Abstract
Mif2p is the budding-yeast orthologue of the mammalian centromere-binding protein CENP-C. We have mapped domains of Saccharomyces cerevisiae Mif2p and studied the phenotyptic consequences of their deletion. Using chromatin immunoprecipitation (ChIP) and electrophoretic mobility shift assays, we have further shown that Mif2p binds in the CDEIII region of the budding-yeast centromere, probably in close spatial association with Ndc10p. Moreover, ChIP experiments show that Mif2p recruits to yeast kinetochores a substantial subset of inner and outer kinetochore proteins, but not the Ndc80 or Spc105 complexes. We have determined the crystal structure of the C-terminal, dimerization domain of Mif2p. It has a “cupin” fold, extremely similar both in polypeptide chain conformation and in dimer geometry to the dimerization domain of a bacterial transcription factor. The Mif2p dimer seems to be part of an enhanceosome-like structure that nucleates kinetochore assembly in budding yeast.
INTRODUCTION
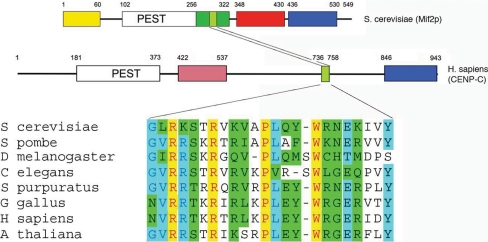
Mif2p is the budding-yeast orthologue of mammalian CENP-C, an essential, inner kinetochore centromere (CEN)-binding protein (Earnshaw and Rothfield, 1985; Saitoh et al., 1992; Brown, 1995; Meluh and Koshland, 1995; Yang et al., 1996). Recognition of this relationship was an early clue that yeast kinetochores, which assemble on “point centromeres” of roughly 150 base pairs, are structurally similar to higher eukaryotic kinetochores (Meluh and Koshland, 1995), which assemble on much longer “regional” centromeres, megabases in length (Pluta et al., 1990). The strongest conservation across Mif2p-CENP-C homologues from all eukaryotes lies in a short (25-residue) “CENP-C signature motif” that is roughly in the middle of the Mif2p polypeptide chain (Figure 1). A C-terminal domain of ∼100 residues is also broadly conserved.
Figure 1.
Domain organization of the Mif2p and CENP-C polypeptide chains. Top, the Mif2p polypeptide chain. Yellow, green, red, and blue blocks indicate regions of distinct function (assigned, in part, through results described herein). The two regions recognizably conserved in human CENP-C (middle) are a short “CENP-C signature sequence” (light green) and a C-terminal domain (blue) shown here to be a dimerizing element. The pink box in the human CENP-C diagram represents the presumptive DNA-binding domain. Various signature-motif sequences are shown in the bottom panel.
The budding-yeast centromere is composed of three elements, conserved among all chromosomes: CDEI, a nonessential eight-base pair palindrome; CDEII, an essential 78- to 86-base pair AT-rich sequence; and CDEIII, an imperfect palindrome with an ∼24 base-pair “core” and a less well conserved CDEII-distal sequence of 50–60 bp (Fitzgerald-Hayes et al., 1982). CDEI binds the helix-turn-helix protein, Cbf1p, which also functions as a transcription factor in other contexts (Hemmerich et al., 2000). CDEIII binds the four-protein CBF3 complex to form a structure essential for all subsequent steps in assembly of a yeast kinetochore (Lechner and Carbon, 1991; Doheny et al., 1993; Strunnikov et al., 1995), which comprises ∼60 unique protein subunits (McAinsh et al., 2003; Westermann et al., 2007). Chromatin immunoprecipitation experiments show that CEN binding by Mif2p depends on active CBF3 and on the CENP-A orthologue, Cse4p, and that it is sensitive to mutations in CDEI and CDEIII, leading to the conclusion that Mif2p might associate with Cbf1p, Cse4p, and CBF3 (Meluh and Koshland, 1997; Westermann et al., 2003). Although binding of Mif2p to CDEII was suggested initially (Brown et al., 1993; Meluh and Koshland 1995), Mif2p associates in vivo with a CEN construct that contains CDEIII (with CBF3 bound) but that lacks both CDEI and CDEII (Meluh and Koshland, 1997; Ortiz et al., 1999). Thus, CDEIII, rather than CDEII, would seem to be the critical sequence for kinetochore recruitment of Mif2p.
Mif2p orthologues are required for recruitment of many additional kinetochore components (Oegema et al., 2001; Cheeseman et al., 2004; Liu et al., 2006; Kwon et al., 2007). Mif2p is therefore both an integral part of the inner kinetochore and a critical subunit for assembling the entire kinetochore superstructure. We report here cellular, biochemical, and structural characterization of the domain organization of Mif2p. A region (residues 256–549) that includes the CENP-C signature motif and a likely DNA-binding domain is important for normal cell growth. Deletion N-terminal to this essential segment or deletion of a conserved C-terminal domain results in slow-growing, temperature-sensitive cells. High-resolution chromatin immunoprecipitation (ChIP) experiments show that Mif2p associates in vivo with CDEIII. Moreover, expression and purification of various fragments of the protein allow us to demonstrate that residues 256–549 form a DNA-binding dimer, which associates strongly with an A:T-rich region of CDEIII, consistent with the predicted presence of an “AT-hook” in the DNA-binding region of many yeast Mif2p homologues (Brown, 1995; Lanini and McKeon, 1995; Talbert et al., 2004). CDEII also provides stretches of A:T-rich DNA, but consistent with recently published reports (Camahort et al., 2007; Furuyama and Biggins, 2007; Mizuguchi et al., 2007; Stoler et al., 2007), our experiments show that this region is occupied instead by a Cse4p-containing nucleosomal assembly. Residues 438–549 at the C terminus of the Mif2p polypeptide chain create a dimerization domain. We have determined the crystal structure of this domain and established that it has a “cupin” fold, with striking structural similarity to the dimeric C-terminal domain of Hth-3, a putative transcription factor from Vibrio cholerae. Thus, an essential component of all eukaryotic kinetochores is structurally related to a bacterial gene regulatory protein. We integrate structural and biochemical information into a schematic picture of the centromere-proximal elements of a yeast kinetochore.
MATERIALS AND METHODS
Strains and Constructs
See Supplemental Table for details.
Analysis of Mif2p Truncation Phenotypes
An S288C haploid yeast strain was generated in which a chromosomal deletion of the endogenous MIF2 gene was rescued by MIF2 expressed from a URA3-CEN plasmid (to create pMIF2:URA3 mif2::KANr). Deletion constructs under the control of the endogenous MIF2 promoter were then integrated at LEU2 in these cells, and the ability of deletions to substitute for wild-type (wt) MIF2 tested by counterselection for pMIF2:URA3 on 5-fluoroorotic acid. Plates were incubated overnight at 30 or 37°C and photographed.
Expression and Purification of Mif2p Fragments
DNA encoding residues 256–549, 365–549, or 365–530 of Mif2p was amplified using the polymerase chain reaction (PCR) and inserted into the pET28a(+) plasmid (Novagen, Madison, WI) downstream of a cleavable, N-terminal hexahistidine tag. The fragment was overexpressed in Rosetta pLysS cells (Novagen), induced with 1 mM isopropyl β-d-thiogalactoside at an optical density of 0.6, and purified from the bacterial lysate by nickel-nitrilotriacetic acid chromatography (QIAGEN, Valencia, CA), by using 2 ml resin/l culture. The purified protein was concentrated in an Ultracel-10-kDa centrifugal filter (Millipore, Billerica, CA) to a volume of 1 ml and further purified by size exclusion chromatography on a 16/60 Superdex S75 column (GE Healthcare, Piscataway, NJ) equilibrated with 10 mM HEPES, pH 7.5, 800 mM NaCl (fragment 256–549), or 250 mM NaCl (fragment 365–549).
Crystallization of Mif2p(365-530) and x-Ray Structure Determination
Mif2p(365-530) crystallizes from 3 to 4% polyethylene glycol (PEG) 3000, 70–100 M Li2SO4, or Na2SO4, 100 mM imidazole, pH 8.0, in space group P213 (a = 106.0 Å), with a dimer in the asymmetric unit. For flash freezing, the mother liquor was changed stepwise to 25% glycerol, 6% PEG 3000, with other components as in the initial crystallization. We identified derivatives by using a native gel to test binding with a number of compounds (Boggon and Shapiro 2000). Of the compounds that altered protein mobility, two (K2OsCl6 and di-m-iodobis(ethylenediamine) diplatinum [PIP]) could be soaked into the Mif2p(365-530) crystals without causing damage or nonisomorphism. We collected multiwavelength data sets from these two derivatives at beamline 8.2.2 at the Advanced Light Source and processed the data using HKL2000 (Otwinowski and Minor, 1997). The Os sites were located using SOLVE (Terwilliger and Berendzen, 1997); phases from this derivative were used to locate the Pt sites in the PIP derivative. Multiple isomorphous replacement refinement yielded an interpretable map using structure factors to 3.2-Å resolution.
Electron density for one of the two chains in the asymmetric unit was better defined than for the other chain. We initially built the clearest two strands of each molecule with the program O (Jones et al., 1991) and calculated a rotation matrix for noncrystallographic symmetry (NCS) averaging by using coordinates for residues in these strands and the NCS-related heavy-atom positions. We created a suitable mask and carried out iterative NCS averaging and solvent flattening, using the program DM (Cowtan and Main, 1998; CCP4). A model for residues 437–530 was built into this map and refined, with cycles of rebuilding, using Refmac (CCP4). The final model has Rcryst and Rfree of 21.0 and 24.2%, respectively; 90% of the residues are in the most favored region of the Ramachandran plot, and the remainder are in allowed regions. Coordinates and structure factors have been deposited in the PDB with accession number 2vpv. Data collection, phasing, and refinement statistics are shown in Table 1.
Table 1.
Data and refinement statistics
| Data collection | Native | PIP | Os MAD λ1 | Os MAD λ2 | Os MAD λ3 |
|---|---|---|---|---|---|
| Wavelength (Å) | 0.97950 | 1.50030 | 1.14083 | 1.14048 | 1.14010 |
| Space group | P213 | P213 | P213 | P213 | P213 |
| Resolution (Å)a | 2.70 (2.80–2.70) 2.90 | (3.00–2.90) 3.10 | (3.21–3.10) 3.10 | (3.21–3.10) 3.20 | (3.34–3.20) |
| Unique observations | 11,184 | 15,323 | 12,290 | 12,098 | 11,793 |
| Redundancya | 5.2 (4.0) | 1.9 (1.6) | 4.3 (2.1) | 4.3 (2.0) | 4.2 (2.4) |
| Completenessa | 99.3 (95.8) | 89.7 (80.6) | 88.3 (57.6) | 86.9 (47.9) | 84.0 (68.8) |
| Rsymb | 5.0 (38.5) | 7.9 (43.3) | 5.7 (36.8) | 6.2 (38.5) | 6.0 (39.7) |
| I/σ(I)a | 26.7 (2.3) | 9.6 (1.5) | 19.5 (1.5) | 18.3 (1.3) | 17.6 (2.1) |
| Refinement | |||||
| Resolution (Å) | 30.0–2.70 | ||||
| Refined residues | 186 | ||||
| Refined water molecules | 29 | ||||
| Rcrystc (%) | 21.0 | ||||
| Rfreed (%) | 24.2 | ||||
| Average B factors (Å2) | |||||
| Mif2p | 56.6 | ||||
| Water molecules | 53.8 | ||||
| R.m.s. deviations from ideal geometry | |||||
| Bond lengths (Å) | 0.012 | ||||
| Bond angles (°) | 1.53 | ||||
| Dihedral angles (°) | 18.2 | ||||
| Improper angles (°) | 1.1 | ||||
a Numbers in parentheses refer to highest resolution shell.
b Rsym = [ΣhΣi Ii(h) − <I(h)>/ΣhΣiIi(h)] × 100, where <I(h)> is the average intensity of I symmetry related observations of reflections with Bragg index h.
c Rcryst = [Σhkl Fo − Fc /Σhkl Fo] × 100, where Fo − Fc are the observed and calculated structure factors.
d Rfree was calculated as for Rcryst, but on 5% of data excluded before refinement.
Chromatin Immunoprecipitation
Chromatin Immunoprecipitation was performed based on the protocol of (Megee et al., 1999), with modifications. Specifically, cells were cross-linked with formaldehyde for 2 h at room temperature (RT) and then lysed using glass beads in a Bio101 FastPrep FP120. Genomic DNA was sonicated 6 × 10 s (on ice between bursts) to an average of 100–300 base pairs (estimated from agarose gel in comparison with X/HaeIII MW ladder) and centrifuged to remove cellular debris. Immunoprecipitations (IPs) were performed using anti-green fluorescent protein (GFP) (GFP-Cse4p and Cbf1p-GFP; Clontech, Mountain View, CA), anti-myc (Ndc10p-myc; Santa Cruz Biotechnology, Santa Cruz, CA), anti-H3 (Abcam, Cambridge, MA), anti-CEP3 or anti-Mif2p (Sorger laboratory), or no antibody (negative control; data not shown). An untagged strain also served as negative control (data not shown). Serial dilutions of recovered DNA and Total DNA (pre-IP genomic) were used to establish the linearity of PCR amplifications. Nonoverlapping pairs of oligonucleotides generated 100- or 150-bp DNA fragments, which span Chromosome IV SGD coordinates 449131–450480 (centered on CENIV CDEI-II-III). (CEOL 307F) 5′ ATTGCAAAATTTAATTGCTTGCAAAA and (CEOL 309R) 5′ TTCTTGAGCGGTTTTATGTTTCGG encompass CDEI-II-III. Sequences for the remaining oligos are available upon request.
Electrophoretic Mobility Shift Assays (EMSAs) or Bandshift Assay
Bandshift/competition probes were based on Saccharomyces cerevisiae CENIII. For each probe, 1 nmol (each) of two complementary oligonucleotides were annealed in 40 μl of 20 mM Tris, pH 8.0, 2 mM MgCl2, and 50 mM NaCl, by heating at 90°C for 10 min and then slowly cooling to room temperature. Samples from each reaction were separated on agarose gels to ensure the presence of a single band and to quantitate the annealed product (ImageQuant software; GE Healthcare). Random 88-bp DNA was generated by PCR from the pUC19, purified and quantitated. Probe DNA was labeled with 32P-dATP using T4 polynucleotide kinase (NEB). To measure DNA-binding affinity, 30-μl reactions containing 0.2–4 μg of recombinant Mif2p or CBF3 Complex, 50 fmol of DNA probe, 9 μg casein, 3 μg of sheared salmon sperm DNA, in 10 mM HEPES (pH 8.0), 6 mM MgCl2, 10% glycerol, and adjusted with 1M KCl to a final concentration of 150 mM were incubated at RT for 45 min and then loaded onto a 4% nondenaturing polyacrylamide bandshift gel (as described in Sorger et al., 1995). Competition experiments were carried out by mixing unlabeled wild-type or random DNA (pUC19) with labeled probe DNA and then adding Mif2p. To measure the ratio of sequence-specific to nonspecific binding, sheared salmon sperm DNA was used as a competitor. The average length of the salmon sperm DNA was 500 base pairs.
RESULTS
Phenotypes of Mif2p Truncations
To evaluate the roles of various protein sequence elements in the Mif2p polypeptide, we designed a series of deletion constructs based on multiple sequence alignments (Figure 1). A S288C haploid yeast strain was generated in which a chromosomal deletion of the endogenous MIF2 gene was rescued by MIF2 expressed from a URA3-CEN plasmid (to create pMIF2:URA3 mif2Δ::KANr). Deletion constructs under the control of the endogenous MIF2 promoter were then integrated at LEU2 in these cells, and the ability of the deletions to substitute for wt MIF2 tested by counterselection for pMIF2:URA3 on 5-fluoroorotic acid. Two of the five regions of Mif2p were found to be essential for function: those between residues 267 and 347, a segment that contains the CENP-C signature box (green; Figure 1), and those between 331 and 438 (red; Figure 2). These regions contain the likely nuclear localization sequence (QRRKKQKK; Nair and Rost, 2005), but we did not explicitly test whether the sequence is essential for correct localization. The red region also contains an AT-hook, PRGRPKK, that is conserved in eight of nine sequenced fungal species with point centromeres and in 19 of 31 fungal species overall (including Schizosaccharomyces pombe). A deletion in the N-terminal region (residues 3–39) had no effect on growth. Deletion of the PEST (Δ103-255) and C-terminal (Δ455-548) regions of the protein resulted in strains that were slow growing and temperature sensitive, with similar phenotypes (large, single-budded cells, indicating mitotic arrest) at the nonpermissive temperature (data not shown).
Figure 2.
Phenotypes of Mif2p domain deletants. (A) amino-acid residue boundaries of domains. (B) Growth at 30°C of yeast expressing various MIF2 deletions as sole copy. Color code corresponds to A. (C) Temperature sensitivity of deletion mutants lacking the N-terminal, PEST, and dimerization domains, respectively. (D) Temperature sensitivity of various point mutations (see text for details). (E) Effects of deleting parts or all of the DNA-binding region (red block in A). The deletion Δ366-436 removes the region between the AT-hook (residues 346–365) and the dimerization domain. For details of each deletion, see Supplemental Table S1.
Expression and Purification of Recombinant Mif2p Fragments
We were unable to express full-length Mif2p in either bacterial or insect cells. N-Terminal truncations of human CENP-C have been shown to yield increased expression levels (Lanini and McKeon, 1995); therefore, we chose to examine expression of truncated Mif2p. Residues 250–549 were selected for expression of a truncated protein based on sequence conservation among 14 fungal Mif2 proteins, which showed an N-proximal PEST region we presumed might destabilize the recombinant product (Figure 1). When fused to a C-terminal hexahistidine tag, the Mr ∼45-kDa fragment Mif2p(250-549) was indeed expressed at high levels in E. coli, and soluble protein could be recovered in good yield from extracts (Supplemental Figure 1A). After purification using a nickel-chelating resin, SDS-polyacrylamide gel electrophoresis (PAGE) also revealed the presence of a slightly more rapidly migrating species, shown by Edman degradation to correspond to Mif2p lacking residues 250–255 (data not shown), and to a species with Mr ∼37 kDa, which had lost the hexahistidine tag (Fig S1A). To identify additional unstructured segments in the Mif2p(250-549) polypeptide, purified protein was subjected to limited proteolysis using trypsin or chymotrypsin. A species with Mr ∼25 kDa was recovered as a stable product of digestion with either protease, and Edman sequencing showed it to correspond to the C-terminal region of Mif2p in both cases (Supplemental Figure 1A). A polypeptide corresponding to the proteolytic endpoint, Mif2p(365-549)His6, was selected for further crystallographic studies, and other fragments were used for DNA binding assays.
Analytical equilibrium ultracentrifugation of bacterially expressed Mif2p(365-549)His6 yielded a molecular mass of 40.6 ± 2.5 kDa (Supplemental Figure 1B); because the predicted mass of a single chain is 19.1 kDa, the protein was clearly dimeric. When SDS-PAGE was performed under reducing and nonreducing conditions, the apparent mass was ∼25 kDa in both cases, demonstrating that disulfide bond formation was not involved in dimerization. Gel filtration suggested a native mass closer to ∼70 kDa, indicating either an elongated structure or the presence of extended “arms”; crystallographic results, presented below, show the latter to be the case. From these biochemical studies, we conclude that the ∼200 C-terminal residues in Mif2p correspond to a well folded domain that forms a stable, noncovalent dimer with extended (and potentially flexible) arms.
Crystallization and Structure Determination of Mif2p(365-530)
While performing a screen to find crystallization conditions for Mif2p(365-549)His6, we found that it was helpful to eliminate the C-terminal 19 residues of Mif2p from the expression construct and to remove the hexahistidine tag (by thrombin cleavage after purification). The resulting species, Mif2p(365-530), crystallized in 4% PEG 3000 at neutral pH in space group P213, with two molecules per asymmetric unit. The structure was determined as described in Materials and Methods, by using multiple isomorphous replacement with multiwavelength anomalous dispersion. The final model included density for residues 437–530 (Figure 3), but residues 365–436 were not visible, implying that they were disordered. Three pairs of chains (each pair defining an asymmetric unit of the P213 U cell) form a ring of six protein monomers (Supplemental Figure 2A), with a crystallographic threefold axis running through its center and a noncrystallographic twofold axis perpendicular to the threefold and intersecting it. One twofold relationship is determined by an extended protein interface; the other, by a tenuous contact that includes a disulfide. The disulfide seems to have formed during crystallization, because the crystals were grown in the absence of reducing agent, and the dimer in solution is noncovalent (see above). Nine water molecules were added in strong electron density, where the capacity to form two hydrogen bonds with groups on the protein was clear. The disordered N-terminal arm extends into the empty region of the crystal delimited by the hexamer packing (Supplemental Figure 2B). The resistance of this arm to both trypsin (Supplemental Figure 1A) and chymotrypsin (data not shown) suggests that it might be a flexibly tethered, folded unit rather than a fully disordered segment of polypeptide chain.
Figure 3.
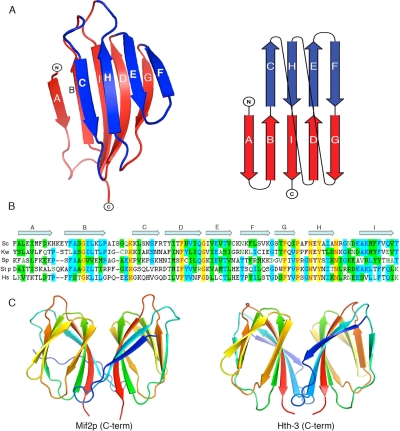
Structure of the Mif2p dimerization domain. (A) Two views of the domain in ribbon representation. The two β-sheets of the cupin domain are in red (ABIDG) and blue (CHEF), respectively. The red sheet is at the dimer interface. On the right is a schematic representation of the strand connectivity in a cupin fold. (B) Amino acid sequence of the Mif2p dimerization domain (residues 438–530) from S. cerevisiae (top line), with secondary-structure elements indicated by arrows. For comparison, sequences of the homologues from Kluyveromyces walti, Schizosaccharomyces pombe, Stongylocentrotus purpuratus, and Homo sapiens (CENP-C) are aligned over the relevant regions. (C) Ribbon representation of the domain dimer (left), compared with the C-terminal domain of Hth-3 from V. cholerae (right).
The C-Terminal Domain of Mif2p Is a Dimeric, β-Jelly Roll
Residues 441–530 of Mif2p fold as a nine-stranded β-jelly roll (Figure 3A), a domain first identified in virus capsids (Harrison et al., 1978) and subsequently in a variety of proteins, including transcription factors, lectins, and kinases (Soisson et al., 1997; Baker et al., 2001; Dunwell et al., 2001; Williams and Westhead, 2002). The jelly roll has come to be called a “cupin” domain, because in many instances it has a barrel-like or cup-like aspect with a ligand-binding site at the open end of the cup (Dunwell et al., 2001). Indeed, multiple sequence alignments have predicted that the C-terminal region of CENP-C proteins might belong to this family (Dunwell et al., 2001). The Mif2p jelly-roll has a five-stranded, antiparallel β-sheet, with strand order ABIDG, and a four-stranded antiparallel sheet with strand order CHEF (Figure 3A). Many conserved residues are glycines or prolines, which break the hydrogen-bonding pattern of individual strands and allow the jelly roll to fold (Dunwell et al., 2001; Williams and Westhead, 2002). The positions of a number of aromatic and aliphatic residues are also conserved. Residues in the ABIDG sheet, which lies at the dimer contact, are better conserved than those in the CHEF sheet.
The ABIDG sheet (Figure 3A) creates an extended dimerization surface (Figure 3C). Conserved, generally nonpolar, residues line the interface. The surface area buried at the dimer interface is 1500 Å2, comparable with the area buried in the complex of an antibody and a protein antigen (Janin and Chothia, 1990) and consistent with the observed stability of the dimer. The area buried at the other twofold contact is only 752 Å2. The thermal parameters of the ABIDG interface residues are lower than those of other surface residues, consistent with their tightly complementary packing. The outer surface of the dimer has no obvious patches of conservation, however, nor does it have prominent insertions into the core cupin fold.
Dimerization In Vivo
Deletion of the C-terminal domain of Mif2p produces cells with a temperature-sensitive growth phenotype (Figure 2). To confirm that the effect is due to failure to dimerize, we introduced mutations Y451S, F452A, and F523A, designed to disrupt the dimer contact (see Supplemental Figure 3). As shown in Figure 2, these mutations indeed impaired cell growth at high temperature, whereas mutation of a conserved residue that lies outside the dimer interface, T488A, had no effect. The severity of our directed mutations is comparable with that of the classic temperature-sensitive mif2-3 mutation (a P505L change), which has previously been subjected to extensive genetic analysis (Brown et al., 1993). P505L should destabilize the conformation of a strand that connects one sheet to another and may destabilize the dimerization domain more generally. Thus, Mif2p dimerization is probably essential for function in vivo.
DNA Binding by Mif2p
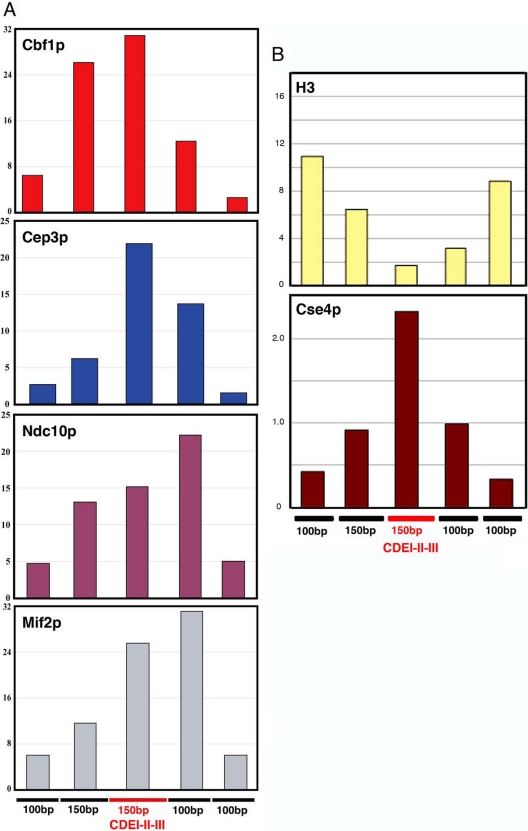
Like its human homologue, CENP-C, Mif2p associates with centromeric DNA (Meluh et al., 1997). CEN binding by Mif2p in vivo requires both CBF3 (Meluh et al., 1997; Ortiz et al., 1999) and Cse4p (Westermann et al., 2003), and quantitative fluorescence microscopy estimates suggest the presence of one Mif2p dimer per CBF3 complex and hence one Mif2p dimer per kinetochore (Joglekar et al., 2006). We carried out high-resolution chromatin immunoprecipitation (ChIP) experiments to determine the approximate location of the Mif2p binding site. Cbf1p and Cep3p served as guides, because their centromeric binding sites are known (Bram and Kornberg 1987; Lechner and Carbon, 1991; Baker and Masison, 1990; Cai and Davis, 1990; Espelin et al., 1997). As shown in Figure 4A (top two panels), Cbf1p maps, as expected, to the CDEI-proximal end of the CENIV CDEI-II-III fragment (red line), whereas Cep3p binds toward the CDEIII-proximal end. The spread of a Gaussian fit to each histogram has a full-width at half maximum of ∼350 base pairs, but the position of the centroid can be determined more precisely, as evident from the distributions in Figure 4 (e.g., Bevington and Robinson, 2002). Ndc10p and Mif2p generate very similar ChIP patterns, both biased toward the CDEIII-proximal side of the centromere (Figure 4A, bottom two panels). The centroid of the Mif2p distribution lies just outside the core CDEI-II-III interval of CENIV, but still within the “extended” (∼80-base pair) region of CDEIII (Espelin et al., 1997). Ndc10p has a closely related distribution, but it also exhibits some tendency to immunoprecipitate the fragment at the opposite end of the CDEI-II-III interval. The relatively large size and extended shape of Ndc10p (Espelin et al., 1997) might account for this pattern. Alternatively, Ndc10p might have a somewhat bimodal binding distribution. Ndc10p is an essential component of the CDEIII-binding, CBF3 complex, but it also can bind CDEII (Espelin et al., 2003). Binding to CDEII in some of the cells (e.g., those cells in G2 that have not yet assembled mature kinetochores) could account for an apparent bimodality. The principal occupant of CDEII is, however, a Cse4p-containing nucleosome (Figure 4B; see below).
Figure 4.
Chromatin immunoprecipitation of CENIV DNA with Cbf1p-GFP, Cep3p, Ndc10p-myc12, and Mif2p (A) and H3 and Cse4p (B). DNA in immunoprecipitated fractions was amplified by PCR with contiguous, nonoverlapping primers spanning CENIV as shown at the bottom of the figures. Bar graphs show the percentage of IP of each PCR fragment relative to total input DNA.
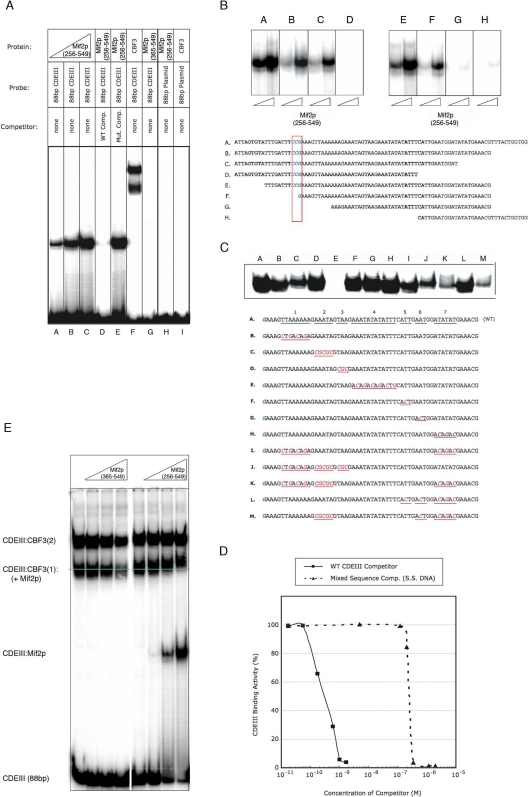
The presence of an AT-hook in Mif2p suggests binding to A:T-rich sequences. Preliminary EMSA demonstrated association of Mif2p(256-549) both with a DNA fragment that included CDEI, -II, and -III (data not shown) and also with an 88-base pair DNA fragment that included only CDEIII (Figure 5A). All binding experiments were performed in the presence of excess random sequence DNA to ensure specificity of the observed interactions. The amount of complex (as indicated by the intensity of the mobility shifted band) on the 88-bp CDEIII probe increased more or less linearly with the concentration of Mif2p, over the full probe concentration range, consistent with our conclusion from the ChIP data (above) that CDEIII contains the principal Mif2p binding site. The addition of excess “cold,” wild-type competitor DNA eliminated the Mif2p-CDEIII binding, whereas addition of DNA with a random sequence did not, further demonstrating specificity of the interaction (Figure 5A). Moreover, we could detect no binding to a radiolabeled 88-base pair negative-control fragment derived from plasmid sequences (Figure 5A). We estimate that Mif2p(256-549) binds wild-type, 88 base-pair CDEIII DNA probes, with a Kd of ∼0.5 nM (Figure 5D).
Figure 5.
Binding of Mif2p to S. cerevisiae CENIV. Recombinant proteins were incubated with radiolabeled DNA fragments, and complexes were resolved on nondenaturing bandshift gels (see Materials and Methods). (A) Binding of Mif2p to CENIV. Recombinant Mif2p(256-549) (lanes A–E), Mif2p(365-549) (lane G), or CBF3 complex (lane F), was mixed with 32P-radiolabeled 88-base pair wild-type CDEIII alone or in addition to unlabeled wild-type 88bp CDEIII (lane D) or 88-bp random plasmid DNA (lane E). Recombinant Mif2p(256-549) (lane H) or CBF3 complex (lane I) was mixed with 32P-radiolabeled 88-base pairs random plasmid DNA. (B and C) DNA requirements for Mif2p binding. Recombinant Mif2p(256-549) was incubated with 32P-radiolabeled DNA probes of indicated sequence and binding evaluated as described in A. The boxed CCG is the conserved binding triplet for one of the Cep3p Zn clusters (cf. Figure 6; McGrew et al., 1986, Ng and Carbon, 1987; Espelin et al., 1997; Bellizzi et al., 2007). (D) Recombinant Mif2p(256-549) was incubated with 32P-radiolabeled 88-bp CDEIII DNA alone, in the presence of increasing amounts of unlabeled 88-bp wild-type CDEIII DNA, or increasing amounts of sonicated salmon sperm DNA. Percentage of binding in the presence of unlabeled WT CDEIII DNA or salmon sperm DNA is shown relative to binding to radiolabeled CDEIII probe alone. (E) Binding of Mif2p to CENIV (88bp CDEIII) in the presence of CBF3. Recombinant Mif2p(365-549) (left lanes) or Mif2p(256-549) (right lanes) was incubated with 32P-radiolabeled 88-bp CDEIII DNA in the presence of CBF3. The two CBF3-shifted bands, CDEIII:CBF3(1) and CDEIII:CBF3(2) contain one and two Ndc10p dimers, respectively. Mif2p(365-549) does not bind DNA, either on its own or with CBF3 bound; Mif2p(256-549) binds both on its own (to excess free DNA, lower shifted band) or together with CBF3 (supershift of CDEIII:CBF3(1) band).
Mif2p(256-549) forms a stable DNA–protein complex with 88-base pair CDEIII DNA; Mif2p(365-549), which lacks the AT-hook motif and the CENP-C signature region, does not (Figure 5A). Mif2p(345-549), which contains the AT-hook but lacks the signature region, binds as strongly as Mif2p(256-549) (data not shown). The requirement of an AT-hook for CEN binding by dimeric Mif2p led us to examine whether the binding site is itself twofold symmetric, with a defined spacing between A:T-rich regions. The data in Figure 5, B and C, suggest that Mif2p binding requires at least 30 base pairs of DNA containing almost exclusively A:T or T:A, but no single sequence within the CDEIII fragment seems to be essential. In particular, the conserved CCG triplet, crucial for CEN binding by CBF3 in vivo and in vitro (McGrew et al., 1986; Hegemann and Fleig, 1993; Espelin et al., 1997), is not important for binding of Mif2p, but the exclusive presence of A:T base pairs seems to be necessary in the sequence that lies between 25 and 35 base pairs to the right of the conserved CCG (Figure 5C, lane E). Overall, the sequence requirements for Mif2p-DNA binding in vitro seem not to depend on single-nucleotide identity, consistent with observations in mammalian cells (Yang et al., 1996; Politi et al., 2002).
We examined the consequences in vivo of deleting or mutating the region between residues 256 and 365 (Figure 2E). The AT-hook did not seem to be essential for viability, but combined deletion of the AT-hook and the sequence linking it to the dimerization domain had a pronounced effect on cell growth. In general, AT-hooks are adjunct DNA-binding elements (Aravind and Landsman, 1998), implying that the region of Mif2p between the AT-hook and the dimerization domain might also contribute to DNA binding. Indeed, this region might be the principal DNA-binding element, because the AT-hook is not absolutely conserved among budding yeast.
Co-occupancy of centromeres with CBF3 may enhance the sequence selectivity of Mif2p. We carried out the EMSA “supershift” experiment reported in Figure 5E to show that CBF3 and Mif2p can together occupy the same DNA fragment. The two bands present in all CBF3-containing lanes represent complexes with one Skp1p:Ctf13p heterodimer, one Cep3p homodimer, and one or two Ndc10p homodimers, respectively (Espelin et al., 1997). Only the band with a single Ndc10p shows a detectable supershift with Mif2p (Figure 5E). Mif2p and (Ndc10p)2 may be mutually exclusive on the second (Ndc10p)2 site, which includes the segment that is altered in sequence E of Figure 5C. Thus, one interpretation of the combined data in Figures 5C, lane E, and 5E is that the complete complex of DNA-binding proteins on a yeast centromere includes one Ndc10p dimer and one Mif2p dimer. It is possible, however, that experimental factors involving in vitro conditions and/or kinetics of binding might have obscured a supershift of the upper band.
We included the variant, H3-like histone Cse4p in our ChIP experiments. The immunoprecipitation histogram in Figure 4B (bottom) has a maximum just between those for Cbf1p and Cep3p, and it is thus centered precisely on CDEII. Moreover, a similar immunoprecipitation experiment with antibodies to H3 shows depletion exactly complementary to Cse4p occupancy (Figure 4B, top). These results, which are consistent with the recently published evidence for a single centromeric nucleosome in budding yeast (Furuyama and Biggins, 2007) and with earlier quantitative fluorescence measurements, suggesting that no more than one nucleosome is present per CBF3 complex (Joglekar et al., 2006), provide additional markers for validating the relative position of Mif2p. Our data also support the notion that the Cse4p-containing nucleosomal assembly binds CDEII.
Role of Mif2p in Kinetochore Assembly
To examine the role of Mif2p in kinetochore assembly, we used ChIP and fluorescence microscopy of GFP-tagged proteins to analyze CEN binding by inner and outer kinetochore proteins in mif2-3 and mif2td degron mutants under permissive (room temperature) and nonpermissive (37°C) conditions (Table 2 and Supplemental Figure 4). As specificity controls, we analyzed kinetochore recruitment of the CBF3 subunit Cep3p, which binds to CEN DNA independently of Mif2p, as well as Mif2p recruitment in ndc10-1 and cse4-1, two mutants in which Mif2p is lost from kinetochores at 37°C. We also analyzed Bim1p-GFP recruitment in mif2-3, because Bim1p requires Mif2p to associate with kinetochores (De Wulf, unpublished). These experiments confirm that CBF3 and Cse4p are required for CEN binding by Mif2p (Table 2). We found that Mif2p function is required to recruit the conserved, inner kinetochore, COMA complex (Ctf19p, Okp1p, Mcm21p, and Ame1p) as well as the conserved outer kinetochore MIND comples (Mtw1p, Nnf1p, Nsl1p, and Dsn1p). The conserved outer kinetochore complexes, Ndc80 (Ndc80p, Nuf2p, Spc24p, and Spc25p) and Spc105 (Spc105p and Kre28p), associate with CEN DNA even under nonpermissive conditions (Table 2). We conclude that although the CBF3 complex is required for the recruitment of all kinetochore proteins (including Cse4p and Mif2p) to budding yeast centromeres, the Cse4p-Mif2p axis seems to be involved in recruiting only a subset. This selective recruitment corresponds to the phenotypes of mutations in proteins recruited by Mif2p and Cse4p. Although CBF3 is required to activate the spindle checkpoint, Cse4p, Mif2p, COMA, and MIND components are not (Gardner et al., 2001, Scharfenberger et al., 2003). Moreover, like mif2-3 and cse4-1, mutants in the COMA and MIND complexes arrest at G2/M with kinetochore attachment (either bipolar or monopolar) established, whereas mutants in the Ndc80 and Spc105 complexes pass through the checkpoint, for which both these complexes are essential (Kiyomitsu et al., 2007; De Wulf, unpublished data). Thus, Mif2p (together with upstream component Cse4p) recruits kinetochore proteins/complexes that contribute to sister chromatid biorientation and force generation in mitosis (e.g., COMA, MIND), proteins that depend on COMA and MIND (e.g., Ctf3), and proteins that protect centromeric cohesin during meiosis I (Iml3p, Chl4p). Kinetochore association by proteins involved in microtubule attachment and spindle checkpoint activity (e.g., the Ndc80p and Spc105p complexes) does not seem to require Mif2p, but only CBF3 (Supplemental Figure 4).
Table 2.
Recruitment to centromeres of various kinetochore components, determined by ChIP and/or fluorescence microscopy in various kinetochore-mutant backgrounds
| Mutant background | Mutated protein (complex) | Analyzed protein (complex) | Temp |
Positive control (Cep3p) |
||
|---|---|---|---|---|---|---|
| 24°C | 37°C | 24°C | 37°C | |||
| cse4–1 | Cse4-Scm3 nucleosome | Mif2p (Mif2p dimer) | +a | −a | +a | +a |
| Okp1p-GFP (COMA) | +a | −a | +a | +a | ||
| Mtw1p-GFP (MIND) | +a | −a | +a | +a | ||
| Ndc80p-GFP (Ndc80 complex) | + | + | + | + | ||
| Spc105p-GFP (Spc105 complex) | + | + | + | + | ||
| mif2–3 | Mif2p dimer | Cse4p-GFP (Cse4p-Scm3p nucleosome) | + | + | n.d.b | n.d. |
| Spc105p-GFP | + | + | + | + | ||
| mif2td | Mif2p dimer | Mtw1p-GFP | +a | −a | n.d. | n.d. |
| Ndc80p-GFP | + | + | n.d. | n.d. | ||
| Ctf19p-GFP (COMA) | +a | −a | n.d. | n.d. | ||
| ndc80–1 | Ndc80 complex | Mif2p-TAP | +c | +c | n.d. | n.d. |
| Spc105p-GFP | + | + | + | + | ||
| spc105–15 | Spc105 complex | Mif2p | + | + | + | + |
| Okp1p-GFP | + | + | + | + | ||
| Mtw1p-GFP | + | + | + | + | ||
| Ndc80p-GFP | + | + | + | + | ||
+, protein localizes to CENIV (ChIP) and/or kinetochores (fluorescence microscopy). Recruitment of all proteins was scored by ChIP, except for Cse4p-GFP in mif2–3 and Mtw1p-GFP, Ndc80p-GFP, and Cftf19p-GFP in the Mif2p degron strain mif2td, for which recruitment was analyzed using wide-field Deltavision fluorescence microscopy (De Wulf et al., 2003). −, protein does not localize to CENIV (ChIP).
a Confirms a previous result reported by Westermann et al. (2003).
b n.d., not determined.
c See Westermann et al. (2003).
Proteins Related to Mif2p
Psi-BLAST searches, using the C-terminal domain sequence of Mif2p as a query, identified a large number of potential relatives in various species. These presumably fall into one or more of the 18 cupin-domain subfamilies tabulated previously (Dunwell et al., 2001). One of the 18 subfamilies contains helix-turn-helix (HTH) transcription factors, with the cupin domain at the C terminus (Aravind and Koonin, 1999). A search with the DALI server (Holm and Sander, 1995) by using the Mif2p dimerization domain as probe uncovers one of the members of this subfamily, Hth-3 from Vibrio cholerae, as a structural homologue. Indeed, Hth-3 contains a dimeric β-jelly roll, with essentially the same dimer interface as Mif2p (Figure 3C). Hth-3 has not been characterized functionally: it is the unpublished product of a structural genomics effort. We can assume that it is indeed a DNA-binding protein, because its HTH domain is convincingly similar to those of known bacterial repressors. Another class of cupin-domain containing transcription factors, in which the DNA-binding domain is C-terminal to the cupin domain, includes AraC (Dunwell et al., 2001). It thus seems that Mif2p is a “borrowed” transcription factor, with a conserved dimerization element present in a number of bacterial DNA-binding proteins.
DISCUSSION
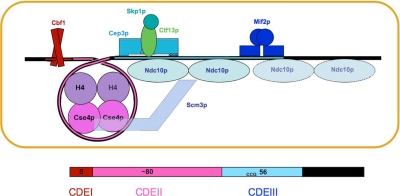
We summarize in Figure 6 our conclusions from the present analysis of Mif2p–centromere interactions, together with the earlier detailed mapping of CBF3 contacts (Espelin et al., 1997) and other published data, as referenced in the figure caption. The DNA-binding properties of Mif2p depend on contacts from an AT-hook and probably from additional structural elements in the region of the molecule between the AT-hook and the dimerization domain. Optimal binding requires an AT-rich DNA segment of ∼30 base pairs. In particular, the sequence between 25 and 35 base pairs to the right of the conserved CCG seems to be essential, at least for in vitro binding to CENIV. This palindromic sequence is appropriate for binding a Mif2p homodimer, but because it is not conserved among all centromeres, the symmetry could be coincidental. One possible binding model would place Mif2p across the major-groove dyad of the palindrome, with the AT-hooks inserted symmetrically into minor grooves one turn apart. Alternatively, other parts of the protein might interact with major-groove sequences one turn apart, with the AT-hooks inserted into the flanking minor grooves. The protein would then cover at least 20–25 base pairs. The AT-rich region of CDEIII to which Mif2p binds tightly is within the extended footprint of Ndc10p, which also binds AT-rich DNA. Some of our data suggest that Mif2p might exclude the second Ndc10p dimer in CBF3, which generates the extended coverage (Espelin et al., 1997). In any case, fully specific association of Mif2p with a centromere probably requires Ndc10p or other components of CBF3, on which Mif2p depends for binding in vivo. The arrangement and overlap of centromeric binding sites for transcription-factor–like proteins recalls the properties of a transcriptional enhancer (Panne et al., 2007), as first suggested over a decade ago (Meluh and Koshland, 1995). In some enhanceosomes, the identity of proteins occupying particular sites can vary with the physiological state of the cell; likewise, in yeast kinetochores it is possible that Ndc10p and Mif2p occupy the same or overlapping sites at different cell cycle stages.
Figure 6.
Diagram summarizing binding data in this article, in previous work on CBF3–centromere interactions (Espelin et al., 1997), and in the literature. CBF3 comprises a Skp1p:Ctf13p heterodimer associated closely with a Cep3p dimer; addition of one or two dimers of Ndc10p generates “core” and “extended” complexes, respectively. The Cse4p nucleosome contains Scm3p, Cse4p, and H4 in 1:1:1 proportion, but it seems to lack H2a and H2b (Mizuguchi et al., 2007). An interaction of Scm3p and Ndc10p has been proposed to be the mechanism by which CBF3 recruits the Cse4p nucleosome (Camahort et al., 2007). We discuss in the text whether adding a second Ndc10p dimer, to form an extended complex, is compatible with recruitment of Mif2p; the possibility, that the two are mutually exclusive is suggested by a white hatching across the second Ndc10p dimer. The guide at the bottom of the figure relates the spatial organization diagrammed in the upper part to the sequence and organization of centromeric DNA in yeast.
What about CDEII? The recent demonstration that the Cse4p-containing nucleosome, recruited jointly by CBF3 and Scm3p, lacks H2A and H2B, suggests that it might cover only ∼80 base pairs of DNA, which corresponds closely to the required length of CDEII in S. cerevisiae (Gaudet and Fitzgerald-Hayes, 1987; Mizuguchi et al., 2007). Moreover, in two budding yeasts with point centromeres, Eremothecium gossypii and Kluyveromyces lactis, CDEII is twice that length and might therefore contain two such tetramer-core nucleosomes (Meraldi et al., 2006). Wrapping around a histone tetramer core would probably exclude Ndc10p and Mif2p from CDEII, even though Ndc10p can associate with empty CDEII in vitro (Espelin et al., 2003).
The parts of Mif2p that are clearly conserved in CENP-C proteins of higher eukaryotes are the CENP-C signature element and the dimerization domain. The presumptive DNA-binding domain lies between these regions in yeast but N-terminal to the CENP-C signature in metazoans and plants. Because DNA-binding regions of many yeast transcription factors (e.g., the Zn2Cys6 clusters of Gal4p and its relatives, including the Cep3p component of CBF3) seem to have evolved divergent structures, and as CBF3 itself is yeast specific, the transposition of the two essential regions is still consistent with conserved overall function.
Mif2p is not required for recruitment of Cse4p, but it is required for centromeric association of the MIND and COMA complexes and of proteins that depend on MIND and COMA for kinetochore localization (e.g., Ctf3 complex, Impl3p, Chl4p; Measday et al., 2002) (Supplemental Figure S4). Although the presence of Mif2p at a centromere leads to recruitment of only a subset of kinetochore components in budding yeast, its orthologues, Cnp3 in fission yeast and CENP-C in animal cells, participate in recruitment of all kinetochore subunits (except for the upstream Cse4p orthologues Cnp1 and CENP-A) (Oegema et al., 2001; Cheeseman et al., 2004; Liu et al., 2006; Kwon et al., 2007). Thus, CENP-C in higher eukaryotes seems to have functions that are shared between Mif2p and CBF3 in budding yeast.
The dimerization domain is necessary for proper Mif2p function, but it is not essential for viability. It is therefore unlikely to be the recruitment structure. Indeed, its surface exhibits no particularly extended patches of conserved residues, as one might expect for a protein association domain. We suggest instead that the conserved CENP-C signature element, present in homologues of every species examined, creates at least part of the contact for the next members of the assembly hierarchy.
Supplementary Material
ACKNOWLEDGMENTS
We thank Nick Larsen for help with data collection; John Bellizzi, Piotr Sliz, and Ben Spiller for advice on crystallographic methods; Jawdat Al-Bassam, for help with analytical ultracentrifugation; Kevin Corbett, for some final adjustments and refinement of the model; Caroline George for help with the kinetochore recruitment experiments; members of Fred Winston's laboratory (Harvard Medical School) for advice on yeast-strain constructs; and JJ Miranda and Ronnie Wei for discussion. P.K.S. acknowledges National Institutes of Health grants GM-51464 and CA-84179. S.C.H. is an Investigator in the Howard Hughes Medical Institute. P.D.W. and K.T.S. acknowledge fellowships from the Charles King Trust.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-03-0297) on August 13, 2008.
REFERENCES
- Aravind L., Koonin D. DNA-binding proteins and evolution of transcription regulation in the archaea. Nucleic Acids Res. 1999;27:4658–4670. doi: 10.1093/nar/27.23.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L., Landsman D. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998;26:4413–4421. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker L. J., Dorocke J. A., Harris R. A., Timm D. E. The crystal structure of yeast thiamin pyrophosphokinase. Structure. 2001;9:539–546. doi: 10.1016/s0969-2126(01)00615-3. [DOI] [PubMed] [Google Scholar]
- Baker R. E., Masison D. C. Isolation of the gene encoding the Saccharomyces cerevisiae centromere-binding protein CP1. Mol. Cell Biol. 1990;10:2458–2467. doi: 10.1128/mcb.10.6.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellizzi J. J., III, Sorger P. K., Harrison S. C. Crystal structure of the yeast inner kinetochore subunit, Cep3p. Structure. 2007;15:1422–1430. doi: 10.1016/j.str.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevington P. R., Robinson D. K. Data Reduction and Error Analysis for the Physical Sciences. New York: McGraw-Hill; 2002. [Google Scholar]
- Boggon T. J., Shapiro L. Screening for phasing atoms in protein crystallography. Structure. 2000;8:R143–R149. doi: 10.1016/s0969-2126(00)00168-4. [DOI] [PubMed] [Google Scholar]
- Bram R. J., Kornberg R. D. Isolation of a Saccharomyces cerevisiae centromere DNA-binding protein, its human homolog, and its possible role as a transcription factor. Mol. Cell Biol. 1987;7:403–409. doi: 10.1128/mcb.7.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. T. Sequence similarities between the yeast chromosome segregation protein Mif2 and the mammalian centromere protein CENP-C. Gene. 1995;160:111–116. doi: 10.1016/0378-1119(95)00163-z. [DOI] [PubMed] [Google Scholar]
- Brown M. T., Goetsch L., Hartwell L. H. MIF2 is required for mitotic spindle integrity during anaphase spindle elongation in Saccharomyces cerevisiae. J. Cell Biol. 1993;123:387–403. doi: 10.1083/jcb.123.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai M., Davis R. W. Yeast centromere binding protein CBF1, of the helix-loop-helix protein family, is required for chromosome stability and methionine prototrophy. Cell. 1990;61:437–446. doi: 10.1016/0092-8674(90)90525-j. [DOI] [PubMed] [Google Scholar]
- Camahort R., Li B., Florens L., Swanson S. K., Washburn M. P., Gerton J. L. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol. Cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Cheeseman I. M., Niessen S., Anderson S., Hyndman F., Yates J. R., 3rd, Oegema K., Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K., Main P. Miscellaneous algorithms for density modification. Acta Crystallogr. D Biol. Crystallogr. 1998;54:487–493. doi: 10.1107/s0907444997011980. [DOI] [PubMed] [Google Scholar]
- De Wulf P., McAinsh A. D., Sorger P. K. Hierarchical assembly of the budding yeast kinetochore form multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doheny K. F., Sorger P. K., Hyman A. A., Tugendreich S., Spencer F., Hieter P. Identification of essential components of the S. cerevisiae kinetochore. Cell. 1993;73:761–774. doi: 10.1016/0092-8674(93)90255-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwell J. M., Culham A., Carter C. E., Sosa-Aguirre C. R., Goodenough P. W. Evolution of functional diversity in the cupin superfamily. Trends Biochem. Sci. 2001;26:740–746. doi: 10.1016/s0968-0004(01)01981-8. [DOI] [PubMed] [Google Scholar]
- Earnshaw W. C., Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- Espelin C. W., Kaplan K. B., Sorger P. K. Probing the architecture of a simple kinetochore using DNA-protein crosslinking. J. Cell Biol. 1997;139:1383–1396. doi: 10.1083/jcb.139.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espelin C. W., Simons K. T., Harrison S. C., Sorger P. K. Binding of the essential Saccharomyces cerevisiae kinetochore protein Ndc10p to CDEII. Mol. Biol. Cell. 2003;14:4557–4568. doi: 10.1091/mbc.E02-08-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M., Clarke L., Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- Furuyama S., Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. USA. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet A., Fitzgerald-Hayes M. Alterations in the adenine-plus-thymine-rich region of CEN3 affect centromere function in Saccharomyces cerevisiae. Mol. Cell Biol. 1987;7:68–75. doi: 10.1128/mcb.7.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner R. D., Poddar A., Yellman C., Tovormina P. A., Monteagudo M. C., Burke D. J. The spindle checkpoint of the yeast Saccharomyces cerevisiae requires kinetochore fuction and maps to the CBF3 domain. Genetics. 2001;157:1493–1502. doi: 10.1093/genetics/157.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S. C., Olson A. J., Schutt C. E., Winkler F. K., Bricogne G. Tomato bushy stunt virus at 2.9 Å resolution. Nature. 1978:368–373. doi: 10.1038/276368a0. [DOI] [PubMed] [Google Scholar]
- Hegemann J. H., Fleig U. N. The centromere of budding yeast. Bioessays. 1993;15:451–460. doi: 10.1002/bies.950150704. [DOI] [PubMed] [Google Scholar]
- Hemmerich P., Stoyan T., Wieland G., Koch M., Lechner J., Diekmann S. Interaction of yeast kinetochore proteins with centromere-protein/transcription factor Cbf1. Proc. Natl. Acad. Sci. USA. 2000;97:12583–12588. doi: 10.1073/pnas.97.23.12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L., Sander C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- Janin J., Chothia C. The structure of protein-protein recognition sites. J. Biol. Chem. 1990;265:16027–16030. [PubMed] [Google Scholar]
- Joglekar A. P., Bouck D. C., Molk J. N., Bloom K. S., Salmon E. D. Molecular architecture of a kinetochore-microtubule attachment site. Nat. Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kiyomitsu T., Obuse C., Yanagida M. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell. 2007;13:663–676. doi: 10.1016/j.devcel.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Kwon M. S., Hori T., Okada M., Fukagawa T. CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol. Biol. Cell. 2007;18:2155–2168. doi: 10.1091/mbc.E07-01-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanini L., McKeon F. Domains required for CENP-C assembly at the kinetochore. Mol. Biol. Cell. 1995;6:1049–1059. doi: 10.1091/mbc.6.8.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner J., Carbon J. A 240 kD multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- Liu S. T., Rattner J. B., Jablonski S. A., Yen T. J. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAinsh A. D., Tytell J. D., Sorger P. K. Structure, function, and regulation of budding yeast kinetochores. Annu. Rev. Cell Dev. Biol. 2003;19:519–539. doi: 10.1146/annurev.cellbio.19.111301.155607. [DOI] [PubMed] [Google Scholar]
- McGrew J., Diehl B., Fitzgerald-Hayes M. Single base-pair mutations in centromere element III cause aberrant chromosome segregation in Saccharomyces cerevisiae. Mol. Cell Biol. 1986;6:530–538. doi: 10.1128/mcb.6.2.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday V., Hailey D. W., Pot I., Givan S. A., Hyland K. M., Cagney G., Fields S., Davis T. N., Hieter P. Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Genes Dev. 2002;16:101–113. doi: 10.1101/gad.949302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee P. C., Mistrot C., Guacci V., Koshland D. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol. Cell. 1999;4:445–450. doi: 10.1016/s1097-2765(00)80347-0. [DOI] [PubMed] [Google Scholar]
- Meluh P. B., Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh P. B., Koshland D. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 1997;11:3401–3412. doi: 10.1101/gad.11.24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., McAinsh A. D., Rheinbay E., Sorger P. K. Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 2006;7:R23. doi: 10.1186/gb-2006-7-3-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi G., Xiao H., Wisniewski J., Smith M. M., Wu C. Nonhistone Scm3 and histones CenH3–H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Nair R., Rost B. Mimicking cellular sorting improves prediction of subcellular localization. J. Mol. Biol. 2005;348:85–100. doi: 10.1016/j.jmb.2005.02.025. [DOI] [PubMed] [Google Scholar]
- Ng R., Carbon J. Mutational and in vitro protein-binding studies on centromere DNA from Saccharomyces cerevisiae. Mol. Cell Biol. 1987;7:4522–4534. doi: 10.1128/mcb.7.12.4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K., Desai A., Rybina S., Kirkham M., Hyman A. A. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz J., Stemmann O., Rank S., Lechner J. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Panne D., Maniatis T., Harrison S. C. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta A. F., Cooke C. A., Earnshaw W. C. Structure of the human centromere at metaphase. Trends Biochem. Sci. 1990;15:181–185. doi: 10.1016/0968-0004(90)90158-8. [DOI] [PubMed] [Google Scholar]
- Politi V., Perini G., Trazzi, Pliss S., A., Raska I., Earnshaw W. C., Della Valle G. CENP-C binds the alpha-satellite DNA in vivo at specific centromere domains. J. Cell Sci. 2002;115:2317–2327. doi: 10.1242/jcs.115.11.2317. [DOI] [PubMed] [Google Scholar]
- Saitoh H., Tomkiel J., Cooke C. A., Ratrie H., 3rd, Maurer M., Rothfield N. F., Earnshaw W. C. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- Scharfenberger M., Ortiz J., Grau N., Janke C., Schiebel E., Lechner J. Nsl1p is essential for the establishment of bipolarity and the localization of the Dam-Duo complex. EMBO J. 2003;22:6584–6597. doi: 10.1093/emboj/cdg636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soisson S. M., MacDougall-Shackleton B., Schleif R., Wolberger C. The 1.6 A crystal structure of the AraC sugar-binding and dimerization domain complexed with D-fucose. J. Mol. Biol. 1997;273:226–237. doi: 10.1006/jmbi.1997.1314. [DOI] [PubMed] [Google Scholar]
- Sorger P. K., Doheny K. F., Hieter P., Kopski K. M., Huffaker T. C., Hyman A. A. Two genes required for the binding of an essential Saccharomyces cerevisiae kinetochore complex to DNA. Proc. Natl. Acad. Sci. USA. 1995;92:12026–12030. doi: 10.1073/pnas.92.26.12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler S., Rogers K., Weitze S., Morey L., Fitzgerald-Hayes M., Baker R. E. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proc. Natl. Acad. Sci. USA. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov A. V., Kingsbury J., Koshland D. CEP3 encodes a centromere protein of Saccharomyces cerevisiae. J. Cell Biol. 1995;128:749–760. doi: 10.1083/jcb.128.5.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P. B., Bryson T. D., Henikoff S. Adaptive evolution of centromere proteins in plants and animals. J. Biol. 2004;3:18. doi: 10.1186/jbiol11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T. C., Berendzen J. Bayesian correlated MAD phasing. Acta Crystallogr. D Biol. Crystallogr. 1997;53:571–579. doi: 10.1107/S0907444997005398. [DOI] [PubMed] [Google Scholar]
- Westermann S., Cheeseman I. M., Anderson S., Yates J. R., 3rd, Drubin D. G., Barnes G. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J. Cell Biol. 2003;163:215–222. doi: 10.1083/jcb.200305100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams A., Westhead D. R. Sequence relationships in the legume lectin fold and other jelly rolls. Protein Eng. 2002;15:771–774. doi: 10.1093/protein/15.10.771. [DOI] [PubMed] [Google Scholar]
- Yang C. H., Tomkiel J., Saitoh H., Johnson D. H., Earnshaw W. C. Identification of overlapping DNA-binding and centromere-targeting domains in the human kinetochore protein CENP-C. Mol. Cell Biol. 1996;16:3576–3586. doi: 10.1128/mcb.16.7.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.