Abstract
The Smc5-Smc6 holocomplex plays essential but largely enigmatic roles in chromosome segregation, and facilitates DNA repair. The Smc5-Smc6 complex contains six conserved non-SMC subunits. One of these, Nse1, contains a RING-like motif that often confers ubiquitin E3 ligase activity. We have functionally characterized the Nse1 RING-like motif, to determine its contribution to the chromosome segregation and DNA repair roles of Smc5-Smc6. Strikingly, whereas a full deletion of nse1 is lethal, the Nse1 RING-like motif is not essential for cellular viability. However, Nse1 RING mutant cells are hypersensitive to a broad spectrum of genotoxic stresses, indicating that the Nse1 RING motif promotes DNA repair functions of Smc5-Smc6. We tested the ability of both human and yeast Nse1 to mediate ubiquitin E3 ligase activity in vitro and found no detectable activity associated with full-length Nse1 or the isolated RING domains. Interestingly, however, the Nse1 RING-like domain is required for normal Nse1-Nse3-Nse4 trimer formation in vitro and for damage-induced recruitment of Nse4 and Smc5 to subnuclear foci in vivo. Thus, we propose that the Nse1 RING-like motif is a protein–protein interaction domain required for Smc5-Smc6 holocomplex integrity and recruitment to, or retention at, DNA lesions.
INTRODUCTION
Structural maintenance of chromosomes complexes (SMCs) are essential for genome integrity during the unchallenged cell cycle and play key roles in DNA repair after genotoxic stress (Losada and Hirano, 2005; Nasmyth and Haering, 2005). In eukaryotes, this evolutionary conserved family consists of six members, pairs of which heterodimerize to form the core of high-molecular-weight complexes: cohesin (Smc1-Smc3), condensin (Smc2-Smc4), and Smc5-Smc6 (Losada and Hirano, 2005; Nasmyth and Haering, 2005). Although cohesin and condensin are well characterized, the mechanisms by which Smc5-Smc6 maintains chromosomal integrity during vegetative growth and DNA repair remain largely undefined (Losada and Hirano, 2005; Nasmyth and Haering, 2005).
In yeast, the Smc5-Smc6 heterodimer associates with six non-SMC elements (Nse1–6; Fousteri and Lehmann, 2000; Hazbun et al., 2003; McDonald et al., 2003; Morikawa et al., 2004; Pebernard et al., 2004; Sergeant et al., 2005; Zhao and Blobel, 2005; Pebernard et al., 2006). In humans, Nse1 to Nse4 proteins are clearly conserved, whereas homologues of Nse5 and Nse6 remain unidentified (Hu et al., 2005; Potts and Yu, 2005; Taylor et al., 2008). Hypomorphic mutants of fission yeast Smc6 and Nse1 to Nse4 display severe DNA segregation defects and a terminal elongated phenotype when shifted to restrictive temperature (Fousteri and Lehmann, 2000; McDonald et al., 2003; Pebernard et al., 2004). Budding yeast Smc5-Smc6 binds to repetitive genomic sequences such as rDNA and promotes its faithful segregation at mitosis (Torres-Rosell et al., 2005b). It also localizes to centromeres in a cell cycle–dependent manner, with peak binding at G2/M phase, indicating a potential role in centromere and kinetochore function (Lindroos et al., 2006).
Depending on the nature of the DNA lesion, Smc5-Smc6 appears to play both early and late roles in homologous recombination repair (HRR). In response to ionizing radiation (IR) or enzymatically generated site-specific DNA double-strand breaks (DSBs), Smc5-Smc6 apparently promotes intersister HRR of DSBs, perhaps through local recruitment of the cohesin complex (e.g., Lindroos et al., 2006; Potts et al., 2006). However, after replicative stress and UV-induced DNA damage, Smc5-Smc6 is required either to suppress HRR or to resolve late HRR intermediates that otherwise cause chromosome missegregation at mitosis (Lehmann et al., 1995; Ampatzidou et al., 2006; Miyabe et al., 2006; Pebernard et al., 2006).
The Nse1 and Nse2 subunits of the Smc5-Smc6 complex contain RING finger-like motifs, indicating that these subunits might have catalytic activities (Fujioka et al., 2002; McDonald et al., 2003). The RING finger is a small domain containing eight conserved metal-binding residues (either cysteines or histidines), which coordinate two zinc atoms in a cross-brace conformation (Borden, 2000). Nse2 (Mms21) contains a RING variant called an SP-RING (Hochstrasser, 2001) for which E3 SUMO ligase activity has been confirmed, both in vitro and in vivo (Andrews et al., 2005; Potts and Yu, 2005; Zhao and Blobel, 2005). Although Nse2-dependent E3 SUMO ligase activity is not essential for cellular viability, it promotes key DNA repair functions of Smc5-Smc6 (Andrews et al., 2005; Potts and Yu, 2005; Zhao and Blobel, 2005). Interestingly, Nse2-dependent sumoylation of telomere factors positively modulates HRR at the telomeres of cancer cells that lack telomerase and utilize the HRR-dependent alternative lengthening of telomeres (ALT) pathway (Potts and Yu, 2007).
Nse1 contains a C-terminal RING-like motif, indicating that Nse1 could mediate ubiquitin E3 ligase activity (McDonald et al., 2003). Canonical RING motifs have been categorized as RING-HC and RING-H2 families according to their cysteine/histidine content (C3HC4 and C3H2C3, respectively), also taking into account conservation of spacing between these residues (Saurin et al., 1996; Lorick et al., 1999). Many RING variants with different cysteine/histidine arrangements and spacing but retaining ubiquitin E3 ligase activity have been described, e.g., vRING (C4HC3; Hewitt et al., 2002), RING-C2 (C8, Albert et al., 2002; Aravind et al., 2003; Dodd et al., 2004; Hassink et al., 2005; Stone et al., 2005). Importantly, several proteins that contain RING-type motifs do not support detectable ubiquitin E3 ligase activity. For example, by itself the RING finger protein BMI-1 is not an E3 ligase (Wei et al., 2006). However, when bound to its heterodimeric RING-containing protein partner RING1b, the complex displays robust ubiquitin ligase activity toward histone H2A (Buchwald et al., 2006). Similarly, BRCA1 and BARD1 form a heterodimeric RING-RING heterodimer, whose ubiquitin ligase activity is far greater than the sum of the individual activities (Hashizume et al., 2001). The recently identified SUMO-targeted ubiquitin ligase (STUbL) complexes of yeast are also heterodimeric RING-RING complexes, in which only one subunit is active in ubiquitination assays (Prudden et al., 2007; Perry et al., 2008). Thus, the presence of a RING-like signature is not always predictive of ubiquitin E3 ligase activity.
Here, we functionally characterize the fission yeast Nse1 RING-like motif, analyzing its role in the essential and DNA repair functions of Smc5-Smc6. The Nse1 RING-like motif is distinct from all previously described RING variants, and forms an evolutionary subclass found in Nse1 homologues across species (hereafter referred to as RING in Nse1 homologues; NH-RING). Human and fission yeast Nse1 expressed in and purified from bacteria do not support ubiquitin E3 ligase activity in vitro. To investigate the role of the NH-RING in vivo, we generated a series of Nse1 mutants and demonstrated that the NH-RING is not essential for vegetative growth, but is critical for Smc5-Smc6 DNA repair functions. In addition, we determined that the NH-RING constitutes a protein–protein interaction motif required for stable formation of the Nse1-Nse3-Nse4 heterotrimer. Finally, we find that the NH-RING is required for the formation of Nse4 subnuclear foci after DNA damage induced by methyl methanesulfonate (MMS). Thus, the NH-RING contributes to the DNA damage response structurally, via Nse1-Nse3-Nse4 trimer stabilization and is likely required for Smc5-Smc6 holocomplex recruitment/accumulation at sites of DNA damage.
MATERIALS AND METHODS
Strains and Fission Yeast Methods
Standard fission yeast culture and genetics methods were used as described previously (Moreno et al., 1991). All strains are ura4-D18 leu1-32 unless otherwise stated: PR100, h+; PR109, h−; NBY1395, h+ nse1C197A:flag:kanMx6; NBY1262, h− nse1C199A:myc:kanMx6; NBY1265, h− nse1C197A/C199A:myc:kanMx6; NBY1396, h+ nse1C219A:flag:kanMx6; NBY1397, h+ nse1ΔRING:flag:kanMx6; NBY1512, gar1:mRFP:KanMx6 nse1-1:myc:KanMx6; NBY1513, gar1:mRFP:KanMx6 nse1ΔRING:flag:KanMx6; NBY128, mus81::KanMx6, h+; NBY1941, mus81::KanMx6 nse1ΔRING:flag:KanMx6; NBY202, rqh1::Ura4+ h+; NBY527K, nse2-1:myc:KanMx6 h+; NBY952, rhp51::Ura4+; NBY1421, rhp51::Ura4+ nse1ΔRING:flag:KanMx6; NBY1471, nse1wt:flag:KanMx6 nse4:GFP:KanMx6; NBY1472, nse1ΔRING:flag:KanMx6 nse4:GFP:KanMx;NBY480, Smc5-GFP:KanMx6 h+; NBY1975, smc5-GFP:KanMx6 nse1ΔRING:flag:KanMx6. For spot assays, cells were grown in YES medium at 25°C until they reached exponential phase, before being spotted on YES plates supplemented with the indicated drugs. Fivefold dilutions from a starting 200 cells/μl concentration were performed before spotting. Plates were incubated at the indicated temperature for 2–3 d before scanning. In kill-curve assays, 500 cells of each strain were plated on YES and irradiated with the indicated UV dose using a UV Stratalinker-18000 (Stratagene, La Jolla, CA). Results are represented as the percentage of surviving colonies on UV-treated plates relative to the full amount of colonies on nonirradiated plates. Each condition was tested in triplicate.
Homology Modeling
Searching with fission yeast Nse1 sequence (Uniprot ID: Q53EK2) for homologues, using PSI-BLAST (Altschul et al., 1997) and SAM-T06 (Karplus et al., 1998) revealed significant sequence similarity to the Homo sapiens Nse1 homologue, whose RING domain structure has been experimentally determined by nuclear magnetic resonance in structural genomics studies (2CT0.pdb). For comparative modeling, the sequence alignment of fission yeast Nse1 to its structural homologue was optimized by iterative multiple sequence alignment methods (Burke et al., 1999), meta-server fold recognition (Kurowski and Bujnicki, 2003; Wallner et al., 2007) and manual adjustment. These sequence alignments were performed using the BioEdit (Ibis Therapeutics, Carlsbad, CA) and CLUSTALW alignment software. A fission yeast Nse1 comparative model was generated using Modeler 8v2 (Sali and Blundell, 1993). Model validation steps included the ADIT Validation Server at the protein databank (http://www.rcsb.org/pdb/index.html).
Generation of Endogenous Nse1 NH-RING Mutants
The full Nse1:myc:kanMx6 cassette was amplified from its endogenous genomic locus and cloned into a TOPO vector (Invitrogen, Carlsbad, CA) to give pSPG-135. Mutations were generated within Nse1 open reading frame (ORF) previously cloned into a TOPO vector by classical PCR or by site-directed mutagenesis using the Quick-Change XLII kit (Stratagene). Primer sequences used in these clonings are available upon request. The Nse1 ORF in pSGP-135 was replaced by mutation-containing ORFs between PvuII and XhoI sites, and resulting mutant Nse1:myc:kanMx6 cassettes were used for gene replacement of endogenous Nse1 genomic locus after transformation of wild-type Schizosaccharomyces pombe PR100 (h+) or PR109 (h−) cells according to Bahler et al. (1998). Transformants were selected on YES+G418, and sequencing confirmed the mutations.
Expression of Recombinant S. pombe Proteins and Interaction Assays in Insect Cells
Cloning and Sf9 expression techniques for Nse1, Nse3, and Nse4 were described elsewhere (Pebernard et al., 2006). Nse1 point and deletion RING mutant ORFs obtained as above were subcloned into the pSGP-75 vector (pFASTBAC-GST-PreScission-Nse1) to give pSGP-124, 138–142, and 158 (pFASTBAC-GST-PreScission-Nse1C216A, C197A, C199A, C197A/C199A, C219A, C216S/C219A, and ΔRING, respectively). Presence of mutations was confirmed by sequencing, and baculoviruses expressing these recombinant proteins were produced as previously described (Pebernard et al., 2006). Depending on the experiment performed, the baculovirus combinations used to coinfect Sf9 cells are described in the figure legends. Purification method consisted of a glutathione S-transferase (GST) pulldown followed by elution by PreScission protease cleavage (GE Healthcare, Waukesha, WI), and is described in details in Pebernard et al. (2006).
In Vitro Ubiquitination Assays
Standard reaction mixture (20 μl) contained 50 mM Tris-HCl, pH 7.5, 4 mM ATP, 10 mM MgCl2, 0.2 mM CaCl2, 1 mM DTT, 1–2 mM ubiquitin (bovine erythrocytes, recombinant, Sigma-Aldrich, St. Louis, MO) or GST-ubiquitin, 100 nM E1 ubiquitin-activating enzyme (yeast, Boston Biochem, Cambridge, MA), 1 μM of the indicated E2-conjugating enzyme (UbcH2, 6xHis-UbcH3, UbcH5a, UbcH5b, UbcH5c, UbcH8, UbcH9, 6xHis-UbcH10, UbcH13/Uev1a, human recombinant, Boston Biochem), and ∼1–2 μg bacteria- or SF9-produced complex containing the indicated proteins as ubiquitin E3 ligases. Reactions were incubated for 30 min at 30°C, stopped by adding 20 μl 2× SDS sample buffer, and subjected to SDS/PAGE followed by immunoblotting using the indicated antibodies.
Immunoblotting
Proteins denatured in 2× SDS sample buffer (1% SDS, 5% glycerol, 0.1% bromophenol blue, 40 mM Tris-HCl, pH 6.8, 1/10 [vol/vol] β-mercaptoethanol) were resolved on 8–12% SDS/PAGE gels, except for in vitro ubiquitination assays where 4–20% Tris-glycine gels (Invitrogen) were used. Gels were transferred on Immobilon-P membrane (Millipore, Billerica, MA), blocked in 5% milk in Tris-saline buffer with 0.3% Tween-20, and probed with mouse mAb raised against hemagglutinin (H; 12CA5; BABCo, Richmond, CA), FLAG (anti-FLAG M2; Sigma-Aldrich, Saint Louis, MO), or myc (9E10; BABCo) epitopes, or against full-length ubiquitin (UBI-1; Zymed, San Francisco, CA). The peroxidase antiperoxidase reagent (PAP; Sigma-Aldrich) was used for detection of TAP-tagged proteins. After amplification by probing with a secondary horseradish peroxidase–conjugated rabbit anti-mouse antibody, the protein signal was revealed using ECL reagent (Pierce, Rockford, IL) on autoradiography films (Genesee, San Diego, CA). When indicated, Western blots were stripped for reprobing using stripping buffer (0.2 M glycine, pH 2.5, 1% SDS).
Yeast Two-Hybrid Assays
Yeast two-hybrid assays were performed as described previously (Prudden et al., 2007). Briefly, the indicated cDNAs were amplified by PCR and cloned in frame with the Gal4-activation domain (AD), in a yeast expression vector (pAS404). Resulting vectors were integrated at the TRP locus to give stable recombinant strains, which were then transformed with plasmids containing either the Gal4-binding domain (DBD) alone or fused in frame with cDNAs as indicated. Transformed strains were selected on dextrose minus Trp and Leu (D-WL), grown at 32°C, and spotted at an OD = 0.6 onto the indicated media. Specific interactions were exposed by spotting the strains on media without His and complemented with 10–20 mM 3-amino-1,2,4-triazole (3-AT).
Microscopy
Cells were grown in liquid EMM medium (Qbiogene, Carlsbad, CA) supplemented with leucine, uracil, arginine, and histidine (LUAH) and sterilized by filtration at 25 or 32°C for at least 36 h before treatment. For MMS treatment, cells were then diluted to OD = 0.6, treated with 0.03% MMS (vol/vol), and grown for 6h at 25°C (for optimal fluorescence). Cells were then pelleted and kept on ice in the same medium for live cell imaging. Pictures were taken on a Nikon Eclipse E800 microscope (Melville, NY) using a Photometrics Quantix CDD camera (Woburn, MA) and processed with the IP lab spectrum P (BD Biosciences, Rockville, MD) and Adobe Photoshop CS software (San Jose, CA). A minimum of 300 cells were counted in two to three independent experiments, but for figure purposes only one representative experiment is shown.
RESULTS
Sequence Analysis and Comparative Modeling of Fission Yeast Nse1 RING-like Domain
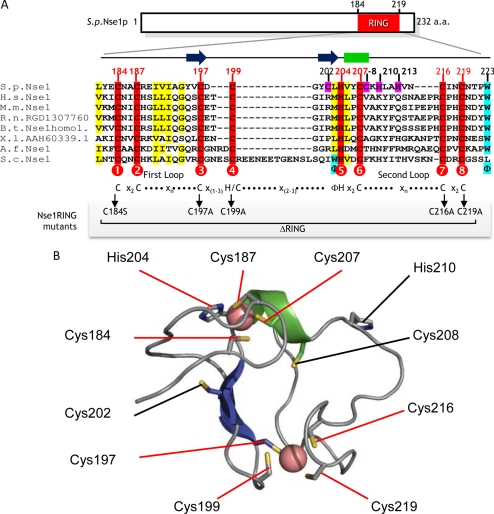
Nse1 is a nuclear ∼27-kDa protein conserved from yeast to humans that contains a RING-like motif at its C-terminus, which we call an NH-RING (McDonald et al., 2003). RING motifs contain a series of eight cysteine or histidine residues, with variable spacing, which act as ligands to chelate two Zn2+ ions in a cross-brace conformation (Borden, 2000). Zn2+ binding is essential for both folding and function of RING domains (Borden, 2000). Conventional RING motifs have a C3HC4 consensus, whereas the closely related PHD motif (plant homology domain) contains a C4HC3 consensus (Borden and Freemont, 1996). Interestingly, the Nse1 NH-RING has a “PHD-like” consensus (C4HC3) but lacks some key features of a bona fide PHD domain, including a conserved aromatic residue in the second loop (Figure 1A; Aravind et al., 2003). The solution structure of the human Nse1 NH-RING domain has been determined, confirming that the sequence adopts a RING-like cross-brace structure (protein databank code: 2CT0.pdb). Mammalian Nse1 sequence homologues readily align to human Nse1 in our structure-based sequence alignment (Figure 1A): RING domains usually respect a C1(X2)C2(X9–39)C3(X1–3)H/C4(X2–3)C5(X2)C6(X4–48)C7(X2)C8 consensus, and the two Zn2+ atoms are coordinated by two groups of ligands in positions 1, 2, 5, and 6 and in positions 3, 4, 7, 8, respectively. Ligand spacing in the mammalian and Xenopus NH-RINGs, as well as those in the human Nse1 structure, conform to this consensus (Figure 1A). Interestingly, the budding yeast NH-RING contains a much larger insertion between C4 and H5, in addition to its general sequence divergence from mammalian sequences. The fission yeast NH-RING (184–219aa) also shows sequence divergence from mammalian sequences and several additional cysteine and histidine residues occur in the sequence at Zn2+ binding regions (Figure 1A, pink boxes). Therefore, we conducted comparative modeling of the fission yeast NH-RING structure, using the human NH-RING structure as a template, to aid model-based mutagenesis studies. For example, in the fission yeast NH-RING residues C197 and C199 or C202 are potential zinc ligands. Comparative modeling strongly indicates that residue C197 is the C3 and residue C199 the C4. This region differs with a one residue spacer, rather than a two-amino acid spacing observed in the mammalian homologues and in most C4HC3 RING sequences (Aravind et al., 2003; Dodd et al., 2004; Hassink et al., 2005), but still conforms to a more general consensus sequence of C3(X1–3)H/C4. Moreover, a model with C202 as the C4 chelating residue would disrupt both the conserved central β-sheet of the human NH-RING structure and the second Zn2+ binding site. The fission yeast NH-RING comparative model and its alignment to homologues are depicted in Figure 1. Based on this model, mutations of critical NH-RING cysteines and a full deletion of the region between C1 and C8 (ΔRING) were introduced into Nse1 for in vivo and in vitro analysis.
Figure 1.
Structural analysis of the Nse1 RING-like motif. (A) A multiple sequence alignment between fission yeast Nse1 (SPCC550.05), human NSMCE1 (NP_659547), mouse NSMCE1 (AAH49558), rat RGD1307760, bovine NSMCE1 (NP_001030483), Xenopus MGC68739 (AAH60339), Aspergillus Nse1 (XP_748562), and budding yeast YDR288W (NP_013107) is depicted. Conserved cysteines are shaded in red. Alternate potential Zn2+ ligands of fission yeast Nse1 NH-RING are shaded in pink. Other conserved residues of interest are shaded in yellow and blue. Numbered circles indicate the eight predicted Zn2+ ligand positions in the interspecies Nse1 NH-RING consensus. The C4HC3 RING consensus, with expected spacing, is shown below this alignment, as well as the two loop positions. A shaded box below indicates the positions of the ligands mutated in this study, as well as the region deleted in the ΔRING mutant. Φ, conserved hydrophobic residue. (B) The fission yeast Nse1 homology model. Comparative modeling studies based on human Nse1 structure (2CT0.pdb) suggest that part of the fission yeast Nse1 sequence forms a RING-like structure, comprised of a central β-sheet (two strands, in blue), a short α-helix (in green) and two binding sites for Zn2+ ions (pink spheres). The positions of the central β-sheets and α-helix are also represented on the alignment in A for better understanding. The Cysteine (Cys) residues and a Histidine (His) residue that chelate Zn2+ are highlighted with red line labels. Cys202, Cys208, and His210 (highlighted with black line labels) are present in the structure, but sequence analysis and modeling studies indicate that they are not situated at the Zn2+ coordination sites. This model includes Nse1 residues Leu181 to Ile229.
Nse1 NH-RING Mutations Uncouple the Essential and DNA Repair Roles of Smc5-Smc6
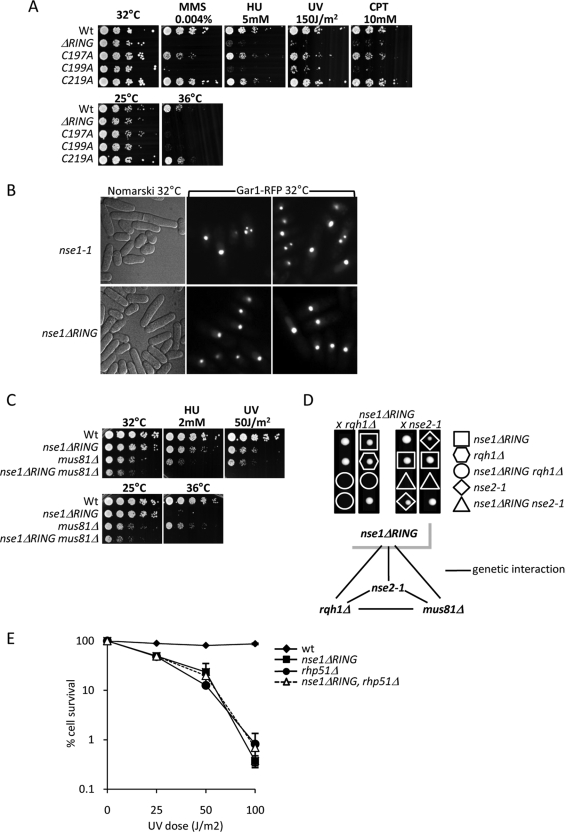
Fission yeast lacking Nse1 (nse1Δ) form microcolonies that cease growing and display extreme cell elongation after a few divisions due to activation of the DNA damage checkpoint (McDonald et al., 2003). As observed for other Smc5-Smc6 subunits, cells hypomorphic for Nse1 display aberrant chromosome segregation including “cut” phenotypes and DNA stretched along the mitotic spindle axis (Lehmann et al., 1995; Boddy et al., 2003; McDonald et al., 2003; Morikawa et al., 2004; Pebernard et al., 2004; Sergeant et al., 2005). To determine the contribution of the NH-RING to Smc5-Smc6 functions, we replaced the endogenous nse1+ locus with an nse1-myc:KanMx6 cassette that carried either point mutations or a full deletion of the NH-RING. We generated and characterized strains with NH-RING point mutations C197A, C199A, C197A/C199A, and C219A, and an in-frame RING deletion (ΔRING; Figure 1A). The ΔRING, C197A, C199A, and C197A/C199A mutants were mildly temperature sensitive but grew normally at 25°C (Figure 2A and data not shown). At 32°C the NH-RING mutants grew with almost wild-type kinetics but displayed noticeable heterogeneity in cell length, indicating activation of the DNA damage checkpoint (Figure 2A and data not shown). The C219A mutant was not temperature sensitive (Figure 2A). Overall, this analysis demonstrates that the NH-RING is not required for the essential roles of the Smc5-Smc6 complex during the unchallenged cell cycle.
Figure 2.
The Nse1 NH-RING domain is required for DNA repair. (A) Spot assays showing temperature and genotoxic stress sensitivities of the indicated Nse1 NH-RING mutants. Serial dilutions of the indicated strains were spotted onto rich media with or without the indicated doses of DNA-damaging agents. Plates were grown at the indicated temperature for 3–5 d. (B) nse1ΔRING cells properly segregate their DNA. Live cell microscopy showing the RFP-tagged nucleolar marker Gar1 segregation in nse1-1 and nse1ΔRING cells. Phase constrast (DIC or Nomarski) shows these mutants cell length when grown at 32°C. (C) nse1ΔRING and mus81Δ are synthetically sick mutations. Serial dilutions of the indicated strains were spotted as in A. Combined nse1ΔRING and mus81Δ mutations show additive growth defects and hypersensitivity to genotoxic stress when compared with single mutants. (D) Genetic interactions for nse1ΔRING. Top, tetrad dissections showing synthetic lethality of combined nse1ΔRING and rqh1Δ or nse2-1 mutations. Genetic backgrounds of resulting strains are represented by the various shapes circling each colony. Bottom, summary of genetic interactions (synthetic sickness or lethality) between nse1ΔRING and the indicated alleles. (E) rhp51Δ and nse1ΔRING display similar sensitivities to UV and are not additive. Kill curves showing the percentage of cell survival for each indicated strain, which equals the number of colony forming after exposure to increasing UV doses versus the number of colonies forming on untreated plates.
In addition to its essential role(s) in chromosome segregation, Smc5-Smc6 plays key roles in the cellular response to DNA damage (Lehmann et al., 1995; McDonald et al., 2003; Pebernard et al., 2004, 2006; Andrews et al., 2005; Sergeant et al., 2005). Therefore, we determined the contribution of the NH-RING to cellular survival after exposure to DNA damaging agents (Figure 2A). The Nse1 ΔRING mutant was hypersensitive to low concentrations of MMS, HU, and high doses of UV (Figure 2A). It was also mildly sensitive to high concentrations of the topoisomerase I poison camptothecin (CPT, Figure 2A). Cells with the Nse1 C199A point mutation exhibited similar sensitivities to these genotoxic agents as Nse1 ΔRING cells (Figure 2A). Although the C197A mutant is temperature sensitive, it is only mildly sensitive to this array of DNA damaging agents (Figure 2A). Finally, Nse1 C219A is neither temperature nor damage sensitive (Figure 2A). Although most nullifying mutations in RING motifs comprise simultaneous loss of two zinc ligands, we mutated each ligand singly to rigorously test their individual contributions to NH-RING function. Thus, observed variation in phenotypic severity between the single NH-RING point mutations is anticipated. Where other local nonzinc coordinating intradomain contacts exist, loss of these single zinc ligands will be better tolerated in the NH-RING structure. Indeed, the RING-related SP-RING and U-box domains adopt the RING fold, but lack one or all of the zinc coordinating sites, respectively, which are replaced by hydrogen-bonding networks (see Ohi et al., 2003 and protein databank code: 2yu4.pdb). On the basis of these data, we propose that the Nse1 NH-RING contributes to the DNA repair functions of the Smc5-Smc6 holocomplex.
We also compared nucleolar segregation phenotypes of the Nse1 ΔRING versus hypomorphic nse1-1 mutant cells at 32°C (McDonald et al., 2003). In nse1-1 mutant cells, aberrant chromatin structures accumulate at elevated growth temperatures (McDonald et al., 2003). Following the red fluorescent protein (RFP)-tagged nucleolar marker Gar1 by fluorescence microscopy, we discovered that nse1-1 nucleoli segregated aberrantly at 32°C (Figure 2B), which is reminiscent of the rDNA segregation defects observed in Saccharomyces cerevisiae SMC5-SMC6 hypomorphic mutants (Torres-Rosell et al., 2005a,b). Although heterogeneous in length (Figure 2B, left panels), Nse1 ΔRING cells did not display nucleolar segregation defects (Figure 2B, right panels). This result clearly differentiates the Nse1 ΔRING mutant phenotype from other hypomorphic mutations in Nse1 and further indicates DNA repair-specific roles for the Nse1 NH-RING.
To further characterize the DNA repair pathways facilitated by the Nse1 NH-RING, we tested for genetic interactions between Nse1 ΔRING and key DNA repair factors. The Nse1 ΔRING mutation was synthetic lethal in combination with a deletion of Rqh1 (human BLM) and synthetic sick when combined with a deletion of the Holliday junction endonuclease Mus81 (Figure 2, C and D). These genetic interactions indicate a role for the Nse1 NH-RING in stabilizing and/or processing stalled and damaged replication forks, a function ascribed to the Smc5-Smc6 holocomplex (Pebernard et al., 2004, 2006; Branzei et al., 2006; Lindroos et al., 2006; Murray and Carr, 2008). Defects in Smc5-Smc6 function cause toxic DNA structures to accumulate after UV irradiation in a manner dependent on the HRR machinery (Branzei et al., 2006; Pebernard et al., 2006). To test whether such structures contribute to the UV sensitivity of Nse1 ΔRING cells, we analyzed the genetic interaction between a deletion of the HRR protein Rhp51 (RAD51) and Nse1 ΔRING (Figure 2E). UV sensitivities were comparable for Nse1 ΔRING and rhp51Δ single mutant cells and Nse1 ΔRING mutant UV sensitivity was not additive with that of rhp51Δ, indicating that Nse1 acts in an Rhp51-dependent repair pathway (Figure 2E). Interestingly, the UV sensitivity of cells lacking the Smc5-Smc6 subunit Nse6 (nse6Δ) is due to pathological Rhp51-dependent recombination (Pebernard et al., 2006).
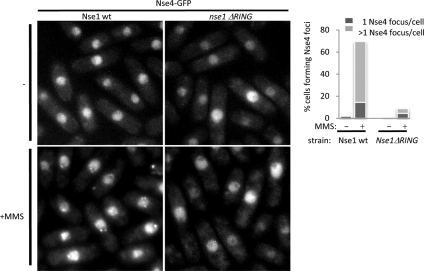
Formation of MMS-induced Nse4 Foci Is Nse1 NH-RING-dependent
We observe that GFP-tagged subunits of the Smc5-Smc6 complex, including Nse1, Nse3, Nse4, and Smc5, form multiple subnuclear foci upon MMS treatment (Figure 3 and our unpublished observations). Having determined that the Nse1 NH-RING domain is crucial for the Smc5-Smc6–mediated response to DNA damage, we asked whether Nse1 ΔRING cells would be defective in MMS-induced Nse4-GFP focus formation. On MMS treatment, Nse4-GFP localized to multiple foci in wild-type but not Nse1 ΔRING cells (Figure 3). A similar effect on Smc5-GFP focus formation was observed, although Smc5-GFP focus formation is more difficult to visualize because of higher background fluorescence in the nucleus (Supplementary Figure S1). Because Nse3 tagged with GFP is hypomorphic, it was not possible to combine Nse3-GFP with the Nse1 ΔRING mutation (our unpublished observations). Overall, these data indicate that the Nse1 NH-RING promotes the localization or retention of Nse4 and thus, likely the Smc5-Smc6 holocomplex, at DNA damage sites upon genotoxic stress.
Figure 3.
The Nse1 NH-RING domain is required for damage-induced Smc5-Smc6 localization. Left, live cell microscopy monitoring endogenously tagged Nse4-GFP localization in untreated (−) or MMS-treated (+MMS) at 25°C, in wild-type or nse1ΔRING backgrounds. Right, percentage of cells showing one or more than one Nse4-GFP focus on the left panels were scored.
Assessment of the Nse1 NH-RING as an Ubiquitin E3 Ligase
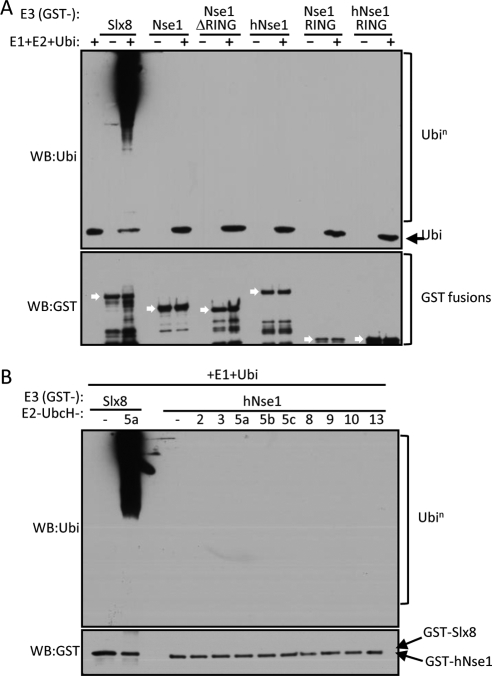
Based on the presence of a RING-like domain, close relatives of which often catalyze ubiquitination, we proposed that Nse1 might support ubiquitin E3 ligase activity (McDonald et al., 2003). Therefore, we tested this possibility using in vitro assays that are standard for demonstrating such activity. Recombinant full-length GST-Nse1 fusion proteins were expressed in and purified from bacteria. A reaction using the bona fide Slx8 ubiquitin E3 ligase (GST-Slx8) was used as a positive control for our experimental conditions (Figure 4A; Prudden et al., 2007). We assayed both fission yeast and human Nse1, because the human NH-RING is more related to a canonical RING domain (Figure 1). In addition to full-length proteins, we assayed just the NH-RING domains of both human and fission yeast Nse1, which often reveals ubiquitin ligase activity in bona fide E3 proteins (Figure 4A). The control reaction using GST-Slx8 as E3 ligase produced polyubiquitin chains in a very efficient manner; however, none of the GST-Nse1 fusions catalyzed polyubiquitin chain formation under the same reaction conditions (Figure 4A). This experiment was performed in the presence of the potent and processive E2-conjugating enzyme UbcH5a, which nonetheless might not be a cognate E2 for Nse1. Therefore, we tested the E3 activity of human GST-Nse1 in the presence of a broad panel of human E2-conjugating enzymes (Figure 4B). Again, Nse1 did not promote ubiquitination in the presence of any E2 tested, indicating that Nse1 NH-RING might not be an ubiquitin E3 ligase (see Discussion).
Figure 4.
Human and fission yeast Nse1 proteins and their NH-RINGs do not mediate ubiquitin E3 ligase activity in vitro. (A) In vitro ubiquitination assay testing GST fusions of fission yeast or human (h) Nse1 full-length, ΔRING, or RING-only constructs, as putative ubiquitin E3 ligases. GST-Slx8 is used as a positive control for E3-mediated polyubiquitin chain formation. WB, Western blot; Ubi, ubiquitin. White arrows in the bottom panel show the position of each GST fusion used. (B) Human Nse1 GST fusion was tested as ubiquitin E3 ligase with a panel of E2-conjugating enzymes. A reaction containing GST-Slx8 and the E2 enzyme UbcH5a was used as a positive control.
We considered that because Nse1 is normally in complex with Nse3 and Nse4, these interactions might affect Nse1 ubiquitin E3 ligase activity. Using Nse1-Nse3 dimers or Nse1-Nse3-Nse4 trimers purified form Sf9 insect cells, we were able to detect weak ubiquitin E3 ligase activity associated with these complexes in vitro (Supplementary Figure S2). This activity was detected using UbcH5 E2 proteins, which had failed to promote ubiquitination with GST-Nse1 (Supplementary Figure 2C). Notably, the E3 activity detected was independent of the Nse1 NH-RING domain, as a dimer containing Nse3 and Nse1 ΔRING or Nse1 NH-RING point mutants still catalyzed polyubiquitin chain formation (Supplementary Figure 2D). Furthermore, no activity was detected for Nse1-Nse3 dimers coexpressed in bacteria (Supplementary Figure 2E). Hence, we conclude that the Nse1 NH-RING does not support in vitro ubiquitin E3 ligase activity. The slight NH-RING-independent activity detected using Sf9-produced complexes is likely due either to a contaminant activity pulled down with the complex or to the presence of Nse3 (see Discussion). Future studies centered on Nse3 will help clarify this issue.
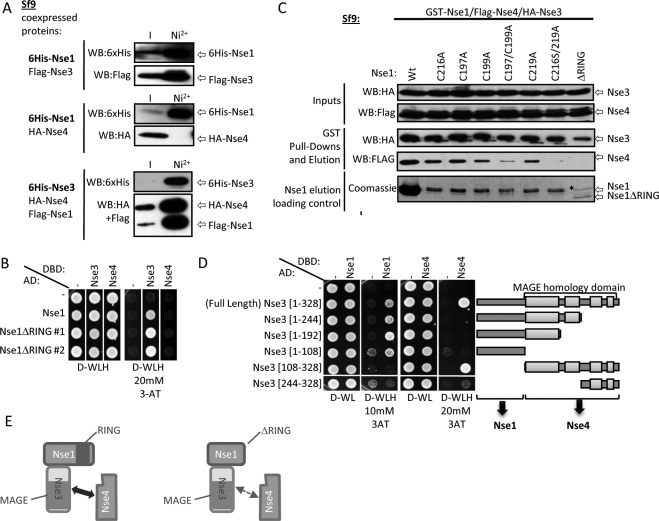
The Nse1 NH-RING Domain Stabilizes the Nse1-Nse3 Interaction with Nse4
Nse1, Nse3, and Nse4 form a stable heterotrimer, of which the Nse4 subunit bridges the Smc5 and Smc6 head domains in the holocomplex (Sergeant et al., 2005; Palecek et al., 2006; Pebernard et al., 2006). Nse1 and Nse3 stably dimerize, whereas Nse1 and Nse4 do not interact directly (Figure 5A). Thus, Nse3 contacts both Nse1 and Nse4 to form a stable trimer (Figure 5A and Sergeant et al., 2005; Pebernard et al., 2006). The Nse1-Nse3 interaction is not perturbed by deletion of Nse1 NH-RING as tested by yeast two-hybrid assay, which is consistent with a previous report (Figure 5B and Sergeant et al., 2005). We hypothesized that the Nse1 NH-RING might contribute to, or stabilize, contacts between the Nse1-Nse3 dimer and Nse4. To test this, we assessed the capacity of Nse1 NH-RING mutants to interact with Nse3 and Nse4 by coprecipitating these proteins from coinfected Sf9 insect cell extracts. This system was effectively used in our laboratory to dissect contacts between subunits of the Smc5-Smc6 complex, which is not possible in vivo (Pebernard et al., 2006). We first used GST-PreScission-Nse1 fusions as baits to inquire whether HA-Nse3 and Flag-Nse4 coprecipitated with Nse1 NH-RING mutants (Figure 5C). After GST purification, Nse1 and coprecipitating proteins were eluted by cleavage with the PreScission protease (Figure 5C). All Nse1 NH-RING single-point mutants interacted with Nse3 and Nse4, albeit to a lesser extent than wild-type Nse1. Notably, the Nse1 ΔRING and double zinc ligand mutants (C197, 199A and C216S, C219A) still bound Nse3 but interacted very weakly or not at all with Nse4, indicating that the Nse1 NH-RING is critical for Nse4 but not Nse3 binding (Figure 5C). It is also noteworthy that there is more wild-type Nse1 visible than any of the NH-RING mutants after purification. Despite this, Nse1 NH-RING point mutants coprecipitate equivalent amounts of Nse3 as wild-type Nse1. This likely reflects the capacity of Nse1 to homodimerize, which is a common feature of RING finger proteins that can multimerize through RING-RING interactions (Kentsis et al., 2002a,b).
Figure 5.
Analysis of Nse1-Nse3-Nse4 trimer formation in the presence of Nse1 NH-RING mutations. (A) Nse1 interaction with Nse4 is bridged by Nse3. Protein interaction assays were performed in Sf9 insect cells coinfected with the indicated protein-expressing baculoviruses, using Nickel (Ni2+) purification columns and Western blotting for copurifying proteins. Nse1 can interact with Nse3 but not with Nse4 in the absence of Nse3, whereas the full trimer can be pulled down when all three proteins are coexpressed. (B) Nse1 NH-RING is not required for interaction with Nse3. Yeast two-hybrid interaction assays were performed using the indicated strains and spotted on media without (D-WLH) or with (D-WLH + 3-AT) selection to test for direct interaction between the expressed proteins. Nse1 does not interact with Nse4 in the absence of Nse3, but interacts with Nse3 in an NH-RING–independent manner. −, empty vector control; AD, Gal4 activation domain; DBD, Gal4 DNA-binding domain. (C) The Nse1 NH-RING domain is required for robust Nse1-Nse3 trimerization with Nse4 in vitro. Sf9 insect cells were simultaneously infected with baculoviruses to coexpress the indicated proteins. Complexes were purified by GST pulldown, eluted by PreScission Protease cleavage, and detected by Western blotting against the HA and Flag tags. * Nonspecific band appearing in every purification, slightly larger than Nse1 full-length. (D) Distinct Nse3 domains are required to contact Nse1 and Nse4. The indicated yeast two-hybrid strains were performed as in B using the indicated strains. Nse3 protein domains are illustrated on the right. Regions of high sequence homology between species are represented by light gray boxes. (E) The Nse1 NH-RING domain optimizes Nse1-Nse3-Nse4 trimer formation. Schematics summarizing interaction analyses. Nse1 and Nse3 N-termini interact together independently of Nse1 NH-RING. Nse3 MAGE domain interacts with Nse4 independently of Nse1. Nse1 NH-RING is required for stable trimer formation, as an Nse1 ΔRING mutant (right panel) loses contact with Nse4, but not with Nse3.
The Nse3 MAGE Homology Domain Interacts with Nse4
Little is known about the involvement of Nse3 structural domains in Nse1-Nse3-Nse4 trimer formation. The Nse3 structure has not been determined but sequence analysis identified a conserved MAGE homology domain, which covers around two thirds of the protein (Figure 5D; Pebernard et al., 2004). We used the yeast two-hybrid assay to map Nse3 domains required for interaction with Nse1 and Nse4. Constructs expressing the Gal4 AD fused to Nse3 full-length, or various N- and C-terminal deletion mutants, were introduced in a yeast two-hybrid strain and tested for their interaction with full-length Nse1 and Nse4 proteins fused to the Gal4 DNA-binding domain (Figure 5D, DBD). We established that the minimal Nse3 region required for interaction with Nse1 is a disordered N-terminal 108-amino acid region (Figure 5D). On the contrary, the entire Nse3 MAGE domain is necessary for proper interaction with Nse4 (Figure 5D). Altogether, our analyses suggest that the Nse1 NH-RING motif, specifically in context of the Nse1-Nse3 dimer, promotes stable contact of Nse1-Nse3 with Nse4. Thus, Nse1 NH-RING mutations might lead to partial loss of conformation of the Nse1-Nse3-Nse4 trimer (e.g., Figure 5E), causing the observed NH-RING mutant phenotypes, including mislocalization of Nse4 upon DNA damage.
DISCUSSION
Nse1 and Nse2 are unique among the non-SMC subunits of all SMC complexes, in that they contain motifs predictive of catalytic functions in the ubiquitin and SUMO posttranslational modifier pathways, respectively (Fujioka et al., 2002; McDonald et al., 2003; Sergeant et al., 2005). Nse2 has been confirmed as a SUMO E3 ligase, with critical roles in DNA repair (Andrews et al., 2005; Potts and Yu, 2005; Zhao and Blobel, 2005). We have now executed a detailed structure-function analysis of the unique RING-like domain of Nse1 (NH-RING). Strikingly, despite the essential nature of Nse1, the NH-RING motif is dispensable for cellular viability. However, cells lacking a functional Nse1 NH-RING are hypersensitive to genotoxic stress and temperature sensitive for growth. Furthermore, NH-RING mutant cells are dependent on the key DNA repair factors Rqh1 (human BLM) and Mus81-Eme1 for viability. Given the replication-associated roles of Rqh1 and Mus81-Eme1 (Boddy et al., 2000; Doe et al., 2000; Roseaulin et al., 2008), these genetic interactions underscore the intimate relationship between Smc5-Smc6 and replication fork stabilization/processing (Ampatzidou et al., 2006; Pebernard et al., 2006; Murray and Carr, 2008).
Because the Nse1 NH-RING is related to RING domains of ubiquitin E3 ligases, we tested for such activity in vitro, but did not detect E3 ligase activity associated with either the human or the fission yeast NH-RING domain. We utilized a large panel of E2-conjugating enzymes in conjunction with the human Nse1 NH-RING, none of which yielded detectable activity. Thus, under standard conditions used to assay RING-dependent E3 ligase activity (e.g., Slx8, Figure 4), neither full-length Nse1 nor the NH-RING alone stimulates detectable ubiquitination. Although our in vitro data imply that Nse1 is not an ubiquitin E3 ligase, we do not exclude the possibility that the Smc5-Smc6 complex contains such activity in vivo. For example, it is possible that we have not coupled Nse1 with its cognate E2-conjugating enzyme in vitro. Alternatively, as for a number of RING-type E3 ubiquitin ligases, Nse1 might require heterodimerization with another RING-containing factor to be active (Hashizume et al., 2001; Buchwald et al., 2006; Prudden et al., 2007). However, in our extensive purifications of the Smc5-Smc6 holocomplex we have identified no candidates for such a cofactor (McDonald et al., 2003; Pebernard et al., 2004, 2006). Because Nse1 normally functions in a heterotrimeric subcomplex of Smc5-Smc6 (Sergeant et al., 2005; Pebernard et al., 2006), we also tested the possibility that Nse1 needs to be complexed with Nse3 and Nse4 to be active. The Nse1-Nse3 dimer produced in insect cells displayed weak but reproducible ubiquitin E3 ligase activity in our in vitro ubiquitination assays. Notably, this activity was independent of the Nse1 NH-RING and thus might be due to contaminant proteins copurifying from insect cells, as no activity was detected for Nse1-Nse3 dimer produced in bacteria. However, it is conceivable that Nse3 may constitute a new class of ubiquitin E3 ligase supported by its structurally undefined melanoma antigen (MAGE) or necdin-like domain (Barker and Salehi, 2002; Pebernard et al., 2004; Sergeant et al., 2005). In this scenario, the MAGE domain could bind the E2 and the Nse1 NH-RING could function in substrate recognition. Further studies centered on Nse3, including structural analyses, are required to distinguish between these possibilities.
The Nse1 NH-RING does not conform to previous RING-like signatures but shows features of several, including the PHD domain, i.e., C4HC3 configuration (see above and Bienz, 2006). PHD domains constitute protein interaction interfaces for many nuclear and chromatin associated factors, including methylated histone H3 (Bienz, 2006). Interestingly, we find that the Nse1 NH-RING domain is required for stable trimerization of the Nse1-Nse3-Nse4 subcomplex, of the Smc5-Smc6 holocomplex. Nse1 dimerizes with Nse3 (NH-RING independent), and this dimer then binds avidly to Nse4 (Sergeant et al., 2005; Pebernard et al., 2006). An Nse1-Nse3 dimer that lacks the NH-RING does not bind as strongly to Nse4, making the Nse1-Nse3-Nse4 trimer labile. Thus, we propose that the Nse1 NH-RING forms a “latch” for high-affinity Nse4 binding when Nse1 is complexed with Nse3. It will be necessary to determine the crystal structure of the Nse1-Nse3-Nse4 trimer to elucidate how dimerization between Nse1 and Nse3 induces Nse4 binding by the NH-RING, which may involve an allosteric effect.
Notably, we determined that the Nse1 NH-RING domain is required for the localization/enrichment of Nse4 and Smc5 in subnuclear foci after DNA damage. Because there is no evidence for extensive pools of Nse4 and Smc5 that are not part of the octameric holocomplex (McDonald et al., 2003; Andrews et al., 2005; Sergeant et al., 2005; Pebernard et al., 2006), we propose this result applies to the entire complex. A defect in accumulation of the Smc5-Smc6 complex at DNA damage sites is an economical explanation for the observed DNA damage hypersensitivities of NH-RING mutant cells. The Nse1 NH-RING domain, like some PHD domains, might have affinity for histones with certain tail modifications, perhaps those induced by DNA damage. Alternatively, in vivo Nse1 might promote monoubiquitination of a component in the Smc5-Smc6 complex leading to focal accumulation of the complex, similar to that seen after FANCD2-mediated monoubiquitination in the Fanconi anemia DNA repair complex (see Huang and D'Andrea, 2006 and references therein).
Supplementary Material
ACKNOWLEDGMENTS
We thank John Prudden, Jenny Scorah, and Matthias Kaeser for critical reading of the manuscript, and all members of the Scripps Cell Cycle group for constant support and encouragement. S.P. was supported by a fellowship from the Swiss Cancer League (KLS-01525-02-2004). This study was funded by National Institutes of Health (NIH) Grant GM068608 awarded to M.N.B. and NIH Grant CA104660 to J.A.T.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-02-0226) on July 30, 2008.
REFERENCES
- Albert T. K., Hanzawa H., Legtenberg Y. I., de Ruwe M. J., van den Heuvel F. A., Collart M. A., Boelens R., Timmers H. T. Identification of a ubiquitin-protein ligase subunit within the CCR4-NOT transcription repressor complex. EMBO J. 2002;21:355–364. doi: 10.1093/emboj/21.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ampatzidou E., Irmisch A., O'Connell M. J., Murray J. M. Smc5/6 is required for repair at collapsed replication forks. Mol. Cell. Biol. 2006;26:9387–9401. doi: 10.1128/MCB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews E. A., Palecek J., Sergeant J., Taylor E., Lehmann A. R., Watts F. Z. Nse2, a component of the Smc5–6 complex, is a SUMO ligase required for the response to DNA damage. Mol. Cell. Biol. 2005;25:185–196. doi: 10.1128/MCB.25.1.185-196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L., Iyer L. M., Koonin E. V. Scores of RINGS but no PHDs in ubiquitin signaling. Cell Cycle. 2003;2:123–126. doi: 10.4161/cc.2.2.335. [DOI] [PubMed] [Google Scholar]
- Bahler J., Wu J. Q., Longtine M. S., Shah N. G., McKenzie A., 3rd, Steever A. B., Wach A., Philippsen P., Pringle J. R. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Barker P. A., Salehi A. The MAGE proteins: emerging roles in cell cycle progression, apoptosis, and neurogenetic disease. J. Neurosci. Res. 2002;67:705–712. doi: 10.1002/jnr.10160. [DOI] [PubMed] [Google Scholar]
- Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem. Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Boddy M. N., Lopez-Girona A., Shanahan P., Interthal H., Heyer W. D., Russell P. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 2000;20:8758–8766. doi: 10.1128/mcb.20.23.8758-8766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy M. N., Shanahan P., McDonald W. H., Lopez-Girona A., Noguchi E., Yates I. J., Russell P. Replication checkpoint kinase Cds1 regulates recombinational repair protein Rad60. Mol. Cell. Biol. 2003;23:5939–5946. doi: 10.1128/MCB.23.16.5939-5946.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden K. L. RING domains: master builders of molecular scaffolds? J. Mol. Biol. 2000;295:1103–1112. doi: 10.1006/jmbi.1999.3429. [DOI] [PubMed] [Google Scholar]
- Borden K. L., Freemont P. S. The RING finger domain: a recent example of a sequence-structure family. Curr. Opin. Struct. Biol. 1996;6:395–401. doi: 10.1016/s0959-440x(96)80060-1. [DOI] [PubMed] [Google Scholar]
- Branzei D., Sollier J., Liberi G., Zhao X., Maeda D., Seki M., Enomoto T., Ohta K., Foiani M. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006;127:509–522. doi: 10.1016/j.cell.2006.08.050. [DOI] [PubMed] [Google Scholar]
- Buchwald G., van der Stoop P., Weichenrieder O., Perrakis A., van Lohuizen M., Sixma T. K. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 2006;25:2465–2474. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D. F., et al. An iterative structure-assisted approach to sequence alignment and comparative modeling. Proteins Suppl. 1999;3:55–60. doi: 10.1002/(sici)1097-0134(1999)37:3+<55::aid-prot8>3.3.co;2-2. [DOI] [PubMed] [Google Scholar]
- Dodd R. B., Allen M. D., Brown S. E., Sanderson C. M., Duncan L. M., Lehner P. J., Bycroft M., Read R. J. Solution structure of the Kaposi's sarcoma-associated herpesvirus K3 N-terminal domain reveals a novel E2-binding C4HC3-type RING domain. J. Biol. Chem. 2004;279:53840–53847. doi: 10.1074/jbc.M409662200. [DOI] [PubMed] [Google Scholar]
- Doe C. L., Dixon J., Osman F., Whitby M. C. Partial suppression of the fission yeast rqh1(-) phenotype by expression of a bacterial Holliday junction resolvase. EMBO J. 2000;19:2751–2762. doi: 10.1093/emboj/19.11.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fousteri M. I., Lehmann A. R. A novel SMC protein complex in Schizosaccharomyces pombe contains the Rad18 DNA repair protein. EMBO J. 2000;19:1691–1702. doi: 10.1093/emboj/19.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka Y., Kimata Y., Nomaguchi K., Watanabe K., Kohno K. Identification of a novel non-structural maintenance of chromosomes (SMC) component of the SMC5-SMC6 complex involved in DNA repair. J. Biol. Chem. 2002;277:21585–21591. doi: 10.1074/jbc.M201523200. [DOI] [PubMed] [Google Scholar]
- Hashizume R., Fukuda M., Maeda I., Nishikawa H., Oyake D., Yabuki Y., Ogata H., Ohta T. The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J. Biol. Chem. 2001;276:14537–14540. doi: 10.1074/jbc.C000881200. [DOI] [PubMed] [Google Scholar]
- Hassink G., et al. TEB4 is a C4HC3 RING finger-containing ubiquitin ligase of the endoplasmic reticulum. Biochem. J. 2005;388:647–655. doi: 10.1042/BJ20041241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazbun T. R., et al. Assigning function to yeast proteins by integration of technologies. Mol. Cell. 2003;12:1353–1365. doi: 10.1016/s1097-2765(03)00476-3. [DOI] [PubMed] [Google Scholar]
- Hewitt E. W., Duncan L., Mufti D., Baker J., Stevenson P. G., Lehner P. J. Ubiquitylation of MHC class I by the K3 viral protein signals internalization and TSG101-dependent degradation. EMBO J. 2002;21:2418–2429. doi: 10.1093/emboj/21.10.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. SP-RING for SUMO: new functions bloom for a ubiquitin-like protein. Cell. 2001;107:5–8. doi: 10.1016/s0092-8674(01)00519-0. [DOI] [PubMed] [Google Scholar]
- Hu B., Liao C., Millson S. H., Mollapour M., Prodromou C., Pearl L. H., Piper P. W., Panaretou B. Qri2/Nse4, a component of the essential Smc5/6 DNA repair complex. Mol. Microbiol. 2005;55:1735–1750. doi: 10.1111/j.1365-2958.2005.04531.x. [DOI] [PubMed] [Google Scholar]
- Huang T. T., D'Andrea A. D. Regulation of DNA repair by ubiquitylation. Nat. Rev. Mol. Cell Biol. 2006;7:323–334. doi: 10.1038/nrm1908. [DOI] [PubMed] [Google Scholar]
- Karplus K., Barrett C., Hughey R. Hidden Markov models for detecting remote protein homologies. Bioinformatics. 1998;14:846–856. doi: 10.1093/bioinformatics/14.10.846. [DOI] [PubMed] [Google Scholar]
- Kentsis A., Gordon R. E., Borden K. L. Control of biochemical reactions through supramolecular RING domain self-assembly. Proc. Natl. Acad Sci. USA. 2002a;99:15404–15409. doi: 10.1073/pnas.202608799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentsis A., Gordon R. E., Borden K. L. Self-assembly properties of a model RING domain. Proc. Natl. Acad Sci. USA. 2002b;99:667–672. doi: 10.1073/pnas.012317299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurowski M. A., Bujnicki J. M. GeneSilico protein structure prediction meta-server. Nucleic Acids Res. 2003;31:3305–3307. doi: 10.1093/nar/gkg557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann A. R., Walicka M., Griffiths D. J., Murray J. M., Watts F. Z., McCready S., Carr A. M. The rad18 gene of Schizosaccharomyces pombe defines a new subgroup of the SMC superfamily involved in DNA repair. Mol. Cell. Biol. 1995;15:7067–7080. doi: 10.1128/mcb.15.12.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindroos H. B., Strom L., Itoh T., Katou Y., Shirahige K., Sjogren C. Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol. Cell. 2006;22:755–767. doi: 10.1016/j.molcel.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Lorick K. L., Jensen J. P., Fang S., Ong A. M., Hatakeyama S., Weissman A. M. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad Sci. USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A., Hirano T. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev. 2005;19:1269–1287. doi: 10.1101/gad.1320505. [DOI] [PubMed] [Google Scholar]
- McDonald W. H., Pavlova Y., Yates J. R., 3rd, Boddy M. N. Novel essential DNA repair proteins Nse1 and Nse2 are subunits of the fission yeast Smc5-Smc6 complex. J. Biol. Chem. 2003;278:45460–45467. doi: 10.1074/jbc.M308828200. [DOI] [PubMed] [Google Scholar]
- Miyabe I., Morishita T., Hishida T., Yonei S., Shinagawa H. Rhp51-dependent recombination intermediates that do not generate checkpoint signal are accumulated in Schizosaccharomyces pombe rad60 and smc5/6 mutants after release from replication arrest. Mol. Cell. Biol. 2006;26:343–353. doi: 10.1128/MCB.26.1.343-353.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Morikawa H., Morishita T., Kawane S., Iwasaki H., Carr A. M., Shinagawa H. Rad62 protein functionally and physically associates with the smc5/smc6 protein complex and is required for chromosome integrity and recombination repair in fission yeast. Mol. Cell. Biol. 2004;24:9401–9413. doi: 10.1128/MCB.24.21.9401-9413.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J. M., Carr A. M. Smc5/6, a link between DNA repair and unidirectional replication? Nat. Rev. Mol. Cell Biol. 2008;9:177–182. doi: 10.1038/nrm2309. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Haering C. H. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- Ohi M. D., Vander Kooi C. W., Rosenberg J. A., Chazin W. J., Gould K. L. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat. Struct. Biol. 2003;10:250–255. doi: 10.1038/nsb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palecek J., Vidot S., Feng M., Doherty A. J., Lehmann A. R. The Smc5-Smc6 DNA repair complex. bridging of the Smc5-Smc6 heads by the KLEISIN, Nse4, and non-Kleisin subunits. J. Biol. Chem. 2006;281:36952–36959. doi: 10.1074/jbc.M608004200. [DOI] [PubMed] [Google Scholar]
- Pebernard S., McDonald W. H., Pavlova Y., Yates J. R., 3rd, Boddy M. N. Nse1, Nse2, and a novel subunit of the Smc5-Smc6 complex, Nse3, play a crucial role in meiosis. Mol. Biol. Cell. 2004;15:4866–4876. doi: 10.1091/mbc.E04-05-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pebernard S., Wohlschlegel J., McDonald W. H., Yates J. R., 3rd, Boddy M. N. The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5-Smc6 complex. Mol. Cell. Biol. 2006;26:1617–1630. doi: 10.1128/MCB.26.5.1617-1630.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J. J., Tainer J. A., Boddy M. N. A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem. Sci. 2008;33:201–208. doi: 10.1016/j.tibs.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Potts P. R., Porteus M. H., Yu H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 2006;25:3377–3388. doi: 10.1038/sj.emboj.7601218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts P. R., Yu H. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol. Cell. Biol. 2005;25:7021–7032. doi: 10.1128/MCB.25.16.7021-7032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts P. R., Yu H. The SMC5/6 complex maintains telomere length in ALT cancer cells through SUMOylation of telomere-binding proteins. Nat. Struct. Mol. Biol. 2007;14:581–590. doi: 10.1038/nsmb1259. [DOI] [PubMed] [Google Scholar]
- Prudden J., Pebernard S., Raffa G., Slavin D. A., Perry J. J., Tainer J. A., McGowan C. H., Boddy M. N. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089–4101. doi: 10.1038/sj.emboj.7601838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roseaulin L., Yamada Y., Tsutsui Y., Russell P., Iwasaki H., Arcangioli B. Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J. 2008;27:1378–1387. doi: 10.1038/emboj.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A., Blundell T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Saurin A. J., Borden K. L., Boddy M. N., Freemont P. S. Does this have a familiar RING? Trends Biochem. Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- Sergeant J., Taylor E., Palecek J., Fousteri M., Andrews E. A., Sweeney S., Shinagawa H., Watts F. Z., Lehmann A. R. Composition and architecture of the Schizosaccharomyces pombe Rad18 (Smc5–6) complex. Mol. Cell. Biol. 2005;25:172–184. doi: 10.1128/MCB.25.1.172-184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S. L., Hauksdottir H., Troy A., Herschleb J., Kraft E., Callis J. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 2005;137:13–30. doi: 10.1104/pp.104.052423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor E. M., Copsey A. C., Hudson J. J., Vidot S., Lehmann A. R. Identification of the proteins, including MAGEG1, that make up the human SMC5–6 protein complex. Mol. Cell. Biol. 2008;28:1197–1206. doi: 10.1128/MCB.00767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Rosell J., Machin F., Aragon L. Smc5-Smc6 complex preserves nucleolar integrity in S. cerevisiae. Cell Cycle. 2005a;4:868–872. doi: 10.4161/cc.4.7.1825. [DOI] [PubMed] [Google Scholar]
- Torres-Rosell J., Machin F., Farmer S., Jarmuz A., Eydmann T., Dalgaard J. Z., Aragon L. SMC5 and SMC6 genes are required for the segregation of repetitive chromosome regions. Nat. Cell Biol. 2005b;7:412–419. doi: 10.1038/ncb1239. [DOI] [PubMed] [Google Scholar]
- Wallner B., Larsson P., Elofsson A. Pcons.net: protein structure prediction meta server. Nucleic Acids Res. 2007;35:W369–374. doi: 10.1093/nar/gkm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J., Zhai L., Xu J., Wang H. Role of Bmi1 in H2A ubiquitylation and Hox gene silencing. J. Biol. Chem. 2006;281:22537–22544. doi: 10.1074/jbc.M600826200. [DOI] [PubMed] [Google Scholar]
- Zhao X., Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl Acad Sci. USA. 2005;102:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.