antibody, and YY1 interaction with HDAC5 was detected using antibodies to GAL4 or FLAG. The only YY1 construct that did not interact with HDAC5 lacked region 174–200 (arrow). Cells transfected with HDAC5-MYC or HDAC5-MYC and CMV-GAL4 constructs were used as controls for the immunoprecipitation experiments. (B) NRVMs were transfected using the Amaxa system. Overexpression of the YY1 del 174–200 FLAG construct results in expression of the protein in NRVMs as detected by Western blot using the FLAG antibody. (C) Gene expression was analyzed by RT-PCR. Results were compared with vehicle-treated cells transfected with pcDNA, defined as 100%. White bars, control; black bars, PE treatment. (D) NRVMs were transfected with siRNA for HDAC5 by using the Amaxa system. Down-regulation of HDAC5 was detected by Western blot using the HDAC5 antibody. HDAC4 expression was not changed. (E) Gene expression was analyzed by RT-PCR. Results of YY1 infection in siRNA control transfected cells were compared with control-infected cells transfected with control siRNA, defined as 100%. Results of YY1 infection in siRNA HDAC% transfected cells were compared with control-infected cells transfected with HDAC5 siRNA, defined as 100%.
Down-Regulation of HDAC5 Results in a Switch of YY1 Function in the Regulation of Gene Expression from Transcription Repressor to Transcription Activator
To test whether HDAC5 is required for YY1 to function as a transcription repressor NRVMs were transfected with a HDAC5-specific siRNA. As shown in Figure 8D, expression of HDAC5, but not of the closely related HDAC4, is down-regulated in cells transfected with HDAC5 siRNA. We next tested YY1 regulation of gene expression in cells where HDAC5 was down-regulated. As shown in Figure 8E down-regulation of HDAC5 resulted in YY1 up-regulation of βMyHC, ANF, and BNP, all genes previously shown to be repressed by YY1. Skeletal α-actin was still repressed by YY1, suggesting that the mechanism of YY1 repression of skeletal α-actin is not mediated by HDAC5.
DISCUSSION
In the work presented here, we show that YY1 is a repressor of fetal gene expression in untreated and α-AR–stimulated cardiac cells through interaction with HDAC5. HDACs promote deacetylation of histones, which tightens the chromatin structure around genes and results in transcription repression at those sites (Courey and Jia, 2001). Our results suggest that overexpression of YY1 in cardiac cells results in interaction with and retention of HDAC5 in the nucleus and subsequent repression of fetal genes even when myocytes are stimulated with PE treatment. The retention of HDAC5 in the nucleus likely results from an interaction between YY1 and HDAC5 that prevents phosphorylation of HDAC5 in response to PE stimulation; however, it is also possible that YY1 regulates expression of a kinase or a phosphatase that, in turn, blocks HDAC5 phosphorylation and nuclear export. Because HDAC5 has been shown to be phosphorylated by protein kinase D (PKD) in response to PE stimulation (Vega et al., 2004), we hypothesized that PKD expression may be regulated by YY1, but our results show that PKD levels were unchanged in cells overexpressing YY1 (Supplemental Figure 1S). It remains possible that there are other factors that are important for regulating HDAC5 localization and that these factors are regulated by YY1. Our results also show that YY1 is important for regulating protein synthesis in response to PE (Figure 2). Interestingly YY1 stimulates sarcomeric organization in the absence of PE. It is possible that YY1, through its function as a polycomb group protein (see below), accelerates signaling events that are involved in the organization of the sarcomere. Further experiments would be necessary to determine whether YY1 affects other signaling pathways.
An interaction between YY1 and HDAC5 may lead to the transcriptional repression of fetal genes by causing the recruitment of HDAC5 to the promoter regions of these genes. Consistent with this model, the down-regulation of YY1 by siRNA is sufficient to induce the expression of the fetal gene isoforms. When levels of YY1 are synthetically reduced, HDAC5 would have no mechanism to specifically interact to these promoters and transcription of fetal genes would increase. This is evidenced by the lack of further increase in gene expression in response to PE treatment. Furthermore, the YY1 deletion construct that lacks interaction with HDAC5 functions as transcription activator, suggesting that the ability of YY1 to act as a repressor is dependent on HDAC5 interaction.
These data suggest that YY1 is critically important to the maintenance of normal, adult gene expression patterns. By acting to repress fetal genes, YY1 effectively prevents a regression to fetal programs and promotes the continued terminal differentiation of cardiac cells. Consistent with this idea that YY1 acts as a global regulator of differentiation, YY1 has been identified as a homologue of polycomb group (PcG) proteins and has been shown to be involved in development (Atchison et al., 2003). During the development/differentiation process, there is a fundamental mechanistic need to maintain key transcription patterns throughout the development and lifetime of an organism. PcG proteins have been shown to be an essential component of the maintenance of transcription repression in development and differentiation. PcG proteins can repress transcription by generating chromatin structures that are refractory to gene expression (reviewed in Levine et al., 2004).
ChIP assays done with an anti-YY1 antibody indicate that the interaction of YY1 with the endogenous fetal promoters is unchanged in response to PE treatment, suggesting that the activity of YY1 is not simply a result of YY1 binding to these promoters. Instead, the activity of YY1 at these promoters likely results from an interaction with other factors that bind to and/or modify YY1. Alternatively, retention of HDAC5 in the nucleus of cells overexpressing YY1 is likely to promote transcription repression not only through interaction with YY1 on the promoter region of the various genes, but also through interaction with other transcription factors known to be important in the regulation of these genes, i.e., MEF2. In heart cells, the interaction of YY1 with HDAC5 seems to be critical for the controlled repression of fetal genes. However, the factors that mediate the affinity of YY1 for HDAC5 would also be critical for the proper regulation of development in the mammalian system. In experiments with undifferentiated and differentiated H9C2 cells, YY1 acts as a transcription repressor only in differentiated cells (Sucharov et al., 2006).
Last, we show that HDAC5 is necessary for YY1 function as a transcription repressor of the fetal isoforms in cardiac cells. YY1 has been shown to interact with class I HDACs in HeLa cells and this interaction occurs in two regions of YY1, the 170–200 and 261–333 (Yao et al., 2001). YY1 interaction with HDAC5 occurs through the 170–200 but not the 261–333 region (Figure 8A). It is possible that other domains of YY1 are important for interaction with class II HDACs. However, our results suggest that deletion of the 170–200 region is the only one required for the interaction to occur. Although we showed that HDAC5 is necessary for YY1 to function as a transcription repressor, it is possible that in cardiac cells interaction of YY1 and class I HDACs is important for regulation of a subset of genes. Interestingly, repression of skeletal α-actin seems to be independent of HDAC5, suggesting that other mechanisms are involved in YY1 function as a repressor. Changes in gene expression in response to HDAC5 down-regulation are part of different study (Dockstader and Sucharov, unpublished data). The ability of YY1 to control fetal gene expression does not seem to be limited to an interaction with HDAC5. YY1 regulation of the adult isoforms of gene expression, αMyHC and SERCA, is the opposite of what is seen for the fetal isoforms. YY1 up-regulates expression of these genes independently of its interaction with HDAC5, and repression of these genes in response to PE is also independent of the YY1–HDAC5 interaction (Figure 8C). However, in response to down-regulation of HDAC5, αMyHC gene expression does not change in cells overexpressing YY1, whereas SERCA levels are up-regulated (Figure 8E). This suggests that regulation of the adult isoforms by YY1 is mediated by factors other than HDAC5. Down-regulation of HDAC5 did not have an effect on YY1 repression of skeletal α-actin, whereas deletion of the region that interacts with HDAC5 resulted in up-regulation of skeletal α-actin gene expression by YY1. This suggests that YY1 down-regulation of skeletal α-actin is likely due to interaction with a different HDAC. We had shown previously that YY1 represses human αMyHC promoter activity and gene expression (Sucharov et al., 2003; Mariner et al., 2005). In the previous experiments, NRVMs were infected or transfected for only 48 h, whereas the cells were infected for 72 h (24 h of infection previous to treatment and 48 h of PE treatment) in the experiments shown here. Future experiments in transgenic animal will allow us to further investigate these phenomena.
As a result of the work described here, we postulate that the increase in YY1 expression is a protective mechanism of the cell to prevent further increases in gene expression. We have recently shown that YY1 expression is up-regulated in human heart cells, and in a transgenic model of hypertrophic cardiomyopathy (Sucharov et al., 2003). Future in vivo studies will allow us to gain a better understanding of YY1 function during hypertrophy and heart failure. Our results show that YY1 is an important factor in preventing cardiac hypertrophy through a mechanism that involves inhibition of HDAC5 nuclear export.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Michael Atchison and Ed Seto for the YY1 deletion constructs. Finally, we thank Dr. Aristidis Moustakas for the YY1-GFP virus. This study was funded by National Institutes of health grants 2R01 HL-48013, R01 HL-48013–10S1, and 1K01 HL-088708-01 and was facilitated by the infrastructure and resources provided by the National Institutes of Health CFAR Core grant P30 AI27767. T. M. is affiliated with Gilead Colorado, Inc. This work has been facilitated by the infrastructure and resources by the NIH CFAR core grant P30AI27767.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-12-1217) on July 16, 2008.
REFERENCES
- Abraham W. T., et al. Coordinate changes in myosin heavy chain isoform gene expression are selectively associated with alterations in dilated cardiomyopathy phenotype. Mol. Med. 2002;8:750–760. [PMC free article] [PubMed] [Google Scholar]
- Atchison L., Ghias A., Wilkinson F., Bonini N., Atchison M. L. Transcription factor YY1 functions as a PcG protein in vivo. EMBO J. 2003;22:1347–1358. doi: 10.1093/emboj/cdg124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla S. S., Robitaille L., Nemer M. Cooperative activation by GATA-4 and YY1 of the cardiac B-type natriuretic peptide promoter. J. Biol. Chem. 2001;276:11439–11445. doi: 10.1074/jbc.M100208200. [DOI] [PubMed] [Google Scholar]
- Bushmeyer S., Park K., Atchison M. L. Characterization of functional domains within the multifunctional transcription factor, YY1. J. Biol. Chem. 1995;270:30213–30220. doi: 10.1074/jbc.270.50.30213. [DOI] [PubMed] [Google Scholar]
- Bushmeyer S. M., Atchison M. L. Identification of YY1 sequences necessary for association with the nuclear matrix and for transcriptional repression functions. J. Cell. Biochem. 1998;68:484–499. [PubMed] [Google Scholar]
- Calalb M. B., Polte T. R., Hanks S. K. Tyrosine phosphorylation of focal adhesion kinase at sites in the catalytic domain regulates kinase activity: a role for Src family kinases. Mol. Cell. Biol. 1995;15:954–963. doi: 10.1128/mcb.15.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caretti G., Di Padova M., Micales B., Lyons G. E., Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey A. J., Jia S. Transcriptional repression: the long and the short of it. Genes Dev. 2001;15:2786–2796. doi: 10.1101/gad.939601. [DOI] [PubMed] [Google Scholar]
- Emter C. A., McCune S. A., Sparagna G. C., Radin M. J., Moore R. L. Low-intensity exercise training delays onset of decompensated heart failure in spontaneously hypertensive heart failure rats. Am. J. Physiol. Heart. Circ. Physiol. 2005;289:H2030–H2038. doi: 10.1152/ajpheart.00526.2005. [DOI] [PubMed] [Google Scholar]
- Han S., Lu J., Zhang Y., Cheng C., Han L., Wang X., Li L., Liu C., Huang B. Recruitment of histone deacetylase 4 by transcription factors represses interleukin-5 transcription. Biochem J. 2006;400:439–448. doi: 10.1042/BJ20061085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison B. C., Roberts C. R., Hood D. B., Sweeney M., Gould J. M., Bush E. W., McKinsey T. A. The CRM1 nuclear export receptor controls pathological cardiac gene expression. Mol. Cell. Biol. 2004;24:10636–10649. doi: 10.1128/MCB.24.24.10636-10649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Galvin K. M., See R. H., Eckner R., Livingston D., Moran E., Shi Y. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 1995a;9:1188–1198. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- Lee J. S., See R. H., Galvin K. M., Wang J., Shi Y. Functional interactions between YY1 and adenovirus E1A. Nucleic Acids Res. 1995b;23:925–931. doi: 10.1093/nar/23.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. C., Shi Y., Schwartz R. J. Displacement of BrdUrd-induced YY1 by serum response factor activates skeletal alpha-actin transcription in embryonic myoblasts. Proc. Natl. Acad. Sci. USA. 1992;89:9814–9818. doi: 10.1073/pnas.89.20.9814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine S. S., King I. F., Kingston R. E. Division of labor in polycomb group repression. Trends Biochem. Sci. 2004;29:478–485. doi: 10.1016/j.tibs.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Liu Q., Merkler K. A., Zhang X., McLean M. P. Prostaglandin F2alpha suppresses rat steroidogenic acute regulatory protein expression via induction of Yin Yang 1 protein and recruitment of histone deacetylase 1 protein. Endocrinology. 2007;148:5209–5219. doi: 10.1210/en.2007-0326. [DOI] [PubMed] [Google Scholar]
- Lompre A. M., Schwartz K., d'Albis A., Lacombe G., Van Thiem N., Swynghedauw B. Myosin isoenzyme redistribution in chronic heart overload. Nature. 1979;282:105–107. doi: 10.1038/282105a0. [DOI] [PubMed] [Google Scholar]
- Luke M. P., Sui G., Liu H., Shi Y. Yin Yang 1 physically interacts with Hoxa11 and represses Hoxa11-dependent transcription. J. Biol. Chem. 2006;281:33226–33232. doi: 10.1074/jbc.M606584200. [DOI] [PubMed] [Google Scholar]
- Maass A., Grohe C., Kubisch C., Wollnik B., Vetter H., Neyses L. Hormonal induction of an immediate-early gene response in myogenic cell lines–a paradigm for heart growth. Eur. Heart J. 1995;16(suppl C):12–14. doi: 10.1093/eurheartj/16.suppl_c.12. [DOI] [PubMed] [Google Scholar]
- MacLellan W. R., Brand T., Schneider M. D. Transforming growth factor-beta in cardiac ontogeny and adaptation. Circ. Res. 1993;73:783–791. doi: 10.1161/01.res.73.5.783. [DOI] [PubMed] [Google Scholar]
- Mariner P. D., Luckey S. W., Long C. S., Sucharov C. C., Leinwand L. A. Yin Yang 1 represses alpha-myosin heavy chain gene expression in pathologic cardiac hypertrophy. Biochem. Biophys. Res. Commun. 2005;326:79–86. doi: 10.1016/j.bbrc.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Patten M., Hartogensis W. E., Long C. S. Interleukin-1beta is a negative transcriptional regulator of alpha1-adrenergic induced gene expression in cultured cardiac myocytes. J. Biol. Chem. 1996;271:21134–21141. doi: 10.1074/jbc.271.35.21134. [DOI] [PubMed] [Google Scholar]
- Patten M., Wang W., Aminololama-Shakeri S., Burson M., Long C. S. IL-1 beta increases abundance and activity of the negative transcriptional regulator yin yang-1 (YY1) in neonatal rat cardiac myocytes. J. Mol. Cell Cardiol. 2000;32:1341–1352. doi: 10.1006/jmcc.2000.1169. [DOI] [PubMed] [Google Scholar]
- Sucharov C. C., Langer S. J., Bristow M. R., Leinwand L. A. Shuttling of HDAC5 in H9C2 cells regulates YY1 function through CaMKIV/PKD and PP2A. Am. J. Physiol. Cell Physiol. 2006;291:C1029–C1037. doi: 10.1152/ajpcell.00059.2006. [DOI] [PubMed] [Google Scholar]
- Sucharov C. C., Mariner P., Long C., Bristow M., Leinwand L. Yin Yang 1 is increased in human heart failure and represses the activity of the human alpha-myosin heavy chain promoter. J. Biol. Chem. 2003;278:31233–31239. doi: 10.1074/jbc.M301917200. [DOI] [PubMed] [Google Scholar]
- Thomas M. J., Seto E. Unlocking the mechanisms of transcription factor YY 1, are chromatin modifying enzymes the key? Gene. 1999;236:197–208. doi: 10.1016/s0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- Vega R. B., Harrison B. C., Meadows E., Roberts C. R., Papst P. J., Olson E. N., McKinsey T. A. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol. Cell. Biol. 2004;24:8374–8385. doi: 10.1128/MCB.24.19.8374-8385.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waspe L. E., Ordahl C. P., Simpson P. C. The cardiac beta-myosin heavy chain isogene is induced selectively in alpha 1-adrenergic receptor-stimulated hypertrophy of cultured rat heart myocytes. J. Clin. Invest. 1990;85:1206–1214. doi: 10.1172/JCI114554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y. L., Yang W. M., Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 2001;21:5979–5991. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. L., McKinsey T. A., Chang S., Antos C. L., Hill J. A., Olson E. N. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.