Abstract
Centromeric (CEN) chromatin is placed under mechanical tension and stretches as kinetochores biorient on the mitotic spindle. This deformation could conceivably provide a readout of biorientation to error correction mechanisms that monitor kinetochore–spindle interactions, but whether CEN chromatin acts in a tensiometer capacity is unresolved. Here, we report observations linking yeast Topoisomerase II (Top2) to both CEN mechanics and assessment of interkinetochore tension. First, in top2-4 and sumoylation-resistant top2-SNM mutants CEN chromatin stretches extensively during biorientation, resulting in increased sister kinetochore separation and preanaphase spindle extension. Our data indicate increased CEN stretching corresponds with alterations to CEN topology induced in response to tension. Second, Top2 potentiates aspects of the tension checkpoint. Mutations affecting the Mtw1 kinetochore protein activate Ipl1 kinase to detach kinetochores and induce spindle checkpoint arrest. In mtw1top2-4 and mtw1top2-SNM mutants, however, kinetochores are resistant to detachment and checkpoint arrest is attenuated. For top2-SNM cells, CEN stretching and checkpoint attenuation occur even in the absence of catenation linking sister chromatids. In sum, Top2 seems to play a novel role in CEN compaction that is distinct from decatenation. Perturbations to this function may allow weakened kinetochores to stretch CENs in a manner that mimics tension or evades Ipl1 surveillance.
INTRODUCTION
Accurate chromosome segregation requires that sister kinetochores connect to microtubules from opposing poles of the mitotic spindle, a process known as biorientation. Misaligned kinetochore attachments can also occur, and they must be resolved to prevent lethal chromosome segregation errors or the formation of aneuploid cells. Syntelic attachments arise when sister kinetochores connect to microtubules from the same spindle pole; merotelic attachments result if a single kinetochore attaches to both poles. The Aurora-B kinases, complexed with conserved INCENP/Sli15 and Survivin/Bir1 subunits of the chromosomal passenger complex, are key regulators of the error correction machinery that resolves such inappropriate attachments (reviewed in Ruchaud et al., 2007). Aurora-B (Ipl1 in budding yeast) facilitates error correction through two mechanisms. First, these kinases destabilize defective kinetochore–microtubule interactions by phosphorylating kinetochore-associated proteins. Second, Ipl1/Aurora-B plays a role in activating the spindle assembly checkpoint (SAC), both in response to syntelic attachments as well as mutations that affect the kinetochore or sister chromatid cohesion. Checkpoint activation may occur indirectly because Ipl1/Aurora-B creates unoccupied kinetochores (Pinsky et al., 2006) or directly through phosphorylation of SAC regulators (King et al., 2007).
The manner in which Ipl1/Aurora-B distinguishes such a wide range of spindle lesions is not well understood. With the exception of merotelic interactions (Cimini, 2007), those attachments that are targeted for error correction generally tend to be characterized by a reduction in the mechanical tension generated by pulling bioriented kinetochores toward opposite spindle poles. Ipl1/Aurora-B has thus been suggested to control a tension checkpoint, implying a mechanism to monitor tension (Biggins and Murray, 2001). But whether tension is sensed directly, or transduced into an alteration of the kinetochore-microtubule interface that allows kinetochores to become resistant to Ipl1/Aurora-B, is a matter of debate (Pinsky and Biggins, 2005; Sandall et al., 2006). As one facet of this, it has long been recognized that elastic deformation of chromatin underlying and/or adjacent to the kinetochore (collectively denoted here as CEN chromatin) could provide a mechanical readout of biorientation (McIntosh, 1991; Shelby et al., 1996). In yeast, for example, kinetochores are drawn apart to half the length of the preanaphase spindle by bipolar force, accompanied by a localized dissolution of CEN cohesion and partial unraveling of separated CEN fibers (He et al., 2000; Pearson et al., 2001; Eckert et al., 2007; Ocampo-Hafalla et al., 2007). Recent evidence suggests these separated regions fold back to form a cruciform structure that behaves as an integrated tensile unit, stretching and relaxing as kinetochores oscillate on the spindle (Yeh et al., 2008). Ipl1/Aurora-B could be coupled to CEN deformation in a variety of ways. In one model, stretching of CEN fibers and/or components of the kinetochore under tension has been proposed to separate Ipl1/Aurora-B within the inner CEN from the kinetochore substrates involved in detachment (Tanaka et al., 2002; Cimini, 2007). Overall, these considerations suggest the mechanical resistance of CEN chromatin to deformation could be an important parameter influencing the activity of error correction mechanisms.
Several observations indicate DNA Topoisomerase II (Topo II or Top2 in yeast) may be one factor that that is connected to Ipl1/Aurora-B in the tension checkpoint. First, in vertebrates Topo II accumulates and exhibits robust activity within mitotic CEN chromatin (reviewed in Porter and Farr, 2004). Sumoylation of Topo II (or possibly other substrates) plays an as yet undefined role in controlling Topo II localization to CENs (Azuma et al., 2005; Diaz-Martinez et al., 2006; Dawlaty et al., 2008). Treatments that block this modification also perturb sister chromatid separation during anaphase, suggesting Topo II CEN targeting helps resolve a final population of topological intertwinings (catenates) so sister CENs can disjoin efficiently. Second, in both yeast and vertebrates, Topo II inhibition can restore chromatid attachment to the spindle and silence the tension checkpoint in cohesin mutants, an effect that may be attributable to unresolved catenates substituting for cohesin in generating tension (Dewar et al., 2004; Vagnarelli et al., 2004; Toyoda and Yanagida, 2006). Third, perturbations to Topo II can in some cases activate Ipl1/Aurora-B–dependent preanaphase arrest (Andrews et al., 2006; Diaz-Martinez et al., 2006). This has suggested Ipl1/Aurora-B may be involved in monitoring some aspect of Topo II function, but the basis for this surveillance has remained unclear.
In this report, we find that budding yeast Top2 also plays an unanticipated role in helping Ipl1 enforce the tension checkpoint. Specifically, mutations in the Mtw1 kinetochore protein lead to weakened microtubule attachments that are disconnected by Ipl1. In temperature-sensitive top2-4 mutants and top2-SNM mutants that prevent Top2 SUMO modification, however, these defective attachments are stabilized. In the case of top2-SNM mutants it is possible to show that stabilization of attachment does not occur through catenates restoring tension to mtw1 kinetochores. Rather, the tension checkpoint defects associated with top2 mutants correspond more closely with a requirement for Top2 in preventing excessive CEN stretching as sister kinetochores are pulled toward opposite spindle poles. These results suggest the ability of CEN chromatin to resist spindle traction may indeed be connected to tension assessment mechanisms. In this view, sumoylation promotes an aspect of Top2 function that maintains CEN compaction against poleward force. Perturbations to this function could allow weakened kinetochores to stretch CEN chromatin into a configuration that confers resistance to Ipl1 detachment activity.
MATERIALS AND METHODS
Yeast Strain Construction and Culture Conditions
All strains were derived from W303-related CRY1 (MATa his3-11,15 leu2-3112 trp1-1 ura3-1 ade2-1 can1-100) by using standard genetic techniques. CEN4-GFP comprises 256 copies of the Lac operator targeted 705 base pairs to the right side of CEN4 CDEIII (Bachant et al., 2002). Saccharomyces cerevisiae were cultured in yeast extract/peptone/dextrose (YPD) or synthetic complete media. Cultures for microscopy were supplemented with 50 μg/ml adenine. For G1 synchronization, overnight cultures were diluted into fresh media supplemented with 10 μg/ml α-factor (Bio-Synthesis, Lewisville, TX) for 2–3 h; bar1 strains were treated with 1 μg/ml. Cells were washed in H2O and released in fresh media at an appropriate temperature. Efficient release at 37°C required addition of 0.1 mg/ml Pronase (P-6911; Sigma-Aldrich, St. Louis, MO). For α-factor add-back, cells were released into YPD, pH 3.9, and α-factor was restored (20 μg/ml) when ∼80% of the cells exhibited a small budded morphology. Nocodazole (Sigma-Aldrich) was dissolved in dimethyl sulfoxide at 10 mg/ml and used at a final concentration of 15 μg/ml in YPD.
Strains for in situ CEN excision were constructed as follows. A 1408-base pair SpeI-XhoI fragment from YCp50 containing CEN4 was cloned into pLITMUS29 (pJBN137). A 450-base pair AvrII fragment containing CEN4 CDE I-II-III elements on pJBN137 was replaced with a 3576-base pair loxP(CEN1:kanMX4)loxP cassette to form pJBN150. The elements on this cassette include two loxP sites from pSE934, a CEN1 sequence originating from a 1769-base pair HindIII fragment from YRp14/ARS1/CEN (Hieter et al., 1985), chromosome I (base pairs 150402–152170), and a 1561-base pair NotI fragment from pFA6a/kanMX4 containing kanMX4 (Wach et al., 1994). To replace CEN4 with loxP(CEN1:kanMX4)loxP, a CRY1 TRP1-GFP strain was transformed with a 4522-base pair fragment derived from pJBN150 by SpeI-XhoI digestion. Transformants were selected for G418 resistance, and CEN4 replacement was verified by the appearance of a novel 4534-base pair XhoI fragment on Southern blots by using a 1408-base pair CEN4 SpeI-XhoI fragment isolated from pJBN137 to prepare radiolabeled probes. Standard genetic crosses were used to transfer loxCENlox to other strains. To induce CEN excision, loxP(CEN1:kanMX4)loxP strains were transformed with pBS49 (pGAL-Cre CEN/ARS/URA3; Sauer, 1987) and processed into galactose media.
Microscopy
For Pds1-Myc immunofluorescence, ∼5 × 106 cells were fixed in 3.7% formaldehyde for 2–4 h and spheroplasted (15 min; 37°C) in 0.5 ml of 1.2 M sorbitol, 100 mM KPO4, pH 7.5, containing 1 μl of β-mercaptoethanol (β-ME) and 5 μl of 5 mg/ml Zymolyase 100-T (MP Biomedicals, Irvine, CA). Spheroplasts were resuspended in 0.5 ml of phosphate-buffered saline (PBS), 0.1% Triton X-100 (PBS: 150 mM NaCl, 50 mM KPO4, pH 7.5); this permeabilization step was essential for Pds1-Myc staining. Staining was performed by sequential incubation in 50 μl of a 1:100 dilution of 9E10 (Covance Research Products, Princeton, NJ) (diluted in PBS; 1–2 h; room temperature) and 50 μl of a 1:250 dilution of fluorescein isothiocyanate (FITC)-conjugated α-mouse antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA). Stained specimens were washed into PBS and spotted onto 0.1% poly-lysine–coated slides. For α-tubulin immunofluorescence, cells were fixed 4–15 h, permeabilized in 70% ethanol, spheroplasted 1 h, and stained using a 1:50 dilution of YOL1/34 (Accurate Chemical & Scientific, Westbury, NY) and a 1:100 dilution of FITC-conjugated α-rat antibodies (Sigma-Aldrich).
For budding analyses, SPC42-GFP measurements, and 4,6-diamidino-2-phenylindole (DAPI) staining, cells were fixed in 3.7% formaldehyde for 3 min and washed into PBS. DAPI staining (1 μg/ml) was performed in mounting medium (VECTASHIELD; Vector Laboratories, Burlingame, CA). For TRP1-GFP (GFP, green fluorescent protein) separation/segregation kinetics, cells were fixed in 0.05% NaN3 and 2.5 mM EDTA, washed into STOP solution (0.9% NaCl, 1 mM NaN3, and 10 mM EDTA), and stored at 4°C. Visualization of CEN4-GFP and Ask1-GFP was performed using live cell mounts on a thermal stage (Tokai Hit, Shizuoka-ken, Japan), a 100× Plan Apo 1.4 numerical aperture objective and a number 4 neutral density filter. Spindle length and Ask1-GFP measurements were performed using MetaMorph software (Molecular Devices, Sunnyvale, CA). Live cell imaging was performed as described previously (Bachant et al., 2005). The statistical significance of differences between strains in the distribution of CEN stretching lengths, interkinetochore distance, and preanaphase spindle extension were evaluated using Student's t test.
Chromosome Loss Assays
Cells from single colonies cultured to maintain CFIII (Ura− or Ade−) were resuspended in H2O, inoculated into YPD at low density, and grown to saturation. Cell density was determined at the beginning and end of the culture by using a hemacytometer, and the number of cell divisions (n) was calculated as [log2(final cell number) − log2(initial cell number)]. CFIII loss was monitored either by plating a defined number of cells onto 5′-fluoroorotic acid media or by plating ∼750 cells onto YPD and dividing the number of solid red colonies by total colony number. Using these values, the rate of CFIII loss per division (p) was determined using the equation p = 1 − e(1/n)ln(Rn/R0), where R0 and Rn are the fraction of cells containing CFIII after 0 and n generations under nonselective conditions, respectively (Murakami et al., 1995). The significance of loss rate or frequency differences between strains was assessed using Student's t test.
Minichromosome Catenation and CEN Topoisomer Analysis
For analysis of minichromosome catenation, DNA was prepared from 10 ml of OD600 0.8 cultures synchronized as appropriate for the experiment. Cells were spheroplasted (1 h; 37°C) in 0.6 ml of 0.9 M sorbitol, 100 mM EDTA, a 1:500 dilution of β-ME, and 0.3 mg/ml Zymolyase 20T (MP Biomedicals). Spheroplasts were resuspended in 0.6 ml of TE (20 mM Tris, pH 8.0, and 2 mM EDTA) and lysed (65°C; 30 min) by adding 240 μl of 250 mM EDTA, 0.5 M Tris base, and 2.5% SDS. Then, 400 μl of 5 M KOAC was added to the solution, which was incubated on ice for 1 h and clarified at 13.5K rpm (15 min; 4°C). Nucleic acids were precipitated using isopropanol, resuspended in 500 μl of TE containing 50 μg/ml RNase, and incubated for 1 h at 37°C. After a second precipitation (adding 20 microliters SM NaCl), DNA was dissolved in 40 μl of TE. Then, 7 μl of the sample was loaded onto wide (2.3-cm) well format 20- by 20-cm 0.6% agarose gels containing ethidium bromide (0.5 μg/ml) and electrophoresed in 1× TAE overnight at 50-V constant voltage. Southern transfer was carried out under alkaline conditions onto Hybond-XL (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) membranes. Radiolabeled probes to detect both p3847 and pRS416 were synthesized using a 1549-base pair ScaI/PvuII fragment from pRS416. Hybridization was performed in RapidHyb (GE Healthcare) according to manufacturer's instructions. For analysis of CEN loop-out topoisomers, DNA was prepared from 50 ml of saturated cultures that were refed and processed as described in the legend for Figure 5. Genomic DNA was prepared as described for catenation samples, scaling up as appropriate for the larger cultures. The final DNA prep was resuspended in 200 μl of TE, and 50 μg of each sample was separated on 25- × 20 cm 1% agaraose gels in TAE containing an appropriate concentration of chloroquine in both the gel and running buffer. Electrophoresis and Southern transfer was carried out as described for catenation samples, but electrophoresis time was extended to 24 h, and ethidium bromide was omitted from both the gel and running buffer. The use of wide (2.3 cm) wells and extended electrophoretic separation proved essential for resolution of linking number variants. CEN topoisomers were visualized with radiolabeled probes prepared from a 1087-base pair DNA fragment amplified from pFA6a-kanMX4 (Wach et al., 1994) by using primers JB.101 5′-CCGAACATAAAGCGGCCGCGGTAAGGAAAAGACTCACGTTTCGAGGC and JB.102 5′-ATCGATGAATTCGAGCTCGTTTTCGACACTGGATGGC.
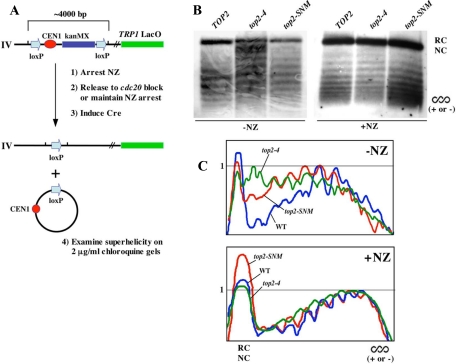
Figure 5.
Topoisomer distribution of loxCENlox excision products. (A) Schematic detailing the loxCENlox cassette and experimental procedure. (B) cdc20-1 (TWY286), cdc20-1top2-4 (TWY258), and cdc20-1top2-SNM (TWY291) strains harboring loxCENlox and pGAL-Cre were arrested in nocodazole at 23°C for 3 h and then washed into galactose media in the presence (+) or absence (−) of nocodazole (NZ) at 35°C. Cells were harvested after 4 h. Genomic DNA preparations were fractionated on 2 μg/ml chloroquine gels and analyzed by Southern blotting to visualize topoisomers for loxCENlox excision products. Chloroquine induces positive supercoiling, and the resulting topoisomers will therefore resolve on these one-dimensional gels as a distribution of negatively wound, relaxed, or positively wound linking number variants depending on the initial winding state of the excised CEN circle. In all these samples, there are also likely to be nicked forms of the excision product resulting from sample preparation that will migrate as completely relaxed molecules. SC; position of either (+) or (−) supercoiled forms. RC/NC; relaxed/nicked circle. (C) To more readily compare samples, the gel panels shown in B were analyzed by densitometry to create graphical representations of topoisomer distributions. The most prominent topoisomer band was assigned a relative gray scale value of 1, and the graphs show overlays of the normalized profiles. Blue, cdc20TOP2 (denoted on graphs as WT); green, cdc20top2-4; red, cdc20top2-SNM.
To create the CEN topoisomers profiles shown in Figure 5 and Supplemental Figure 4, digitized images of Southern autoradiograms were analyzed using the plot profile features of NIH ImageJ software (http://rsb.info.nih.gov/ij/), ensuring all gray scale values were in the nonsaturated range. Y coordinate values were normalized such that the highest topoisomer band for each sample (not the nicked/relaxed circle form) was assigned a relative gray scale value of 1. This allowed the profile plots to be overlaid directly in a graphical representation.
Protein Analysis
Protein extracts for immunoblotting were prepared using mechanical lysis in 20% trichloroacetic acid as described previously (Bachant et al., 2002).
RESULTS
Sister Chromatid Separation and Segregation in top2-SNM Mutants
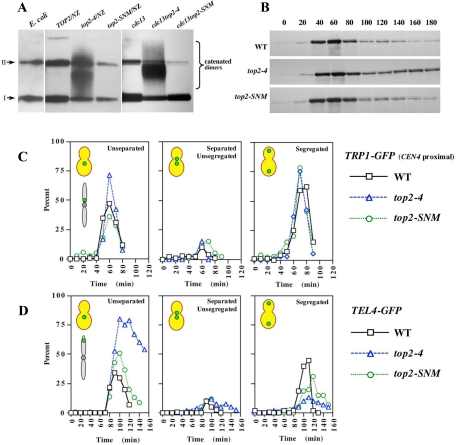
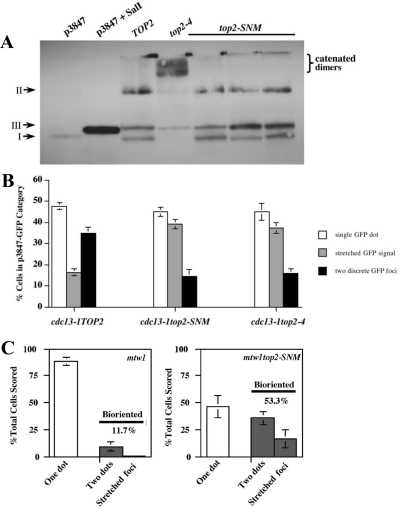
Accumulating evidence suggests Topo II sumoylation promotes sister chromatid decatenation and chromosome segregation in vertebrate cells (Azuma et al., 2003, 2005; Diaz-Martinez et al., 2006; Dawlaty et al., 2008), but whether this modification played an analogous role in yeast Top2 function was unclear. To evaluate functions associated with Top2 sumoylation, we therefore initially compared the catenation status of pRS416, a topologically closed circular minichromosome, in top2-SNM and top2-4 mutants. The top2-SNM allele consists of mutations within the noncatalytic C-terminal domain that abolish SUMO modification (Bachant et al., 2002; Takahashi et al., 2006). top2-4 is a well-characterized temperature-sensitive allele (DiNardo et al., 1984; Holm et al., 1985). Because the force of the spindle has been proposed to help resolve catenates (Holm, 1994), we isolated pRS416 from mitotic cells in the presence or absence of a spindle. Wild-type (WT), top2-SNM, and top2-4 cells were arrested using nocodazole to depolymerize microtubules, whereas cdc13-1, cdc13-1top2-4, and cdc13-1top2-SNM cells were arrested in the presence of a spindle at a cdc13-1 mitotic block. cdc13-1 induces telomeric DNA damage and activates mitotic arrest through the DNA damage checkpoint (Garvik et al., 1995). Consistent with previous studies (Koshland and Hartwell, 1987), we found pRS416 was completely decatenated under both arrest conditions in WT cells, whereas catenated dimers accumulated in top2-4 strains. Similar to WT controls, only supercoiled and nicked forms of pRS416 were observed in top2-SNM mutants (Figure 1A).
Figure 1.
Chromosome decatenation and segregation in top2-SNM Mutants. (A) WT (TOP2; JBY772), top2-4 (JBY775), top2-SNM (TWY179), cdc13-1 (TWY181), cdc13-1top2-4 (TWY185), and cdc13top2-SNM (TWY183) strains harboring pRS416 were released from α-factor at 35°C either in the presence (CDC13 strains) or absence (cdc13 strains) of nocodazole. After 3 h, pRS416 electrophoretic mobility was evaluated by Southern blotting. A bacterial plasmid preparation (E. coli) was included as a control. I and II denote supercoiled and nicked circular plasmid species. (B) WT (JBY649), top2-4 (JBY1523) and top2-SNM (JBY1520) PDS1-MYC strains were arrested in G1 with α-factor, released at 35°C, and α-factor was restored after budding to rearrest cells in G1 after completion of mitosis. Pds1-myc was analyzed by immunoblotting. (C) WT (MNY37-8B), top2-4 (MNY37-24A), and top2-SNM (MNY44-3D) TRP1-GFP strains were released from α-factor at 37°C and rearrested following completion of mitosis. The percentage of budded cells displaying unseparated TRP1-GFP foci (first graph), separated but unsegregated TRP1-GFP foci (second graph) and segregated TRP1-GFP foci (third graph) was determined at each time point. (D) WT (MNY46-3C), top2-4 (MNY33-7C), and top2-SNM (MNY46-9C) TEL4-GFP strains were processed and analyzed as described in C.
Although minichromosomes are decatenated before anaphase, Top2 must act at the metaphase–anaphase transition to resolve catenates that persist on endogenous chromosomes. This decatenation activity is essential for sister chromatids to disjoin completely (Holm et al., 1989). In particular, it has been shown that CENs separate normally in top2-4 cells, whereas telomeric regions fail to disjoin, suggesting catenates may roll down chromosome arms as sisters are pulled apart (Spell and Holm, 1994; Bhalla et al., 2002). To determine whether Top2 SUMO modification facilitated sister separation, we examined chromatid disjunction at CEN-proximal (TRP1-GFP; 13kb from CEN4) and telomere-proximal (TEL4-GFP; 94kb from right end of chromosome 4) loci in top2-4 and top2-SNM strains. As evaluated by degradation of the anaphase inhibitor Pds1, top2-4 and top2-SNM cells entered anaphase with similar kinetics to WT controls, although a fraction of Pds1 was stabilized in the top2-4 strain (Figure 1B). As anticipated, WT, top2-4 and top2-SNM cells displayed similar kinetics of separation and segregation at the CEN proximal locus (Figure 1C), whereas top2-4 mutants were largely blocked for separation at telomeres (Figure 1D). DAPI staining revealed the block to telomere disjunction was accompanied by extensive chromosome bridging (Supplemental Figure 1). In comparison, top2-SNM mutants also exhibited a slight delay in telomere separation (Figure 1D) and a transient accumulation of stretched anaphase chromosomes (Supplemental Figure 1). However, chromatids seemed to eventually disjoin completely and segregate to spindle poles. Consistent with this, top2-SNM cells did not exhibit an increased rate of chromosome missegregation (Supplemental Figure 2A). Furthermore, deletion of RAD52, required for repair of DNA double-strand breaks, did not affect top2-SNM survival (data not shown) or chromosome loss (Supplemental Figure 2A), arguing transient stretching does not cause chromosome breakage. Overall, these results indicate Top2 sumoylation is dispensable for decatenating small minichromosomes but makes a minor contribution to the timing of chromatid disjunction on endogenous chromosomes. top2-SNM mutants may therefore experience a subtle perturbation to chromosome decatenation. If so, this perturbation that does not obviously interfere with chromosome segregation accuracy.
CEN Tensioning in top2-SNM and top2-4 Mutants
Having evaluated whether Top2 sumoylation contributes to the overall program of anaphase chromosome separation and segregation, we next focused on possible roles for this modification in CEN chromatin dynamics (here, we use the term CEN chromatin to encompass both the CEN locus and adjacent periCEN regions). We felt this might be informative because in vertebrates sumoylation seems to preferentially promote Topo II function at CENs (Diaz-Martinez et al., 2006; Dawlaty et al., 2008). CEN dynamics during kinetochore biorientation have been characterized extensively in yeast, primarily using GFP tags integrated immediately adjacent to the kinetochore (Goshima and Yanagida, 2000; He et al., 2000; Tanaka et al., 2000; Pearson et al., 2001). These studies have shown that sister CENs are locally pulled apart as sister kinetochores attain bipolar attachment, accompanied by stretching of separated CEN chromatin. This initial distension, however, is quickly followed by a localized recompaction of the separated chromatid fibers.
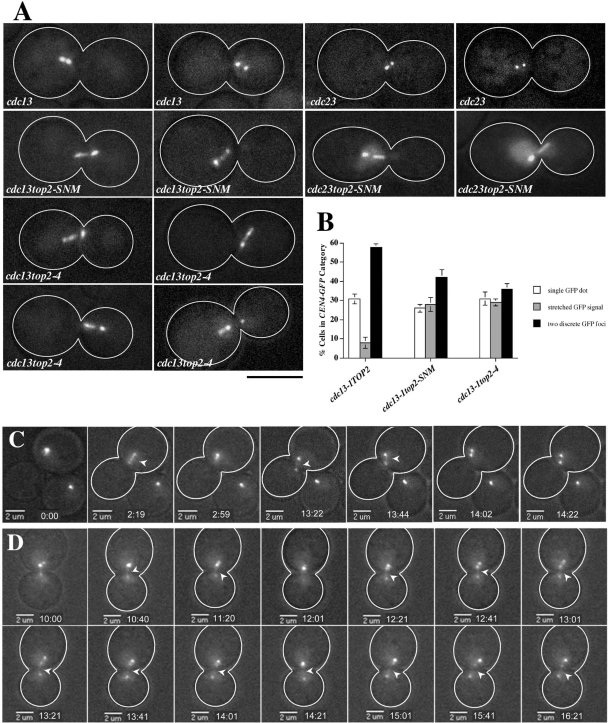
As a first step in analyzing CEN dynamics, we examined CEN morphology in cycling top2-SNM mutants. Twenty-three percent of medium/large-budded top2-SNM cells exhibited a filamentous CEN4-GFP morphology distinct from the compact, spherical foci typically associated with bipolar attachment; a similar morphology was only observed in 7% of WT controls (Supplemental Figure 3, A and B). Direct measurement suggested CEN4-GFP was indeed drawn into a more extended configuration in top2-SNM cells (Supplemental Figure 3C). Importantly, our previous work indicated top2 mutants remain largely proficient for chromatid cohesion at CEN-proximal chromosomal regions (Bachant et al., 2002). Thus, the extension of CEN chromatin in top2 mutants is likely to reflect increased chromatid stretching rather than increased chromatid separation, and we therefore refer to this extension as a CEN stretching phenotype. To ensure we were not visualizing CEN stretching in early anaphase, we examined CEN morphology in metaphase-arrested cdc13top2-SNM and cdc13top2-4 strains. Compared with cdc13 controls, both top2 mutants exhibited an approximately threefold increase in cells exhibiting CEN4-GFP stretching, corresponding with a reduction in cells with two discrete CEN4-GFP foci (Figure 2, A and B). This was not a consequence of cdc13-1 DNA damage, as a similar type of stretching occurred in top2-SNM mutants blocked in metaphase by inactivating the anaphase-promoting complex (APC; Figure 2A).
Figure 2.
CEN4-GFP Stretching in top2-4 and top2-SNM Mutants. (A) cdc13-1 (JBY607), cdc23-1 (JBY686), cdc13-1top2-SNM (JBY1100), cdc23-1top2-SNM (JBY1633), and cdc13-1top2-4 (JBY845) CEN4-GFP strains were arrested at 32°C for 2.5 h before imaging. Micrographs illustrate bioriented CEN4-GFP foci in cdc13 and cdc23 cells and CEN4-GFP stretching in top2 mutants. Bar, 5 μm. (B) cdc13-1 (JBY607; 3 experiments), cdc13-1top2-SNM (JBY1100; 3 experiments), and cdc13-1top2-4 (JBY845, JBY846; 2 experiments each) CEN4-GFP cells were processed for microscopy as in A and scored using the indicated categories. Graphs depict the average and SD for the combined experiments. (C) Selected frames showing a cdc13TOP2 cell exhibiting CEN4-GFP stretching and separation into two compact foci. Numbers, elapsed time; arrows, CEN4-GFP stretching into a filamentous configuration. (D) Sequential frames illustrating cdc13top2-SNM CEN4-GFP stretching.
CEN dynamics were further characterized by imaging cells over a 20-min period (Supplemental Videos 1–4). In cdc13TOP2 cells, bioriented CEN foci remained close together, and stretching events were of short duration. For example, Figure 2C depicts stretched CEN4-GFP chromatin that coalesces into a single GFP focus after 40 s. A second stretching event occurs that quickly resolves into two compact, spherical foci. In cdc13top2-SNM cells it proved difficult to visualize stretched CEN fibers for extended periods without photobleaching. However, based on successfully imaging five cells for a complete 20-min sequence, stretching of CEN4-GFP arrays seemed more persistent in top2-SNM cells (≥6 min for the cell in Figure 2D), suggesting recompaction to form closely juxtaposed foci occurred less efficiently.
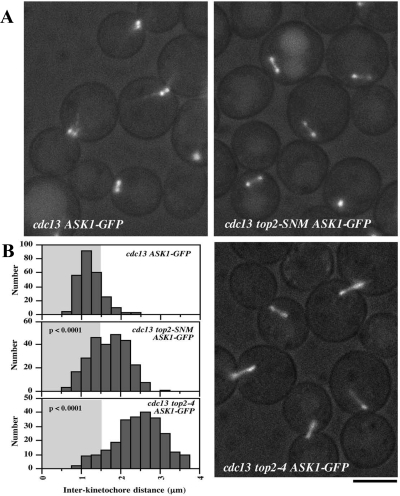
A notable feature of the above-mentioned analysis was that CEN chromatin displayed an extent of stretching during biorientation in top2-SNM mutants that was comparable to the catalytically inactive top2-4 allele. This suggested to us that sumoylation might be promoting a functional role for Top2 in endowing CEN fibers with the tensile properties to resist spindle traction. We reasoned that if this were true we would expect to observe an increased distance between bioriented kinetochores in both top2 mutants, a parameter frequently used to evaluate the magnitude of interkinetochore tension. To test this we visualized the distribution of bioriented kinetochore clusters in cdc13top2-SNM and cdc13top2-4 cells using Ask1-GFP, a component of the Dam1 kinetochore complex (Li et al., 2002). This analysis revealed kinetochores were in fact pulled further apart in both top2 strains with mean distances of Ask1-GFP separation of 1.24 ± 0.34, 1.69 ± 0.47, and 2.46 ± 0.62 μm, respectively, in TOP2, top2-SNM and top2-4 strains (Figure 3). Consistent with the normal chromosome loss rate of top2-SNM cells (Supplemental Figure 2A), increased kinetochore separation was not accompanied by obvious perturbations to kinetochore alignment. However, chromosome loss was slightly (∼2-fold) but significantly (p = 0.0002) increased in Δmad2top2-SNM strains (Supplemental Figure 2B), indicating the SAC contributes to chromosome segregation fidelity in top2-SNM cells. This suggests there may be infrequent kinetochore alignment problems in the top2-SNM mutant.
Figure 3.
Interkinetochore distance in top2-SNM and top2-4 Mutants. (A) cdc13-1 (JBY1483), cdc13-1top2-SNM (JBY1485) and cdc13-1top2-4 (JBY1694) ASK1-GFP cells were arrested for 2.5 h at 35°C and visualized by fluorescence microscopy. Bar, 5 μm. (B) cdc13-1, cdc13-1top2-SNM, and cdc13-1top2-4 ASK1-GFP cells were arrested and visualized as in A. For each strain, the distance between Ask1-GFP foci was measured for ≥250 cells. Histograms show the distribution of these measurements. p values indicate the significance of the differences between the cdc13-1top2-SNM and cdc13-1top2-4 distributions compared with the cdc13-1 control.
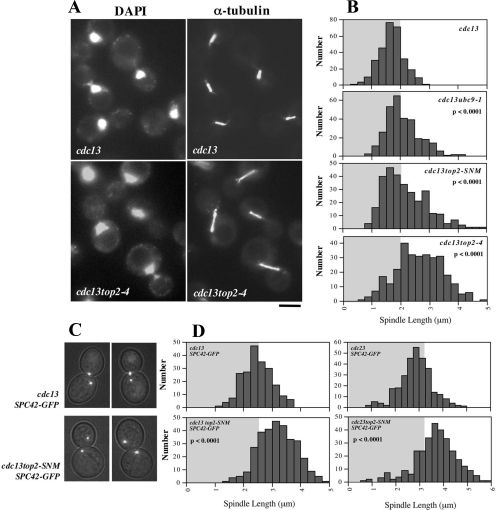
As a second metric for tension, we compared preanaphase spindle lengths in cdc13 (mean length, 1.69 ± 0.42 μm), cdc13top2-SNM (2.17 ± 0.76 μm), cdc13top2-4 (2.83 ± 0.73 μm), and cdc13ubc9-1 (2.08 ± 0.59 μm) strains by using α-tubulin immunofluorescence (Figure 4, A and B) and in cdc13 (mean pole separation, 2.49 ± 0.50 μm), cdc13top2-SNM (3.20 ± 0.67 μm), cdc23 (2.87 ± 0.70 μm), and cdc23top2-SNM (3.67 ± 0.91 μm) cells by visualizing GFP-labeled spindle poles (Figure 4, C and D). Both methods of measurement indicated a statistically significant increase in spindle length in top2 mutants. cdc13ubc9-1 mutants, defective for SUMO conjugation, displayed similar spindle extension to cdc13top2-SNM cells. To summarize the results in this section, we observed that CEN chromatin is more prone to stretch in top2-4 and top2-SNM strains, corresponding with increased sister kinetochore separation during biorientation and an accompanying increase in the length of the preanaphase spindle.
Figure 4.
Preanaphase Spindle Length in top2-4 and top2-SNM Mutants. (A) cdc13-1 (JBY432) and cdc13-1top2-4 (JBY626) strains were shifted to 35°C for 3 h and processed for α-tubulin immunofluorescence and DAPI staining. Bar; 5 μm. (B) Spindle length was measured in ≥300 cdc13-1 (JBY607), cdc13-1ubc9-1 (JBY1009), cdc13top2-SNM (JBY1100), and cdc13-1top2-4 (JBY846) cells arrested as in A. Histograms show the distribution of spindle lengths. (C) cdc13-1 (TWY001) and cdc13-1top2-SNM (TWY004) SPC42-GFP strains were arrested at 32°C for 3 h before imaging. (D) cdc13-1, cdc13-1top2-SNM, cdc23-1 (JBY1289), and cdc23-1top2-SNM (JBY1601) SPC42-GFP strains were arrested as in C, and the distance between Spc42-GFP foci was evaluated in ≥300 cells. In B and D, p values indicate the significance of differences between the indicated data sets compared with either cdc13-1 or cdc23-1 controls.
CEN Superhelicity in top2 Mutants
Current evidence suggests at least three possible functions for Top2 within CEN chromatin. First, a primary function for Topo II at CENs, at least in vertebrates, seems to be untangling sister CENs as cells transition into anaphase (Downes et al., 1991; Shamu and Murray, 1992; Clarke et al., 1993; Azuma et al., 2005; Diaz-Martinez et al., 2006; Dawlaty et al., 2008). Second, our previous work suggested Top2 might mediate a cohesive linkage between sister CENs that was potentially distinct from catenation. This is because we found that either the top2-SNM allele or Top2 overproduction could partially suppress a CEN proximal cohesion defect (Bachant et al., 2002). Third, Top2 has been implicated in yeast chromosome condensation (Vas et al., 2007). Of these functions, it was not readily apparent why increased winding or linkage between sister CENs would cause kinetochores to be pulled further apart during biorientation. In contrast, a defect in maintaining CEN compaction seemed a reasonable hypothesis to account for the top2 CEN stretching phenotype. There is at present no means to assay higher order coiling within CEN chromatin. However, Top2 has recently been shown to be more proficient than Top1 at relaxing torsional strain on chromatin templates (Salceda et al., 2006), and it seemed possible that biorientation might induce alterations to superhelical winding as CEN fibers were placed under tension. Based on this reasoning, we decided to test whether Top2 might be required to modulate intrachromatid topology at CENs.
To do this, we constructed strains in which CEN4 was replaced with a CEN cassette flanked by loxP repeats, allowing excision of a circular CEN DNA molecule after Cre production under control of a galactose-inducible promoter (Figure 5A). cdc20-1, cdc20-1top2-4, and cdc20-1top2-SNM strains harboring loxCENlox were arrested in nocodazole and released into galactose media at a nonpermissive temperature for the cdc20-1 and top2-4 alleles. Under these conditions, spindle assembly and biorientation could proceed, but cells were prevented from going into anaphase by inactivation of the Cdc20 APC subunit. Genomic DNA was then fractionated on 2 μg/ml chloroquine gels to analyze CEN topoisomers. All three strains showed a similar distribution of linking number variants in the absence of tension (+NZ; Figure 5, B and C, and Supplemental Figure 4). In comparison, when kinetochores were allowed to biorient at the cdc20 block (−NZ), we observed CEN loop-out products from cdc20top2-4 cells migrating as a noticeably more relaxed set of topoisomers compared with cdc20TOP2 controls. By analyzing these samples under a range of chloroquine concentrations, it seemed as though CEN circles were comparatively overwound (more positively supercoiled) in top2-4 cells (Supplemental Figure 5). Alterations to CEN topology were less apparent in cdc20top2-SNM strains. However, by overlaying profiles of topoisomer distributions (Figure 5C and Supplemental Figure 4) it did seem that there was also a shift toward more relaxed linking number variants in cdc20top2-SNM samples. Thus, based on this assay, it seems that perturbations to Top2 are associated with alterations to CEN topology induced in response to tension.
Top2 Promotes Tension Checkpoint Arrest and Chromatid Detachment in Mtw1 Mutants
A particularly important question in our analysis concerned the functional significance of the role we had uncovered for Top2 in CEN tensioning. In particular, because chromatids bioriented and segregated properly in top2-SNM cells, this aspect of Top2 function did not seem to contribute markedly to chromosome stability, at least under normal conditions. As elaborated in the Introduction, however, the tensile properties of CEN chromatin may be one factor influencing Ipl1/Aurora B targeting of kinetochore–spindle attachment errors. Based on this idea, we considered the possibility that a function for Top2 in the ability of CEN chromatin to resist deformation by the spindle might become important when Ipl1 was forced to evaluate a partial reduction of bipolar tension.
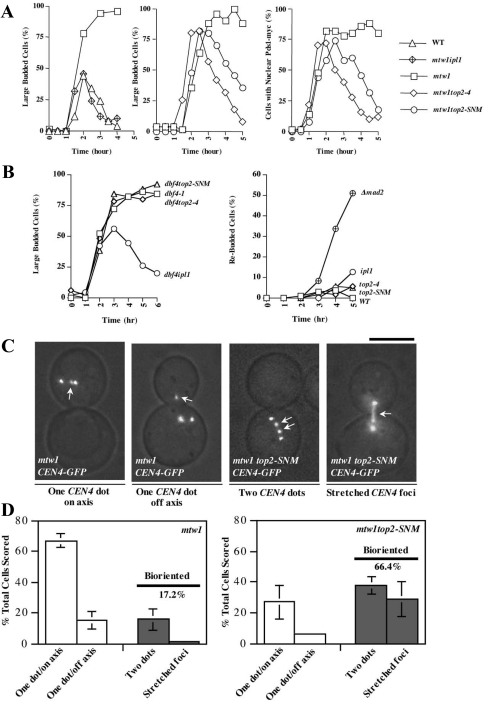
Mutations affecting the kinetochore protein Mtw1 have been shown to activate Ipl1 to detach chromatids from the spindle and induce SAC arrest (Pinsky et al., 2003), presumably because kinetochores attach to microtubules in a weakened configuration that leads to reduced tension. Accordingly, mtw1-1top2-4, and mtw1-1top2-SNM strains, along with WT, mtw1-1 and mtw1-1ipl1-321 controls were released from G1 at 37°C, and SAC arrest was monitored by determining the fraction of large-budded cells and cells with nuclear Pds1 staining. mtw1 cells arrested, whereas mtw1ipl1 cells progressed through mitosis with similar kinetics to WT controls, indicating a complete checkpoint defect (Figure 6A). In comparison, mtw1top2-4 and mtw1top2-SNM exhibited an attenuated response where SAC arrest diminished over 5 h.
Figure 6.
Tension checkpoint integrity in top2 mutants. (A) Each graph shows a separate experiment where WT (JBY649), mtw1-1 (JBY1544), mtw1-1ipl1-321 (SBY1724), mtw1-1top2-4 (JBY1560) or mtw1-1top2-SNM (JBY1553) PDS1-MYC strains were released from a G1 arrest (α-factor) at 37°C. α-factor was restored after budding to restore G1 arrest in cells that completed mitosis. The percentage of large budded cells (left and middle graphs) or cells displaying nuclear Pds1-myc immunofluorescence staining (right graph) was determined at each time point. (B) Left graph. WT (Y300), dbf4-1 (TWY211), dbf4-1ipl1-321 (JBY1643), dbf4-1top2-4 (JBY1640), and dbf4-1top2-SNM (JBY1644) strains were released from a G1 block at 36°C and α-factor was restored after budding. The percentage of large-budded cells was determined at each time point. Right graph. WT (Y300), Δmad2 (JBY554), ipl1-321 (YM311), top2-4 (JBY335), and top2-SNM (JBY1452) strains were released from α-factor into nocodazole-containing media at 36°C. The percentage of rebudded cells was determined at each time point. A rebudded cell is a large budded cell that has sent out one or more additional buds, indicating a failure in SAC arrest. (C) Micrographs illustrating CEN4-GFP alignment categories. Arrows mark CEN4-GFP; unmarked foci are Spc42-GFP. Bar, 5 μm. (D) mtw1-1 SPC42-GFP CEN4-GFP (TW194; 3 experiments, TWY199; 1 experiment) and mtw1-1top2-SNM SPC42-GFP CEN4-GFP (TWY195, 196, 197, 200; 1 experiment each) strains were shifted to 37°C for 2 h before microscopy. Using Spc42-GFP to mark spindle poles, large budded cells that had not undergone spindle extension were scored into the indicated categories. Graphs depict the mean and SD for the combined experiments; in total, 506 and 514 cells were evaluated for mtw1-1 and mtw1top2-SNM strains.
If top2-SNM interferes with how the mtw1 defect is perceived by Ipl1, kinetochore–spindle connections should be stabilized in mtw1top2-SNM strains. Indeed, the frequency of mtw1 cells exhibiting bipolar CEN4-GFP attachments increased in mtw1top2-SNM cells at 37°C, associated with cells exhibiting stretched CEN4-GFP (Figure 6, C and D). Three criteria were used to address whether this restoration of attachment corresponded with improved chromosome segregation. First, the presence of the top2-SNM allele did not suppress mtw1 temperature sensitivity (data not shown). Second, mtw1 cells exhibited a slight increase in chromosome loss at a semipermissive temperature of 34°C; this was not significantly reduced in mtw1top2-SNM strains (Supplemental Figure 6A). Third, at 35.5°C mtw1 cells arrested only transiently, making it possible to evaluate the frequency of cells with two TRP1-GFP foci after completion of mitosis. A partial reduction in this segregation error was in fact observed in mtw1top2-SNM mutants (Supplemental Figure 6B). Overall, however, the data suggest improved kinetochore attachment in mtw1top2-SNM cells is not sufficient to restore chromosome segregation. This is similar to what has been documented in mtw1ipl1 mutants; one possibility is that even though mtw1 kinetochores are not being actively disconnected they are unable to remain attached to the spindle during anaphase (Pinsky et al., 2003).
Ipl1 is also required to activate the SAC in strains defective for the initiation of DNA replication or chromatid cohesion, but not when unoccupied kinetochores are generated directly using nocodazole (Biggins and Murray, 2001). We therefore examined SAC arrest under these conditions in top2 mutants. We observed that replication defective top2-4dbf4-1 and top2-SNMdbf4-1 strains arrested similarly to dbf4-1 controls, whereas ipl1-321dbf4-1 mutants failed to arrest (Figure 6B). Thus, Top2 is not required for tension checkpoint arrest if bipolar attachment is completely circumvented. In keeping with this, top2-4 and top2-SNM mutants seemed largely proficient for SAC arrest after treatment with nocodazole (Figure 6B). Previous experiments have established that inactivating top2-4 silences the tension checkpoint in strains deficient for chromatid cohesion (Dewar et al., 2004). To determine whether the top2-SNM allele behaved similarly, cohesin-deficient scc1-73top2-4, and scc1-73top2-SNM mutants were analyzed as described for mtw1top2 strains. We observed scc1top2-4 and scc1top2-SNM cells proceeded through mitosis more rapidly than scc1 controls (Supplemental Figure 7), suggesting SAC arrest was less robust in both top2 alleles.
Restoration of Chromatid Attachment in mtw1top2-SNM Mutants in the Absence of Catenation
In the case of scc1top2-4 mutants, inactivation of Top2 has been proposed to silence the tension checkpoint because failure to resolve catenates provides an alternative means of linking sister chromatids and restoring tension (Dewar et al., 2004). Because top2-SNM mutants seemed to have a subtle defect in decatenating sister chromatids (Figure 1D), it was possible that checkpoint override in scc1top2-SNM mutants occurred through a similar mechanism. This brought into question the basis for the attenuated checkpoint response of mtw1top2 strains; specifically, could trapped catenates restore tension to weakened mtw1 kinetochores? To address this issue unambiguously, we examined catenation, CEN morphology under tension, and chromatid attachment to the spindle in mtw1top2-SNM cells harboring p3847, a GFP-tagged circular minichromosome containing 10.7-kb of CEN4 flanking DNA (Tanaka et al., 1999). Previous studies have shown that replicated minichromosomes remain paired as sister chromatids until the metaphase to anaphase transition and are joined by cohesive linkages that are capable of supporting tension (Guacci et al., 1994; Megee and Koshland, 1999). Even with this relatively large minichromosome (∼20 kb) catenated forms of p3847 were not observed in cdc13 and cdc13top2-SNM cells, but they were readily detected in cdc13top2-4 strains (Figure 7A; similar results were obtained in nocodazole-arrested cells; data not shown). Compared with endogenous CEN4, p3847 exhibited an increased tendency to adopt a filamentous configuration in cdc13TOP2 cells during biorientation (Figures 7B and 2B). Nonetheless, the frequency of cells displaying a stretched chromatin morphology was clearly elevated in cdc13top2-4 and cdc13top2-SNM transformants, again corresponding with a reduction in separated, compact GFP foci. Finally, we examined p3847 attachment to the spindle in mtw1 and mtw1top2-SNM mutants (Figure 7C). After a shift to the nonpermissive temperature, the majority of mtw1 cells showed a single GFP dot. In contrast, mtw1top2-SNM strains showed a higher percentage of cells with two foci and stretched GFP-labeled chromatin, suggesting a restoration of bipolar attachment. We conclude that kinetochore–microtubule interactions can be stabilized in mtw1top2-SNM mutants even in the absence of catenation between sister chromatids.
Figure 7.
Analysis of minichromosome catenation, elasticity, and spindle attachment. (A) cdc13-1 (TOP2; JBY1461), cdc13-1top2-4 (JBY1462), and cdc13-1top2-SNM (JBY1454, 1455, 1456) strains harboring p3847 were released from α-factor at 35°C. After 3 h, p3847 electrophoretic mobility was evaluated by Southern blotting. Supercoiled (p3847) and linearized (p3847 + SalI) p3847 prepared from E. coli were included as controls. I, II, and III, respectively, denote supercoiled, nicked circular, and linear plasmid species. (B) cdc13-1 (JBY1466, JBY1467; 2 experiments each), cdc13-1top2-SNM (JBY1473, JBY1474, JBY1471; 1 experiment each) and cdc13-1top2-4 (JBY1476; 3 experiments) p3847 transformants were arrested at 34°C for 2.5 h before imaging. Minichromosome stretching was evaluated using the indicated categories. Graphs depict the mean and SD for the combined experiments. (C) mtw1-1 (TWY259; 3 experiments, TWY260; 3 experiments) and mtw1-1top2-SNM (TWY264; three experiments, TWY265; three experiments) strains harboring p3847 were either released from α-factor or shifted as asynchronous cell culture to 37°C for 2 h before microscopy to visualize the GFP-tagged p3847 minichromosome. Large-budded cells were scored into the indicated categories. Graphs depict the mean and SD for the combined experiments; in total, 604 and 686 cells were evaluated for mtw1-1 and mtw1top2-SNM strains.
DISCUSSION
In vertebrates, disruption of PIASγ or RanBP2, two SUMO ligases implicated in Topo II sumoylation, interferes with Topo II localization to CENs and chromatid separation in anaphase (Azuma et al., 2005; Diaz-Martinez et al., 2006; Dawlaty et al., 2008). It is not known whether Top2 is targeted to yeast CENs in an equivalent manner. However, Top2-SUMO fusion proteins can exhibit robust cross-linking to CEN DNA (Takahashi et al., 2006) or accumulate in the nucleolus (Takahashi and Strunnikov, 2007), indicating sumoylation may also regulate yeast Top2 trafficking. In this report, we used the top2-SNM allele to specifically examine functions associated with Top2 SUMO modification, comparing top2-SNM strains to catalytically inactive top2-4 mutants. The two main phenotypes we observed were a tendency for CENs to adopt a drawn out, filamentous appearance as kinetochores bioriented on the spindle and a defect in sustaining tension checkpoint arrest in kinetochore- and cohesin-defective strains. As discussed below, these observations suggest a novel role for Top2 in maintaining tensile CENs that seems to be connected to enforcing aspects of the tension checkpoint.
Top2 Function in CEN Tensioning
Based on increased CEN stretching, interkinetochore distance, and preanaphase spindle extension, a main conclusion from this study is that Top2 acts in some capacity to allow CENs to resist the force of the spindle. There are two aspects to this. First, because increased stretching occurs in both top2-SNM and top2-4 mutants the simplest interpretation is that sumoylation promotes Top2 function in limiting CEN expansion, although we cannot exclude that different defects are responsible for CEN stretching in top2-4 and top2-SNM strains. Second, this aspect of Top2 function seems to be separable from decatenation, which has been considered the main CEN function for TopoII. Most directly, as evaluated using a topologically closed minichromosome, increased stretching occurred in top2-SNM cells even in the absence of catenation.
One hypothesis to explain our data are that Top2 modulates CEN tensioning by reinforcing a compact CEN structure. This might be in keeping with the observation that treating vertebrate cells with the TopoII inhibitor ICRF-193 disorganizes CEN chromatin (Rattner et al., 1996). At present, we can only speculate as to how Top2 might mediate CEN compaction. Because Top2 binds DNA as a dimer, Top2 could act in a structural role to stabilize DNA loops (Aguilar et al., 2005), potentially facilitating coiling within the paired CEN foci that form during biorientation.
Top2 could also maintain CEN compaction in its capacity as a topoisomerase. Our CEN excision experiments are consistent with this, because they reveal top2 mutants display altered CEN superhelicity when chromatids are placed under tension. This may suggest Top2 is required to relax topological stress induced during biorientation, although it is also possible that perturbations to Top2 are a direct cause of aberrant changes to CEN topology. We can envision at least two ways in which torsional strain might arise as CENs are placed under tension. In vitro pulling experiments have shown that DNA overwinds when stretched (Gore et al., 2006), and it is therefore possible that tension could sufficiently distort the DNA helix to alter topology. Alternatively, CEN chromatin unravels extensively during biorientation, potentially to a point where nucleosomes are displaced as a means to dissipate tension (He et al., 2000; Pearson et al., 2001). This would be expected to release superhelical turns constrained by the nucleosomes. Interestingly, a recent study has shown there is in fact a relationship between nucleosome packaging and CEN elasticity, with reduced nucleosome density producing exacerbated CEN stretching and spindle extension (Bouck and Bloom, 2007), reminiscent of top2 mutants.
Finally, a role for Top2 in CEN compaction could be mediated through a functional interaction with the condensin complex. Condensin can reconfigure DNA topology in a Topo II-dependent manner (Stray et al., 2005) and has been suggested to localize to yeast CENs (Bachellier-Bassi et al., 2008). Condensin has also been implicated in preventing CEN expansion under tension (Oliveira et al., 2005; Gerlich et al., 2006; Yong-Gonzalez et al., 2007).
Top2 Function in the Tension Checkpoint
Topo II inactivation silences the SAC and stabilizes chromatid-spindle attachment in cohesin-deficient cells, probably reflecting the ability of unresolved catenates to restore tension (Dewar et al., 2004; Vagnarelli et al., 2004; Toyoda and Yanagida, 2006). In our experiments, restoration of chromatid-spindle attachments in mtw1top2-SNM mutants did not require catenation between sister CENs and was not accompanied by suppression of the underlying mtw1 kinetochore defect. Thus, if top2-SNM causes an increase in interkinetochore tension that compensates for mtw1 kinetochores, this presumably occurs through an as yet unknown mechanism. Such an increase in tension would also be counterintuitive to kinetochores being pulled further apart in top2-SNM strains. One scenario, however, is that perturbations to kinetochore structure or geometry in top2-SNM or top2-4 mutants (potentially associated with alterations to CEN chromatin structure) could make it more difficult for Ipl1 to phosphorylate kinetochore substrates or could allow kinetochore reattachment to occur more efficiently.
A distinct view, prompted by tensiometer models invoking CEN stretching as a factor controlling Ipl1/Aurora B targeting of kinetochores, is that in mtw1top2 mutants weakened kinetochores can distend CENs into a configuration that mimics tension. The observation that Top2 is not required for Ipl1 to respond to monopolar attachments in replication-defective dbf4-1 mutants is consistent with this; in the absence of bipolar force it presumably becomes unnecessary to calibrate CEN resistance to spindle pulling. By extension, Top2 would not be expected to play a role in resolving syntelic attachments, explaining the normal chromosome loss rate of top2-SNM strains. Further analysis of Ipl1 localization and activity in top2 mutants may help shed light on whether there actually is a mechanistic coupling between CEN stretching and stabilization of mtw1 kinetochore attachments.
A model in which Top2 enforces the sensitivity of the tension checkpoint would seem to contrast with reports indicating perturbations to Topo II activate metaphase arrest through pathways that overlap extensively, if are not identical to, the SAC (Mikhailov et al., 2002; Skoufias et al., 2004; Clarke et al., 2006; Diaz-Martinez et al., 2006; Toyoda and Yanagida, 2006). The signal(s) that elicits this arrest has been controversial, with one suggestion being unresolved catenates activate a Topo II-responsive checkpoint (Skoufias et al., 2004; Clarke et al., 2006). Although this has remained unclear, metaphase delay after Topo II inhibition has recently been shown to be abolished by the Aurora B inhibitor ZM447439 (Diaz-Martinez et al., 2006). Similarly, in budding yeast a new group of top2 alleles has been identified that delay anaphase in an Ipl1-dependent manner (Andrews et al., 2006). If Topo II influences both CEN decatenation and compaction, perturbations to Topo II could have variable outcomes with respect to Ipl1/Aurora B signaling. For example, depleting Topo II in vertebrate cells where sister CENs seems to be tightly linked by catenation has been reported to lead to a shortened metaphase interkinetochore distance (Spence et al., 2007), which could possibly simulate a reduction in tension. In contrast, if sister CENs were not extensively joined by catenates, or if it were possible to specifically perturb Topo II CEN compaction activity, CEN fibers might distend more easily into a configuration that conferred resistance to the tension checkpoint. Such a framework may prove useful for analyzing what is turning out to be a complex set of interactions between Topo II, CEN mechanics, and the tension branch of the SAC.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. S. Biggins, S. Elledge, P. Hieter, A. Murray, and K. Nasmyth for generously providing strains and reagents. We also thank Drs. S. Biggins, K. Bloom, D. Clarke, C. Nugent, and members of the Bachant laboratory for useful comments on the manuscript and insightful discussions. This work was supported by National Institutes of Health grant GM-66190 (to J. B.).
Abbreviations used:
- APC
anaphase promoting complex
- CEN
centromere
- GFP
green fluorescent protein
- NZ
nocodazole
- SAC
spindle assembly checkpoint
- Topo II
DNA topoisomerase II
- WT
wild type.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-05-0547) on August 13, 2008.
REFERENCES
- Aguilar C., Davidson C., Dix M., Stead K., Zheng K., Hartman T., Guacci V. Topoisomerase II suppresses the temperature sensitivity of Saccharomyces cerevisiae pds5 mutants, but not the defect in sister chromatid cohesion. Cell Cycle. 2005;4:1294–1304. doi: 10.4161/cc.4.9.1997. [DOI] [PubMed] [Google Scholar]
- Andrews C. A., Vas A. C., Meier B., Gimenez-Abian J. F., Diaz-Martinez L. A., Green J., Erickson S. L., Vanderwaal K. E., Hsu W. S., Clarke D. J. A mitotic topoisomerase II checkpoint in budding yeast is required for genome stability but acts independently of Pds1/securin. Genes Dev. 2006;20:1162–1174. doi: 10.1101/gad.1367206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y., Arnaoutov A., Anan T., Dasso M. PIASy mediates SUMO-2 conjugation of Topoisomerase-II on mitotic chromosomes. EMBO J. 2005;24:2172–2182. doi: 10.1038/sj.emboj.7600700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y., Arnaoutov A., Dasso M. SUMO-2/3 regulates topoisomerase II in mitosis. J. Cell Biol. 2003;163:477–487. doi: 10.1083/jcb.200304088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachant J., Alcasabas A., Blat Y., Kleckner N., Elledge S. J. The SUMO-1 isopeptidase Smt4 is linked to centromeric cohesion through SUMO-1 modification of DNA topoisomerase II. Mol. Cell. 2002;9:1169–1182. doi: 10.1016/s1097-2765(02)00543-9. [DOI] [PubMed] [Google Scholar]
- Bachant J., Jessen S. R., Kavanaugh S. E., Fielding C. S. The yeast S phase checkpoint enables replicating chromosomes to bi-orient and restrain spindle extension during S phase distress. J. Cell Biol. 2005;168:999–1012. doi: 10.1083/jcb.200412076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachellier-Bassi S., Gadal O., Bourout G., Nehrbass U. Cell cycle-dependent kinetochore localization of condensin complex in Saccharomyces cerevisiae. J. Struct. Biol. 2008;162:248–259. doi: 10.1016/j.jsb.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Bhalla N., Biggins S., Murray A. W. Mutation of YCS4, a budding yeast condensin subunit, affects mitotic and nonmitotic chromosome behavior. Mol. Biol. Cell. 2002;13:632–645. doi: 10.1091/mbc.01-05-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S., Murray A. W. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck D. C., Bloom K. Pericentric chromatin is an elastic component of the mitotic spindle. Curr. Biol. 2007;17:741–748. doi: 10.1016/j.cub.2007.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D. Detection and correction of merotelic kinetochore orientationby Aurora B and its partners. Cell Cycle. 2007;6:1558–1564. doi: 10.4161/cc.6.13.4452. [DOI] [PubMed] [Google Scholar]
- Clarke D. J., Johnson R. T., Downes C. S. Topoisomerase II inhibition prevents anaphase chromatid segregation in mammalian cells independently of the generation of DNA strand breaks. J. Cell Sci. 1993;105:563–569. doi: 10.1242/jcs.105.2.563. [DOI] [PubMed] [Google Scholar]
- Clarke D. J., Vas A. C., Andrews C. A., Diaz-Martinez L. A., Gimenez-Abian J. F. Topoisomerase II checkpoints: universal mechanisms that regulate mitosis. Cell Cycle. 2006;5:1925–1928. doi: 10.4161/cc.5.17.3200. [DOI] [PubMed] [Google Scholar]
- Dawlaty M. M., Malureanu L., Jeganathan K. B., Kao E., Sustmann C., Tahk S., Shuai K., Grosschedl R., van Deursen J. M. Resolution of sister centromeres requires RanBP2-mediated SUMOylation of topoisomerase IIalpha. Cell. 2008;133:103–115. doi: 10.1016/j.cell.2008.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar H., Tanaka K., Nasmyth K., Tanaka T. U. Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature. 2004;428:93–97. doi: 10.1038/nature02328. [DOI] [PubMed] [Google Scholar]
- Diaz-Martinez L. A., Gimenez-Abian J. F., Azuma Y., Guacci V., Gimenez-Martin G., Lanier L. M., Clarke D. J. PIASgamma is required for faithful chromosome segregation in human cells. PLoS ONE. 2006;1:e53. doi: 10.1371/journal.pone.0000053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo S., Voelkel K., Sternglanz R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc. Natl. Acad. Sci. USA. 1984;81:2616–2620. doi: 10.1073/pnas.81.9.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes C. S., Mullinger A. M., Johnson R. T. Inhibitors of DNA topoisomerase II prevent chromatid separation in mammalian cells but do not prevent exit from mitosis. Proc. Natl. Acad. Sci. USA. 1991;88:8895–8899. doi: 10.1073/pnas.88.20.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert C. A., Gravdahl D. J., Megee P. C. The enhancement of pericentromeric cohesin association by conserved kinetochore components promotes high-fidelity chromosome segregation and is sensitive to microtubule-based tension. Genes Dev. 2007;21:278–291. doi: 10.1101/gad.1498707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvik B., Carson M., Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich D., Hirota T., Koch B., Peters J. M., Ellenberg J. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr. Biol. 2006;16:333–344. doi: 10.1016/j.cub.2005.12.040. [DOI] [PubMed] [Google Scholar]
- Gore J., Bryant Z., Nollmann M., Le M. U., Cozzarelli N. R., Bustamante C. DNA overwinds when stretched. Nature. 2006;442:836–839. doi: 10.1038/nature04974. [DOI] [PubMed] [Google Scholar]
- Goshima G., Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- Guacci V., Hogan E., Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J. Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Asthana S., Sorger P. K. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 2000;101:763–775. doi: 10.1016/s0092-8674(00)80888-0. [DOI] [PubMed] [Google Scholar]
- Hieter P., Mann C., Snyder M., Davis R. W. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985;40:381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- Holm C. Coming undone: how to untangle a chromosome. Cell. 1994;77:955–957. doi: 10.1016/0092-8674(94)90433-2. [DOI] [PubMed] [Google Scholar]
- Holm C., Goto T., Wang J. C., Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985;41:553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- Holm C., Stearns T., Botstein D. DNA topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Mol. Cell. Biol. 1989;9:159–168. doi: 10.1128/mcb.9.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E. M., Rachidi N., Morrice N., Hardwick K. G., Stark M. J. Ipl1p-dependent phosphorylation of Mad3p is required for the spindle checkpoint response to lack of tension at kinetochores. Genes Dev. 2007;21:1163–1168. doi: 10.1101/gad.431507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D., Hartwell L. H. The structure of sister minichromosome DNA before anaphase in Saccharomyces cerevisiae. Science. 1987;238:1713–1716. doi: 10.1126/science.3317838. [DOI] [PubMed] [Google Scholar]
- Li Y., Bachant J., Alcasabas A. A., Wang Y., Qin J., Elledge S. J. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 2002;16:183–197. doi: 10.1101/gad.959402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J. R. Structural and mechanical control of mitotic progression. Cold Spring Harb. Symp. Quant. Biol. 1991;56:613–619. doi: 10.1101/sqb.1991.056.01.070. [DOI] [PubMed] [Google Scholar]
- Megee P. C., Koshland D. A functional assay for centromere-associated sister chromatid cohesion. Science. 1999;285:254–257. doi: 10.1126/science.285.5425.254. [DOI] [PubMed] [Google Scholar]
- Mikhailov A., Cole R. W., Rieder C. L. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr. Biol. 2002;12:1797–1806. doi: 10.1016/s0960-9822(02)01226-5. [DOI] [PubMed] [Google Scholar]
- Murakami S., Yanagida M., Niwa O. A large circular minichromosome of Schizosaccharomyces pombe requires a high dose of type II DNA topoisomerase for its stabilization. Mol. Gen. Genet. 1995;246:671–679. doi: 10.1007/BF00290712. [DOI] [PubMed] [Google Scholar]
- Ocampo-Hafalla M. T., Katou Y., Shirahige K., Uhlmann F. Displacement and re-accumulation of centromeric cohesin during transient pre-anaphase centromere splitting. Chromosoma. 2007;116:531–544. doi: 10.1007/s00412-007-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira R. A., Coelho P. A., Sunkel C. E. The condensin I subunit Barren/CAP-H is essential for the structural integrity of centromeric heterochromatin during mitosis. Mol. Cell. Biol. 2005;25:8971–8984. doi: 10.1128/MCB.25.20.8971-8984.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. G., Maddox P. S., Salmon E. D., Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 2001;152:1255–1266. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky B. A., Biggins S. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 2005;15:486–493. doi: 10.1016/j.tcb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Pinsky B. A., Kung C., Shokat K. M., Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat. Cell Biol. 2006;8:78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- Pinsky B. A., Tatsutani S. Y., Collins K. A., Biggins S. An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev. Cell. 2003;5:735–745. doi: 10.1016/s1534-5807(03)00322-8. [DOI] [PubMed] [Google Scholar]
- Porter A. C., Farr C. J. Topoisomerase II: untangling its contribution at the centromere. Chromosome Res. 2004;12:569–583. doi: 10.1023/B:CHRO.0000036608.91085.d1. [DOI] [PubMed] [Google Scholar]
- Rattner J. B., Hendzel M. J., Furbee C. S., Muller M. T., Bazett-Jones D. P. Topoisomerase II alpha is associated with the mammalian centromere in a cell cycle- and species-specific manner and is required for proper centromere/kinetochore structure. J. Cell Biol. 1996;134:1097–1107. doi: 10.1083/jcb.134.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W. C. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Salceda J., Fernandez X., Roca J. Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA. EMBO J. 2006;25:2575–2583. doi: 10.1038/sj.emboj.7601142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandall S., Severin F., McLeod I. X., Yates J. R., 3rd, Oegema K., Hyman A., Desai A. A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell. 2006;127:1179–1191. doi: 10.1016/j.cell.2006.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B. Functional expression of the cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1987;7:2087–2096. doi: 10.1128/mcb.7.6.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamu C. E., Murray A. W. Sister chromatid separation in frog egg extracts requires DNA topoisomerase II activity during anaphase. J. Cell Biol. 1992;117:921–934. doi: 10.1083/jcb.117.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelby R. D., Hahn K. M., Sullivan K. F. Dynamic elastic behavior of alpha-satellite DNA domains visualized in situ in living human cells. J. Cell Biol. 1996;135:545–557. doi: 10.1083/jcb.135.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias D. A., Lacroix F. B., Andreassen P. R., Wilson L., Margolis R. L. Inhibition of DNA decatenation, but not DNA damage, arrests cells at metaphase. Mol. Cell. 2004;15:977–990. doi: 10.1016/j.molcel.2004.08.018. [DOI] [PubMed] [Google Scholar]
- Spell R. M., Holm C. Nature and distribution of chromosomal intertwinings in Saccharomyces cerevisiae. Mol. Cell. Biol. 1994;14:1465–1476. doi: 10.1128/mcb.14.2.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence J. M., Phua H. H., Mills W., Carpenter A. J., Porter A. C., Farr C. J. Depletion of topoisomerase IIalpha leads to shortening of the metaphase interkinetochore distance and abnormal persistence of PICH-coated anaphase threads. J. Cell Sci. 2007;120:3952–3964. doi: 10.1242/jcs.013730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer F., Gerring S. L., Connelly C., Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990;124:237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stray J. E., Crisona N. J., Belotserkovskii B. P., Lindsley J. E., Cozzarelli N. R. The Saccharomyces cerevisiae Smc2/4 condensin compacts DNA into (+) chiral structures without net supercoiling. J. Biol. Chem. 2005;280:34723–34734. doi: 10.1074/jbc.M506589200. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Strunnikov A. In vivo modeling of polysumoylation uncovers targeting of Topoisomerase II to the nucleolus via optimal level of SUMO modification. Chromosoma. 2007;117:189–198. doi: 10.1007/s00412-007-0137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Yong-Gonzalez V., Kikuchi Y., Strunnikov A. SIZ1/SIZ2 control of chromosome transmission fidelity is mediated by the sumoylation of topoisomerase II. Genetics. 2006;172:783–794. doi: 10.1534/genetics.105.047167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Cosma M. P., Wirth K., Nasmyth K. Identification of cohesin association sites at centromeres and along chromosome arms. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Fuchs J., Loidl J., Nasmyth K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2000;2:492–499. doi: 10.1038/35019529. [DOI] [PubMed] [Google Scholar]
- Tanaka T. U., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M. J., Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Toyoda Y., Yanagida M. Coordinated requirements of human topo II and cohesin for metaphase centromere alignment under Mad2-dependent spindle checkpoint surveillance. Mol. Biol. Cell. 2006;17:2287–2302. doi: 10.1091/mbc.E05-11-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli P., Morrison C., Dodson H., Sonoda E., Takeda S., Earnshaw W. C. Analysis of Scc1-deficient cells defines a key metaphase role of vertebrate cohesin in linking sister kinetochores. EMBO Rep. 2004;5:167–171. doi: 10.1038/sj.embor.7400077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vas A. C., Andrews C. A., Kirkland Matesky K., Clarke D. J. In vivo analysis of chromosome condensation in Saccharomyces cerevisiae. Mol. Biol. Cell. 2007;18:557–568. doi: 10.1091/mbc.E06-05-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A., Brachat A., Pohlmann R., Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in. Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Yeh E., Haase J., Paliulis L. V., Joglekar A., Bond L., Bouck D., Salmon E. D., Bloom K. S. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr. Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong-Gonzalez V., Wang B. D., Butylin P., Ouspenski I., Strunnikov A. Condensin function at centromere chromatin facilitates proper kinetochore tension and ensures correct mitotic segregation of sister chromatids. Genes Cells. 2007;12:1075–1090. doi: 10.1111/j.1365-2443.2007.01109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.