Abstract
In this work, we show several previously unknown features of p120-catenin in a cadherin–catenin complex that are critical for our understanding of cadherin-based adhesion and signaling. We show that in human epithelial A-431 cells, nearly all p120 molecules engage in high-affinity interaction with E-cadherin–catenin complexes located at the cellular surface. p120 is positioned in proximity to α-catenin in the complex with cadherin. These findings suggest a functional cooperation between p120 and α-catenin in cadherin-based adhesion. A low level of cadherin-free p120 molecules, in contrast, could facilitate p120-dependent signaling. Finally, we present compelling evidence that p120 is a key linker cementing the E-cadherin–catenin complex with the transmembrane protease γ-secretase. The cell–cell contact location of this supercomplex makes it an important candidate for conducting different signals that rely on γ-secretase proteolytic activity.
INTRODUCTION
A complete understanding of the mechanism of cadherin adhesion, its regulation, and its impact on intracellular signaling is very important because malfunctions in the cadherin system can lead to severe consequences, including tumor cell metastasis and invasion. Nevertheless, our knowledge of these processes remains very circumstantial. Several conflicting models for cadherin adhesive homodimerization have been proposed (reviewed in Troyanovsky, 2005). Many potential but understudied mechanisms such as cadherin clustering (Kusumi et al., 1999; Gumbiner, 2005), cadherin endocytosis (Yap et al., 2007), modulation of the cadherin dimerization site (Troyanovsky et al., 2007), and proteolytic cadherin ectodomain shedding (Ii et al., 2006) have been suggested to regulate adherens junction strength and stability. Finally, although extensive cross-talk between cadherin adhesion and various signaling processes has been demonstrated previously (Chen and Gumbiner, 2006; Halbleib and Nelson, 2006), its exact molecular mechanisms remain to be studied.
One of the most direct strategies for elucidating the function and regulation of cadherin is a detailed examination of cadherin-binding proteins. It is known that a C-terminal domain of the intracellular cadherin tail forms a complex with β-catenin. This binding diverts β-catenin away from the Wnt-signaling cascade, in which it is a key player. In the complex with cadherin, β-catenin is an important structural element; it anchors the actin-binding protein α-catenin (Provost and Rimm, 1999; Weis and Nelson, 2006). This interaction is critical because α-catenin depletion severally damages cadherin adhesion and actin cytoskeleton (Capaldo and Macara, 2007). Another cadherin partner, p120-catenin (p120), belongs to the same arm-repeat protein family as β-catenin (Anastasiadis and Reynolds, 2000). p120 directly binds to the binding site located at the juxtamembrane portion of the E-cadherin tail. Similar to β-catenin, p120 has an important function outside of adherens junctions. Its cadherin-free pool is thought to regulate the activity of Rho-related GTPases RhoA and Rac1. Binding to cadherin abolishes p120 signaling potential. By unknown mechanisms, this binding shields cadherin from the cell degradation machinery, thereby regulating the cadherin expression level (Kowalczyk and Reynolds, 2004). In contrast to β-catenin, however, no proteins interacting with p120 in the cadherin–catenin complex have been identified. Several fundamental questions regarding cadherin-p120 interactions remain unanswered. For example, the ratio between cadherin-bound and cadherin-unbound forms of p120 is not fully known. Immunoprecipitation experiments indicate that only a small fraction of p120 is complexed with cadherin, whereas cell fractionation experiments suggest that nearly all p120 molecules are present in adherens junctions (for references, see Anastasiadis and Reynolds, 2000). Another remarkable member of the cadherin–catenin complex is presenilin 1 (PS1). This protein constitutes a catalytic portion of the transmembrane protease γ-secretase, which in addition to PS1 also contains three other transmembrane proteins, nicastrin, APH-1, and PEN-2 (Spasic and Annaert, 2008). These four proteins are folded into the transmembrane barrel mediating the regulated intramembrane proteolysis of many cell surface receptors, including amyloid precursor protein, Notch, Ephrin, and many others. Whether the entire γ-secretase or only PS1 alone associates with cadherin is not yet known. It was reported that the hydrophilic loop of PS1 binds directly to the juxtamembrane region of E-cadherin, thereby blocking cadherin-p120 interactions (Baki et al., 2001). However, it was also shown that the same PS1 region directly interacts with the arm domain of different catenins: β-catenin and close p120 relatives, δ-catenin, and p0071 (Zhou et al., 1997; Murayama et al., 1998; Levesque et al., 1999; Stahl et al., 1999). These observations suggest a possible structural flexibility for cadherin–PS1 complexes. In theory, interactions with cadherin may regulate γ-secretase activity, its substrate specificity and localization. Together, the structural data demonstrate that cadherin is a midfielder that couples cell–cell adhesion, cell motility, and key signaling pathways. Detailed structural analysis of the cadherin–catenin complex is required to understand the role of cadherin in orchestrating such divers cellular activities.
This work addresses several unanswered questions about the structure of cadherin–catenin complex. Using blue native polyacrylamide gel electrophoresis (BN PAGE) and protein cross-linking, we showed that nearly all p120 molecules engage in high-affinity interaction with cell surface-located E-cadherin. We found that in the cadherin–catenin complex, p120 is positioned in proximity to α-catenin. Furthermore, we developed a new structural model for the cadherin–PS1 complex. We obtained compelling evidence that a pool of cell surface-located E-cadherin–catenin complex associates with the entire γ-secretase complex and that p120 is a molecular linker between these two complexes.
MATERIALS AND METHODS
Cell Culture and Antibodies
A-431 (human epidermoid carcinoma) and SW48 (human colon carcinoma) cells were cultured in DMEM containing 10% fetal calf serum. All plasmid-transfected stable clones of A-431 have been described previously (Chitaev and Troyanovsky, 1998; Troyanovsky et al., 2003). Immunofluorescence microscopy of the cells was performed as described in Troyanovsky et al. (2003). The following mouse antibodies were used: anti-E-cadherin (C20820), anti-p120 (pp120), and anti-β-catenin (all from BD Biosciences, San Jose, CA); anti-α-catenin and anti-E-cadherin SHE78–7 (Zymed Laboratories, South San Francisco, CA); anti-myc 9E10 (Covance Research Products, Princeton, NJ); anti-presenilin 1 C terminus (Millipore Bioscience Research Reagents, Temecula, CA), and anti-p120 (clone 6H11, Santa Cruz Biotechnology, Santa Cruz, CA). Rat monoclonal anti-presenilin1 N terminus (Millipore Bioscience Research Reagents); polyclonal rabbit anti-myc and goat anti-nicastrin (both from Santa Cruz Biotechnology); rabbit anti-nicastrin (Sigma-Aldrich, St. Louis, MO); and rabbit anti-p0071 (kindly provided by Dr. A. Kowalczyk, Emory University School of Medicine) were also used.
Small Interfering RNA (siRNA) and siRNA Transfection
The nicastrin siRNA (GAC CAC UCU GGU GCC UUC CAU AAC A), two p120 siRNAs (AUA CUA CGC UGG UCA UCC UCU AGCC, p120 Oligo#1; and UAU UGU UGG CAA CAU UGG GAG CUG C, p120 Oligo#2) and three negative control siRNAs (low, medium, and high GC) were obtained from Invitrogen (Carlsbad, CA). Three hours before transfections with siRNAs, the cells were plated on 6-cm dishes at a density of 105 cells per dish. Transfection was performed according to Mirus protocol using TransIT-siQuest transfection reagent (Mirus Bio, Madison, WI). The next day, the cells were replated and assayed 48 or 64 h after transfection.
Blue Native Polyacrylamide Gel Electrophoresis, Cross-Linking, Immunodepletion, and Immunoprecipitation
The BN PAGE was performed according to the manufacturer's protocol (Native-PAGE Novex Bis-Tris Gel system; Invitrogen). In brief, a 2-d-old confluent culture in 3-cm tissue culture dish was washed in phosphate-buffered saline (PBS) and extracted in 0.5 ml of lysis buffer (50 mM Bis-Tris, pH 7.4, 50 mM NaCl, and 1 mM EDTA) with 1% of either of following detergents: Digitonin (Calbiochem, San Diego, CA), dodecyl maltoside (DDM), or Triton X-100. Digitonin was prepared as 5% stock solution in water and was added to the buffer immediately before use. In some cases, 40 μM protease inhibitor 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF) (Calbiochem) and phosphatase inhibitor cocktail (1 mM Na3VO4, 10 mM NaF, and 2 mM β-glycerophosphate) were added. After 10-min (14,000 rpm at 4°C) centrifugation, the lysates were mixed 1:1 with gel loading buffer and placed on the gel. For cross-linking, 100-μl lysate aliquots after centrifugation were combined with 30 μl of freshly prepared 1 mM cross-linker BM[PEO]3 (spacer arm, 14.7 Å) and incubated for 10 min on ice. The reaction was stopped by the addition of dithiothreitol (DTT) (final concentration, 1 mM). Cells in culture were cross-linked by 10-min incubation with cysteine-specific cross-linker Bis-maleimidohexane (BMH) (spacer arm, 16.1 Å) at 4°C. For immunodepletion, the cell lysates (100 μl) were incubated (2 h at 4°C) with protein A-Sepharose preloaded with corresponding antibodies. The protein A-Sepharose was then removed by centrifugation, and the lysates were cross-linked as described above and used for BN PAGE. Cells for immunoprecipitation were lysed either in 1% Triton X-100 or 1% digitonin-containing immunoprecipitation (IP) lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM EDTA, and 0.5 mM AEBSF) and then subjected to immunoprecipitation as described previously (Troyanovsky et al., 2003). Proteomic analyses of the immunoprecipitates were done in the Proteomics Core Facility of the Washington University School of Medicine (St. Louis, MO). Proteins for these analyses were separated using Tris-acetate gel system (Invitrogen).
Biotinylation of Cell Surface Proteins
The cells of one 6-cm dish reaching near confluent growth was washed with ice-cold PBS containing 0.2 mM CaCl2 (PBS-Ca). The plate was incubated at 4°C with 2 ml of 0.5 mg/ml sulfo-NHC-LC-biotin (Pierce Chemical, Rockford, IL) in PBS-Ca for 10 min. The reaction was stopped by washing the cells in ice-cold glycine solution (200 mM glycine and 200 mM Tris, pH 7.5).
RESULTS
Nearly All p120 Molecules Are Complexed with Cadherin in A-431 Cells
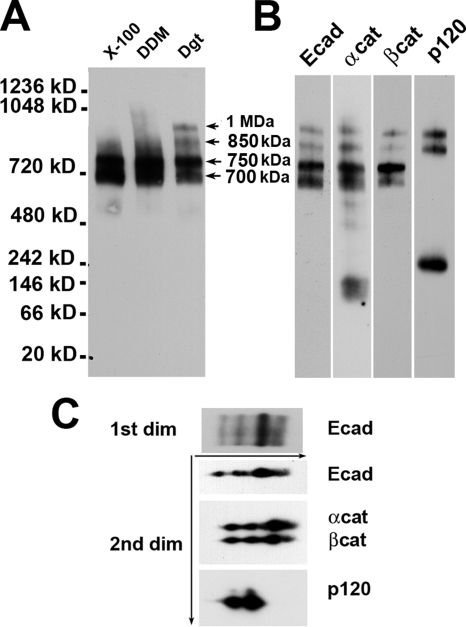
To study the molecular organization of the cadherin-catenin complexes using BN PAGE, we first selected the nonionic detergent most appropriate for this analysis. Figure 1A shows that 1% Triton X-100 or 1% DDM extracted two E-cadherin–containing complexes (700 and 750 kDa). In addition to these two complexes, digitonin lysates exhibited two high-molecular-weight complexes (1 MDa and 850 kDa). The specific feature of these two complexes is that they both contained p120 (Figure 1B). All four complexes were also positive for β-catenin and α-catenin staining. Regardless of the antibody used for detection, the ratios of the Western blot signals from all four complexes were equal in intensity. This indicates that they all have the same stoichiometry of the major components E-cadherin and α- and β-catenins. Even the longest exposure of the Western blots did not produce a signal that may have corresponded to the monomeric E-cadherin or β-catenin. In contrast, both α-catenin and p120 have noticeable pools of low-molecular-weight complexes, which may represent either corresponding monomers, homodimers, or low-molecular-weight complexes with unknown proteins.
Figure 1.
Native gel electrophoresis detects four major cadherin–catenin complexes in A-431 cell lysates. (A) Two-day-old confluent cultures of A-431 cells were lysed with the same volume of the native gel sample buffer containing 1% Triton X-100 (lane X-100), 1% DDM (DDM), or 1% digitonin (Dgt). Identical amounts of the resulting cell lysates were analyzed by BN PAGE and anti-E-cadherin antibody. Note that only the digitonin lysate contained two high-molecular-weight cadherin complexes, 850 kDa and 1 MDa (arrows). (B) A-431 digitonin cell lysate was probed for anti-E-cadherin (Ecad), α-catenin (αcat), β-catenin (βcat), and p120 (p120) antibodies. Note that only the 850-kDa and 1-MDa complexes contained p120. (C) Digitonin cell lysate was subjected to BN PAGE (1st dim). A gel strip from the first dimension was then loaded onto a second-dimension (2nd dim) SDS gel. The resulting gel was analyzed using anti-E-cadherin (Ecad), α-catenin (αcat) plus β-catenin (βcat) and p120 (p120) antibodies.
To test whether such diversity of E-cadherin complexes was caused by proteolytic degradation of one of the complex components, the complexes were analyzed by two-dimensional (2D) native/SDS-PAGE. Western blot analyses of 2D gels showed that E-cadherin and all three catenins have the expected molecular masses (Figure 1C). Together, these biochemical data suggested that the differences between four E-cadherin–containing complexes present in digitonin cell lysates of A-431 cells are caused by the differences in unidentified proteins and/or in overall complex conformations.
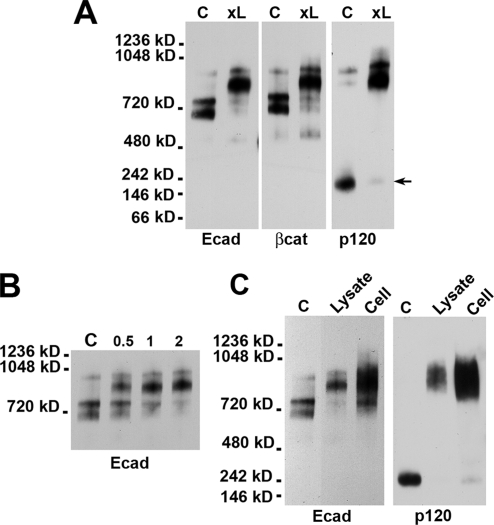
To further characterize these four complexes, A-431 digitonin cell lysate was cross-linked using the cysteine-specific homobifunctional cross-linker BM[PEO]3. Surprisingly, this procedure dramatically increased the yield of both p120-containing complexes (Figure 2A). This increase was accompanied with a corresponding decrease in the amounts of both 700 and 750 kDa p120-deficient complexes as well as with a nearly complete depletion of the presumably free p120 pool (Figure 2A, arrow). There was also a slight decrease in the mobility of the 1-MDa complex in the gel. The kinetics of this cross-linking reaction at 4°C was extremely rapid; the reaction took only 2 min to complete (Figure 2B). Such fast and very efficient cross-linking of p120 to the rest of the cadherin–catenin complex suggests that the majority of p120 was originally present in the complex but dissociated from it during in vitro manipulations. Because the efficiency of cross-linking reactions even upon optimal location of the corresponding reactive groups cannot be 100%, these data suggest that the majority of p120 molecules in the cells are complexed with cadherin. To test whether the same E-cadherin–p120 complexes are present in living cells, A-431 cells were first cross-linked by the membrane-permeable cross-linker BMH and then analyzed by BN PAGE (Figure 2C, lane cell). It showed that nearly all p120 and E-cadherin molecules in living cells were present in the 850 kDa–1 MDa complexes.
Figure 2.
Cross-linking stabilizes E-cadherin–p120 interactions. (A) Before loading the native gel, A-431 cell lysate was cross-linked by BM[PEO]3. The control (C) and cross-linked (xL) samples were analyzed by Western blotting by the anti-E-cadherin (Ecad), anti-β-catenin (βcat), and anti-p120 (p120) antibodies. Note that cross-linking strongly increased the amounts of cadherin–p120 complexes and resulted in the nearly complete disappearance of a cadherin-unbound form of p120 (arrow). (B) Digitonin cell lysates were incubated with BN[PEO]3 for 0, 0.5, 1, or 2 min at 4°C. The reaction was blocked by 1 mM DTT. Note the very fast kinetics of the cross-linking reaction, indicating that p120 and E-cadherin are in the same complex. (C) An intact monolayer of A-431 cells was cross-linked by BMH (Cell) and compared with uncross-linked (C) or cross-linked by BM[PEO]3 (Lysate) digitonin cell lysate. Other abbreviations are as indicated in A. Note that both cross-linking reactions produced similarly sized complexes.
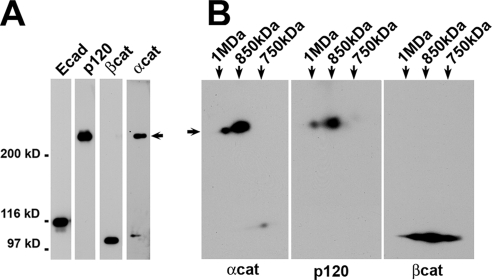
To determine the partner of p120 in BM[PEO]3 cross-linking reaction, digitonin cell lysate after cross-linking was immunoprecipitated using anti-E-cadherin SHE78-7 antibody. The resulting immunoprecipitate was analyzed by SDS-PAGE. This experiment showed that BM[PEO]3 specifically cross-linked p120 to α-catenin, producing a p120–α-catenin heterodimer (Figure 3A, arrow). Neither β-catenin nor E-cadherin participated in this reaction. Furthermore, two-dimensional native/SDS-PAGE showed that both 850 kDa and 1 MDa complexes have identical p120-α–catenin cross-linked dimers (Figure 3B). Such specific cross-linking of p120 to α-catenin suggests that these two proteins are closely apposed in the E-cadherin–catenin complex.
Figure 3.
p120 and α-catenin are a short distance apart. (A) Digitonin cell lysate was cross-linked by BM[PEO]3, immunoprecipitated with SHE78-7 monoclonal antibody (mAb), and then examined by SDS-PAGE and Western blotting. Note that the only cross-linking product was a p120–α-catenin dimer (arrow). (B) The sample was prepared as described in A, but then it was subjected to two-dimensional native/SDS-PAGE as described in Figure 1C. The resulting 2D gels were analyzed using α-catenin (αcat), β-catenin (βcat), and p120 (p120) antibodies. Note that the p120-α–catenin cross-link was present in both the 1-MDa and 850-kDa complexes.
Cadherin–p120 Complex Is Very Stable in Digitonin-containing Buffers
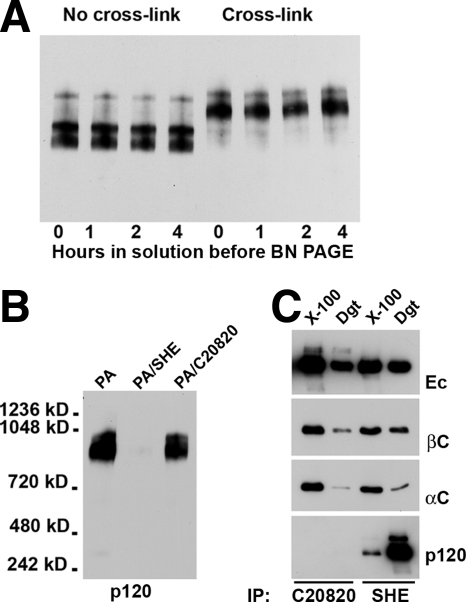
The dramatic increase in the amounts of the E-cadherin–p120 complexes caused by BM[PEO]3 raised a new question: Are the cadherin-p120 bonds so weak that 1% digitonin destroyed most of them in the absence of the cross-linker, or do these bonds become unstable only during BN PAGE? In the latter case, the bonds could be destabilized due to protein–Coomassie G-250 interactions. To answer this question, the digitonin cell lysates were first stored for different times (from 1 to 4 h) at room temperature and an aliquot of each lysate was then cross-linked. Figure 4A shows that the amounts of the cadherin complexes were independent from the store period. This experiment indicated clearly that the cadherin–p120 interactions were destabilized during electrophoresis.
Figure 4.
Stability of cadherin–p120 interactions. (A) Before being placed on the native gel, digitonin cell lysates were stored at room temperature for different times (as indicated below the lanes). A portion of each lysate was then cross-linked. The gel was stained by an anti-E-cadherin antibody. Note that the amounts of the cadherin complexes were independent of storage periods. (B) Three aliquots of the same digitonin lysate were depleted either with protein A-Sepharose alone (PA) or in combination with SHE78-7 (PA/SHE) or C20820 (PA/C20820) anti-cadherin antibodies. The gel was stained by an anti-p120 antibody. Note that antibody C20820 could not deplete p120-containing complexes, whereas SHE78-7 antibody completely removed p120 from the lysate. (C) Triton X-100 (X-100) or digitonin (Dgt) cell lysates were immunoprecipitated either by anti-cadherin C20820 or SHE78–7 antibodies (indicated below the gels). The resulting immunoprecipitates were analyzed for the presence of E-cadherin (Ec), β-catenin (βcat), α-catenin (αcat), and p120 (p120). Note that the antibody C20820 was unable to immunoprecipitate the cadherin–p120 complex. Also note that the yield of the E-cadherin–p120 complex was significantly higher in the digitonin than in Triton X-100 lysates.
To test the stability of E-cadherin–p120 interactions by the alternative approach, we studied whether E-cadherin could be immunodepleted by the anti-E-cadherin antibody C20820, whose epitope is positioned very close to the p120 binding site of E-cadherin (Chitaev and Troyanovsky, 1998). The antibody SHE78-7, recognizing the amino-terminal domain of E-cadherin (Laur et al., 2002) was used as a control. Three aliquots of the same digitonin cell lysate were first incubated either with protein A-agarose alone or in combination with one of these two antibodies. The proteins remaining in solution were then cross-linked by BM[PEO]3 and subjected to BN PAGE. Notably, only SHE78-7 but not the C20820 antibody immunodepleted the lysate for the cadherin–p120 complex (Figure 4B). This result indicated that the cadherin–p120 interactions are stable enough to protect the C20820 epitope from antibody binding. In the control experiment, these two antibodies were used for conventional immunoprecipitation (Figure 4C). Both of the antibodies precipitated E-cadherin and equally well coimmunopreciptated α- and β-catenins from both Triton X-100 and digitonin cell lysates. However, only the SHE78-7 antibody was able to coimmunoprecipitate p120. As expected, the amount of the coimmunoprecipitated p120 was much higher in digitonin lysates, substantiating our previous conclusion that Triton X-100 weakened p120–cadherin interactions.
1-MDa Cadherin Complex Contains γ-Secretase
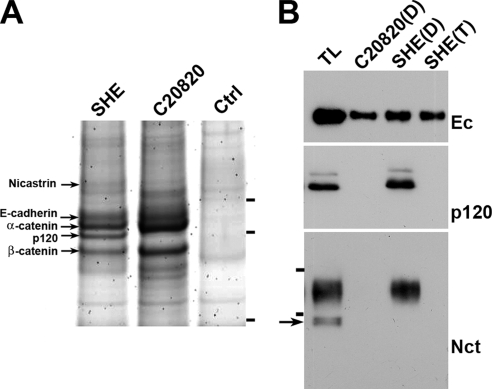
To clarify the composition of cadherin complexes in digitionin lysates, they were precipitated by SHE78-7 or C20820 antibodies and analyzed by SDS-PAGE and mass spectrometry. This work revealed that in addition to catenins, SHE78-7 antibody coimmunoprecipitated 140-kDa protein nicastrin (Figure 5A) that is a large transmembrane component of γ-secretase. An anti-nicastrin antibody confirmed the presence of nicastrin in SHE78-7 but not in C20820 immunoprecipitates (Figure 5B). Importantly, only a completely processed form of nicastrin was found in the complex with E-cadherin. It was also notable that the Triton X-100 lysis buffer completely destroyed both cadherin–nicastrin and cadherin–p120 interactions (Figure 5B).
Figure 5.
Nicastrin is a component of the E-cadherin–catenin complex. (A) Two anti-E-cadherin immunoprecipitates obtained from digitonin A-431 cell lysates by using SHE78-7 (SHE) or C20820 (C20820) mAbs and a control immunoprecipitate (without an added antibody) were subjected to SDS-PAGE and developed using silver staining. The names of proteins identified by mass spectrometry are indicated. Molecular weight markers (from top to bottom: 116, 97.4, and 66 kDa) are shown by horizontal bars. Note the high amount of p120 in the SHE78-7 immunoprecipitate and its complete absence in C20820. (B) A-431 cells were lysed in digitonin lysate buffer and the total lysate (TL) or its C20820 (C20820-D) or SHE78-7 (SHE-D) immunoprecipitates were analyzed by Western blotting. For comparison the SHE78-7 immunoprecipitate obtained from A-431 cells by using Triton X-100–containing buffer was also studied (SHE-T). The blots were stained for E-cadherin (Ec), p120 (p120), and nicastrin (Nct). Horizontal bars in the Nct blot indicate molecular weight markers for 116 and 97.4 kDa. The arrow shows the nicastrin precursor, which was absent in the coimmunoprecipitates. Note that in the immunoprecipitates obtained using C20820 mAb or Triton X-100, neither p120 nor nicastrin were detected.
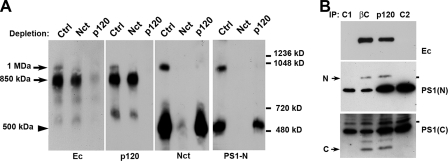
Next, we determined which of the four cadherin complexes detected by BN PAGE contained nicastrin. Western blotting with an anti-nicastrin antibody revealed that digitonin lysates exhibited two nicastrin complexes (Figure 6A, lane Ctrl). The majority of nicastrin was present in the 500-kDa complex, which was much smaller than any of the cadherin complexes. However, the minor 1-MDa nicastrin complex exactly comigrated with the 1-MDa cadherin–p120 complex. The same two complexes (500 kDa and 1 MDa) were also detected by an antibody against PS1, another critical γ-secretase component. As expected, nicastrin depletion using anti-nicastrin antibody (lane Nct) completely removed both nicastrin–PS1 complexes from the digitonin lysate. Concomitantly, this manipulation specifically eliminated the 1-MDa but not the 850-kDa cadherin–p120 complex. A reciprocal p120-depletion exclusively removed the 1-MDa nicastrin–PS1 complex, leaving the 500-kDa complex intact. This experiment unequivocally showed that the 1-MDa complex contains all four proteins—E-cadherin, p120, nicastrin, and PS1. This suggested that the 1-MDa complex consists of two functional elements, an E-cadherin–catenin complex bound to a γ-secretase complex. This interpretation is corroborated further by the fact that this supercomplex contains fully processed amino and carboxy-terminal PS1 fragments because they were both coimmunoprecipitated by the β-catenin or p120 antibodies (Figure 6B).
Figure 6.
Nicastrin and PS1 are components of the 1-MDa E-cadherin–catenin complex. (A) Three aliquots of the digitonin A-431 cell lysate were treated first either with protein A-agarose alone (Ctrl) or in combination with rabbit anti-nicastrin (Nct) or anti-p120 (p120) antibodies. They were then cross-linked by BM[PEO]3 and analyzed by BN PAGE. Blots were stained for E-cadherin (Ec), p120 (p120), nicastrin (Nct), or N-terminal epitope of PS1 (PS1-N). Molecular weight markers (from top to bottom: 1236, 1048, 720, and 480 kDa) are shown by horizontal bars. Note that anti-nicastrin depletion also removed the 1-MDa cadherin complex (arrow) but not the 850-kDa complex. Depletion for p120 results in reciprocal depletion of the 1-MDa nicastrin/PS1-containing complex. The band representing the cadherin-free form of γ-secretase (arrowhead, 500 kDa) is unchanged. (B) Digitonin cell lysates were immunoprecipitated either with anti-β-catenin (IP: βc) or with anti-p120 (IP: p120) antibodies. The resulting immunoprecipitates, as well as the corresponding anti-β-catenin (C1) and anti-p120 (C2) antibodies were analyzed for N-terminal and C-terminal PS1 epitopes. Note that both antibodies coimmunoprecipitated completely processed NTF (N) and CTF (C) PS1 fragments.
p120 Is an Essential Element of the Cadherin–γ-Secretase Supercomplex
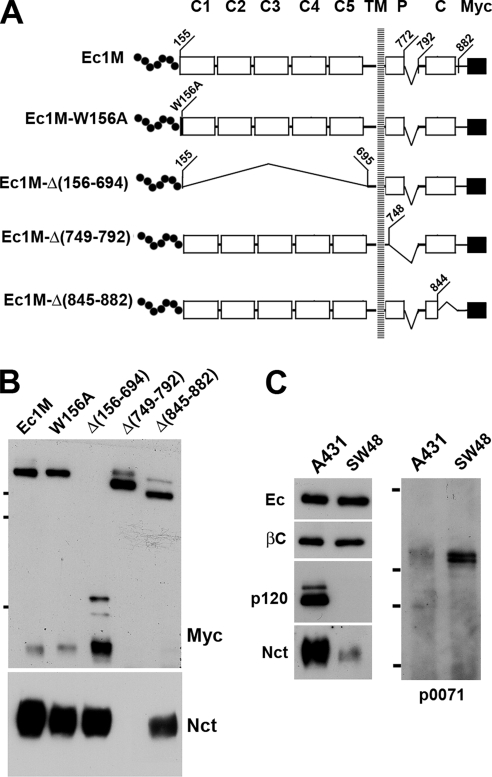
To further characterize the structure of the cadherin-γ–secretase supercomplex, we first determined which E-cadherin region is responsible for γ-secretase binding. Using an anti-myc coimmunoprecipitation assay, we studied A-431 cells expressing several myc-tagged E-cadherin mutants (see the mutant maps in Figure 7A). The mutant Ec1M-W156A contained a point mutation of Trp156, a critical residue for cadherin homodimerization. Three other mutants, Ec1M-Δ(156-694), Ec1M-Δ(749-792), and Ec1M-Δ(845-882), contained extensive deletions in the ectodomain, a p120-binding site, and a β-catenin-binding site, respectively. Remarkably, only the p120-binding site mutant Ec1M-Δ(749-792) was completely unable to coimmunoprecipitate nicastrin (Figure 7B). This observation suggested two mutually exclusive possibilities: 1) the p120-binding site of E-cadherin is also a binding site for γ-secretase or 2) p120 is a linker between γ-secretase and E-cadherin.
Figure 7.
P120 is a critical protein in the cadherin–γ-secretase supercomplex. (A) Schematic representation of the myc-tagged E-cadherin mutants. Signal and precursor peptides (dotted line), five extracellular cadherin-like repeats (C1–C5), the p120-binding site (P), the β-catenin-binding domain (C), and the myc epitope (solid square) are indicated. Deletions are depicted by brackets. Numbers show positions of the corresponding amino acids. (B) A-431 cells stably expressing different cadherin mutants (indicated above the lanes) were lysed with the digitonin-containing buffer and immunoprecipitated with an anti-myc antibody. The resulting immunoprecipitates were analyzed for myc (Myc) and for nicastrin (Nct). Note that only the mutant Ec1M-Δ(749-792) lacking the p120-binding site could not coprecipitate nicastrin. (C) Digitonin lysates of A-431 (A431) or p120-deficient SW48 cells were immunopreciptated using the SHE78-7 antibody. The amounts of the immunoprecipitates loaded on each lane were first equalized for E-cadherin (Ec) and then analyzed for β-catenin (βc), p120 (p120), nicastrin (Nct), and p0071 (p0071).
Neither one of the two previously reported models of cadherin–PS1 interactions is consistent with our data. By one model, PS1 interacts with cadherin via β-catenin (Murayama et al., 1998). Contrary to this model, our experiments with cadherin deletion mutants clearly showed that β-catenin is not required for E-cadherin–γ-secretase interactions. According to the second model, PS1 competes with p120 for the same binding site located within the juxtamembrane E-cadherin region (Baki et al., 2001). However, our experiments unequivocally showed a presence of both p120 and nicastrin/PS1 in the same E-cadherin complex. Still, it was possible that PS1 and p120 independently bind to the same E-cadherin region and that the binding of one of them weakened the interactions of another. Such weak interaction might not have been detected in the previous work. We checked this possibility using two different strategies, analyzing p120-deficient SW48 cells and RNA interference (RNAi).
It was shown that SW48 cells express only a negligible amount of p120, which, in addition, has an extended deletion of its C-terminus (Ireton et al., 2002). According to the competition model, the nearly complete lack of p120 in SW48 cells should promote the formation of the E-cadherin-γ-secretase complex. Coimmunoprecipitation experiments with these cells using an anti-E-cadherin antibody, however, revealed just the opposite—E-cadherin binds much less nicastrin in SW48 than in A-431 cells (Figure 7C). However, despite the p120-deficiency, SW48 cells exhibited a minor but detectable amount of the E-cadherin–nicastrin complex. We proposed that this complex in SW48 cells is formed by the recruitment of close p120 relatives p0071 or ARVCF and/or a p120 C-terminally truncated mutant. Western blotting of anti-cadherin immunoprecipitates indeed showed the presence of the E-cadherin–p0071 and E-cadherin–p120 mutant complexes in SW48 cells (Figure 7C; data not shown).
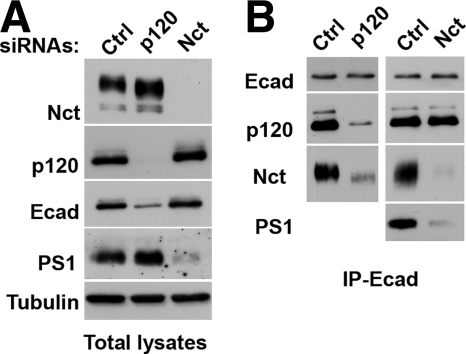
The alternative strategy we applied to study the roles of p120 and nicastrin in the structure of the cadherin-γ-secretase complex was RNAi. A-431 cells were depleted for nicastrin or p120 by using specific siRNAs. The nearly complete depletion of nicastrin by this strategy changed neither the overall cadherin/catenin levels nor cadherin–catenin coimmunoprecipitation (Figure 8). The only notable change associated with nicastrin depletion was a strong reduction of the PS1 level (Figure 8). This observation clearly shows that γ-secretase or its components are not essential for E-cadherin–catenin complex formation and that the absence of γ-secretase cannot increase the binding of p120 to E-cadherin as would be the case by the competition model. In contrast to the depletion of nicastrin, p120 depletion, in agreement with published observations (Ireton et al., 2002), produced a dramatic effect on cell–cell adhesion due to the strong decrease in the total E-cadherin level (Figure 8A). Neither one of our two p120 siRNAs, however, was able to change nicastrin or PS1 expression. To test whether p120 deficiency in A-431 cells abolished cadherin–γ-secretase association, the corresponding anti-E-cadherin immunoprecipitates were equalized first on the E-cadherin level and then probed for p120 and nicastrin. Figure 8B shows that the amount of both proteins in the complex with E-cadherin were equally reduced (Figure 8), confirming that p120 is critical for E-cadherin–γ-secretase binding.
Figure 8.
Nicastrin and p120 depletions by using siRNAs. A-431 cells were transfected with control (Ctrl), p120 (p120), or nicastrin (Nct) siRNAs, and the total digitonin cell lysates (A) or the corresponding anti-E-cadherin SHE78-7 immunoprecipitates (B) were analyzed by Western blotting for nicastrin (Nct), p120 (p120), E-cadherin (Ecad), PS1-NTF (PS1), and tubulin (tubulin). The loading amounts of the immunoprecipitates were equalized for E-cadherin. Note that p120 depletion down-regulated E-cadherin expression and strongly reduced the amount of nicastrin associated with E-cadherin. The depletion of nicastrin, in turn, abolished the expression level of PS1 but did not change the yield of p120 in the immunoprecipitate.
Cadherin-γ–Secretase Supercomplex Is Exposed on the Cell Surface
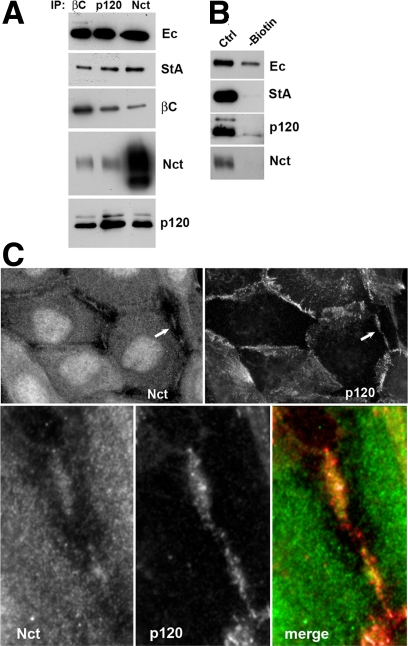
To study the subcellular localization of this complex, A-431 cells were first surface biotinylated and then immunoprecipitated using β-catenin, nicastrin, or p120 antibodies (Figure 9A). All three immunoprecipitates were normalized on the level of E-cadherin. The following streptavidin staining showed that all three immunoprecipitates contained approximately the same amount of biotinylated E-cadherin, indicating that the cadherin–γ-secretase supercomplex is exposed on the cell surface. In an alternative experiment the surface-biotinylated proteins were first removed from the lysate by streptavidin-agarose. The remaining E-cadherin, most of which obviously derived from the intracellular E-cadherin pool, was then immunoprecipitated. Figure 9B shows that only a negligible fraction of the intracellular E-cadherin pool associates with nicastrin or p120. Additional evidence that the cadherin-γ–secretase complex is exposed on the cell surface was provided by anti-nicastrin immunostaining of A-431 cells that revealed a presence of nicastrin in the cell–cell contacts (Figure 9C, Nct). Superimposing of nicastrin and p120 staining (Figure 9C, bottom row) showed, however, that only a limited amount of nicastrin is present in the adherens junctions. The majority of the protein was localized in proximity to those structures.
Figure 9.
Cadherin–γ-secretase supercomplex is present on the cell surface within the cell–cell contact regions. (A) Three identical cultures of A-431 cells were surface biotinylated and their digitonin lysates were immunoprecipitated using β-catenin, p120, or nicastrin antibodies (IP: βc, p120, and Nct, respectively, indicated above the lanes). The resulting immunoprecipitates were first equilibrated for E-cadherin (Ec) and then analyzed for the biotinylated form of E-cadherin by using streptavidin-conjugated peroxidase (StA) and for β-catenin (βc), nicastrin (Nct), or p120 (p120) by using specific antibodies. Note that nicastrin and p120 antibodies precipitated approximately the same amounts of the surface-labeled E-cadherin. (B) Digitonin lysate of surface-biotinylated A-431 cells was split into two equal portions. One portion (-biotin) was then depleted for biotinylated proteins using streptavidin-agarose. This and the second untreated portion (ctrl) were immunoprecipitated using the anti-cadherin SHE78-7 antibody, and the precipitates were analyzed for E-cadherin (Ec), biotin (StA), p120 (p120), and nicastrin (nct). Note that the remaining intracellular nonbiotinylated cadherin interacted only negligibly with p120 and nicastrin. (C) Double immunofluorescence microscopy of A-431 cells stained with rabbit anti-nicastrin (nct) and mouse anti-p120 (p120) antibodies. Higher magnification of the selected region (indicated by arrows) is shown on the bottom row. Note that p120 and nicastrin distribution only partially corresponded to one another. Also note that the anti-nicastrin antibody produced nuclear and cytosolic staining in addition to cell–cell contacts. At least part of this staining is unspecific because it was not eliminated by nicastrin depletion using RNAi (see experiments depicted in Figure 8). In contrast, cell–cell contact anti-nicastrin staining was abolished by nicastrin siRNA.
DISCUSSION
In this work, we examined two E-cadherin complexes. One complex contained the arm-repeat protein p120. The other complex had, in addition, the transmembrane protease γ-secretase. Each of these three proteins—E-cadherin, p120, and γ-secretase—is vital for vertebrate development. This study provides several missing and very important links in our knowledge of these two complexes.
First, we obtained compelling biochemical evidence that a majority of p120 molecules in cells is present in the cadherin-bound form. Furthermore, our data suggest that the interaction between E-cadherin and p120 is of high affinity because the anti-cadherin antibody C20820, which epitope is masked by these interactions, fails to destroy the cadherin–p120 complex. Together, our data support a previous suggestion (Anastasiadis and Reynolds, 2000) that the low yield of the cadherin–p120 complex obtained in regular coimmunoprecipitation assays is caused by the complex's instability in Triton X-100–containing buffers. Our findings are important because the low level of the cadherin-free form of p120, in conjunction with the high-affinity of cadherin–p120 interactions, enables p120-dependent signaling. Local and transient fluctuations in the cadherin-free pool of p120, which apparently mediates p120 signaling, is much easier to achieve once its background level is low. A cadherin-free form of p120 is thought to conduct signaling events either through Rho family GTPases (Anastasiadis et al., 2000, Grosheva et al., 2001, Noren et al., 2000), or by binding to cortactin (Boguslavsky et al., 2007) and transcriptional repressor Kaiso (Daniel and Reynolds, 1999; Park et al., 2006). This cadherin-free form of p120 can be generated by two processes—by preventing p120 degradation (Park et al., 2006) or by disassembly of the cadherin–p120 complex. The latter event has been noted upon cadherin internalization (Xiao et al., 2005) or during carcinogenesis (Thoreson and Reynolds, 2002; Bellovin et al., 2005; Taniuchi et al., 2005). It is possible, therefore, that in normal cells the equilibrium between p120-free and p120-bound form of E-cadherin is tightly regulated. Additional studies of these regulatory mechanisms are very important. Our observation that the cadherin–p120 complex is stable in digitonin-containing buffers establishes a foundation for these future studies.
Our data also indicate that p120 is closely positioned to α-catenin in the E-cadherin–catenin complex. This conclusion is based on the observation that cysteine-specific homobifunctional cross-linker BM[PEO]3, whose spacer arm is just 14.7 Å, selectively and efficiently cross-links p120 to α-catenin. Our previous structural studies of cadherin dimers by using cysteine-specific cross-linking reaction showed that this reaction is highly dependent on the corresponding intercysteine distance (Troyanovsky et al., 2003). Because of such selectivity and the low abundance of cysteines in proteins (each catenin, for example, exhibits ∼10 cysteines, whereas cadherin has no cysteines in the intracellular region), the absence of the cross-linked adducts upon cysteine cross-linking is meaningless, but their formation is a clear indication that two proteins are very close in the protein complex. The intimate positioning of α-catenin and p120 in the complex suggests that p120 can influence the α-catenin conformation, thereby controlling α-catenin binding activities, binding to actin filaments in particular. Indeed, some published experiments support this hypothesis. For example, it was shown that the deletion of the p120 binding site of cadherin 11 stabilizes interactions between adherens junctions and microfilaments (Kiener et al., 2006) or that p120 deficiency increases the severity of morphogenetic defects caused by a hypomorphic mutation in the α-catenin gene of Caenorhabditis elegans (Pettitt et al., 2003). Therefore, although the exact mechanisms and significance of p120–α-catenin associations in the cadherin complex remain to be identified, our data suggests the possibility that p120 can directly modulate α-catenin-actin interactions.
The third important finding of our work is the uncovering of the γ-secretase component nicastrin in the cadherin–catenin complex. In previous studies, another γ-secretase component, PS1, has been detected in the complex with cadherin (Georgakopoulos et al., 1999). But neither the exact composition of this complex nor its abundance was determined. The protein–protein interactions in this complex also remained confusing because PS1 has the potential to interact with different components of the cadherin complex—cadherin itself, β-catenin, δ-catenin, p0071, and ARVCF (Zhou et al., 1997; Murayama et al., 1998; Levesque et al., 1999; Stahl et al., 1999; Baki et al., 2001). Which of these interactions, if any, occur in living cells has not been entirely clear. Finally, no information was available in previous studies on whether the full-size γ-secretase or only PS1 is associated with cadherin. Our work sheds light on the structure of cadherin–PS1 complex. We present compelling evidence that digitinonin cell lysates contain a 1-MDa complex consisting of E-cadherin, all three catenins (α- and β-catenins and p120), and at least two γ-secretase proteins, nicastrin and PS1. Furthermore, our coimmunoprecipitation data showed that the complex has mature forms of both nicastrin and PS1, suggesting that they derive from the full-size γ-secretase. Together, our data suggest that the 1-MDa complex, which we refer as the cadherin–γ-secretase supercomplex, consists of two relatively independent elements, cadherin–catenin and γ-secretase complexes. Remarkably, it is the only one multiprotein complex, detectable by BN PAGE, that contains γ-secretase in association with any other proteins in A-431 cells (as well as in HaCat cells; data not shown). This supercomplex is minor, however, relative to the cadherin-free γ-secretase, which, in complete agreement with previous BN PAGE data (Edbauer et al., 2002; Hansson et al., 2004; Sato et al., 2007), runs in the native gel as 500-kDa protein. Because both γ-secretase and cadherin complexes are in large excess relative to the cadherin–γ-secretase supercomplex, one may envision a specific process regulating supercomplex formation. Our experiments with γ-secretase inhibitors (data not shown) demonstrated that proteolytic activity of this enzyme does not regulate the amounts of the supercomplex. Lithium chloride, a potent inhibitor of glycogen synthase kinase 3β, which has been suggested to regulate cadherin–PS1 interactions (Uemura et al., 2007), also was unable to change the level of the supercomplex in our experiments. Biotinylation and immunofluorescence microscopy indicated that the majority of this supercomplex is located on the cell surface, in the cell–cell contact region in particular. However, the complex seems not only to reside in adherens junctions, but also to be present around these structures. It is possible that supercomplex assembly and the targeting of γ-secretase to the cell surface are mediated by related mechanisms. The clarification of these mechanisms is a very important direction for future studies.
Finally, we obtained several strong evidence that p120 is a key structural element in this supercomplex: Namely, we found that 1) the supercomplex contains both p120 and γ-secretase; 2) the deletion of the p120-binding site of E-cadherin abolishes cadherin binding to both nicastrin and p120; 3) Triton X-100 similarly eliminates both bindings; 4) the antibody C20820, against the p120-binding site of E-cadherin, can coimmunoprecipitate neither p120 nor nicastrin; and 5) the low level of p120 either in SW48 cells or after p120 depletion in A-431 cells by using RNAi strongly reduced the amount of the cadherin–γ-secretase supercomplex. Its residual amount in p120-dificient SW48 cells is apparently caused by the presence of a close p120 relative, p0071. The most likely interpretation of these observations is that p120 is a molecular linker connecting cadherin-catenin and γ-secretase complexes into the supercomplex. This conclusion is further supported by the previous observations that PS1 directly interacts with arm repeat domain of p120 protein family members (Zhou et al., 1997; Murayama et al., 1998; Levesque et al., 1999; Stahl et al., 1999). Additional work, which is currently underway in our laboratory, is required to fully understand the molecular organization of the supercomplex.
At present, it is also difficult to assess the functional significance of the cadherin–γ-secretase supercomplex. Previous works suggested that PS1–cadherin interactions function in cell–cell adhesion (Georgakopoulos et al., 1999). However, the nearly complete elimination of γ-secretase by nicastrin depletion in our experiments had no obvious effect on cadherin distribution in A-431 cells. One of the possible functions of this supercomplex, which we will test in the upcoming works, is its role in targeting of γ-secretase to the cell–cell contacts. Such targeting might be very important because one of the γ-secretase functions is a cleavage of Notch and Eph receptors upon its activation (Selkoe and Kopan, 2003; Georgakopoulos et al., 2006). Because Notch and Eph receptors activation occurs at cell–cell contacts, such targeting might be important for the efficient functioning of this signaling pathway.
ACKNOWLEDGMENTS
We thank P. Gilmore, J. Malone, and R. Townsend (Proteomics Core Facility, Washington University Medical School) for performing proteomic analyses of our samples. We are grateful to Drs. C. Gottardi and A. Kowalczyk for providing SW48 cell line and anti-p0071 antibody. We thank L. Cornelious (Washington University Medical School) for valuable support. The work has been supported in part by grant AR44016–04 from the National Institutes of Health.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-04-0394) on July 16, 2008.
REFERENCES
- Anastasiadis P. Z., Reynolds A. B. The p120 catenin family: complex roles in adhesion, signaling and cancer. J. Cell Sci. 2000;113:1319–1334. doi: 10.1242/jcs.113.8.1319. [DOI] [PubMed] [Google Scholar]
- Anastasiadis P. Z., Moon S. Y., Thoreson M. A., Mariner D. J., Crawford H. C., Zheng Y., Reynolds A. B. Inhibition of RhoA by p120 catenin. Nat. Cell Biol. 2000;2:637–644. doi: 10.1038/35023588. [DOI] [PubMed] [Google Scholar]
- Baki L., et al. Presenilin-1 binds cytoplasmic epithelial cadherin, inhibits cadherin/p120 association, and regulates stability and function of the cadherin/catenin adhesion complex. Proc. Natl. Acad. Sci. USA. 2001;98:2381–2386. doi: 10.1073/pnas.041603398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellovin D. I., Bates R. C., Muzikansky A., Rimm D. L., Mercurio A. M. Altered localization of p120 catenin during epithelial to mesenchymal transition of colon carcinoma is prognostic for aggressive disease. Cancer Res. 2005;65:10938–10945. doi: 10.1158/0008-5472.CAN-05-1947. [DOI] [PubMed] [Google Scholar]
- Boguslavsky S., Grosheva I., Landau E., Shtutman M., Cohen M., Arnold K., Feinstein E., Geiger B., Bershadsky A. p120 catenin regulates lamellipodial dynamics and cell adhesion in cooperation with cortactin. Proc. Natl. Acad. Sci. USA. 2007;104:10882–10887. doi: 10.1073/pnas.0702731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capaldo C. T., Macara I. G. Depletion of E-cadherin disrupts establishment but not maintenance of cell junctions in Madin-Darby canine kidney epithelial cells. Mol. Biol. Cell. 2007;18:189–200. doi: 10.1091/mbc.E06-05-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Gumbiner B. M. Crosstalk between different adhesion molecules. Curr. Opin. Cell Biol. 2006;18:572–578. doi: 10.1016/j.ceb.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Chitaev N. A., Troyanovsky S. M. Adhesive but not lateral E-cadherin complexes require calcium and catenins for their formation. J. Cell Biol. 1998;142:837–846. doi: 10.1083/jcb.142.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J. M., Reynolds A. B. The catenin p120(ctn) interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol. Cell. Biol. 1999;19:3614–3623. doi: 10.1128/mcb.19.5.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edbauer D., Winkler E., Haass C., Steiner H. Presenilin and nicastrin regulate each other and determine amyloid beta-peptide production via complex formation. Proc. Natl. Acad. Sci. USA. 2002;99:8666–8671. doi: 10.1073/pnas.132277899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakopoulos A., et al. Presenilin-1 forms complexes with the cadherin/catenin cell-cell adhesion system and is recruited to intercellular and synaptic contacts. Mol. Cell. 1999;4:893–902. doi: 10.1016/s1097-2765(00)80219-1. [DOI] [PubMed] [Google Scholar]
- Georgakopoulos A., Litterst C., Ghersi E., Baki L., Xu C., Serban G., Robakis N. K. Metalloproteinase/Presinilin1 processing of ephrinB regulates EphB-induced Src phosphorylation and signaling. EMBO J. 2006;25:1242–1252. doi: 10.1038/sj.emboj.7601031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosheva I., Shtutman M., Elbaum M., Bershadsky A. D. p120 catenin affects cell motility via modulation of activity of Rho-family GTPases: a link between cell-cell contact formation and regulation of cell locomotion. J. Cell Sci. 2001;114:695–707. doi: 10.1242/jcs.114.4.695. [DOI] [PubMed] [Google Scholar]
- Gumbiner B. M. Regulation of cadherin-mediated adhesion in morphogenesis. Nat. Rev. Mol. Cell Biol. 2005;6:622–634. doi: 10.1038/nrm1699. [DOI] [PubMed] [Google Scholar]
- Hansson C. A., et al. Nicastrin, presenilin, APH-1, and PEN-2 form active gamma-secretase complexes in mitochondria. J. Biol. Chem. 2004;279:51654–51660. doi: 10.1074/jbc.M404500200. [DOI] [PubMed] [Google Scholar]
- Halbleib J. M., Nelson W. J. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 2006;20:3199–3214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- Ireton R. C., et al. A novel role for p120 catenin in E-cadherin function. J. Cell Biol. 2002;159:465–476. doi: 10.1083/jcb.200205115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ii M., Yamamoto H., Adachi Y., Maruyama Y., Shinomura Y. Role of matrix metalloproteinase-7 (matrilysin) in human cancer invasion, apoptosis, growth, and angiogenesis. Exp. Biol. Med. 2006;231:20–27. doi: 10.1177/153537020623100103. [DOI] [PubMed] [Google Scholar]
- Kiener H. P., Stipp C. S., Allen P. G., Higgins J. M., Brenner M. B. The cadherin-11 cytoplasmic juxtamembrane domain promotes alpha-catenin turnover at adherens junctions and intercellular motility. Mol. Biol. Cell. 2006;17:2366–2376. doi: 10.1091/mbc.E05-08-0745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk A. P., Reynolds A. B. Protecting your tail: regulation of cadherin degradation by p120-catenin. Curr. Opin. Cell Biol. 2004;16:522–527. doi: 10.1016/j.ceb.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Kusumi A., Suzuki K., Koyasako K. Mobility and cytoskeletal interactions of cell adhesion receptors. Curr. Opin. Cell Biol. 1999;11:582–590. doi: 10.1016/s0955-0674(99)00020-4. [DOI] [PubMed] [Google Scholar]
- Laur O. Y., Klingelhöfer J., Troyanovsky R. B., Troyanovsky S. M. Both the dimerization and immunochemical properties of E-cadherin EC1 domain depend on Trp(156) residue. Arch. Biochem. Biophys. 2002;400:141–147. doi: 10.1006/abbi.2002.2774. [DOI] [PubMed] [Google Scholar]
- Levesque G., et al. Presenilins interact with armadillo proteins including neural-specific plakophilin-related protein and beta-catenin. J. Neurochem. 1999;72:999–1008. doi: 10.1046/j.1471-4159.1999.0720999.x. [DOI] [PubMed] [Google Scholar]
- Murayama M., Tanaka S., Palacino J., Murayama O., Honda T., Sun X., Yasutake K., Nihonmatsu N., Wolozin B., Takashima A. Direct association of presenilin-1 with beta-catenin. FEBS Lett. 1998;433:73–77. doi: 10.1016/s0014-5793(98)00886-2. [DOI] [PubMed] [Google Scholar]
- Noren N. K., Liu B. P., Burridge K., Kreft B. p120 catenin regulates the actin cytoskeleton via Rho family GTPases. J. Cell Biol. 2000;150:567–580. doi: 10.1083/jcb.150.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. I., Ji H., Jun S., Gu D., Hikasa H., Li L., Sokol S. Y., McCrea P. D. Frodo links Dishevelled to the p120-catenin/Kaiso pathway: distinct catenin subfamilies promote Wnt signals. Dev. Cell. 2006;11:683–695. doi: 10.1016/j.devcel.2006.09.022. [DOI] [PubMed] [Google Scholar]
- Pettitt J., Cox E. A., Flett A., Broadbent I. D., Hardin J. The Caenorhabditis elegans p120 catenin homologue, JAC-1, modulates cadherin-catenin function during epidermal morphogenesis. J. Cell Biol. 2003;162:15–22. doi: 10.1083/jcb.200212136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost E., Rimm D. L. Controversies at the cytoplasmic face of the cadherin-based adhesion complex. Curr. Opin. Cell Biol. 1999;11:567–572. doi: 10.1016/s0955-0674(99)00015-0. [DOI] [PubMed] [Google Scholar]
- Sato T., Diehl T. S., Narayanan S., Funamoto S., Ihara Y., De Strooper B., Steiner H., Haass C., Wolfe M. S. Active gamma-secretase complexes contain only one of each component. J. Biol. Chem. 2007;282:33985–33993. doi: 10.1074/jbc.M705248200. [DOI] [PubMed] [Google Scholar]
- Selkoe D., Kopan R. Notch and Presenilin: regulated intramembrane proteolysis links development and degeneration. Annu. Rev. Neurosci. 2003;26:565–597. doi: 10.1146/annurev.neuro.26.041002.131334. [DOI] [PubMed] [Google Scholar]
- Spasic D., Annaert W. Building {gamma}-secretase–the bits and pieces. J. Cell Sci. 2008;121:413–420. doi: 10.1242/jcs.015255. [DOI] [PubMed] [Google Scholar]
- Stahl B., Diehlmann A., Südhof T. C. Direct interaction of Alzheimer's disease-related presenilin 1 with armadillo protein p0071. J. Biol. Chem. 1999;274:9141–9148. doi: 10.1074/jbc.274.14.9141. [DOI] [PubMed] [Google Scholar]
- Taniuchi K., Nakagawa H., Hosokawa M., Nakamura T., Eguchi H., Ohigashi H., Ishikawa O., Katagiri T., Nakamura Y. Overexpressed P-cadherin/CDH3 promotes motility of pancreatic cancer cells by interacting with p120ctn and activating rho-family GTPases. Cancer Res. 2005;65:3092–3099. doi: 10.1158/0008.5472.CAN-04-3646. [DOI] [PubMed] [Google Scholar]
- Thoreson M. A., Reynolds A. B. Altered expression of the catenin p120 in human cancer: implications for tumor progression. Differentiation. 2002;70:583–589. doi: 10.1046/j.1432-0436.2002.700911.x. [DOI] [PubMed] [Google Scholar]
- Troyanovsky S. M. Cadherin dimers in cell-cell adhesion. Eur. J. Cell Biol. 2005;84:225–233. doi: 10.1016/j.ejcb.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Troyanovsky R. B., Sokolov E., Troyanovsky S. M. Adhesive and lateral E-cadherin dimers are mediated by the same interface. Mol. Cell. Biol. 2003;23:7965–7972. doi: 10.1128/MCB.23.22.7965-7972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanovsky R. B., Laur O., Troyanovsky S. M. Stable and unstable cadherin dimers: mechanisms of formation and roles in cell adhesion. Mol. Biol. Cell. 2007;18:4343–4352. doi: 10.1091/mbc.E07-01-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura K., Kuzuya A., Shimozono Y., Aoyagi N., Ando K., Shimohama S., Kinoshita A. GSK3beta activity modifies the localization and function of presenilin 1. J. Biol. Chem. 2007;282:15823–15832. doi: 10.1074/jbc.M610708200. [DOI] [PubMed] [Google Scholar]
- Weis W. I., Nelson W. J. Re-solving the cadherin-catenin-actin conundrum. J. Biol. Chem. 2006;281:35593–35597. doi: 10.1074/jbc.R600027200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K., Garner J., Buckley K. M., Vincent P. A., Chiasson C. M., Dejana E., Faundez V., Kowalczyk A. P. p120-Catenin regulates clathrin-dependent endocytosis of VE-cadherin. Mol. Biol. Cell. 2005;16:5141–5151. doi: 10.1091/mbc.E05-05-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap A. S., Crampton M. S., Hardin J. Making and breaking contacts: the cellular biology of cadherin regulation. Curr. Opin. Cell Biol. 2007;19:508–514. doi: 10.1016/j.ceb.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Liyanage U., Medina M., Ho C., Simmons A. D., Lovett M., Kosik K. S. Presenilin 1 interaction in the brain with a novel member of the Armadillo family. Neuroreport. 1997;8:2085–2090. doi: 10.1097/00001756-199705260-00054. [DOI] [PubMed] [Google Scholar]