Abstract
Efg1 is essential for hyphal development and virulence in the human pathogenic fungus Candida albicans. How Efg1 regulates gene expression is unknown. Here, we show that Efg1 interacts with components of the nucleosome acetyltransferase of H4 (NuA4) histone acetyltransferase (HAT) complex in both yeast and hyphal cells. Deleting YNG2, a subunit of the NuA4 HAT module, results in a significant decrease in the acetylation level of nucleosomal H4 and a profound defect in hyphal development, as well as a defect in the expression of hypha-specific genes. Using chromatin immunoprecipitation, Efg1 and the NuA4 complex are found at the UAS regions of hypha-specific genes in both yeast and hyphal cells, and Efg1 is required for the recruitment of NuA4. Nucleosomal H4 acetylation at the promoters peaks during initial hyphal induction in an Efg1-dependent manner. We also find that Efg1 bound to the promoters of hypha-specific genes is critical for recruitment of the Swi/Snf chromatin remodeling complex during hyphal induction. Our data show that the recruitment of the NuA4 complex by Efg1 to the promoters of hypha-specific genes is required for nucleosomal H4 acetylation at the promoters during hyphal induction and for subsequent binding of Swi/Snf and transcriptional activation.
INTRODUCTION
Candida albicans has emerged as one of the most prevalent opportunistic fungal pathogens in humans. It causes mucosal as well as systemic candidiasis, especially in immunocompromised patients. C. albicans can undergo reversible morphogenetic transitions between budding yeast, pseudohyphal, and hyphal growth forms. Its unique ability to switch from yeast to hyphal growth in response to various signals is essential for its pathogenicity.
Central to the yeast-to-hypha transition is the transcription factor Efg1, a member of the Asm1p, Phd1p, Sok2p, Efg1p, and StuAp (APSES) family of fungal proteins that regulate cellular differentiation in ascomycetes. Members of this family share a conserved DNA binding domain. Efg1 is an essential regulator of hyphal development, chlamydospore formation, and white-opaque phenotypic switching in C. albicans (Lo et al., 1997; Stoldt et al., 1997; Sonneborn et al., 1999; Srikantha et al., 2000; Zordan et al., 2007). It is suggested to function downstream of the cAMP/protein kinase A (PKA) pathway in hyphal development (Sonneborn et al., 2000; Bockmuhl and Ernst, 2001), and both Efg1 and PKA are required for the induction of hypha-specific genes. However, Efg1 also regulates other genes that are not modulated by the cAMP pathway (Sohn et al., 2003; Doedt et al., 2004; Harcus et al., 2004; Setiadi et al., 2006). Numerous in vitro and in vivo studies with efg1 mutants have demonstrated that Efg1 is important for C. albicans virulence and for the interactions of C. albicans with endothelial and epithelia cells, as well as biofilm formation and catheter infection (Lo et al., 1997; Phan et al., 2000; Dieterich et al., 2002; Lewis et al., 2002; Ramage et al., 2002; Garcia-Sanchez et al., 2004). Despite its importance, molecular mechanisms for how Efg1 regulates gene expression during hyphal development and other processes are still unknown.
In addition to transcription factors, chromatin structure also plays a critical role in the regulation of gene expression (Narlikar et al., 2002). Chromatin structure is regulated by histone modifications and chromatin remodeling. N-terminal histone tail acetylation by histone acetyltransferases (HATs) strongly correlates with active transcription. In Saccharomyces cerevisiae, Gcn5 encodes the catalytic subunit of the HAT complexes Ada2-Gcn5-Ada3 (ADA) and Spt-Ada-Gcn5-Acetyltransferase (SAGA) that acetylate histones H3 and H2B (Grant et al., 1997). Esa1 encodes a member of the MOZ, Ybf2/Sas3, Sas2, and Tip60 (MYST) family of HATs, and it is the catalytic subunit of the nucleosome acetyltransferase of H4 (NuA4) HAT complex that acetylates histones H4 and H2A (Allard et al., 1999). The NuA4 complex contains two functional modules: the recruitment module for targeting Esa1 acetyltransferase activity to specific genomic loci and the nucleosome HAT module that consists of Esa1, Epl1, and Yng2 (Boudreault et al., 2003). A genome-wide study has shown that both Gcn5 and Esa1 are recruited to the promoters of active protein-coding genes with the maximal occupancy at the upstream activation sequences (Robert et al., 2004), consistent with the notion that they are recruited through DNA binding transcription factors (Naar et al., 2001). A few transcription factors have been shown to recruit SAGA or NuA4 to specific promoters (Utley et al., 1998; Bhaumik and Green, 2001; Brown et al., 2001; Larschan and Winston, 2001; Nourani et al., 2004). Thus far, however, functions of specific HATs in Candida albicans have not been demonstrated. The ATP-dependent chromatin remodeling complex Swi/Snf has been shown to be essential for hyphal development in C. albicans (Mao et al., 2006). Deletion of SWI1 or SNF2 blocks hyphal development and the expression of hypha-specific genes under all hyphal induction conditions examined.
ECE1, HWP1, and ALS3 are the first few hypha-specific genes identified (Birse et al., 1993; Hoyer et al., 1998; Staab and Sundstrom, 1998; Zhao et al., 2004). Although the UAS regions for HWP1 and ALS3 have been mapped (Argimon et al., 2007; Kim et al., 2007), how Efg1 regulates their expression has not been elucidated. In this study, we identify an orthologue of the NuA4 subunit as an Efg1 interacting protein in a two-hybrid screen. Deletion analysis suggests that C. albicans has a conserved NuA4 complex and that NuA4 is required for hyphal development and the expression of hypha-specific genes. We demonstrate Efg1 interacts with NuA4, and both are located at the promoters of hypha-specific genes in yeast and hyphae. Importantly, Efg1 is required for the recruitment of NuA4 and an increase in H4 acetylation at the promoters during the yeast-to-hypha transition. Moreover, we show that the Swi/Snf complex binds to promoters of hypha-specific genes only in hyphae and that this binding is mediated by Efg1. Our data suggest that Efg1 functions by recruiting NuA4 to promoters of hypha-specific genes and that this is required for subsequent recruitment of the Swi/Snf complex and transcriptional activation during the yeast-to-hypha transition.
MATERIALS AND METHODS
Strains and Culture Conditions
The C. albicans and S. cerevisiae strains used in this study are listed in Table 1. YPD + 10% bovine serum media were used for hyphal induction.
Table 1.
C. albicans and S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| C. albicans | ||
| SC5314 | Wild type | Fonzi and Irwin (1993) |
| CAI4 | ura3::1 imm434/ura3::1 imm434 | Fonzi and Irwin (1993) |
| BWP17 | ura3::1 imm434/ura3::1 imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | Wilson et al. (1999) |
| CSC1 | ura3::1 imm434/ura3::1 imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG eaf7::ARG4/eaf7::HIS1 | This study |
| CLY1 | ura3::1 imm434/ura3::1 imm434 YNG2/yng2::hisG-URA3-hisG | This study |
| CLY2 | ura3::1 imm434/ura3::1 imm434 YNG2/yng2::hisG | This study |
| CLY3 | ura3::1 imm434/ura3::1 imm434 yng2::hisG/yng2::hisG-URA3-hisG | This study |
| CLY4 | ura3::1 imm434/ura3::1 imm434 yng2::hisG/yng2::hisG | This study |
| HLC52 | ura3::1 imm434/ura3::1 imm434 efg1::hisG/efg1::hisG-URA3-hisG | Lo et al. (1997) |
| HLC54 | ura3::1 imm434/ura3::1 imm434 efg1::hisG/efg1::hisG | Lo et al. (1997) |
| HLY3234 | ura3::1 imm434/ura3::1 imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG EFG1/EFG1-TAP-URA3 | Cao et al. (2006) |
| CLY5 | ura3::1 imm434/ura3::1 imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG EFG1/EFG1-TAP-URA3 RP10::ACT1p-EAF7-13MYCFLAG-HIS1 | This study |
| CLY6 | ura3::1 imm434/ura3::1 imm434 RP10::ACT1p-EAF7-13MYCFLAG-URA3 | This study |
| CLY7 | ura3::1 imm434/ura3::1 imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG EFG1/EFG1-TAP-URA3 RP10::ACT1p-EPL1-13MYCFLAG-HIS1 | This study |
| CLY8 | ura3::1 imm434/ura3::1 imm434 RP10::ACT1p-EPL1-13MYCFLAG-URA3 | This study |
| CLY9 | ura3::1 imm434/ura3::1 imm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG EFG1/EFG1-TAP-URA3 RP10::ACT1p-YNG2-13MYCFLAG-HIS1 | This study |
| CLY10 | ura3::1 imm434/ura3::1 imm434 RP10::ACT1p-YNG2-13MYCFLAG-URA3 | This study |
| CLY11 | ura3::1 imm434/ura3::1 imm434 SNF2/SNF2-13MYCFLAG-URA3 | This study |
| CLY12 | ura3::1 imm434/ura3::1 imm434 EFG1/EFG1-13MYCFLAG-URA3 | This study |
| S. cerevisiae | ||
| EGY48 (p8op-lacZ) | MATα, his3, trp1, LexAop(×6)-leu2, LexAop(×8)-lacZ flo8-1 | Gyuris et al. (1993) |
Yeast Two-Hybrid Screen
The plasmid pEG202-EFG1, which expresses the LexA-Efg1 fusion protein, was used as a bait vector to screen the C. albicans two-hybrid library in S. cerevisiae strain EGY48 (Ni et al., 2004). Among 1 × 107colonies screened, 80 colonies could grow on the minimal medium (SC gal/raff without Ura, His, Trp, Leu), and 44 of them showed a positive response in the β-galactosidase assay. All 44 positives gave light blue color by β-galactosidase assay. Restriction mapping and DNA sequence analysis demonstrated that these 44 inserts represent 12 different genes (data not shown). Three of 44 contained Eaf7.
Plasmid Construction
pBA1-YNG2 for the expression of C. albicans YNG2 in C. albicans was constructed by placing a 0.9-kb polymerase chain reaction (PCR) fragment containing YNG2 coding sequence into the ClaI-KpnI site of plasmid pBA1. Primer 1 and primer 2 were used for PCR amplification.
The full-length coding sequences of EAF7, EPL1, and YNG2 were PCR-amplified (primers 17, 18, 19, 20, 21, 22) and inserted into pJG4-5, generating pJG-EAF7, pJG-EPL1, and pJG-YNG2 for the yeast two-hybrid assay.
EAF7-MYC13, EPL1-MYC13, and YNG2-MYC13.
The EAF7, YNG2, and EPL1 coding sequences were PCR amplified by using primers 3, 4, 5, 6, 7, and 8 (Table 3). The 1.3-kb PCR product of EAF7 was digested with BamHI and MluI and then inserted into the BamHI-MluI sites of pPR671, to produce pPR671-EAF7 (EAF7-13MYC-FLAG-HIS1). The 2.3-kb PCR product of EPL1 and 0.9-kb PCR product of YNG2 were digested with NotI and MluI and then inserted into the NotI-MluI sites of pPR671, to produce pPR671-EPL1 (EPL1-13MYC-FLAG-HIS1) and pPR671-YNG2 (YNG2-13MYC-FLAG-HIS1). The pPR671-EAF7, pPR671-EPL1, and pPR671-YNG2 were digested with StuI to target the integration of the plasmid into the genomic RP10 locus under HIS1 selection. Expression of Eaf7p, Epl1p, and Yng2p under the ACT1 promoter in YPD at 30°C was verified by Western analysis by using an anti c-Myc-conjugated peroxidase antibody (Sigma-Aldrich, St. Louis, MO).
Table 3.
Primers used in this study
| Primer | Sequence | Purpose and features |
|---|---|---|
| 1 | 5′-GTCATCGATATGGATACATCAACTGTACT | pBA1-YNG2 |
| 2 | 5′-GTCGGTACCTACTATACACTCAGTTCTTC | |
| 3 | 5′-GGCGGATCCCATGGCAACTTTGGATGAGAAT | pPR671-EAF7 |
| 4 | 5′-CGACGCGTCGTCTTTTACGACTACTGCGTG | |
| 5 | 5′-CGGCGGCCGCTTGACTGGATATGGCAGCA | pPR671-EPL1 |
| 6 | 5′-CGACGCGTCGTTTACGTGTTCTATTCAACAA | |
| 7 | 5′-ATCGGCGGCCGCCATGGATACATCAACTGTACT | pPR671-YNG2 |
| 8 | 5′-GGTCACGCGTCGGTTCTTCCGTTTCTTTTTTC | |
| 9 | 5′-GTCGGTACCGGATAATAGGGAAACACATG | pYNG2-KO-1 |
| 10 | 5′-GTCGAATTCTTGTCTGTGCTACGTCAATC | |
| 11 | 5′-GTCggATccAGAACTGAGTGTATAGTATG | pYNG2-KO-2 |
| 12 | 5′-GTCTCTAGAGAATATATTGACAACTCGGA | |
| 13 | 5′-GCTGGATCCATGTCTGACGACGATGATGACAATGAC | pPR675 |
| 14 | 5′-GGAGACGCGTCGATCAAAATTTGCTGGTGTAGACTC | |
| 15 | 5′-GCTGGATCCGAGATGGGATTTTGAAATCAG | pPR676 |
| 16 | 5′-GGAGACGCGTCGATGACTGAACTTGGGGTGAT | |
| 17 | 5′-GTCGAATTCATGGCAACTTTGGATGAGAA | pJG-EAF7 |
| 18 | 5′-GTCCTCGAGTCATCTTTTACGACTACTGC | |
| 19 | 5′-GTCGAATTCATGGCAGCAGCACCACCACC | pJG-EPL1 |
| 20 | 5′-GTCCTCGAGTCTCGCCACAGTTGCCTGCC | |
| 21 | 5′-GTCGAATTCATGGATACATCAACTGTACT | pJG-YNG2 |
| 22 | 5′-GTCCTCGAGCATACTATACACTCAGTTCT |
pYNG2-KO.
Two PCR fragments, 0.68 kb (primers 9 and 10), 0.74 kb (primers 11 and 12), and pCUB6 were digested with KpnI-EcoRI, BamHI-XbaI, and BamHI-PstI, respectively, and then the two PCR fragments and a 4.8-kb fragment containing HISG-URA3-HISG cassette from pCUB6 were sequentially ligated to pBluescript SK+ to generate pYNG2-KO. pYNG2-KO was digested with KpnI and SstI and transformed into CAI4 to produce YNG2/yng2 and yng2/yng2 strains. Spontaneous Ura− derivatives were selected on 5-fluoro-orotic acid-containing medium. The disruption was confirmed by Southern blotting.
SNF2-MYC13-URA3, EPL1-MYC13-URA3, and EFG1-MYC13-URA3.
The HIS1 fragment of pPR671 (digested with HindIII) was replaced with the URA3 fragment from pFLAG-ACT1-URA (Umeyama et al., 2002) to generate plasmid pPR673. The NotI-MluI fragment from pPR673 of EPL1 coding sequence was inserted into NotI-MluI site of pPR673 to generate pPR674. A 0.9-kb PCR product (primers 13 and 14) containing the C-terminal SNF2 coding region was inserted into the BamHI-MluI sites of pPR673 to generate pPR675. A 0.8-kb PCR product (primers 15 and 16) containing the C-terminal EFG1 coding region was inserted into the BamHI-MluI sites of pPR673 to generate pPR676. CAI4, HLC54, and CSC1 were transformed with StuI-digested pPR673 or pPR674, respectively, to generate strains as Myc control or strains for Epl1-myc13 fusion proteins. The pPR675 was digested with PstI to target the integration of the plasmid into its own locus of CAI4, HLC54, and CLY4 to express Snf2-myc13 fusion proteins. The pPR676 was digested with SpeI to target the integration of the plasmid into its own locus of CAI4, CSC1, and CLY4 to express Efg1-myc13 fusion proteins.
SC5314 genomic DNA was used as template for all PCR amplifications. All constructs were verified by DNA sequencing. The plasmids used in this study are listed in Table 2. The primers used for PCR amplification are listed in Table 3.
Table 2.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| Library | C. albicans genomic two-hybrid library in pJG4-5 | Ni et al. (2004) |
| pJG4-5 | 2 μ, TRP1-based vector carrying the B42 activation domain | Gyuris et al. (1993) |
| pJG-EAF7 | 1.34-kb full-length C. albicans EAF7 in pJG4-5 | This study |
| pJG-EPL1 | 2.26-kb full-length C. albicans EPL1 in pJG4-5 | This study |
| pJG-YNG2 | 0.9-kb full-length C. albicans YNG2 in pJG4-5 | This study |
| pEG202 | 2 μ, HIS3-based vector carrying the lexA DNA-binding domain | Gyuris et al. (1993) |
| pEG202-EFG1 | 1.6-kb full-length EFG1 in pEG202 | Cao et al. (2006) |
| pCUB6 | Substitution of S. cerevisiae URA3 by C. albicans URA3 in pNKY50 | Fonzi and Irwin (1993) |
| pBluescript SK+ | Apr; ColEI origin | Stratagene (La Jolla, CA) |
| pYNG2-KO | 0.68 kb, 0.74 kb of C. albicans YNG2 and hisG-URA3-hisG in pBluescript SK+ | This study |
| pBA1 | C. albicans ADH1 promoter in BES116 | Cao et al. (2006) |
| pBA1-YNG2 | 0.9-kb full-length C. albicans YNG2 in BA1 | This study |
| pMSCTAP | A codon-optimized TAP-tag in a Bluescript vector that carries C. albicans URA3 | Cao et al. (2006) |
| pFLAG-ACT1-URA | C. albicans ACT1 promoter, FLAG, URA3 | Umeyama et al. (2002) |
| pPR671 | C. albicans ACT1 promoter,13×myc-FLAG, HIS1 | Cao et al. (2006) |
| pPR671-EPL1 | 2.26-kb full-length C. albicans EPL1 in pPR671 | This study |
| pPR671-EAF7 | 1.34-kb full-length C. albicans EAF7 in pPR671 | This study |
| pPR671-YNG2 | 0.9-kb full-length C. albicans YNG2 in pPR671 | This study |
| pPR673 | C. albicans ACT1 promoter, 13×xmyc-FLAG, URA3 | This study |
| pPR674 | 2.26-kb full-length C. albicans EPL1 in pPR673 | This study |
| pPR675 | 0.9kb of SNF2 in pPR673 | This study |
| pPR676 | 0.8kb of EFG1 in pPR673 | This study |
Cell Synchronization and Fluorescence-activated Cell Sorting (FACS) Analysis
Cell synchronization was performed as described previously (Barelle et al., 2003). Cells were patched to YPD and grown at 25°C. After 36 h, cells were taken from the plates and inoculated at 2 × 108 cells/ml into SC medium lacking glucose at 25°C for 24 h and sonicated once. Then, cells were collected and released to YPD medium for FACS analysis. FACS analysis was performed as described previously (Hazan et al., 2002).
DNA Damage Assay
To test cells for hydroxyurea, camptothecin, and methymethane sulfonate (MMS) sensitivity, freshly grown cells at room temperature were serially diluted 10-fold, and 5 μl was spotted onto YPD plate or YPD plate containing 25 mM hydroxyurea, 0.02% methyl methanesulfonate, or 30 μg/ml camptothecin and incubated at 30°C for 56 h. To quantify the drug sensitivities, ∼200 cells were plated on YPD plates or YPD plates containing DNA-damaging agents. After incubation at 30°C for 72 h, the colony-forming units were determined as described previously (Ciudad et al., 2004).
Immunoprecipitation
Protein extraction and immunoprecipitation were performed as described previously (Cao et al., 2006) with modifications. Protein extract containing 10 mg protein were subjected to immunoprecipitation using 60 μl of immunoglobulin G (IgG) agarose bead slurry (GE Healthcare, Chalfont St. Giles, United Kingdom), which was preincubated once with 0.2 mg/ml sheared salmon sperm DNA, 0.5 mg/ml bovine serum albumin in phosphate-buffered saline. Proteins were separated by 7.5% SDS-polyacrylamide gel electrophoresis and transferred to a polyvinylidene difluoride membrane (Hybond; GE Healthcare). After blocking in 5% skim-milk powder in Tris-buffered saline/0.05% Tween 20, a peroxidase-conjugated anti-c-Myc antibody (Sigma- Aldrich) was used to probe for Myc-tagged proteins, which were then detected using the enhanced chemiluminescence system (Pierce Chemical. Rockford, IL).
Chromatin Immunoprecipitations (ChIPs)
Overnight cultures were grown in YPD for ∼6 h at 25°C to an OD600 of 0.8 for yeast growth or in YPD + serum for 0, 15 min, 30 min, 1 h, 2 h, 3 h, or 3.5 h at 37°C for hyphal induction. Cells were formaldehyde cross-linked and lysed using lysis buffer (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 0.1% sodium deoxycholate). ChIPs were performed as described by Pokholok et al. (2005), with modifications. DNA was sheared by sonication three times for 10 s at high power on a Bioruptor, incubating on ice for 20 s between sonication pulses. For precipitation using tandem affinity purification (TAP)-tagged proteins (Figure 3A), 30 μl of IgG-Sepharose was used in 400 μl (∼4 mg of chromatin proteins) of sheared chromatin (∼0.5 kb). For the ChIPs in Figures 3B, 4, and 6, 5 μl of monoclonal Myc antibody (9E11′ Abcam, Cambridge, MA) was used for ∼4 mg of chromatin proteins in an immunoprecipitation volume of 400 μl. For the ChIPs in Figure 5, 5 μl of polyclonal H4 (ab10158; Abcam) or acetylated H4 (Millipore, Billerica, MA) antibodies were used for ∼4 mg of chromatin proteins in an immunoprecipitation volume of 400 μl. DNA derived from the whole cell extracts and immunoprecipitation (IP) eluate was analyzed by PCR or quantitative PCR (qPCR).
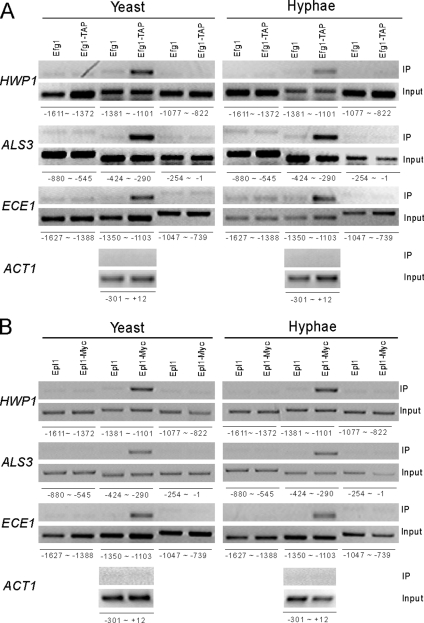
Figure 3.
Efg1 and NuA4 complex are present at the promoters of hypha-specific genes in vivo. (A) ChIP of Efg1 at a specific region of the HWP1, ALS3, and ECE1 promoters in yeast and hyphae. HLY3234 (EFG1-TAP) or SC5314 (no tag control) strains were grown in either YPD at 25°C or YPD + 10% serum at 37°C. Positions of primers for HWP1, ALS3, ECE1, and ACT1 promoters are indicated. (B) ChIP of Epl1 at the promoters of hypha-specific genes. CLY6 (Epl1-Myc) or CAI4+pPR673 (no tag control) strains were grown at 25°C in YPD for yeast growth and at 37°C in YPD + 10% serum for hyphal growth.
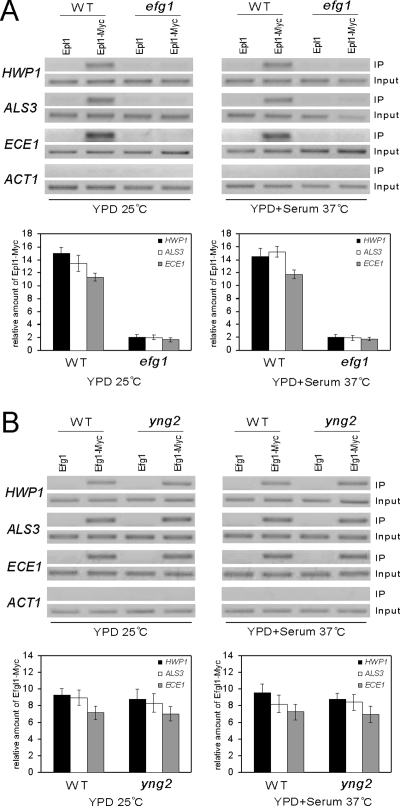
Figure 4.
Recruitment of NuA4 to promoters of hypha-specific genes is Efg1 dependent. (A) Efg1 is essential for NuA4 binding. ChIP of Epl1-myc in wild-type (CAI4) or efg1/efg1 (HLC54) strains. Cells were grown at 25°C in YPD for yeast growth and at 37°C in YPD + 10% serum for hyphal growth. PCR reactions with primers at −1381 to −1101 of HWP1, −424 to −290 of ALS3, −1350 to −1103 of ECE1, and −301 to +12 of ACT1 are shown. ChIP signals were also quantified by real-time PCR. The ADE2 promoter was used as control locus. Epl1-Myc expressing strains were used with nontagged strains as controls. HWP1, ALS3, ECE1, and control locus IP signals (bound/input) in the nontagged strains were subtracted from the values in the Myc-tagged strains. The Epl1-Myc enrichment is then presented as a ratio of HWP1, ALS3, or ECE1 IP versus control locus IP. (B) Efg1 binds the promoters of hypha-specific genes independently of Yng2. ChIP of Efg1-myc in wild-type (CAI4) or yng2/yng2 (CLY4) strains. Cells were grown at 25°C in YPD for yeast growth and at 37°C in YPD + 10% serum for hyphal growth. PCR reactions with primers at −1381 to −1101 of HWP1, −424 to −290 of ALS3, −1350 to −1103 of ECE1 are shown. For qPCR, the ADE2 promoter was used as control locus. IP ratios were calculated and are presented in A.
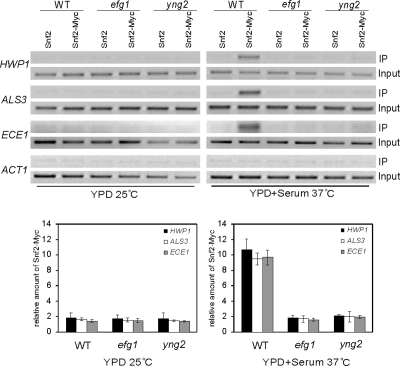
Figure 6.
Hypha-specific recruitment of Swi/Snf complex to the promoters of hypha-specific genes is NuA4 dependent. ChIP of Snf2 in wild-type (CAI4), efg1/efg1 (HLC54), or yng2/yng2 (CLY4) cells grown in YPD at 25°C or induced for 3.5 h in YPD + 10% serum at 37°C. PCR reactions with primers at −1381 to −1101 of HWP1, −424 to −290 of ALS3, −1350 to −1103 of ECE1, and −301 to +12 of ACT1 are shown. The ChIPs of Snf2 were also quantified by real-time PCR. The ADE2 promoter was used as control locus. IP ratios were calculated and are presented in Figure 4.
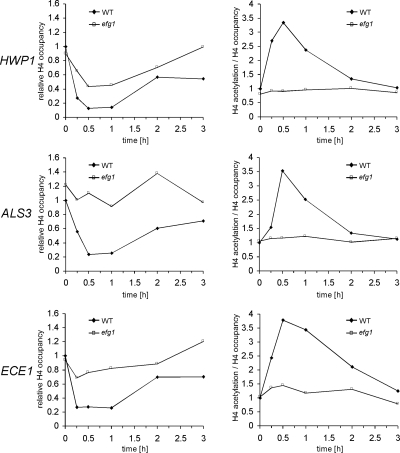
Figure 5.
Nucleosomal H4 acetylation levels at the promoters of hypha-specific genes increase rapidly during initial hyphal induction in wild-type but not in efg1/efg1 strain. Cells of SC5314 (WT) and HLC52 (efg1) were transferred into 10% serum in YPD at 37°C for hyphal induction, and cells were collected at 0 min, 15 min, 30 min, 1 h, 2 h, and 3 h for ChIP with anti-histone H4 and anti-acetylated H4 antibodies. The amounts of HWP1, ALS3, and ECE1 upstream DNA in the ChIP reactions were determined by quantitative PCR with primers at −1381 to −1101 of HWP1, −424 to −290 of ALS3, and −1350 to −1103 of ECE1. Left, levels of H4 (relative H4 occupancy) at these promoters, which are normalized to the respective control DNA at the ADE2 promoter. The 0-min values in the wild-type strain are set to be 1.00. Acetylation levels normalized with respect to H4 levels are presented (right; H4 acetylation/H4 occupancy). They are calculated by dividing the values for acetylated H4 with H4 occupancy values.
RESULTS
Efg1 Interacts with the NuA4 Complex In Vivo
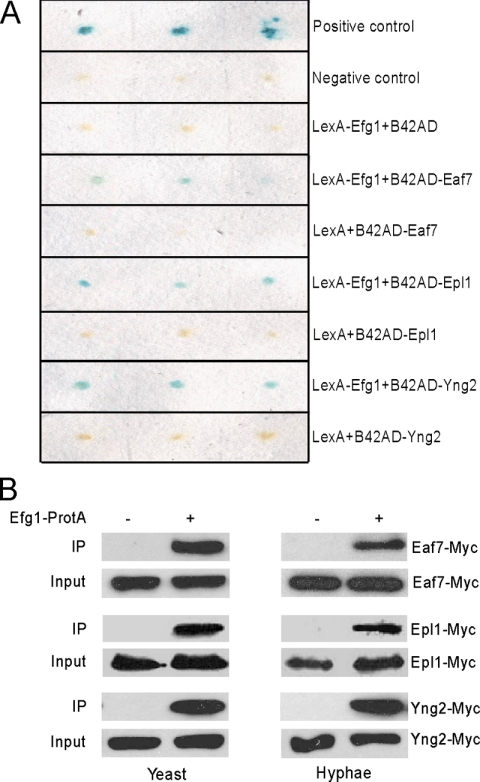
To understand how Efg1 is regulated, we set out to identify Efg1-interacting proteins with a yeast two-hybrid screen. A LexA-Efg1 fusion was used as bait to screen a C. albicans two-hybrid library (Ni et al., 2004) in the yeast S. cerevisiae, taking advantage of the fact that the LexA-Efg1 fusion did not activate the transcription of LexAop-lacZ in yeast (Figure 1A) (Doedt et al., 2004). From the screen, we identified 12 distinct genes whose protein products were able to interact with Efg1. Two of the 12 are likely involved in transcriptional regulation based on the known functions of their orthologues. In this study, we have focused on Orf19.497, which encodes a protein with the highest similarity (45%) to S. cerevisiae Eaf7, a subunit of the NuA4 complex. Based on the Candida Genome Database, orthologues of all but one of the subunits of the S. cerevisiae NuA4 complex exist in C. albicans (Table 4). Hereafter, we refer to these C. albicans genes based on the names of their corresponding S. cerevisiae orthologues.
Figure 1.
Efg1 interacts with NuA4 in yeast and hyphae. (A) Yeast two-hybrid assays. EGY48 (p8op-lacZ) was cotransformed with plasmids as indicated. (B) Coimmunoprecipitation of Efg1 interacts with Eaf7, Epl1, and Yng2 in yeast and hyphae. C. albicans strains CLY5 (Efg1-TAP Eaf7-myc) and CLY6 (Eaf7-myc), CLY7 (Efg1-TAP Epl1-myc) and CLY8 (Epl1-myc), or CLY9 (Efg1-TAP Yng2-myc) and CLY10 (Yng2-myc) were grown at 25°C in YPD for yeast growth and at 37°C in YPD + 10% serum for hyphal growth. Protein lysates were subjected to immunoprecipitation with IgG beads (GE Healthcare), and the precipitated proteins were separated by 7.5% SDS-PAGE and probed with peroxidase-conjugated anti-c-myc (Sigma-Aldrich). As input control, cell lysates were analyzed by Western blotting with the peroxidase-conjugated anti-c-myc.
Table 4.
List of genes coding subunits of NuA4 complex in S. cerevisiae and C. albicans
| NuA4 subunits in S. cerevisiae | NuA4 subunits in C. albicans |
|---|---|
| TRA1 | orf19.139 |
| VID21/EAF1 | VID21/orf19.3077 |
| EPL1 | orf19.7529 |
| SWC4/EAF2 | orf19.7492 |
| ARP4 | orf19.5623 |
| ESA1 | orf19.5416 |
| EAF7 | orf19.497 |
| EAF3 | orf19.2660 |
| ACT1 | ACT1/orf19.5007 |
| YNG2/EAF4 | NBN1/orf19.878 |
| EAF5 | ? |
| YAF9 | YAF9/orf19.5501 |
| EAF6 | orf19.396 |
To confirm the two-hybrid interaction between Efg1 and Eaf7 and to determine whether other subunits of the NuA4 complex also interact with Efg1, we cloned EAF7, YNG2, and EPL1 into pJG4-5 to generate B42AD fusions. The interactions between LexA-Efg1 and B42AD-Eaf7, B42AD-Epl1, and B42AD-Yng2 were detected in the yeast two-hybrid assay (Figure 1A). The three B42AD fusions did not interact with the control LexA. Therefore, Efg1 could interact with several subunits of the NuA4 complex by two-hybrid assay, possibly mediated through the NuA4 complex of S. cerevisiae. The two-hybrid interaction between Efg1 and Eaf7 was also verified by a coimmunoprecipitation assay in S. cerevisiae (data not shown).
An immunoprecipitation experiment in C. albicans was performed to determine whether Efg1 interacts with the NuA4 complex in vivo. Eaf7, Epl1, and Yng2 were tagged at theirs C terminus with myc, and Eaf7-myc, Epl1-myc, Yng2-myc were expressed from the ACT1 promoter in a strain containing Efg1-TAP (Cao et al., 2006). Immunoprecipitation of Efg1-TAP with IgG beads was able to pull down all three Myc-tagged proteins from both yeast and hyphal cells (Figure 1B). The interaction was specific because the interaction was not detectable in the control strain that carried only Eaf7-myc, Epl1-myc, or Yng2-myc. Therefore, Efg1 can interact with the NuA4 complex in vivo, and the interaction is not regulated in connection to growth form.
The NuA4 HAT Activity Is Required for Hyphal Development and the Induction of Hypha-specific Genes
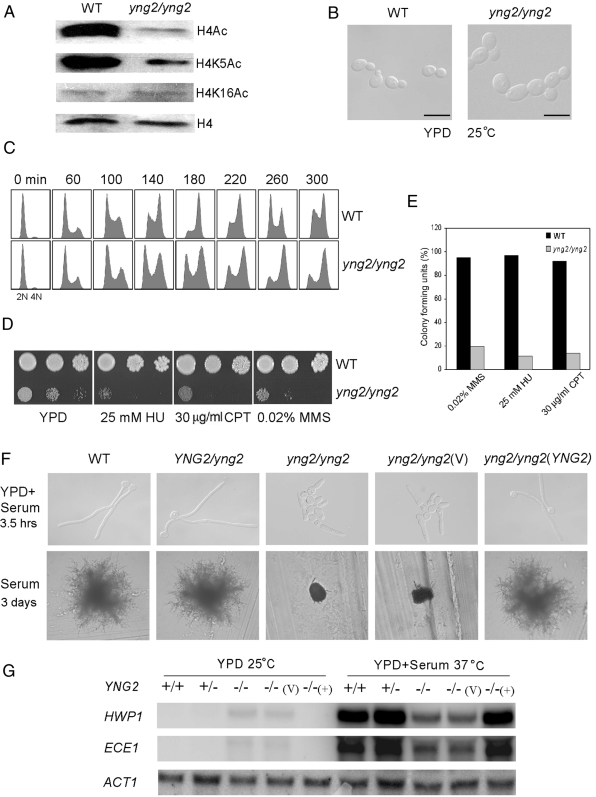
To study the functions of the NuA4 complex in C. albicans, we constructed an yng2/yng2 mutant strain. Among the three subunits of the NuA4 HAT module (Piccolo NuA4) in S. cerevisiae, Esa1 and Epl1 are essential for cell growth. Yng2 is not essential, but the NuA4 HAT activity in Yng2-deficient cells is approximately fivefold lower than in wild-type cells (Loewith et al., 2000; Choy et al., 2001). Therefore, we decided to delete YNG2 in C. albicans. Successful deletion of YNG2 in C. albicans was confirmed by Southern analysis (data not shown). Next, we performed Western analysis to examine the acetylation level of H4 in cells lacking YNG2. The yng2/yng2 cells exhibited a significant decrease in the level of acetylation at histone H4 (Figure 2A). Acetylation was dramatically reduced at H4K5, but not at H4K16 (Figure 2A), probably due to the existence of redundant HATs for H4K16 as in S. cerevisiae (Kimura et al., 2002; Suka et al., 2002). These data demonstrate that NuA4 HAT activity is significantly lower in yng2/yng2 cells compared with wild-type cells. In addition to its function in transcriptional activation as a HAT, NuA4 also plays a role on DNA repair and cell cycle progression in S. cerevisiae (Clarke et al., 1999; Bird et al., 2002; Choy and Kron, 2002). Similar to the yng2 mutant of S. cerevisiae (Loewith et al., 2000; Choy et al., 2001; Choy and Kron, 2002), C. albicans yng2/yng2 cells also showed general growth and cell cycle defects, and they were sensitized to DNA damage. They grew much slower than wild-type cells (170- and 80-min doubling times during log phase, respectively, in YPD at 30°C), and they displayed swollen and multibudded cell morphology (Figure 2B). FACS analysis showed that yng2/yng2 mutant cells delayed in G2/M. Most wild-type cells completed DNA synthesis within 180 min (Figure 2C, top), and progressed through mitosis, whereas most yng2/yng2 cells completed S phase by ∼260 min (Figure 2C, bottom). Moreover, yng2/yng2 mutant cells exhibited sensitivity to DNA damage induced by hydroxyurea (HU), camptothecin (CPT), or MMS (Figure 2D). This sensitivity was not because of growth defect of the mutant. As shown in Figure 2E, the viability of yng2/yng2 cells is at least fourfold lower than that of wild-type cells when known amounts of cells were individually plated on the plates containing HU, CPT or MMS. Therefore, C. albicans Yng2 is a functional homologue of S. cerevisiae Yng2.
Figure 2.
The NuA4 complex is necessary for hyphal development and full expression of hypha-specific genes in C. albicans. (A) Western analyses of histone H4 acetylation levels in wild-type strain SC5314 and yng2/yng2 strain (CLY3), grown in YPD at 25°C, by using antibodies that specifically recognize acetylated H4 (H4Ac; Millipore), H4 acetylated at lysine residues 5 (H4K5Ac; Abcam), 16 (H4K16Ac; Millipore), and histone H4 (H4; Abcam). (B) Cell morphology of yng2/yng2 strain (CLY3) in comparison with wild-type (SC5314) in YPD medium at 25°C. Bars, 10 μm. (C) yng2/yng2 cells display a G2/M delay. Wild-type and yng2/yng2 yeast cells were synchronized in G1 by carbon starvation in minimal medium and then released into YPD at 30°C. Aliquots were harvested at intervals for flow cytometry. (D) yng2/yng2 cells are sensitive to DNA damage. The wild-type and yng2/yng2 mutants were treated with DNA-damaging agents. Cells were serially diluted 10-fold and spotted onto plates. Photographs were taken after 56 h of growth at 30°C. (E) Quantification of the drug sensitivities presented in two dimensions. About 200 cells were plated on YPD plates or YPD plates containing DNA-damaging agents, and they were incubated at 30°C for 72 h. Colony-forming units are calculated as a ratio of the colonies formed in drug-containing plates versus YPD plates. Results are mean of three experiments. (F) yng2/yng2 cells (CLY3) are defective in hyphal development at 37°C in YPD + 10% serum for 3.5 h (top) or on a solid serum-containing medium for 3 d (bottom). (G) Northern analyses show defective induction of hypha-specific genes in yng2/yng2 mutants. Cells were grown in YPD + 10% serum at 37°C for 3.5 h or grown in YPD at 25°C for 6 h and collected for RNA extraction and Northern analyses. Strains shown in C and D are WT (SC5314, +/+), YNG2/yng2 (CLY1, +/−), yng2/yng2 (CLY3, −/−), yng2/yng2 (V) [CLY4 + pBA1, −/−(v)], yng2/yng2 (YNG2) [CLY4 + pBA1-YNG2, −/−(+)].
Deletion of YNG2 caused a profound defect in hyphal development. On serum-containing solid media, the wild-type strains generated florid hyphal colonies, whereas yng2/yng2 strains formed only a few filaments, and colonies remained predominantly smooth (Figure 2F). The defect in hyphal formation associated with yng2/yng2 mutants was also observed in YPD + 10% serum liquid media at 37°C, where most yng2/yng2 cells stayed as swollen, multibudded cells, except for some cells, which were elongated at the tip (Figure 2F). As a control, wild-type cells developed true hyphae. Reintroduction of wild-type YNG2 in yng2/yng2 mutant restored yeast growth in YPD and hyphal growth in serum media.
Consistent with the defect in hyphal morphogenesis, the yng2/yng2 mutants were also defective in the expression of hypha-specific genes. ECE1 and HWP1 were highly induced in wild-type cells under hyphal inducing conditions and were undetectable under yeast growth condition. Compared with the wild-type strain, the levels of ECE1 and HWP1 expression in the yng2/yng2 mutant were dramatically lower under hyphal inducing condition (Figure 2G). These results indicate that H4 acetylation by the NuA4 complex is important for hyphal morphogenesis and the induction of hypha-specific genes. We also observed low levels of HWP1 and ECE1 expression in yng2/yng2 mutant cells under yeast growth conditions. This derepression in the expression of hypha-specific genes in yeast growth could be an indirect result of yng2 deletion or an indication of an inhibitory role of NuA4 in transcription.
Among the 10 non-Piccolo NuA4 subunits in S. cerevisiae, Eaf7, together with Eaf5, may form a subcomplex that mediates only a subset of the cellular functions of NuA4 (Mitchell et al., 2008). To determine whether Eaf7 is required for hyphal development in C. albicans, we deleted two copies of EAF7 by PCR-based homologous recombination as described previously (Wilson et al., 1999). Deletion of EAF7 has no apparent effect on histone H4 acetylation (Supplemental Figure 1A). Therefore, Eaf7 is not required for NuA4 catalytic activity, consistent with the situation in S. cerevisiae (Krogan et al., 2004). The eaf7/eaf7 mutants also have no detectable defects in growth and hyphal development (Supplemental Figure 1, B and C), which suggests that the Eaf7 is not involved in hyphal regulation and as in S. cerevisiae, it is likely required for a distinct subset of NuA4 cellular functions (Mitchell et al., 2008).
Efg1 and the NuA4 Complex Are Present at the UAS Regions of Hypha-specific Genes in Both Yeast and Hyphal Cells
Efg1 can interact with the NuA4 complex in vivo and both are necessary for the induction of hypha-specific genes. To determine whether they are directly involved in the induction of hypha-specific genes, ChIP was performed to determine whether a TAP-tagged Efg1 and a myc-tagged Epl1 are present at the promoters of hypha-specific genes. Cross-linked chromatin from isogenic strains containing either Efg1-TAP or no tag (as a control) was precipitated with IgG-Sepharose beads, and cross-linked chromatin of either Epl1-myc or no tag (as a control) was precipitated with anti-myc and protein-G beads. The precipitated DNA was analyzed by PCR at ≈250 base pairs intervals across the whole promoters of hypha-specific genes. ACT1 served as a negative control as in previously published ChIP experiments (Tebarth et al., 2003). As shown in Figure 3A, a specific region from each of the hypha-specific promoters, −1350 to −1103 for ECE1, −1381 to −1101 for HWP1, and −424 to −290 for ALS3, was specifically detected from the Efg1-TAP ChIP compared with the control ChIP or the ACT1 promoter. Other regions of the same hypha-specific promoters were not detected in the Efg1-TAP ChIP. Among them, surrounding regions were shown in the Figure 3A. Importantly, the Efg1-bound regions on the HWP1 and ALS3 promoters coincide exactly with the recently mapped regions essential for the induction of HWP1 and ALS3 (Argimon et al., 2007; Kim et al., 2007). Interestingly, Efg1 was detected at the promoters in both yeast and hyphal states (Figure 3A). ChIP with Epl1-myc showed that Epl1 binds to the same regions as Efg1 on the hypha-specific promoters (Figure 3B). Again, the promoter binding of Epl1 was detected under both yeast and hyphal growth conditions, and no increase in bound Epl1 was detected during hyphal induction.
Efg1 Is Required for the Recruitment of the NuA4 Complex to Hypha-specific Promoters
Because both Efg1 and NuA4 bind to the same regions in the hypha-specific promoters, it is possible that Efg1 binds to cis-elements in the regions and recruits NuA4. To test this possibility, we performed ChIP with Epl1-myc in an efg1/efg1 mutant, and in a wild-type strain as a positive control. PCR of Epl1-myc–precipitated DNA detected the presence of hypha-specific promoters ECE1, HWP1, and ALS3 in the wild-type control, but not in the efg1/efg1 strain (Figure 4A). We then performed real-time PCR to quantify Epl1-Myc binding. The quantitative PCR revealed that the Epl1-Myc binding on HWP1, ALS3, and ECE1 promoters in efg1/efg1 mutant cells is at least sixfold lower than that in wild-type cells under both yeast and hyphal growth conditions (Figure 4A). Therefore, the binding of NuA4 to hyphal promoters is dependent on Efg1. In contrast to efg1/efg1 cells, Epl1 binding to hypha-specific promoters was not affected by deletion of EAF7 gene (Supplemental Figure 1D), suggesting that Eaf7 is not necessary for the Efg1-mediated recruitment of NuA4 complex to hyphal promoters, consistent with the result that Eaf7 is not required for hyphal development (Supplemental Figure 1C).
Because NuA4 binding to hypha-specific promoters is Efg1 dependent, we performed reciprocal ChIP of Efg1 in yng2/yng2 mutants. Efg1 was tagged with myc at its C terminus, and the Efg1-myc fusion was expressed from its endogenous promoter at the EFG1 locus. ChIP experiments were performed with Efg1-myc in wild-type strain and yng2/yng2 mutants. The Efg1-myc precipitated DNA were detected by PCR and analyzed by quantitative PCR. Both PCR and quantitative PCR results, shown in Figure 4B, indicate that deleting YNG2 has no obvious effect on Efg1-Myc binding at the UAS regions of HWP1, ALS3, or ECE1 under either growth condition. Therefore, Efg1 binding at hyphal promoters is independent of NuA4 HAT activity.
Nucleosomal H4 Acetylation at the Hypha-specific Promoters Peaks during Initial Hyphal Induction in an Efg1-dependent Manner
Genome-wide mapping of nucleosome acetylation in S. cerevisiae has shown a strong correlation between transcriptional activation and acetylation levels of nucleosomal H3 and H4 targeted by Gcn5 and Esa1 (Pokholok et al., 2005). Because our results indicate that Efg1 recruits the NuA4 complex to the promoters of hypha-specific genes in both repressed and activated states, we wanted to determine whether H4 acetylation at those promoters is regulated during the yeast-to-hypha transition. ChIP experiments were performed using anti-histone H4 and anti-acetylated H4 antibodies with wild-type cells after 0 min, 15 min, 30 min, 1 h, 2 h, and 3 h of serum induction at 37°C. The levels of H4 or acetylated H4 were monitored by real-time PCR and compared with those of the control ADE2 promoter, the expression of which does not change in response to hyphal induction. The time courses of relative levels of H4 occupancy and acetylation are shown in Figure 5. The ChIP signal of histone H4 at hypha-specific promoters decreased rapidly after the onset of induction. After 30 min of hyphal induction, histone H4 occupancy reached the lowest level, with an estimated 80% reduction compared with the zero time point. This is consistent with the notion that transcriptional activation correlates with reduced nucleosome occupancy (Pokholok et al., 2005). Nucleosomal H4 acetylation levels were calculated with respect to the H4 levels at the promoters. We found that the acetylation levels of H4 on the hypha-specific promoters peaked at ∼30-min after serum induction (3.34-fold at the HWP1 promoter, 3.54-fold at the ALS3 promoter, and 3.79-fold at the ECE1 promoter relative to time 0). Even after 15 min of induction, H4 acetylation levels at the HWP1 and ECE1 promoters had already increased significantly. But in comparison, the increase in H4 acetylation at the ALS3 promoter was slower, which is consistent with slower induction dynamics of the ALS3 gene than that for ECE1 and HWP1 genes. After the initial peak, the H4 acetylation level decreased slowly to the initial level at the zero time point. The increase in H4 acetylation was hypha-specific, because H4 acetylation levels at these promoters remained almost unchanged when cells were transferred into YPD at 25°C for 15 min or 30 min for yeast growth (data not shown).
We repeated the same ChIP experiments with anti-histone H4 and anti-acetylated H4 antibodies in efg1/efg1 cells followed by real-time PCR. We found that the H4 acetylation levels remained largely unchanged during hyphal induction for all three hypha-specific promoters (Figure 5). These data indicate that Efg1, together with NuA4, may play an important role in transcriptional activation during the very initial stage of hyphal induction.
NuA4 Action Is Required for Hypha-specific Recruitment of the Swi/Snf Complex to the Promoters of Hypha-specific Genes
Our previous study has shown that, like NuA4, the Swi/Snf chromatin remodeling complex is also required for hyphal development in C. albicans. Because there are clear instances in vivo where sequence-specific DNA binding proteins recruit HATs and chromatin remodelers in a temporal order during the process of gene activation (Cosma et al., 1999; Agalioti et al., 2000), it is conceivable that Efg1 recruitment of NuA4 could play a role on recruiting Swi/Snf complex to the promoters of hypha-specific genes. To determine whether Snf2, which is a component of Swi/Snf complex, can bind to promoters of hypha-specific genes, we tagged Snf2 with myc at its C terminus and expressed the Snf2-myc fusion from the endogenous SNF2 promoter. ChIP experiments with Snf2-myc in the wild-type strain showed a specific enrichment of Snf2 at the UAS regions of all three hypha-specific promoters under hyphal growth conditions but not under yeast growth condition (Figure 6). Such enrichment was not observed at other regions of hyphal promoters under both yeast and hyphal growth conditions (data not shown). In contrast to the wild-type strain, Snf2 was not detected at the UAS regions of the promoters in the efg1/efg1 and yng2/yng2 strains, even after 3.5 h under the same inducing condition (Figure 6). To confirm the PCR result, a real-time PCR was performed to quantify Snf2 binding at hypha-specific promoters. The Snf2 binding in efg1/efg1 and yng2/yng2 mutants were approximately fivefold lower than that in wild-type strain under hyphal growth conditions (Figure 6). This result suggests that although Efg1 binding to the promoters is not regulated by the yeast to hyphae transition, Efg1 and NuA4 at the promoters of yeast cells is essential for the recruitment of the Swi/Snf complex and transcriptional activation during hyphal induction.
DISCUSSION
C. albicans Has a Conserved NuA4 Complex and NuA4 Is Required for the Induction of Hypha-specific Genes
SAGA and NuA4 are the two major HAT complexes that contribute to gene activation across the genome in yeast (Robert et al., 2004). Like SAGA, NuA4 is an evolutionary conserved HAT complex from yeast to human (Doyon and Cote, 2004). The NuA4 complex was initially purified biochemically from S. cerevisiae by following its characteristic nucleosomal H4 and H2A HAT activity. The yeast NuA4 complex contains 13 stably associated proteins. Of the 13 components of yeast NuA4, 12 homologous proteins are found in the human NuA4 complex. C. albicans contains orthologues of all 12 NuA4 components conserved from yeast to human. Eaf5 of the yeast NuA4 complex has no corresponding orthologue in the C. albicans genome, and it is also the only one not present in the human NuA4 complex.
The functionality of the C. albicans orthologues as components of NuA4 is confirmed here by studying the YNG2 deletion. Yng2 is a subunit of Piccolo NuA4, which enables Esa1 to acetylate nucleosomal H4. Without Yng2, Esa1 alone can only acetylate free histones, and therefore, nucleosomal H4 acetylation is impaired (Nourani et al., 2001; Selleck et al., 2005). Similar to the loss of Yng2 in S. cerevisiae, we find that NuA4 HAT activity in C. albicans yng2/yng2 cells is dramatically lower than that of wild-type cells, suggesting that Yng2 functions as a subunit of Piccolo NuA4 in C. albicans. In addition to its role on NuA4 HAT activity, Yng2 is also responsible for cell cycle progression and DNA damage response. Similar to the yng2 mutant of S. cerevisiae (Loewith et al., 2000; Choy et al., 2001; Choy and Kron, 2002), C. albicans yng2/yng2 cells also display general growth and cell cycle defects, and they are sensitized to DNA damage. Therefore, C. albicans Yng2 is a functional homologue of S. cerevisiae Yng2. NuA4 is a transcriptional coactivator in S. cerevisiae (Allard et al., 1999; Galarneau et al., 2000). Here, we show that NuA4 is at the promoters of hypha-specific genes, and YNG2 deletion causes a profound defect in the induction of hypha-specific genes. Therefore, NuA4 in C. albicans also functions as a transcriptional coactivator.
In S. cerevisiae, the non-Piccolo NuA4 subunit Eaf7 is not required for NuA4 HAT activity, and it seems to form a subcomplex with Eaf5 that is involved in a subset of cellular functions of NuA4, possibly through the recruitment of NuA4 to distinct chromatin loci (Krogan et al., 2004; Mitchell et al., 2008). In this study, we find that Eaf7 is not required for NuA4 HAT activity in C. albicans (Supplemental Figure 1A). It is also not involved in hyphal development (Supplemental Figure 1C), and it is not required for recruitment of NuA4 at the promoters of hyphal genes. So, like its homologue in S. cerevisiae, Eaf7 is responsible only for a subset of the cellular functions of NuA4 in C. albicans.
Efg1 Interacts with NuA4 In Vivo and Recruits NuA4 to Promoters of Hypha-specific Genes
It has been well demonstrated that HATs are targeted to promoter regions by sequence-specific activators (Naar et al., 2001). But, there have been very few sequence-specific activators for recruitment of NuA4 described to date, and mechanisms of NuA4 recruitment in vivo are mostly elusive. The recruitment of NuA4 to ribosomal protein genes is believed to depend on Rap1 (Reid et al., 2000) and to the PHO5 gene by direct interaction with the Pho2 homeodomain factor (Nourani et al., 2004). In this report, we show that NuA4 is recruited by Efg1 to the promoters of the hypha-specific genes ECE1, HWP1, and ALS3. Both Efg1 and the NuA4 subunit Epl1 are located at the same UAS regions of these genes, and Epl1 binding is dependent on Efg1. So, Efg1 is essential for the induction of hypha-specific genes, probably because it is required for the recruitment of NuA4 to these promoters.
Efg1 may also recruit NuA4 to other promoters. Hypha-specific genes are not the only genes affected in efg1 mutants as determined by genome-wide transcription analyses (Sohn et al., 2003; Doedt et al., 2004; Harcus et al., 2004; Cao et al., 2006; Setiadi et al., 2006), and Efg1 also regulates other cellular processes and development (Lo et al., 1997; Stoldt et al., 1997; Sonneborn et al., 1999; Srikantha et al., 2000; Zordan et al., 2007). Phd1, a member of the APSES family in S. cerevisiae, binds to DNA nonspecifically in vitro (Ho et al., 2006), but it binds specific promoters in vivo (Borneman et al., 2006); therefore, Phd1 is thought to use additional cofactors to achieve sequence-specific binding in yeast (Ho et al., 2006). Efg1 shares the same conserved DNA binding domain with Phd1, so Efg1 probably also functions with different cofactors to regulate different processes. This is reminiscent of another NuA4 binding factor, Pho2, which uses combinatorial control with three transcriptional factors in the transcriptional activation of genes in different pathways (Bhoite et al., 2002).
We identified the function of Efg1 as a sequence-specific recruiter of NuA4 from a two-hybrid screen with Efg1 as the bait. Initially, Eaf7, a subunit in the recruitment module of the NuA4 complex, was identified from the screen. Then, Epl1 and Yng2 of Piccolo NuA4 also gave a similar positive interaction with Efg1 when tested in two-hybrid assay. The interaction between Efg1 and NuA4 was further verified in vivo by coimmunoprecipitation of Efg1 with Eaf7, Epl1, and Yng2. Our data, however, do not address which subunits of NuA4 interact with Efg1 or whether the interaction with the NuA4 complex is direct. All three NuA4 subunits examined gave equally a weak interaction with Efg1 by yeast two-hybrid assay.
Hypha-induced Nucleosomal H4 Acetylation at the Promoters of Hypha-specific Genes
Nucleosomal H4 acetylation levels at the promoters of hypha-specific genes peak shortly after hyphal induction, and this increase in H4 acetylation is specific to hyphal induction. Efg1 is required for the recruitment of NuA4 to the promoters in yeast growth, which is a prerequisite for the increase in nucleosomal H4 acetylation during hypha induction. However, Efg1 alone is not sufficient to achieve this hyphal-induced transcriptional activation. Based on a one-hybrid assay set up for the LexADB-Efg1 fusion protein in C. albicans, Efg1 is suggested to function as an inhibitor, and this inhibitory activity is not regulated during hyphal induction (Doedt et al., 2004). Therefore, the increase in nucleosomal H4 acetylation is likely promoter-specific, and it is probably a net effect from a few hyphal regulators bound at the promoters. Flo8 is a potential cofactor of Efg1 at the promoters of hypha-specific genes as the two regulators interact with each other and share a common set of target genes (Cao et al., 2006). Negative regulators of the hyphal-specific transcription, such as Nrg1 and Sfl1 (Braun et al., 2001; Murad et al., 2001; Garcia-Sanchez et al., 2005; Kadosh and Johnson, 2005; Bauer and Wendland, 2007; Li et al., 2007), are also potential candidates that can function to repress the transcription during yeast growth by keeping H4 acetylation levels low. Future experiments need to address the identity and mechanisms of how these regulators are induced in response to extracellular stimuli and regulate hypha-specific nucleosomal H4 acetylation.
Efg1-mediated Recruitment of NuA4 to Promoters of Hypha-specific Genes Is Required for Subsequent Swi/Snf Binding and Transcriptional Activation uring Hyphal Induction
The ATP-dependent chromatin remodeling complex Swi/Snf is essential for hyphal development and the induction of hypha-specific genes (Mao et al., 2006). In this study, we show the Swi/Snf complex is recruited to the promoters of hyphal genes specifically during hyphal development. Furthermore, Efg1-mediated recruitment of the NuA4 complex to the promoters in yeast cells is likely a prerequisite for Swi/Snf binding during hyphal induction. Therefore, NuA4 at the hypha-specific promoters is responsible for the rise in nucleosomal H4 acetylation during initial hyphal induction, which is essential for later Swi/Snf binding and transcriptional activation. This sequential recruitment of different chromatin remodeling complexes by sequence-specific regulators to specific promoters is reminiscent to that found for the induction of PHO5 and SUC2 in S. cerevisiae. The NuA4 complex is recruited to the PHO5 gene by direct interaction with Pho2 (Nourani et al., 2004). This targeting is a prerequisite for chromatin remodeling and activation of transcription. For the induction of the glucose-regulated SUC2 gene, Esa1 is found at the promoter before induction and is necessary for maximal binding of Swi/Snf during gene activation (Geng and Laurent, 2004). Mechanisms for the recruitment of the Swi/Snf complex to hypha-specific promoters during hyphal induction are not clear. But a previous study has shown that stable promoter occupancy by Swi/Snf in vivo can occur in the absence of transcription activators. This requires the bromodomain of Swi2/Snf2 and nucleosome acetylation by either the SAGA or NuA4 HAT complexes (Hassan et al., 2002). This case illustrates a functional link between HAT complexes and the Swi/Snf chromatin remodeling complex and provides a mechanistic basis for the ordered recruitment of these complexes.
In conclusion, our data demonstrate that Efg1 recruits the NuA4 complex to promoters of hypha-specific genes under yeast growth conditions. This is likely to be required for peaking of nucleosomal H4 acetylation levels during initial hyphal induction and for subsequent Swi/Snf binding and transcriptional activation.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Chinese 863 grant 2006AA02Z178 and Chinese Academy of Sciences grant KSCX2-YW-R-107 (to J. C.) and by National Institutes of Health grant GM-55155 (to H. L.).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-02-0173) on August 6, 2008.
REFERENCES
- Agalioti T., Lomvardas S., Parekh B., Yie J., Maniatis T., Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- Allard S., Utley R. T., Savard J., Clarke A., Grant P., Brandl C. J., Pillus L., Workman J. L., Cote J. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 1999;18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argimon S., et al. Developmental regulation of an adhesin gene during cellular morphogenesis in the fungal pathogen Candida albicans. Eukaryot. Cell. 2007;6:682–692. doi: 10.1128/EC.00340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barelle C. J., Bohula E. A., Kron S. J., Wessels D., Soll D. R., Schafer A., Brown A. J., Gow N. A. Asynchronous cell cycle and asymmetric vacuolar inheritance in true hyphae of Candida albicans. Eukaryot. Cell. 2003;2:398–410. doi: 10.1128/EC.2.3.398-410.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer J., Wendland J. Candida albicans Sfl1 suppresses flocculation and filamentation. Eukaryot. Cell. 2007;6:1736–1744. doi: 10.1128/EC.00236-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik S. R., Green M. R. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 2001;15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhoite L. T., Allen J. M., Garcia E., Thomas L. R., Gregory I. D., Voth W. P., Whelihan K., Rolfes R. J., Stillman D. J. Mutations in the pho2 (bas2) transcription factor that differentially affect activation with its partner proteins bas1, pho4, and swi5. J. Biol. Chem. 2002;277:37612–37618. doi: 10.1074/jbc.M206125200. [DOI] [PubMed] [Google Scholar]
- Bird A. W., Yu D. Y., Pray-Grant M. G., Qiu Q., Harmon K. E., Megee P. C., Grant P. A., Smith M. M., Christman M. F. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- Birse C. E., Irwin M. Y., Fonzi W. A., Sypherd P. S. Cloning and characterization of ECE1, a gene expressed in association with cell elongation of the dimorphic pathogen Candida albicans. Infect. Immun. 1993;61:3648–3655. doi: 10.1128/iai.61.9.3648-3655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockmuhl D. P., Ernst J. F. A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics. 2001;157:1523–1530. doi: 10.1093/genetics/157.4.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman A. R., Leigh-Bell J. A., Yu H., Bertone P., Gerstein M., Snyder M. Target hub proteins serve as master regulators of development in yeast. Genes Dev. 2006;20:435–448. doi: 10.1101/gad.1389306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreault A. A., Cronier D., Selleck W., Lacoste N., Utley R. T., Allard S., Savard J., Lane W. S., Tan S., Cote J. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev. 2003;17:1415–1428. doi: 10.1101/gad.1056603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun B. R., Kadosh D., Johnson A. D. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 2001;20:4753–4761. doi: 10.1093/emboj/20.17.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. E., Howe L., Sousa K., Alley S. C., Carrozza M. J., Tan S., Workman J. L. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science. 2001;292:2333–2337. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- Cao F., Lane S., Raniga P. P., Lu Y., Zhou Z., Ramon K., Chen J., Liu H. The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans. Mol. Biol. Cell. 2006;17:295–307. doi: 10.1091/mbc.E05-06-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy J. S., Kron S. J. NuA4 subunit Yng2 function in intra-S-phase DNA damage response. Mol. Cell. Biol. 2002;22:8215–8225. doi: 10.1128/MCB.22.23.8215-8225.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy J. S., Tobe B. T., Huh J. H., Kron S. J. Yng2p-dependent NuA4 histone H4 acetylation activity is required for mitotic and meiotic progression. J. Biol. Chem. 2001;276:43653–43662. doi: 10.1074/jbc.M102531200. [DOI] [PubMed] [Google Scholar]
- Ciudad T., Andaluz E., Steinberg-Neifach O., Lue N. F., Gow N. A., Calderone R. A., Larriba G. Homologous recombination in Candida albicans: role of CaRad52p in DNA repair, integration of linear DNA fragments and telomere length. Mol. Microbiol. 2004;53:1177–1194. doi: 10.1111/j.1365-2958.2004.04197.x. [DOI] [PubMed] [Google Scholar]
- Clarke A. S., Lowell J. E., Jacobson S. J., Pillus L. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 1999;19:2515–2526. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosma M. P., Tanaka T., Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- Dieterich C., Schandar M., Noll M., Johannes F. J., Brunner H., Graeve T., Rupp S. In vitro reconstructed human epithelia reveal contributions of Candida albicans EFG1 and CPH1 to adhesion and invasion. Microbiology. 2002;148:497–506. doi: 10.1099/00221287-148-2-497. [DOI] [PubMed] [Google Scholar]
- Doedt T., Krishnamurthy S., Bockmuhl D. P., Tebarth B., Stempel C., Russell C. L., Brown A. J., Ernst J. F. APSES proteins regulate morphogenesis and metabolism in Candida albicans. Mol. Biol. Cell. 2004;15:3167–3180. doi: 10.1091/10.1091/mbc.E03-11-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y., Cote J. The highly conserved and multifunctional NuA4 HAT complex. Curr. Opin. Genet. Dev. 2004;14:147–154. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Fonzi W. A., Irwin M. Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarneau L., Nourani A., Boudreault A. A., Zhang Y., Heliot L., Allard S., Savard J., Lane W. S., Stillman D. J., Cote J. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell. 2000;5:927–937. doi: 10.1016/s1097-2765(00)80258-0. [DOI] [PubMed] [Google Scholar]
- Garcia-Sanchez S., Aubert S., Iraqui I., Janbon G., Ghigo J. M., d'Enfert C. Candida albicans biofilms: a developmental state associated with specific and stable gene expression patterns. Eukaryot. Cell. 2004;3:536–545. doi: 10.1128/EC.3.2.536-545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sanchez S., Mavor A. L., Russell C. L., Argimon S., Dennison P., Enjalbert B., Brown A. J. Global roles of Ssn6 in Tup1- and Nrg1-dependent gene regulation in the fungal pathogen, Candida albicans. Mol. Biol. Cell. 2005;16:2913–2925. doi: 10.1091/mbc.E05-01-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng F., Laurent B. C. Roles of SWI/SNF and HATs throughout the dynamic transcription of a yeast glucose-repressible gene. EMBO J. 2004;23:127–137. doi: 10.1038/sj.emboj.7600035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P. A., et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Gyuris J., Golemis E., Chertkov H., Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- Harcus D., Nantel A., Marcil A., Rigby T., Whiteway M. Transcription profiling of cyclic AMP signaling in Candida albicans. Mol. Biol. Cell. 2004;15:4490–4499. doi: 10.1091/mbc.E04-02-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A. H., Prochasson P., Neely K. E., Galasinski S. C., Chandy M., Carrozza M. J., Workman J. L. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell. 2002;111:369–379. doi: 10.1016/s0092-8674(02)01005-x. [DOI] [PubMed] [Google Scholar]
- Hazan I., Sepulveda-Becerra M., Liu H. Hyphal elongation is regulated independently of cell cycle in Candida albicans. Mol. Biol. Cell. 2002;13:134–145. doi: 10.1091/mbc.01-03-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho S. W., Jona G., Chen C. T., Johnston M., Snyder M. Linking DNA-binding proteins to their recognition sequences by using protein microarrays. Proc. Natl. Acad. Sci. USA. 2006;103:9940–9945. doi: 10.1073/pnas.0509185103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer L. L., Payne T. L., Bell M., Myers A. M., Scherer S. Candida albicans ALS3 and insights into the nature of the ALS gene family. Curr. Genet. 1998;33:451–459. doi: 10.1007/s002940050359. [DOI] [PubMed] [Google Scholar]
- Kadosh D., Johnson A. D. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol. Biol. Cell. 2005;16:2903–2912. doi: 10.1091/mbc.E05-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Wolyniak M. J., Staab J. F., Sundstrom P. A 368-base-pair cis-acting HWP1 promoter region, HCR, of Candida albicans confers hypha-specific gene regulation and binds architectural transcription factors Nhp6 and Gcf1p. Eukaryot. Cell. 2007;6:693–709. doi: 10.1128/EC.00341-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A., Umehara T., Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 2002;32:370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- Krogan N. J., et al. Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl. Acad. Sci. USA. 2004;101:13513–13518. doi: 10.1073/pnas.0405753101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E., Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis R. E., Lo H. J., Raad I. I., Kontoyiannis D. P. Lack of catheter infection by the efg1/efg1 cph1/cph1 double-null mutant, a Candida albicans strain that is defective in filamentous growth. Antimicrob. Agents Chemother. 2002;46:1153–1155. doi: 10.1128/AAC.46.4.1153-1155.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Su C., Mao X., Cao F., Chen J. Roles of Candida albicans Sfl1 in hyphal development. Eukaryot. Cell. 2007;6:2112–2121. doi: 10.1128/EC.00199-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo H. J., Kohler J. R., DiDomenico B., Loebenberg D., Cacciapuoti A., Fink G. R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90:939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- Loewith R., Meijer M., Lees-Miller S. P., Riabowol K., Young D. Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol. Cell. Biol. 2000;20:3807–3816. doi: 10.1128/mcb.20.11.3807-3816.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Cao F., Nie X., Liu H., Chen J. The Swi/Snf chromatin remodeling complex is essential for hyphal development in Candida albicans. FEBS Lett. 2006;580:2615–2622. doi: 10.1016/j.febslet.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Mitchell L., Lambert J. P., Gerdes M., Al-Madhoun A. S., Skerjanc I. S., Figeys D., Baetz K. Functional dissection of the NuA4 histone acetyltransferase reveals its role as a genetic hub and that Eaf1 is essential for complex integrity. Mol. Cell. Biol. 2008;28:2244–2256. doi: 10.1128/MCB.01653-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murad A. M., et al. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 2001;20:4742–4752. doi: 10.1093/emboj/20.17.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naar A. M., Lemon B. D., Tjian R. Transcriptional coactivator complexes. Annu. Rev. Biochem. 2001;70:475–501. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- Narlikar G. J., Fan H. Y., Kingston R. E. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- Ni J., Gao Y., Liu H., Chen J. Candida albicans Cdc37 interacts with the Crk1 kinase and is required for Crk1 production. FEBS Lett. 2004;561:223–230. doi: 10.1016/S0014-5793(04)00172-3. [DOI] [PubMed] [Google Scholar]
- Nourani A., Doyon Y., Utley R. T., Allard S., Lane W. S., Cote J. Role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex. Mol. Cell. Biol. 2001;21:7629–7640. doi: 10.1128/MCB.21.22.7629-7640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourani A., Utley R. T., Allard S., Cote J. Recruitment of the NuA4 complex poises the PHO5 promoter for chromatin remodeling and activation. EMBO J. 2004;23:2597–2607. doi: 10.1038/sj.emboj.7600230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan Q. T., Belanger P. H., Filler S. G. Role of hyphal formation in interactions of Candida albicans with endothelial cells. Infect. Immun. 2000;68:3485–3490. doi: 10.1128/iai.68.6.3485-3490.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokholok D. K., et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Ramage G., VandeWalle K., Lopez-Ribot J. L., Wickes B. L. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol. Lett. 2002;214:95–100. doi: 10.1111/j.1574-6968.2002.tb11330.x. [DOI] [PubMed] [Google Scholar]
- Reid J. L., Iyer V. R., Brown P. O., Struhl K. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell. 2000;6:1297–1307. doi: 10.1016/s1097-2765(00)00128-3. [DOI] [PubMed] [Google Scholar]
- Robert F., Pokholok D. K., Hannett N. M., Rinaldi N. J., Chandy M., Rolfe A., Workman J. L., Gifford D. K., Young R. A. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell. 2004;16:199–209. doi: 10.1016/j.molcel.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selleck W., Fortin I., Sermwittayawong D., Cote J., Tan S. The Saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex requires the Enhancer of Polycomb A domain and chromodomain to acetylate nucleosomes. Mol. Cell. Biol. 2005;25:5535–5542. doi: 10.1128/MCB.25.13.5535-5542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiadi E. R., Doedt T., Cottier F., Noffz C., Ernst J. F. Transcriptional response of Candida albicans to hypoxia: linkage of oxygen sensing and Efg1p-regulatory networks. J. Mol. Biol. 2006;361:399–411. doi: 10.1016/j.jmb.2006.06.040. [DOI] [PubMed] [Google Scholar]
- Sohn K., Urban C., Brunner H., Rupp S. EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Mol. Microbiol. 2003;47:89–102. doi: 10.1046/j.1365-2958.2003.03300.x. [DOI] [PubMed] [Google Scholar]
- Sonneborn A., Bockmuhl D. P., Gerads M., Kurpanek K., Sanglard D., Ernst J. F. Protein kinase A encoded by TPK2 regulates dimorphism of Candida albicans. Mol. Microbiol. 2000;35:386–396. doi: 10.1046/j.1365-2958.2000.01705.x. [DOI] [PubMed] [Google Scholar]
- Sonneborn A., Tebarth B., Ernst J. F. Control of white-opaque phenotypic switching in Candida albicans by the Efg1p morphogenetic regulator. Infect. Immun. 1999;67:4655–4660. doi: 10.1128/iai.67.9.4655-4660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantha T., Tsai L. K., Daniels K., Soll D. R. EFG1 null mutants of Candida albicans switch but cannot express the complete phenotype of white-phase budding cells. J. Bacteriol. 2000;182:1580–1591. doi: 10.1128/jb.182.6.1580-1591.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staab J. F., Sundstrom P. Genetic organization and sequence analysis of the hypha-specific cell wall protein gene HWP1 of Candida albicans. Yeast. 1998;14:681–686. doi: 10.1002/(SICI)1097-0061(199805)14:7<681::AID-YEA256>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Stoldt V. R., Sonneborn A., Leuker C. E., Ernst J. F. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suka N., Luo K., Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 2002;32:378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- Tebarth B., Doedt T., Krishnamurthy S., Weide M., Monterola F., Dominguez A., Ernst J. F. Adaptation of the Efg1p morphogenetic pathway in Candida albicans by negative autoregulation and PKA-dependent repression of the EFG1 gene. J. Mol. Biol. 2003;329:949–962. doi: 10.1016/s0022-2836(03)00505-9. [DOI] [PubMed] [Google Scholar]
- Umeyama T., Nagai Y., Niimi M., Uehara Y. Construction of FLAG tagging vectors for Candida albicans. Yeast. 2002;19:611–618. doi: 10.1002/yea.863. [DOI] [PubMed] [Google Scholar]
- Utley R. T., Ikeda K., Grant P. A., Cote J., Steger D. J., Eberharter A., John S., Workman J. L. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- Wilson R. B., Davis D., Mitchell A. P. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 1999;181:1868–1874. doi: 10.1128/jb.181.6.1868-1874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Oh S. H., Cheng G., Green C. B., Nuessen J. A., Yeater K., Leng R. P., Brown A. J., Hoyer L. L. ALS3 and ALS8 represent a single locus that encodes a Candida albicans adhesin; functional comparisons between Als3p and Als1p. Microbiology. 2004;150:2415–2428. doi: 10.1099/mic.0.26943-0. [DOI] [PubMed] [Google Scholar]
- Zordan R. E., Miller M. G., Galgoczy D. J., Tuch B. B., Johnson A. D. Interlocking transcriptional feedback loops control white-opaque switching in Candida albicans. PLoS Biol. 2007;5:e256. doi: 10.1371/journal.pbio.0050256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.