Abstract
In addition to its central role in energy production, oxygen has pervasive regulatory actions. Hypoxia (oxygen limitation) triggers the shutdown of major cellular processes, including gene expression. We carried out a genome-wide RNA interference (RNAi) screen in Drosophila S2 cells for functions required to down-regulate translation during hypoxia. RNAi knockdown of specific genes allowed induction of a green fluorescent protein (GFP) reporter gene and continued protein synthesis during hypoxia. Among the identified genes, Tsc1 and Tsc2, which together form the tuberose sclerosis complex that negatively regulates target of rapamycin (TOR) kinase, gave an especially strong effect. This finding is consistent with the involvement of TOR in promoting translation. Another gene required for efficient inhibition of protein translation during hypoxia, the protein tyrosine phosphatase 61F (Ptp61F), down-regulates TOR activity under hypoxia. Lack of Ptp61F or Tsc2 improves cell survival under prolonged hypoxia in a TOR-dependent manner. Our results identify Ptp61F as a novel modulator of TOR activity and suggest that its function during hypoxia contributes to the down-regulation of protein synthesis.
INTRODUCTION
Oxygen is a pivotal component in metabolism and physiology. Damage caused by limited oxygen supply, as occurs in stroke and myocardial infarction, is one of the leading causes of death in the United States. Limited oxygen availability, called hypoxia, is also a key feature of the microenvironment in solid tumors, and tumor cell survival of hypoxia is important in progression to malignancy.
When a shortfall in oxygen is modest, cells and organisms can adjust their metabolism and physiology to compensate, a response that includes adjustments in gene expression mediated by hypoxia-inducible factor (HIF) (Semenza, 2007). However, when reductions in oxygen levels are more severe, cells cannot continue to function, and survival depends on inducing quiescence. Indeed, organisms that tolerate severe hypoxia do so by entering a state resembling suspended animation, in which energy-consuming activities are curtailed. The embryos of many organisms, including Danio rerio (Padilla and Roth, 2001), Caenorhabditis elegans (Padilla et al., 2002), and Drosophila melanogaster (Wingrove and O'Farrell, 1999; DiGregorio et al., 2001; Teodoro and O'Farrell, 2003), instantly enter such a state under severe hypoxia. Our laboratory showed that abrupt loss of oxygen causes inhibition of DNA replication, gene transcription and protein translation, cell cycle arrest in either interphase or metaphase, and stabilization of mRNA and proteins in Drosophila embryos (Wingrove and O'Farrell, 1999; DiGregorio et al., 2001; Teodoro and O'Farrell, 2003).
Similar hypoxia responses have been observed in mammalian cell culture systems. Among them, hypoxia-induced inhibition of protein translation has been studied in detail (Liu and Simon, 2004; Wouters et al., 2005; Koumenis, 2006; Koumenis and Maxwell, 2006; Liu et al., 2006). Hypoxia engages several regulatory pathways that include documented indirect inputs into a translational activity by HIF and more direct inputs from three regulatory kinases: PKR-like ER kinase, AMP-dependent kinase (AMPK), and TOR kinase. However, the signaling path of particular importance in modulating translation during hypoxia is not clear. Because genome-wide RNAi screening provides an unbiased identification of the genes making an important contribution to a response, it can help define the major regulatory pathway. To pursue such an analysis, we demonstrated that Drosophila S2 cells exhibit hypoxia-induced suppression of translation, and we developed an RNAi screen for the relevant genes. Our findings have focused our attention on a pathway of hypoxia regulation of TOR that is distinct from that emphasized in previous studies.
TOR kinase is of great interest because it is a central positive regulator of normal and oncogenic growth (Neufeld, 2003; Oldham and Hafen, 2003; Bhaskar and Hay, 2007). TOR kinase activates translation by inhibiting translational inhibitors. For example, TOR kinase phosphorylates 4E-binding protein (4E-BP), blocking its inhibitory association with a key translation initiation factor, eukaryotic initiation factor (eIF) 4E (Beretta et al., 1996; von Manteuffel et al., 1996). TOR also phosphorylates and activates S6 kinase, p70S6K, which in turn phosphorylates the ribosomal protein S6, promotes ribosome biogenesis, and acts indirectly to prevent eEF2 suppression (Chung et al., 1992; Kuo et al., 1992; Price et al., 1992; Wang et al., 2000, 2001; Browne and Proud, 2004; Connolly et al., 2006). Thus, regulation of TOR is a central axis of translational control.
Dissection of the growth inputs into TOR activity has revealed contributors to its regulation. Tsc1 and Tsc2 are tumor suppressor genes in humans: their inactivation gives rise to nonmalignant, yet damaging growths in tuberous sclerosis patients. Studies in Drosophila revealed that these genes have a central role in the down-regulation of TOR that is conserved to other organisms (Pan et al., 2004; Martin and Hall, 2005). Together, Tsc1 and Tsc2 form a complex, TSC, which acts as a GTPase-activating protein whose function reduces the active form of Ras homolog enriched in brain (Rheb), a small GTPase that activates TOR (Garami et al., 2003; Inoki et al., 2003; Saucedo et al., 2003; Stocker et al., 2003; Zhang et al., 2003). Insulin promotes growth by activating a phosphatidylinositol 3-kinase (PI3-kinase)–Akt pathway that suppresses TSC function to allow TOR to act (Potter et al., 2001; Tapon et al., 2001; Inoki et al., 2002).
Hypoxia promotes TSC function and hence down-regulates TOR. Two modes of hypoxia regulation of TSC have previously been identified. HIF promotes additional gene expression of two related activators of TSC that are called Scylla and Charybdis in Drosophila and Redd1 and Redd2 in mammals (Reiling and Hafen, 2004; Sofer et al., 2005). In addition, AMPK phosphorylates and activates Tsc2 to provide a HIF-independent inhibitory input into TOR activity (Corradetti et al., 2004).
In this report, we asked how hypoxia induces its responses by using inhibition of protein translation as a readout in Drosophila S2 cells. We recently developed an insect cell culture system that encompasses features of the aforementioned animal models' responses to hypoxia, including inhibition of protein translation. Using this culture system, we can focus on the fundamental features of the oxygen sensing and signal transduction without all the attendant physiological responses that complicate studies in the whole animal. More importantly, in Drosophila tissue culture cells, long double-stranded RNAs stimulate RNAi with a remarkable efficiency. Here, we report a genome-wide RNAi screen that identified genes involved in the down-regulation of protein synthesis under hypoxia.
MATERIALS AND METHODS
Cell Culture, RNAi, and Hypoxia Treatment
Drosophila S2 cells and S2 cells harboring a GFP reporter (GFP-Relish) under the metallothioneine promoter (pMT) were cultured in Schneider's Drosophila medium (GIBCO) supplemented with 10% fetal bovine serum (heat inactivated), penicillin, and streptomycin. We added double-stranded RNA (dsRNA) (final concentration, ∼10 μg/ml) to 2∼4 × 105/ml cells. Cells were incubated with dsRNA for 4 d before hypoxia treatment. Hypoxia was achieved by displacing oxygen in a hypoxia chamber with purified N2 gas whose flow was controlled by an oxygen sensor (Proox model 110; Reming Bioinstruments, Redfield, NY). For rapamycin treatment, cells were treated with rapamycin (final concentration, 50 nM; R0395, Sigma-Aldrich, St. Louis, MO) before being placed in the hypoxia chamber.
Genome-wide RNAi Screening
Generation of a dsRNA library has been described previously (Foley and O'Farrell, 2004). dsRNA (final concentration, ∼10 μg/ml) was added to each well of a 96-well plate containing 200 μl of S2 cells (2∼4 × 105/ml) harboring a GFP reporter (GFP-Relish; Foley and O'Farrell, 2004) under the control of the metallothionein promoter. After 4 d of incubation at 25°C, CuSO4 was added to cells at 400 μM (final concentration), and cells were placed in the hypoxia chamber (0.5% O2) overnight. Cells were removed from the chamber, immediately fixed (2% formaldehyde for 5 min), washed with PBS, and screened for an induced GFP signal. dsRNA for candidates were synthesized and tested again for their influence on induction of GFP as detected by microscopy and Western blot analysis.
Double-Strand RNA Production
The polymerase chain reaction (PCR)-generated DNA templates used for dsRNA production had T7 RNA promoter sequences (TAATACGACTCACTATAGGGAGACCACGGGCGGGT) at both ends and specific exonic sequences internally as described previously (Echard et al., 2004; Foley and O'Farrell, 2004). Transcription of such library templates gave dsRNA products. We made an additional DNA template for Ptp61F, which does not overlap in sequence with the Ptp61F DNA template in the library. The efficiency of dsRNA treatment was examined either by reverse transcription (RT)-PCR or Western blotting analysis.
For RT-PCR, total mRNA was extracted using TRIzol (Invitrogen, Carlsbad, CA). cDNA from either 1 or 5 μg total mRNA was made using Superscript First Strand synthesis system for RT-PCR kit (Invitrogen). Two different amounts of input cDNA were used for PCR to show that the signal is not saturated and that band intensity reflects input level of corresponding mRNA. Ptp61F-A was cloned into pENTR-3C as a BamHI–EcoRV PCR fragment and subsequently moved into a Gateway destination vector containing a copper inducible (pMT) promoter.
Western Blot Analysis
Cells were collected by centrifugation at 3000 rpm for 5 min, washed once with PBS (phosphate buffered solution), and lysed with lysis buffer (1% SDS, 100 mM EDTA in PBS with protease inhibitor cocktail [Complete; Roche Diagnostics, Indianapolis, IN]) for 30 min. Cell lysates were heated for 5 min at 95°C and centrifuged for 15 min at top speed (14,000 rpm). Protein concentration was measured using a DC protein assay kit (Bio-Rad, Hercules, CA), and equal amounts of proteins were loaded for SDS-polyacrylamide gel electrophoresis (PAGE). Proteins separated by SDS-PAGE were transferred to polyvinylidene difluoride membrane (Immobilon; Millipore, Billerica, MA) and analyzed by Western blotting. Anti-GFP antibody (1:2000, MMS-118P) was purchased from BAbCO (Richmond, CA). Antibodies against phosphorylated Drosophila ribosomal S6 kinase (T398, 1:1000; #9029), phosphorylated 4E BP1(Thr37/46, 1:1000; #9459), phosphorylated Drosophila Akt (S505, 1:1000; #4054), and Akt (1:1000; #9272) were purchased from Cell Signaling Technology (Danvers, MA). Anti-tubulin (1:5000; T9026) was from Sigma-Aldrich. Anti-phosphotyrosine (clone 4G10; #05-321x) was from Millipore. Antibody against Drosophila insulin receptor was generously provided by Dr. Oscar Puig (Institute of Biotechnology, University of Helsinki, Helsinki, Finland). Horseradish peroxidase-conjugated secondary antibodies were purchased from Jackson ImmunoResearch Laboratories (West Grove, PA).
[35S]-Methionine Incorporation Assay
S2 cells were placed in a hypoxia chamber for the indicated times. [35S]-methionine (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) was added to S2 cells to a final concentration of 50 μCi/ml. Cells were allowed to incorporate [35S]-methionine for 60 min. Cells were lysed by incubation in 1% SDS, 100 mM EDTA in PBS for 30 min. The cell lysate was heated for 5 min at 95°C, followed by centrifugation at 14,000 rpm for 15 min. Equal amounts of solubilized protein were precipitated with trichloroacetic acid. Precipitated protein was transferred to glass microfiber filters (Whatman GF/C; Whatman, Maidstone, United Kingdom) for measurement of [35S]-methionine incorporation in a scintillation counter.
Cell Survival Assay
Cells were treated with dsRNA against various genes for 4 d, and then they were exposed to hypoxia for the indicated times. Sytox Green Nucleic Acid Stain (S-7020; Invitrogen) was added to cells at a final concentration of 10 nM. Cells were analyzed by fluorescence-activated cell sorting (FACS) after incubation at room temperature for 10 min and addition of 800 μl of phosphate-buffered saline (PBS).
RESULTS
Hypoxia Causes Inhibition of Protein Translation in S2 Cells
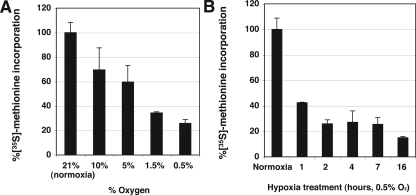
Hypoxia down-regulates protein synthesis in Drosophila embryos and in mammalian tissue culture cells. We examined the effects of hypoxia on translation by assaying [35S]-methionine incorporation by Drosophila S2 cells in culture. Different levels of hypoxia were achieved by admixing an inert gas (nitrogen or argon) and air, and incorporation was measured between the first and second hour after hypoxia exposure. Radiolabel incorporation declined with increasing hypoxia, reaching ∼75% inhibition at 0.5% oxygen (Figure 1A). To assess the speed of onset, we measured [35S]-methionine incorporated during 1-h intervals at different times after exposure of cells to 0.5% oxygen. There was an immediate drop in incorporation (∼60% inhibition) at 1 h followed by continued, slower decline so that at 16 h, there was ∼85% inhibition (Figure 1B). No decline in cell viability occurred during overnight hypoxia (<0.1% O2; Figure 6A). Our results show that, like other cells that have been examined, Drosophila S2 cells exhibit a rapid and dramatic suppression of translation in response to hypoxia.
Figure 1.
Hypoxia induces inhibition of protein translation in Drosophila S2 cells. (A) Protein translation is inhibited in an oxygen concentration-dependent manner. Cells were exposed to the indicated concentration of O2 for 1 h, [35S]-methionine was then added, and incorporation assessed after an additional hour incubation. Counts per minute of [35S]-methionine incorporation were normalized to that in normoxia (100%). (B) Protein translation is rapidly inhibited under hypoxia. Cells were exposed to hypoxia (0.5% O2) for the indicated time. [35S]-Methionine was added during the last hour of the hypoxia treatment.
Figure 6.
Lack of Ptp61F improves cell survival under hypoxia. (A) Cells deficient in Ptp61F displayed enhanced survival under hypoxia compared with control cells (LacI, GFP, or luciferase RNAi). *p < 0.05. (B and C) Inhibition of TORC1 activity abolishes the effect of depletion of Ptp61F (B) or Tsc2 (C) on cell survival under hypoxia. In Figure 6, cells were treated with various dsRNAs for 4 d and exposed to hypoxia treatment for indicated days. Cell death was measured by staining cells with Sytox Green, which only stains dead cells, followed by FACS analysis.
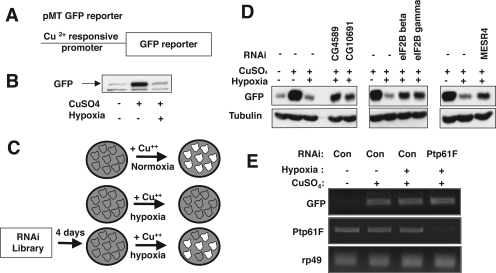
To assess whether hypoxia would block induction of new gene expression, we examined a stable cell line harboring a GFP-tagged transgene expressed under the Cu2+-inducible methallothioneine promoter (Figure 2A). Overnight exposure to CuSO4 induced bright fluorescence in normoxic cells but not hypoxic cells (∼0.5% O2). This hypoxia suppression of GFP expression was readily detected by fluorescence microscopy and by Western blot analysis (Figure 2B and Supplemental Figure S1). In contrast to the protein, induction of GFP mRNA was not reduced by hypoxia (Figure 2E), indicating that the inhibition of GFP expression is mainly due to suppression of translation.
Figure 2.
Genome wide RNAi screening in S2 cells identifies sets of genes required for efficient suppression of translation in response to hypoxia. (A) Cu2+-inducible GFP reporter. (B) The expression of a GFP reporter gene is inhibited under hypoxia. CuSO4 (400 μM final concentration) was added to S2 cells harboring the Cu2+-inducible GFP reporter. Cells were incubated in either normoxia or hypoxia for 16 h. GFP expression levels were measured by Western blotting of cell lysates with an anti-GFP antibody. (C) Scheme of RNAi screening. S2 cells harboring the GFP reporter were treated with dsRNA in a 96-well format and incubated for 4 d. CuSO4 was added to dsRNA-treated cells. Cells were placed under hypoxia (∼0.5% O2) overnight, fixed with 2% formaldehyde, and screened for GFP signal. (D) Western analysis of reporter gene induction shows that RNAi against genes identified in the screen diminishes translational suppression during hypoxia. Cells were treated for 4 d with dsRNA against genes identified in our screen. CuSO4 was added to dsRNA-treated cells. Cells were placed under hypoxia (∼0.5% O2) overnight. GFP expression level was measured by Western blotting of cell lysates with an anti-GFP antibody. Tubulin was detected with anti-tubulin antibody (Sigma-Aldrich) as a loading control. (E) Transcript levels from the GFP reporter were not affected by hypoxia. Cells were treated as described above, and the induction of the GFP reporter under hypoxia and normoxia was measured by RT-PCR.
Genome-wide RNAi Screening Identifies Genes Involved in Inhibition of GFP Induction under Hypoxia
To probe the relevant mechanisms of oxygen sensing and the signaling steps leading to suppression of gene expression, we used a library of 7216 dsRNAs, representing the conserved genes of Drosophila (Foley and O'Farrell, 2004), in an RNAi screen for genes required for suppression of GFP induction in hypoxia. Our starting premise was that the suppression of new gene expression by hypoxia was not merely a passive consequence of cellular energy depletion but represented an active response requiring specific effector functions.
We added dsRNA to S2 cells harboring the GFP reporter and incubated these for 4 d. After 4 d of dsRNA treatment, CuSO4 was added and the cells were placed under hypoxia (∼0.5% O2) overnight (Figure 2C). After this period of induction in hypoxia, cells were fixed and screened for a GFP signal. In the first round of screening, we visually identified wells where the dsRNA allowed substantial GFP induction. We rescreened candidates by the same test, and then assessed the degree of the bypass of the hypoxia-induced inhibition by Western blotting for GFP (Figure 2D).
The genes identified in our screen are summarized in Table 1. We identified sets of genes involved in signaling pathways, including Tsc1, Tsc2, Ptp61F, SH3PX1, and MESR4. Among them, Tsc1 and Tsc2 form a complex (TSC complex), and have been reported previously to be involved in inhibition of protein translation under hypoxia. We also recovered several genes involved in mitochondrial protein transport (Tom40, Tim44) as well as in mitochondrial function (CG4589, CG10691). Some of the candidates are components of translation machinery (eIF2Bβ; Figure 2D) or are known to negatively regulate protein translation (Musashi). We discuss the screen further below, but focus results in this report on the TOR pathway and involvement of Ptp61F in the regulation of translation in hypoxia.
Table 1.
Genes required for hypoxia-induced inhibition of protein translation in Drosophila S2 cells
| CG no. | Gene name | Protein/function |
|---|---|---|
| Protein translational machinery | ||
| CG2677 | eIF2B-β | Initiation factor in protein translation |
| CG8190 | eIF2B-γ | Initiation factor in protein translation |
| CG5099 | msi | Musashi/mRNA processing, negative regulation of translation |
| CG17737 | Translation initiation factor SUI1 | |
| Signal transduction pathways | ||
| CG6147 | Tsc1 | Tsc1/negative regulation of cell growth, size, insulin receptor signaling pathway, organ size |
| CG6975 | gig | Gigas, or Tsc2/negative regulation of cell growth, size, insulin receptor signaling pathway, organ size |
| CG9181 | Ptp61F | Ptp61F/protein tyrosine phosphatase 61F |
| CG6757 | SH3PX1 | SH3PX1/intracellular protein transport, signaling cascade, tyrosine phosphorylated protein that binds to the src homology 2 domain of Dock |
| CG4903 | MESR4 | MESR4/Misexpression suppressor of ras 4 |
| Transcription factor related | ||
| CG32296 | Mrtf | Transcriptional activator activity involved in actin cytoskeleton organization and biogenesis |
| CG16902 | Hr4 | Ligand-dependent nuclear receptor activity, steroid hormone receptor activity |
| CG18389 | Eip93F | Autophagic cell death, ecdysone mediated, positive regulation of transcription |
| CG12690 | CHES-1-like | Checkpoint suppressor homologue. Fork head transcription factor Winged helix repressor DNA binding |
| Mitochondria | ||
| CG4589 | LETM1/calcium ion binding, mitochondrio | |
| CG10691 | l(2)37Cc | Prohibitin/DNA replication, regulation of cell cycle, mitochondria |
| CG5844 | Dodecenoyl-CoA δ-isomerase | |
| CG12157 | Tom40 | Tom40/mitochondrial import, translocase of outer membrane |
| CG11779 | Tim44/mitochondrial import, translocase of inner membrane | |
| CG33066 | Tim9b | Tim9b/mitochondrial import, translocase of, inner membrane |
| Miscellaneous | ||
| CG10688 | Phosphomannomutase activity, manose 6-phosphate/mannose 1-phosphate interconversion in GDP-mannose biosynthesis | |
| CG1559 | Upf1 | RNA helicase activity |
| CG12403 | Vha68-1 | Proton pump, vacuolar ATPase |
| CG12359 | ulp1 | Protein deubiquitination, protein processing, protein targeting, proteolysis and peptidolysis–cys protease |
TSC Complex and Ptp61F Are Required for Efficient Inhibition of Protein Translation under Hypoxia
Consistent with the involvement of TOR in promoting protein translation in other contexts (Ellisen, 2005), addition of the TOR inhibitor rapamycin to normoxic S2 cells suppressed GFP induction as assessed by Western blots at 16 h after induction (Figure 3A, lane 2 vs. lane 4). The identification of Tsc1 and Tsc2 in our screen suggests that down-regulation of TOR contributes to suppression of gene expression during hypoxia. Phosphatase and tensin homologue (PTEN), another negative regulator in the TOR pathway, was not recovered in our screens. However, a direct test of PTEN RNAi revealed that it results in cell death in Drosophila S2 cells, which precluded test of a contribution to the response to hypoxia. In contrast to the dramatically increased GFP accumulation induced during hypoxia after RNAi of Tsc1 or Tsc2 (Figure 3A, lane 3 vs. lanes 6 and 7), HIF-1 knockdown had no detectable effect (Figure 3A, lane 3 vs. lane 5). These results indicate that down-regulation of TOR plays an important role in robust suppression of GFP expression induced under hypoxia and that this regulation is independent of HIF-1.
Figure 3.
Ptp61F is required for efficient inhibition of protein translation under hypoxia. (A) The GFP reporter is induced in cells treated with RNAi against Tsc1, Tsc2, or Ptp61F. Experiments were performed as described in Figure 2D. Rapamycin was added to 50 nM (final concentration) where indicated. (B) The effectiveness of dsRNA against Ptp61F is confirmed by RT-PCR. Cells were treated with indicated dsRNA for 4 d, and depletion of mRNA of indicated genes was examined by RT-PCR. (C) [35S]-Methionine incorporation is decreased by exposure to hypoxia and incorporation in hypoxia is increased by depletion of Tsc2 or Ptp61F. Cells were treated with dsRNA for 4 d, and exposed to hypoxia for 16 h. [35S]-Methionine incorporation was measured as described in Figure 1A. Counts per minute of [35S]-methionine incorporation was normalized to that of control RNAi-treated cells in normoxia (100%). *p < 0.05.
We hypothesized that, in addition to Tsc1 and Tsc2, our screen might identify other negative regulators operating at different points in the signal transduction pathway controlling TOR. One of the genes identified in our screen was of particular interest in this regard. Protein tyrosine phosphatase 61F (Ptp61F) has two homologues in mammals, protein tyrosine phosphatase 1B (PTP-1B) and the related protein TC-PTP. PTP-1B is localized in the membrane and negatively regulates the insulin receptor signaling and leptin signaling in mammals (Seely et al., 1996; Goldstein et al., 1998; Haj et al., 2003). TC-PTP is localized in the nucleus, has a negative input on Janus tyrosine kinase (JAK)/signal transducer and activator of transcription (STAT) pathway in mammals, and is important in immune homeostasis (Simoncic et al., 2002, 2006).
Drosophila Ptp61F has four different splice forms (A, B, C, and D); A and D are bound to membrane and B and C are localized in the nucleus (McLaughlin and Dixon, 1993). Ptp61F interacts with dreadlocks (dock), which has been shown to bind to the insulin receptor (Song et al., 2003), and it was recently reported to be a negative regulator of JAK/STAT pathways (Baeg et al., 2005; Muller et al., 2005). Although its roles in hypoxia have not been reported, we were attracted by the involvement of Ptp61F in pathways of potential relevance: insulin is an upstream regulator of the TOR pathway, and JAK/STAT has been implicated in stress responses.
The dsRNA used in most of our experiments targets all four splice forms. The effectiveness of RNAi knockdown of Ptp61F was confirmed by RT-PCR, and specificity was confirmed by knockdown by using an independent dsRNA targeting nonoverlapping sequences in Ptp61F (Figure 3B) as well as by knockdown of specific isoforms (below). The magnitude of the Ptp61F contribution to hypoxia suppression of induced GFP expression was assessed by Western blot. As seen in Figure 3A, Ptp61F-RNAi increased the level of induced GFP expression in hypoxic cells (Figure 3A, lane 3 vs. lane 8), but to a smaller degree than the knockdown of Tsc1 or Tsc2 (Figure 3A, lanes 6 and 7).
We examined [35S]-methionine incorporation as a measure of effects on translation (Figure 3C). Cells were treated with dsRNA against Tsc2, Ptp61F or control RNAi for 4 d, and then they were exposed to normoxia or hypoxia treatment (<0.5% O2) for 16 h before addition of [35S]-methionine. Although Tsc2 knockdown seemed to cause a slight increase in [35S]-methionine incorporation during normoxia, this effect was not significant. Ptp61F had no evident effect on translation in normoxia. Hypoxia caused a clear suppression of incorporation as described above. Cells treated with dsRNA against Tsc2 or Ptp61F had a significantly higher level of incorporation than cells treated with control dsRNA under hypoxia (1.4–2.5-fold increase; p < 0.05). These results indicate that Ptp61F is required for efficient inhibition of protein translation under hypoxia.
Down-Regulation of TOR Activity under Hypoxia Requires Ptp61F
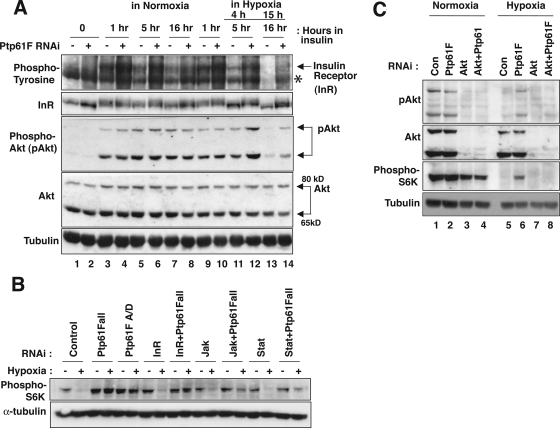
Because mammalian (m)TOR down-regulation contributes to the suppression of translation in mammalian cells, we tested TSC and Ptp61F contributions to down-regulation of TOR during hypoxia in Drosophila S2 cells. We measured TOR activity by examining phosphorylation of two well-characterized TOR substrates, p70S6K and 4E-BP (Figure 4A). A modest level of p70S6K and 4E-BP phosphorylation was observed in control RNAi-treated cells in normoxia, and this phosphorylation was reduced in hypoxia, indicating that TOR activity declined in hypoxia (Figure 4A, lane 1 vs. lane 2). Tsc2 RNAi dramatically increased TOR phosphorylation of 4E-BP in normoxia. This finding suggests that the TSC is active under normal growth conditions and that knockdown of its function results in supraphysiological levels of TOR activity. Tsc2-depleted cells sustained TOR activity under hypoxia, albeit with a slight decrease compared with normoxia (Figure 4A, lanes 1 and 2 vs. lanes 7 and 8). Similarly, Ptp61F-depleted cells sustained TOR activity under hypoxia (Figure 4A, lane 2 vs. lanes 4 and 6); however, Ptp61F RNAi had only a slight effect, if any, on the phosphorylation levels of p70S6K and 4E-BP in normoxia (Figure 4A, lane 1 vs. lanes 3 and 5).
Figure 4.
Ptp61F contributes to inhibition of protein translation under hypoxia by down-regulating TOR activity. (A) Ptp61F is required for down-regulation of TOR activity. Cells were treated with dsRNA against Tsc2 or Ptp61F, exposed to hypoxia (0.5% O2, 16 h), and lysed. Phosphorylated p70S6K (phospho-S6K) and phosphorylated 4E-BP (phospho-4E-BP) were detected by Western blotting with antibodies recognizing phosphorylated forms of these proteins (Cell Signaling Technology). N, normoxia; H, hypoxia. (B) Expression of Ptp61F-A with C-terminal Myc tag restores a decrease in phosphorylation of p70S6K (phospho-S6K) in hypoxia to Ptp61F-A/D–depleted cells. Cells harboring a copper inducible Ptp61F-A-myc gene were treated for 4 d with dsRNA specifically targeting the 3′ UTR of Ptp61F-A/D. Ptp61F-A was induced with copper sulfate (100 μM) for the same period. (C) Inhibition of TOR activity by rapamycin suppresses the effect of Ptp61F depletion on inhibition of protein translation under hypoxia. Cells were treated with dsRNA against Ptp61F for 4 d. Rapamycin was added where indicated, and cells were exposed to hypoxia (0.5% O2 for 16 h). [35S]-Methionine incorporation was measured as described in Figure 1A. Rapa, rapamycin. *p < 0.05.
It has been reported that RNAi using long dsRNA may have off-target effects in Drosophila S2 cells. The selectivity of dsRNA against Ptp61F was demonstrated by rescuing the effects of RNAi by selective expression of Ptp61F. Using a dsRNA complementary to the 3′ untranslated region (UTR) of transcripts encoding Ptp61F membrane isoforms A and D, we were able to selectively knockdown the cognate transcripts (Supplemental Figure S3). Although we were unable to design UTR targeted dsRNAs capable of knocking out isoforms B and C, treatment with the dsRNA specific for the A and D isoforms was sufficient to cause an increased TOR activity during hypoxia (Figure 4B, lanes 5 and 6, and Supplemental Figure S3). Expression of a cDNA that encodes the Ptp61F-A isoform but that lacks the 3′ untranslated region of the endogenous transcript restores Ptp61F-A to Ptp61F-A/D–depleted cells and also restored down-regulation of TOR activity under hypoxia (Figure 4B, lanes 7 and 8). This shows that the effect we observed in cells treated with Ptp61F RNAi is indeed due to Ptp61F depletion rather than an off-target effect, and that Ptp61F functions as a negative regulator of the TOR pathway in hypoxia. Furthermore, these findings show that nonnuclear forms of Ptp61F play an important role in hypoxia down-regulation of TOR activity in Drosophila S2 cells.
Our findings suggest that TOR activity is down-regulated during hypoxia and that this down-regulation makes a substantial contribution to the suppression of translation in hypoxia. To determine whether any consequential residue of TOR function persists during hypoxia, we examined the effect of rapamycin on [35S]-methionine incorporation after 16 h of hypoxia. Rapamycin addition abolished phosphorylation of p70S6K in Drosophila S2 cells (Supplemental Figure S2A) and suppresses [35S]-methionine incorporation in normoxia, but it had little or no effect in hypoxia (Figure 4C, control RNAi). This suggests that TOR activity is effectively suppressed during hypoxia, at least in so far as hypoxia eliminates the contribution of TOR activity to [35S]-methionine incorporation.
Knockdown of Ptp61F had little effect on [35S]-methionine incorporation in normoxia or its suppression by rapamycin (Figure 4C), suggesting that Ptp61F did not substantially alter TOR contributions to translation in normoxia. When Ptp61F-knockdown cells were subjected to hypoxia, rapamycin suppressed the sustained level of [35S]-methionine incorporation to the levels seen in control cells in hypoxia (Figure 4C, bottom). This result shows that knockdown of Ptp61F results in a sustained level of TOR during hypoxia, which promotes the sustained translation.
In a parallel set of experiments, Raptor RNAi was used to inactivate TOR, and its consequence on the ability to induce GFP was examined under normoxia and hypoxia. Raptor forms a complex with TOR and is required for TOR function as a positive regulator of protein translation. Raptor RNAi diminished the ability of Ptp61F RNAi to permit induction of GFP expression during hypoxia (Supplemental Figure S1). These results show that Ptp61F is required for effective down-regulation of TOR during hypoxia and that the failure to down-regulate TOR is sufficient to explain the effect of Ptp61F RNAi on [35S]- methionine incorporation and gene expression under hypoxia.
Ptp61F Governs Persistence of Akt-dependent TOR Activity during Hypoxia
Our results show that Ptp61F negatively regulates TOR activity and that it has a nonredundant role in suppressing TOR function in hypoxia. Work in flies and mammals suggests possible upstream roles of Ptp61F in down-regulating the TOR pathway. PTP-1B, a mammalian homolog of Ptp61F, down-regulates growth factor receptor tyrosine kinase signaling and JAK/STAT signaling (Seely et al., 1996; Goldstein et al., 1998; Zabolotny et al., 2002; Haj et al., 2003). Although JAK/STAT signaling pathway has not been reported to affect the TOR pathway, we tested its possible contribution by knockdown of JAK or STAT. These depletions did not modify the influence of Ptp61F knockdown on persistence of TOR activity during hypoxia (Figure 5B). Receptor tyrosine kinases, such as the insulin receptor, activate Akt-mediated phosphorylation of Tsc2, which alleviates TSC inhibition of TOR (Dan et al., 2002; Inoki et al., 2002). We hypothesized that Ptp61F may inhibit TOR by interfering with growth factor signaling.
Figure 5.
Ptp61F down-regulates phosphorylation of a TOR substrate in hypoxia by suppressing an Akt-dependent signal. (A) Ptp61F is required for efficient down-regulation of insulin signaling under hypoxia. Cells treated with dsRNA against Ptp61F were stimulated with 10 μg/ml bovine insulin for an hour, exposed to normoxia or hypoxia (0.5% O2) for the indicated time, and lysed. Antibodies against phospho-tyrosine (4G10; Millipore), insulin receptor (from O. Puig), phosphorylated Akt, Akt (Cell Signaling Technology), and tubulin (Sigma-Aldrich) were used in Western blot analysis to detect activation of insulin signaling pathway. The star-like symbol indicates a band that stains for phospho-tyrosine that is unrelated to but is close to the insulin receptor band. (B) dsRNAi targeting the insulin receptor, JAK, or STAT does not compromise Ptp61F modulation of phosphorylation of p70S6K (phospho-S6K) under hypoxia. (C) Depletion of Akt inhibits the effect of Ptp61F RNAi on phosphorylated p70S6K under hypoxia. Cells treated with dsRNA against Ptp61F, Akt, or Akt in combination with Ptp61F were exposed to hypoxia (<0.1% O2 for 16 h) or normoxia and phosphorylated p70S6K was detected as described in Figure 4A.
To examine this, S2 cells were stimulated with insulin, and insulin receptor phosphorylation, a marker for its activity, was monitored by Western blotting with a phospho-tyrosine specific antibody as well as by its reduced mobility in SDS-PAGE. We assessed the influence of hypoxia and Ptp61F knockdown on receptor activation (Figure 5A, top two panels). In normoxia, insulin stimulation caused tyrosine phosphorylation and a mobility shift of insulin receptor within 1 h (Figure 5A, compare lanes 1–3 or 9, Supplemental Figure S2B), and this modification persisted with slight reduction for 16 h (Figure 5A, lane 7). When cells were stimulated with insulin for 1 h in normoxia and then exposed to hypoxia (<0.1% O2) for 15 h, the tyrosine-phosphorylated band and the mobility shift of insulin receptor disappeared (Figure 5A, compare lanes 13 and 7), suggesting gradual inactivation of the insulin receptor in hypoxia. Similarly, phosphorylation of Akt, a downstream step in insulin signaling, was almost undetectable after 15 h of hypoxia (Figure 5A, third panel, lane 7 vs. lane 13). These findings suggest that insulin-dependent signaling is gradually down-regulated during hypoxia.
RNAi knockdown of Ptp61F slightly increased total cellular tyrosine phosphorylation in normoxic cells whether or not they were stimulated with insulin (Figure 5A, top, lanes 1–8). After 15 h of hypoxia, Ptp61F-deficient cells sustained a high level of tyrosine phosphorylation and substantially retained the slower migrating form of insulin receptor (Figure 5A, top two panels, lane 13 vs. lane 14). Cells treated with Ptp61F RNAi also displayed sustained Akt phosphorylation (Figure 5A, middle, lane 13 and lane 14) compared with control cells. These results show that Ptp61F contributes importantly to down-regulation of insulin signaling under hypoxia.
Although Ptp61F operates to down-regulate persistence signaling by added insulin after hypoxia, the above-mentioned experiments did not test whether down-regulation of insulin signaling is the relevant factor in the response examined under normal culture conditions. Indeed, under our normal experimental conditions without added insulin, RNAi against insulin receptor did not abolish the effect of Ptp61F RNAi on TOR activity during hypoxia (Figure 5B). Because insulin signaling is not required, Ptp61F must operate on alternative or redundant pathways. The insulin receptor is one many receptor tyrosine kinases (rtk). We considered the possibility that various rtk's might provide redundant TOR activation signal and that Ptp61F is a more general inhibitor of rtk signaling. Supporting this possibility, PTP-1B is known to down-regulate other receptor tyrosine kinases in mammalian cells in addition to insulin receptor.
To test the possible role of generic rtk signaling, we knocked down Akt, a common downstream effector of growth factor signaling pathways. To examine the consequence on TOR activity, we assessed p70S6K phosphorylation (Figure 5C). RNAi of Akt only slightly reduced p70S6K phosphorylation (Figure 5C, lane 1 vs. lane 3), suggesting that both Akt-dependent and independent activities phosphorylate p70S6K. These results suggest that Akt is a significant, but not the sole, activator of p70S6K phosphorylation in normoxic S2 cells.
The knockdown of Akt did not seem to reduce the already extremely low level of p70S6K phosphorylation in hypoxia (Figure 5C, lane 5 vs. lane 7). Thus, Akt dependent TOR activity is somehow eliminated during hypoxia. In contrast, knockdown of Akt reduced the phosphorylation of p70S6K that persists in cells under hypoxia when Ptp61F was knocked down (Figure 5C, lane 6 vs. lane 8), suggesting that a major consequence of reducing Ptp61F is to promote/allow a persistence of Akt-dependent signaling, which activates TOR. These results suggest that Ptp61F acts as a negative regulator of Akt-dependent pathway of TOR regulation whose activity is particularly significant in preventing persistent signaling during hypoxia.
Ptp61F-depleted Cells Are Resistant to Cell Death under Hypoxia
A priori, it is not clear whether inhibition of protein translation during hypoxia will be beneficial or harmful to cell survival. In mammalian cells, lack of Tsc2 prevents efficient down-regulation of translation during hypoxia and allows cells (Tsc2−/− p53−/− mouse embryonic fibroblasts) to grow and proliferate under hypoxia (Brugarolas et al., 2004; Wouters et al., 2005; Kaper et al., 2006). We examined this phenomenon in Drosophila S2 cells and asked whether depletion of Ptp61F has a similar effect on S2 cells under hypoxia.
Cells were treated with control RNAi or Ptp61F RNAi for 4 d, exposed to hypoxia. Cell viability was assessed by FACS after staining with the vital dye Sytox Green, which stains only dead cells. When control RNAi-treated cells were exposed to hypoxia (<0.1% O2), cell viability declined by 3–4 d (Figure 6A). However, Tsc2 or Ptp61F RNAi-treated cells survived better than control RNAi-treated cells under hypoxia.
Because depletion of Tsc2 or Ptp61F impaired the inhibition of TOR activity under hypoxia, we examined whether TOR activity is responsible for survival of these cells. TOR forms two complexes, mTORC1 and mTORC2 (Sarbassov et al., 2004; Martin and Hall, 2005). mTORC1, which includes TOR and Raptor, phosphorylates p70S6K and 4E-BP1 to promote protein translation (Loewith et al., 2002). mTORC1 is under the control of the TSC complex, requires Rheb for its activation, and it is inhibited by rapamycin. We confirmed that RNAi against Rheb or Raptor abolished p70S6K phosphorylation in Drosophila S2 cells in normoxia (Supplemental Figure S4) (Yang et al., 2006). Rheb or Raptor RNAi treatment abolished the effect of Ptp61F on cell survival under hypoxia (Figure 6B), suggesting that TORC1 activity is important in cell survival under prolonged hypoxia treatment in cells lacking Ptp61F. Similarly, the ability of Tsc2 RNAi to improve viability in hypoxia was also eliminated by Raptor or Rheb RNAi (Figure 6C). These data show that TORC1 activity contributes to the increased cell survival under hypoxia that occurs upon Ptp61F or Tsc2 depletion.
DISCUSSION
In several systems, severe and abrupt deprivation of oxygen interferes with new gene expression by suppressing transcription and translation. We show that Drosophila S2 cells exhibit this response, and we used a genome-wide RNAi screen to identify new genes contributing to this poorly understood response to hypoxia. The identified genes outline a signaling system that includes mitochondrial genes and regulators of translation. Here, we focus our attention on the segment of signal transduction in which hypoxia reduces the activity of the TOR kinase, a positive regulator of translation. We chose this focus because of the strong phenotype associated with knockdown of two negative regulators of TOR, Tsc1 and Tsc2, and because TOR is a pivotal regulator of translation. We define the contributions of a tyrosine phosphatase, Ptp61F, to hypoxia responses, and identify it as a novel regulator that is important for the down-regulation of TOR and translation during hypoxia.
TOR Down-Regulation during Hypoxia in Drosophila S2 Cells
Our finding that knockdown of the TSC allows continued protein translation under hypoxia in Drosophila S2 cells is consistent with analyses in both Drosophila and in mammalian cells (Brugarolas et al., 2004; Wouters et al., 2005; Kaper et al., 2006). In normoxic culture conditions, the basal phosphorylation of TOR substrates 4E-BP and p70S6K is low. Nonetheless, knockdown or inhibition of TOR function in normoxic S2 cells revealed that this low activity contributes to protein synthesis under these conditions. Knockdown of Tsc1 or Tsc2 resulted in increase in phosphorylation of TOR substrates to higher levels than we have seen in normal cells in any of our growth conditions. Apparently, without constraining TSC suppression, TORC1 activity can be very high, but the increase in activity was not associated with a substantial increase in protein synthesis (Figure 3). Nonetheless, inhibition of TORC1 suppressed translation (Figures 3A and 4C). This suggests that, under our culture conditions, basal TOR function and a modest level of TOR substrate phosphorylation is nearly saturating in terms of protein synthesis (Hall et al., 2007).
Hypoxia substantially reduced the phosphorylation of TOR substrates, and addition of rapamycin during 16 h in hypoxic conditions caused no further suppression of translation, suggesting that TOR is effectively suppressed during hypoxia. Under these conditions, knockdown of Tsc1 or Tsc2 caused both an increase in phosphorylation of TOR substrates as well as increased growth, protein synthesis, and survival. These increases were, in turn, suppressed by inhibition or knockdown of TORC1, indicating that TORC1 activity is physiologically limiting during hypoxia. Although RNAi to Tsc1 or Tsc2 increased [35S]-methionine incorporation in hypoxia by about twofold, incorporation remained well below the levels in nomoxia. Thus, hypoxia seems to have additional inputs through non-TOR pathway(s) that act in parallel and contribute to hypoxia suppression of translation. Here, we focus on the modulation of TOR and its contribution to the inhibition of translation during hypoxia.
Distinctions between TSC Knockdown and Ptp61F Knockdown
Insulin signaling increases TOR activity (Oldham and Hafen, 2003), and mammalian PTP-1B promotes down-regulation of insulin signaling pathway (Goldstein et al., 1998). Although it has not been demonstrated, these relationships suggest that PTP-1B and its homologue in Drosophila, Ptp61F, might contribute to TOR down-regulation during hypoxia. Indeed, we recovered Ptp61F in our screen, and we show that it does contribute to the down-regulation of TOR and to the suppression of translation during hypoxia. In contrast to the marked up-regulation of TOR-mediated phosphorylation after RNAi of Tsc1 or Tsc2, Ptp61F RNAi had at most a slight effect on the phosphorylation of TOR substrates in normoxic conditions. However, although TOR is effectively suppressed in control cells after 16 h of hypoxia, when Ptp61F is knocked down, substantial TOR function persists during hypoxia, translation continues, and this translation is suppressed by rapamycin. Thus, Ptp61F plays a minor role in down-regulating TOR in normoxia, but it has a significant role during hypoxia.
We show that Ptp61F down-regulates the insulin-signaling pathway during hypoxia, and we suggest that this could represent a more general role in down-regulating receptor tyrosine kinase signaling pathways under hypoxia. To examine the involvement of receptor tyrosine kinase pathways in the absence of insulin, we knocked down Akt, which is a common downstream mediator of receptor tyrosine kinase signaling pathways, and we showed that Ptp61F effects depend on Akt. Thus, Ptp61F suppresses Akt-dependent TOR activity during hypoxia, and we propose that this represents down-regulation of generic receptor tyrosine kinase signaling.
Our findings suggest that Ptp61F is especially effective at suppressing TOR activity during hypoxia. This suggests either that the extant Ptp61F activity only becomes important for TOR function during hypoxia or that its activity is altered during hypoxia, perhaps by localization so that it is targeted to substrates germane to this action. The detailed mechanism awaits further investigation.
Consequence of Ptp61F Action during Hypoxia
At present, the connection between the role of TOR in suppressing translation during hypoxia and its influence on S2 cell survival is unclear. Although our results show that RNAi of either Ptp61F or TSC leads to an increase in cell survival during extended hypoxia, it is not clear whether this is connected to the increased protein translation also induced by these treatments. The improvement in cell survival is not reversed by acute rapamycin treatment, which does page the RNAi-induced increased translation under hypoxia. Similar findings have been seen in Tsc2−/− p53−/− mouse embryonic fibroblast cells (Kaper et al., 2006). Additionally, we found that early knockdown or inhibition of TOR before the onset of hypoxia can negate the survival advantage incurred upon RNAi of Ptp61F or Tsc2 (Figure 6, B and C). Apparently, TOR activity in advance of a hypoxic experience influences survival during the subsequent hypoxia.
There is some doubt that the ability of cells to survive hypoxia in culture is a reliable indicator of the physiological role of hypoxia signaling in the organism. In cell culture, continued growth supported by glycolysis during hypoxia might be considered beneficial to the cells; indeed, these are properties of tumor cells in vivo where the continued growth occurs at the expense of the organism. The tumor suppressor activity of Tsc1/2 and PTEN suggests that one role of the translational down-regulation is to enforce altruistic cell behavior, allowing other hypoxia-sensitive cells to survive. Our findings suggest that Ptp61F down-regulation may have a similar but more focused function. Perhaps inhibition of its activity in vivo may modify hypoxia suppression of translation and influence tumor behavior.
Uncovering a Pathway
In addition to Ptp61F whose action on the TOR pathway we describe here, we have identified several genes that we predict will contribute directly or indirectly to an elaborate signaling process that impinges on TOR and perhaps other regulatory nodes to mediate cellular responses to hypoxia. Although diverse categories of genes are identified in the screen, genes with mitochondrial function and genes with roles in translation are especially enriched. We recovered the regulatory subunits (eIF2Bβ, δ, γ) of eIF2B complex from our screening. Although these may have a regulatory input in eIF2B function during hypoxia to inhibit protein translation, the underlying mechanism needs further investigation. Because mitochondria consume the majority of the oxygen for use in energy generation through oxidative phosphorylation, it is plausible that mitochondria may be the major oxygen sensing organelle. Indeed, mitochondria are required for HIF-1 regulation under mild-to-severe hypoxia but not under anoxia (Guzy et al., 2005; Mansfield et al., 2005). Furthermore, it has been speculated that mitochondrial function provides input in regulating TOR activity (Kim et al., 2002). We found that knockdown of genes encoding the mitochondrial proteins, LETM1 or prohibitin blocks hypoxic down-regulation of TOR (SJL, unpublished data). Our analysis of hypoxia-induced inhibition of translation opens up new avenues to study the signaling process by which the mitochondria communicates with the rest of the cell, a process that is central to cellular physiology and that has major importance to human health.
Supplementary Material
ACKNOWLEDGMENTS
We thank Davide Ruggero, Mark McCleland, Steve Deluca, and O'Farrell laboratory members for reading the manuscript. We thank O. Puig, M. Zeidler, T. Murphy, and R. Vale for providing antibody, plasmids, and Drosophila Gateway vectors. S.-J.L. was supported by an American Heart Association postdoctoral fellowship. R. F was supported by The Jane Coffin Childs Memorial Fund for Medical Research postdoctoral fellowship. This work was supported by National Institutes of Health grant AI-60102.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-03-0265) on July 23, 2008.
REFERENCES
- Baeg G. H., Zhou R., Perrimon N. Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila. Genes Dev. 2005;19:1861–1870. doi: 10.1101/gad.1320705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beretta L., Gingras A. C., Svitkin Y. V., Hall M. N., Sonenberg N. Rapamycin pages the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. EMBO J. 1996;15:658–664. [PMC free article] [PubMed] [Google Scholar]
- Bhaskar P. T., Hay N. The two TORCs and Akt. Dev. Cell. 2007;12:487–502. doi: 10.1016/j.devcel.2007.03.020. [DOI] [PubMed] [Google Scholar]
- Browne G. J., Proud C. G. A novel mTOR-regulated phosphorylation site in elongation factor 2 kinase modulates the activity of the kinase and its binding to calmodulin. Mol. Cell. Biol. 2004;24:2986–2997. doi: 10.1128/MCB.24.7.2986-2997.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugarolas J., Lei K., Hurley R. L., Manning B. D., Reiling J. H., Hafen E., Witters L. A., Ellisen L. W., Kaelin W. G., Jr Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J., Kuo C. J., Crabtree G. R., Blenis J. Rapamycin-FKBP specifically pages growth-dependent activation of and signaling by the 70 kD S6 protein kinases. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- Connolly E., Braunstein S., Formenti S., Schneider R. J. Hypoxia inhibits protein synthesis through a 4E-BP1 and elongation factor 2 kinase pathway controlled by mTOR and uncoupled in breast cancer cells. Mol. Cell. Biol. 2006;26:3955–3965. doi: 10.1128/MCB.26.10.3955-3965.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradetti M. N., Inoki K., Bardeesy N., DePinho R. A., Guan K. L. Regulation of the TSC pathway by LKB 1: evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev. 2004;18:1533–1538. doi: 10.1101/gad.1199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan H. C., et al. Phosphatidylinositol 3-kinase/Akt pathway regulates tuberous sclerosis tumor suppressor complex by phosphorylation of tuberin. J. Biol. Chem. 2002;277:35364–35370. doi: 10.1074/jbc.M205838200. [DOI] [PubMed] [Google Scholar]
- DiGregorio P. J., Ubersax J. A., O'Farrell P. H. Hypoxia and nitric oxide induce a rapid, reversible cell cycle arrest of the Drosophila syncytial divisions. J. Biol. Chem. 2001;276:1930–1937. doi: 10.1074/jbc.M003911200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echard A., Hickson G. R., Foley E., O'Farrell P. H. Terminal cytokinesis events uncovered after an RNAi screen. Curr. Biol. 2004;14:1685–1693. doi: 10.1016/j.cub.2004.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellisen L. W. Growth control under stress: mTOR regulation through the REDD1-TSC pathway. Cell Cycle. 2005;4:1500–1502. doi: 10.4161/cc.4.11.2139. [DOI] [PubMed] [Google Scholar]
- Foley E., O'Farrell P. H. Functional dissection of an innate immune response by a genome-wide RNAi screen. PLoS. Biol. 2004;2:E203. doi: 10.1371/journal.pbio.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garami A., Zwartkruis F. J., Nobukuni T., Joaquin M., Roccio M., Stocker H., Kozma S. C., Hafen E., Bos J. L., Thomas G. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol. Cell. 2003;11:1457–1466. doi: 10.1016/s1097-2765(03)00220-x. [DOI] [PubMed] [Google Scholar]
- Goldstein B. J., Ahmad F., Ding W., Li P. M., Zhang W. R. Regulation of the insulin signalling pathway by cellular protein-tyrosine phosphatases. Mol. Cell. Biochem. 1998;182:91–99. [PubMed] [Google Scholar]
- Guzy R. D., Hoyos B., Robin E., Chen H., Liu L., Mansfield K. D., Simon M. C., Hammerling U., Schumacker P. T. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1:401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Haj F. G., Markova B., Klaman L. D., Bohmer F. D., Neel B. G. Regulation of receptor tyrosine kinase signaling by protein tyrosine phosphatase-1B. J. Biol. Chem. 2003;278:739–744. doi: 10.1074/jbc.M210194200. [DOI] [PubMed] [Google Scholar]
- Hall D. J., Grewal S. S., de la Cruz A. F., Edgar B. A. Rheb-TOR signaling promotes protein synthesis, but not glucose or amino acid import, in Drosophila. BMC Biol. 2007;5:10. doi: 10.1186/1741-7007-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K., Li Y., Xu T., Guan K. L. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K., Li Y., Zhu T., Wu J., Guan K. L. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- Kaper F., Dornhoefer N., Giaccia A. J. Mutations in the PI3K/PTEN/TSC2 pathway contribute to mammalian target of rapamycin activity and increased translation under hypoxic conditions. Cancer Res. 2006;66:1561–1569. doi: 10.1158/0008-5472.CAN-05-3375. [DOI] [PubMed] [Google Scholar]
- Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Koumenis C. ER stress, hypoxia tolerance and tumor progression. Curr. Mol. Med. 2006;6:55–69. doi: 10.2174/156652406775574604. [DOI] [PubMed] [Google Scholar]
- Koumenis C., Maxwell P. H. Low oxygen stimulates the intellect. EMBO Rep; Symposium on hypoxia and development, physiology and disease; 2006. pp. 679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C. J., Chung J., Fiorentino D. F., Flanagan W. M., Blenis J., Crabtree G. R. Rapamycin selectively inhibits interleukin-2 activation of p70 S6 kinase. Nature. 1992;358:70–73. doi: 10.1038/358070a0. [DOI] [PubMed] [Google Scholar]
- Liu L., Cash T. P., Jones R. G., Keith B., Thompson C. B., Simon M. C. Hypoxia-induced energy stress regulates mRNA translation and cell growth. Mol. Cell. 2006;21:521–531. doi: 10.1016/j.molcel.2006.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Simon M. C. Regulation of transcription and translation by hypoxia. Cancer Biol. Ther. 2004;3:492–497. doi: 10.4161/cbt.3.6.1010. [DOI] [PubMed] [Google Scholar]
- Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell. 2002;10:457–468. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- Mansfield K. D., Guzy R. D., Pan Y., Young R. M., Cash T. P., Schumacker P. T., Simon M. C. Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-alpha activation. Cell Metab. 2005;1:393–399. doi: 10.1016/j.cmet.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. E., Hall M. N. The expanding TOR signaling network. Curr. Opin. Cell Biol. 2005;17:158–166. doi: 10.1016/j.ceb.2005.02.008. [DOI] [PubMed] [Google Scholar]
- McLaughlin S., Dixon J. E. Alternative splicing gives rise to a nuclear protein tyrosine phosphatase in Drosophila. J. Biol. Chem. 1993;268:6839–6842. [PubMed] [Google Scholar]
- Muller P., Kuttenkeuler D., Gesellchen V., Zeidler M. P., Boutros M. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature. 2005;436:871–875. doi: 10.1038/nature03869. [DOI] [PubMed] [Google Scholar]
- Neufeld T. P. Body building: regulation of shape and size by PI3K/TOR signaling during development. Mech. Dev. 2003;120:1283–1296. doi: 10.1016/j.mod.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Oldham S., Hafen E. Insulin/IGF and target of rapamycin signaling: a TOR de force in growth control. Trends Cell Biol. 2003;13:79–85. doi: 10.1016/s0962-8924(02)00042-9. [DOI] [PubMed] [Google Scholar]
- Padilla P. A., Nystul T. G., Zager R. A., Johnson A. C., Roth M. B. Dephosphorylation of cell cycle-regulated proteins correlates with anoxia-induced suspended animation in Caenorhabditis elegans. Mol. Biol. Cell. 2002;13:1473–1483. doi: 10.1091/mbc.01-12-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla P. A., Roth M. B. Oxygen deprivation causes suspended animation in the zebrafish embryo. Proc. Natl. Acad. Sci. USA. 2001;98:7331–7335. doi: 10.1073/pnas.131213198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D., Dong J., Zhang Y., Gao X. Tuberous sclerosis complex: from Drosophila to human disease. Trends Cell Biol. 2004;14:78–85. doi: 10.1016/j.tcb.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Potter C. J., Huang H., Xu T. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell. 2001;105:357–368. doi: 10.1016/s0092-8674(01)00333-6. [DOI] [PubMed] [Google Scholar]
- Price D. J., Grove J. R., Calvo V., Avruch J., Bierer B. E. Rapamycin-induced inhibition of the 70-kilodalton S6 protein kinase. Science. 1992;257:973–977. doi: 10.1126/science.1380182. [DOI] [PubMed] [Google Scholar]
- Reiling J. H., Hafen E. The hypoxia-induced paralogs Scylla and Charybdis inhibit growth by down-regulating S6K activity upstream of TSC in Drosophila. Genes Dev. 2004;18:2879–2892. doi: 10.1101/gad.322704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov D. D., Ali S. M., Kim D. H., Guertin D. A., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- Saucedo L. J., Gao X., Chiarelli D. A., Li L., Pan D., Edgar B. A. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat. Cell Biol. 2003;5:566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- Seely B. L., Staubs P. A., Reichart D. R., Berhanu P., Milarski K. L., Saltiel A. R., Kusari J., Olefsky J. M. Protein tyrosine phosphatase 1B interacts with the activated insulin receptor. Diabetes. 1996;45:1379–1385. doi: 10.2337/diab.45.10.1379. [DOI] [PubMed] [Google Scholar]
- Semenza G. L. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci. STKE. 2007;2007:cm8. doi: 10.1126/stke.4072007cm8. [DOI] [PubMed] [Google Scholar]
- Simoncic P. D., Lee-Loy A., Barber D. L., Tremblay M. L., McGlade C. J. The T cell protein tyrosine phosphatase is a negative regulator of janus family kinases 1 and 3. Curr. Biol. 2002;12:446–453. doi: 10.1016/s0960-9822(02)00697-8. [DOI] [PubMed] [Google Scholar]
- Simoncic P. D., McGlade C. J., Tremblay M. L. PTP1B and TC-PTP: novel roles in immune-cell signaling. Can. J. Physiol. Pharmacol. 2006;84:667–675. doi: 10.1139/y06-012. [DOI] [PubMed] [Google Scholar]
- Sofer A., Lei K., Johannessen C. M., Ellisen L. W. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol. Cell. Biol. 2005;25:5834–5845. doi: 10.1128/MCB.25.14.5834-5845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Wu L., Chen Z., Kohanski R. A., Pick L. Axons guided by insulin receptor in Drosophila visual system. Science. 2003;300:502–505. doi: 10.1126/science.1081203. [DOI] [PubMed] [Google Scholar]
- Stocker H., Radimerski T., Schindelholz B., Wittwer F., Belawat P., Daram P., Breuer S., Thomas G., Hafen E. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat. Cell Biol. 2003;5:559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- Tapon N., Ito N., Dickson B. J., Treisman J. E., Hariharan I. K. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell. 2001;105:345–355. doi: 10.1016/s0092-8674(01)00332-4. [DOI] [PubMed] [Google Scholar]
- Teodoro R. O., O'Farrell P. H. Nitric oxide-induced suspended animation promotes survival during hypoxia. EMBO J. 2003;22:580–587. doi: 10.1093/emboj/cdg070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Manteuffel S. R., Gingras A. C., Ming X. F., Sonenberg N., Thomas G. 4E-BP1 phosphorylation is mediated by the FRAP-p70s6k pathway and is independent of mitogen-activated protein kinase. Proc. Natl. Acad. Sci. USA. 1996;93:4076–4080. doi: 10.1073/pnas.93.9.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang X., Proud C. G. Activation of mRNA translation in rat cardiac myocytes by insulin involves multiple rapamycin-sensitive steps. Am. J. Physiol. Heart Circ. Physiol. 2000;278:H1056–H1068. doi: 10.1152/ajpheart.2000.278.4.H1056. [DOI] [PubMed] [Google Scholar]
- Wang X., Li W., Williams M., Terada N., Alessi D. R., Proud C. G. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 2001;20:4370–4379. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingrove J. A., O'Farrell P. H. Nitric oxide contributes to behavioral, cellular, and developmental responses to low oxygen in Drosophila. Cell. 1999;98:105–114. doi: 10.1016/S0092-8674(00)80610-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wouters B. G., van den Beucken T., Magagnin M. G., Koritzinsky M., Fels D., Koumenis C. Control of the hypoxic response through regulation of mRNA translation. Semin. Cell Dev. Biol. 2005;16:487–501. doi: 10.1016/j.semcdb.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Yang Q., Inoki K., Kim E., Guan K. L. TSC1/TSC2 and Rheb have different effects on TORC1 and TORC2 activity. Proc. Natl. Acad. Sci. USA. 2006;103:6811–6816. doi: 10.1073/pnas.0602282103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabolotny J. M., et al. PTP1B regulates leptin signal transduction in vivo. Dev. Cell. 2002;2:489–495. doi: 10.1016/s1534-5807(02)00148-x. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Gao X., Saucedo L. J., Ru B., Edgar B. A., Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat. Cell Biol. 2003;5:578–581. doi: 10.1038/ncb999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.