Abstract
During intraflagellar transport (IFT), the regulation of motor proteins, the loading and unloading of cargo and the turnover of flagellar proteins all occur at the flagellar tip. To begin an analysis of the protein composition of the flagellar tip, we used difference gel electrophoresis to compare long versus short (i.e., regenerating) flagella. The concentration of tip proteins should be higher relative to that of tubulin (which is constant per unit length of the flagellum) in short compared with long flagella. One protein we have identified is the cobalamin-independent form of methionine synthase (MetE). Antibodies to MetE label flagella in a punctate pattern reminiscent of IFT particle staining, and immunoblot analysis reveals that the amount of MetE in flagella is low in full-length flagella, increased in regenerating flagella, and highest in resorbing flagella. Four methylated proteins have been identified in resorbing flagella, using antibodies specific for asymmetrically dimethylated arginine residues. These proteins are found almost exclusively in the axonemal fraction, and the methylated forms of these proteins are essentially absent in full-length and regenerating flagella. Because most cells resorb cilia/flagella before cell division, these data indicate a link between flagellar protein methylation and progression through the cell cycle.
INTRODUCTION
Intraflagellar transport (IFT) was originally discovered in the biflagellate green alga Chlamydomonas (Kozminski et al., 1993b) and is characterized by the rapid, bidirectional movement of IFT particle polypeptides (Cole et al., 1998) and their associated cargo and motor proteins along the length of eukaryotic cilia and flagella. One of the striking observations about the process is that IFT particles move from base to tip at a constant rate without pauses. At the flagellar tip, IFT particles are remodeled (Iomini et al., 2001; Pedersen et al., 2006) and then begin transport back to the cell body. Kinesin-2 (Kozminski et al., 1995) and Osm-3 (Snow et al., 2004) are the motors for anterograde (base to tip) movement, and dynein 2 is the motor for retrograde movement (Gibbons et al., 1994; Pazour et al., 1999; Porter et al., 1999; Signor et al., 1999). However, little is known about 1) how the relative activities of the IFT motors are controlled, 2) how cargo is loaded and unloaded, or 3) how assembly and disassembly of the flagellum are regulated, except that all of these activities occur within a few tenths of a micrometer of the flagellar tip, i.e., where anterograde IFT stops and retrograde IFT begins. We refer to this region of the flagellum and the proteins comprising it as the flagellar tip complex (FTC).
Although morphological structures unique to the FTC were described several decades ago (Dentler and Rosenbaum, 1977; Sale and Satir, 1977; Dentler, 1980), the biochemical composition and function of these structures remain unknown. The following data relevant to the FTC, however, are known: 1) tubulin subunit incorporation into regenerating flagellar microtubules occurs at the flagellar tip (Johnson and Rosenbaum, 1992); 2) full-length flagella continually turn over subunits (Song and Dentler, 2001), and tubulin subunit turnover occurs at the flagellar tip (Marshall and Rosenbaum, 2001); 3) the IFT machinery is required for flagellar growth and maintenance (Kozminski et al., 1995; Piperno et al., 1996); 4) IFT is involved in flagellar length control (Marshall et al., 2005); and 5) IFT returns flagellar components to the cell body for degradation and/or recycling (Iomini et al., 2001). These data suggest that identification of the proteins involved in flagellar assembly and maintenance, in IFT motor regulation and hence IFT directionality, and in cargo loading and unloading will be informed by a molecular and biochemical analysis of the components of the FTC.
To begin such an analysis, we have used a biochemical screen based on difference gel electrophoretic (DIGE) analysis of purified flagella to identify proteins that localize to the FTC. DIGE was used to compare the protein composition of full-length versus regenerating (i.e., short) flagella. Proteins in short flagella that increase in abundance relative to full-length flagella would be potential tip proteins. Such a screen might also be expected to identify flagellar proteins whose abundance in flagella is uniformly increased during regeneration. One protein identified by this screen was the cobalamin (vitamin B12) independent form of methionine synthase (MetE; EC 2.1.1.14). MetE was previously identified in Chlamydomonas as a protein whose gene transcription is up-regulated in gametes (Kurvari et al., 1995). MetE is also a member of the Chlamydomonas flagellar proteome (Pazour et al., 2005). The Chlamydomonas genome also encodes the cobalamin-dependent form of methionine synthase (MetH; EC 2.1.1.13); however, MetH is not found in the flagellar proteome. Chlamydomonas grown in the presence of vitamin B12 preferentially express MetH, not MetE. When cultured in the absence of vitamin B12, the usual laboratory growth conditions for Chlamydomonas, cells express both MetE and MetH (Croft et al., 2005).
Surprisingly, given the hypothesis for the screen used, MetE is not localized to the flagellar tip but rather is distributed along the length of the flagellum. However, the amount of MetE is higher in regenerating flagella compared with control, full-length flagella. What could be the function of MetE in flagella? MetE catalyzes the conversion of homocysteine to methionine, which is then converted to S-adenosyl methionine (SAM) by methionine adenosyltransferase (EC 2.5.1.6), itself a member of the flagellar proteome, indicating a potential requirement for protein methylation during flagellar assembly or disassembly dynamics. Using antibodies to asymmetrically dimethylated arginine residues, we have analyzed flagella under control, regenerating, and resorbing conditions. A very low, basal level of asymmetric dimethyl arginine can be detected in one protein from control or regenerating flagella. By contrast, four axonemal proteins are strongly methylated during the events of flagellar resorption, indicating a role for protein methylation during flagellar disassembly. Protein methylation has long been recognized as an important nuclear event, as histone methylation plays a key role in chromatin structure and transcriptional control. Because cilia and flagella are resorbed before cell division (Bloodgood, 1974; Rieder et al., 1979), the data reported here link progression through the cell cycle to a requirement for protein methylation in a cellular compartment other than the nucleus.
MATERIALS AND METHODS
Cells and Antibodies
Chlamydomonas reinhardtii strain CC125 (wild type, mt+) and fla10ts (fla10-1, strain CC-1919, mt−) were used for the work reported here; these cells are available from the Chlamydomonas Center (http://www.chlamy.org/). Monoclonal antibodies to IFT139 were generously provided by Joel Rosenbaum and Dennis Diener (Yale University). These antibodies were raised using purified IFT particles as the immunogen, followed by selection of cell lines secreting antibodies specific for IFT139 (see Cole et al., 1998). Antibodies to Chlamydomonas MetE (EC 2.1.1.14) were raised in rabbits against a synthetic peptide coupled to keyhole limpet hemocyanin. The peptide used as immunogen (NH3+GC-667AIDRMDADVLTIENSRSD) corresponded to residues 667–684 of Chlamydomonas MetE with the addition of a penultimate N-terminal Cys residue to provide a free sulfhydryl for conjugation to the carrier protein via 3-maleimidobenzoyl-N-hydroxysuccinimide ester (MBS); an N-terminal Gly residue was appended to decrease the chance of dimerization or cross-linking of the peptide via the thiol group. Immunization procedures were carried out by Bio-Synthesis (Lewisville, TX). The specificity of the anti-MetE serum was confirmed in the following manner. First, protein expression driven by full-length MetE cDNA was performed using the reticulocyte lysate-based T7 TNT Quick Coupled Transcription/Translation system (Promega, Madison, WI); an ∼87-kDa polypeptide was detected in the programmed lysate (but not in the control lysate) using the Transcend/Western Blue detection system (Promega) with alkaline phosphatase as the substrate. Next, immunoblot analysis of the proteins synthesized in vitro was performed using the MetE antiserum, which recognized the band at ∼87 kDa in the programmed lysate. Finally, affinity purification of MetE-specific antibodies was performed either with MetE immobilized on nitrocellulose (Talian et al., 1983) or by affinity chromatography using the synthetic peptide covalently bound to Affigel-15 (Bio-Rad, Richmond, CA) according to the manufacturer's instructions. Antibodies to α-tubulin were from Sigma (St. Louis, MO; clone B-5-1-2). Antibodies to methylated arginine (Asym24) were from Millipore (Bedford, MA).
Flagellar Isolation
Cells were grown in tris-acetate-phosphate (TAP) (Gorman and Levine, 1965) at 23°C on a cycle of 14 h of light and 10 h of dark. Flagella, prepared fresh for each experiment from 1.5- or 8-l cultures, were purified by differential centrifugation as previously described (Sloboda and Howard, 2007). To produce subfractions of flagella for more defined analysis, purified flagella were frozen at −80°C, thawed, and centrifuged in a microfuge at top speed for 2 min to produce a freeze-thaw supernatant; the flagella from the freeze-thaw pellet were resuspended and extracted with 0.05% NP-40 for 20 min at room temperature followed by centrifugation to produce a membrane supernatant. The resulting pellet was resuspended to constitute the axoneme fraction. For some experiments, the freeze-thaw step was omitted, and purified flagella were extracted directly with 0.05% NP-40 to produce membrane-matrix and axoneme fractions. To produce a sample of regenerating flagella, cells were deflagellated by pH shock (Witman et al., 1972), allowed to regenerate to a length of 3–4 μm, and then deflagellated again using dibucaine (Witman et al., 1978). To produce a sample of resorbing flagella, cells were incubated in 20 mM sodium pyrophosphate as described previously (Lefebvre et al., 1978) until the length of the flagella had been reduced 3–4 μm; the cells were then deflagellated using dibucaine. Regenerating or resorbing flagella were purified by differential centrifugation as described above.
Differential Interference Contrast Microscopy and Immunofluorescence
Samples were viewed with a Zeiss Axioskop 2 mot plus microscope using a 63×/1.4 NA Plan Apochromatic objective (Thornwood, NY). Images were projected from the microscope to the faceplate of a Hamamatsu ORCA-ER camera via an optivar lens set at a magnification of 2× (Bridgewater, NJ). The microscope, camera, and shutters for the transmitted and epifluorescence pathways were controlled by a dedicated computer running Windows XP (Microsoft, Redmond, WA) and MetaMorph software (Molecular Devices, Sunnyvale, CA). Exposure times varied from 60 ms for differential interference contrast (DIC) to 400 ms for fluorescence. For figure preparation, data files were transferred from the PC via DVD or the Dartmouth computer network to a McIntosh Powerbook G4 computer (Apple, Cupertino, CA). Software was written in MatLab (MathWorks, Natick, MA) to scale the 12-bit images from the ORCA-ER for manipulation on the McIntosh. The MatLab program takes each MetaMorph file, finds the minimum and maximum pixel values, performs a linear scaling of the pixel values to expand them from a range of 0–4095 (12 bit) to a range of 0–65,535 (16 bit), and then rewrites the data to a new 16-bit file for the McIntosh. During this process no original pixel values are discarded. Linearly scaled microscope images were then cropped on the McIntosh using Photoshop CS (Adobe, San Jose, CA) and arranged into composite figures using Illustrator CS2 (Adobe) without further manipulation.
Samples were fixed and processed for immunofluorescence microscopy as previously described (Pedersen et al., 2003), except that the blocking solution was 0.5% BSA, 10% goat serum in phosphate-buffered saline. Secondary antibodies were labeled with Alexa 488, Alexa 568 (Molecular Probes/Invitrogen, Eugene, OR), or Cy3 (Jackson ImmunoResearch Laboratories, West Grove, PA). Samples were mounted in Prolong Gold (Molecular Probes/Invitrogen) before viewing.
Difference Gel Electrophoresis, SDS-Gel Electrophoresis, and Immunoblotting
Difference gel electrophoresis (DIGE; Viswanathan et al.; 2006) was carried out at the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University (New Haven, CT). Total amino acid analysis was conducted to provide an accurate measure of the total protein concentration of each sample. The samples were then labeled with Cy3 or Cy 5 in a reaction mixture containing 400 pmol dye/50 μg protein, which yields a labeling stoichiometry of ∼1 fluor per protein molecule. Complete details of the sample preparation, labeling, and analysis procedures for DIGE can be obtained by going to http://www.gehealthcare.com/ and searching for “DIGE User Manual.” Relative spot intensities on the two-dimensional (2D) gels were determined via computer software (DeCyder from GE Healthcare, Waukesha, WI).
For IFT particle isolation, purified flagella were extracted with 0.05% NP-40 and 10 mM Mg-ATP for 20 min at room temperature (Cole et al., 1998) followed by centrifugation at 12,000 × g for 15 min in a Sorvall HB-4 rotor. The supernatant (the membrane-matrix fraction, containing IFT complex A and B particle polypeptides, among others) was loaded onto an 11-ml, 10–30% sucrose gradient and centrifuged in a Beckman SW41Ti rotor (Fullerton, CA) at 35,000 rpm for 18 h. The tube was removed from the rotor, and 50% sucrose was pumped into the bottom of the tube while collecting 0.5-ml fractions from the top. Sedimentation standards used to interpolate experimental S values were from Sigma: ovalbumin (3.7S), BSA (4.4S), catalase (11.3S), and thyroglobulin (19.4S).
Proteins were analyzed by SDS-PAGE on either 4–10% acrylamide/2–8 M urea gradient gels or 7% gels with a 3% stacking gel according to the buffer formulations of Laemmli (1970) or on 4–20% precast gradient gels (Bio-Rad). Resolved proteins were transferred to nitrocellulose and probed with antibodies as noted in the text, using peroxidase-labeled secondary antibodies and detection via ECL (GE Healthcare).
RESULTS
Using DIGE analysis, we compared the relative abundance of individual proteins in full-length flagella to those in short flagella on the same 2D gel to avoid gel-to-gel differences and artifacts. To do this, Chlamydomonas cells were deflagellated and full-length, long (L) flagella were purified. When the deflagellated cells had regenerated their flagella to a length of 3–4 μm, the cells were again deflagellated and the short (S) flagella were purified. Thus, S flagella were one-third the length of L flagella in this experiment. An aliquot of each sample was subjected to total amino acid analysis to obtain an accurate determination of the protein concentration of the L and S samples. Proteins in the L flagella sample were then solubilized and labeled with the fluorescent dye Cy3, and proteins in the S flagella sample were solubilized and labeled with Cy5. Each sample was labeled at an average stoichiometry of about one fluorochrome per polypeptide. An identical amount of total protein for each sample was resolved simultaneously on the same 2D gel, and individual protein spots were analyzed for relative fluorescent intensities. Proteins in the S sample that increased in spot intensity by a factor of threefold or more relative to the L sample were judged to be potential FTC proteins and analyzed further.
Figure 1 shows the Cy 3 (A) and Cy5 (B) images of a single 2D gel on which have been resolved both L and S flagella protein samples, respectively. Because tubulin is the major component of the axoneme, and because tubulin concentration is constant per unit length of the flagellum, the similar shapes and intensities of the tubulin region (T) from the Cy3 and Cy5 gel images confirmed that an identical quantity of total protein was loaded for each sample. Recall that the S flagella are one-third the length of the L flagella. Thus, assuming essential components of the FTC are always present in L and S samples, protein spots that showed at least a threefold increase in amount in the S sample relative to the L control sample might represent proteins that are specific to the FTC.
Figure 1.

Two-dimensional gel analysis of flagellar proteins. Proteins from long and short flagella samples were fluorescently labeled and then analyzed on the same 2D gel. (A) Cy3 image of a 2D gel showing the proteins from full-length flagella. (B) Cy5 image of the same 2D gel showing the proteins from short, regenerating flagella. The similar shape and intensity of the tubulin (T) region of each image indicates the same amount of total flagellar protein was loaded for each sample. Numbers 1–4 indicate proteins referred to in Results. The small circles with plus signs in the center (two per gel image) were used for image alignment.
Computer software was used to analyze the gel images generated by excitation of each fluorochrome, and this analysis identified several dozen proteins whose spot intensities increased by a factor of three or more in the Cy 5 relative to the Cy 3 image. Twenty-five of these spots were punched out of the 2D gel, and their protein composition was determined by mass spectrometry. Approximately half of these proteins were chloroplast or cytoplasmic contaminants or proteins that could not be clearly identified. Some examples of proteins that increased in relative amounts in the S sample are as follows: Arrow 1 (Figure 1B) points to Oxygen Evolving Enhancer Protein 1 (Mayfield et al., 1989), a chloroplast component; arrow 2 points to C1a-18, a flagellar central pair complex protein (Dutcher et al., 1984; Wargo et al., 2005); arrow 3 points to an unidentified protein; and arrow 4 points to a series of spots identified as MetE (EC 2.1.1.14), which can have multiple isoforms (see below).
MetE, a component of the Chlamydomonas flagellar proteome (Pazour et al., 2005), was previously identified in Chlamydomonas as a protein whose transcript amounts increased several-fold in activated gametes as compared with normal gametes (Kurvari et al., 1995). MetE is the cobalamin (vitamin B12) independent form of methionine synthase that catalyzes the conversion of homocysteine to methionine via transfer of a methyl group from 5-methyltetrahydrofolate. To study flagellar MetE further, we generated a polyclonal antibody directed against a synthetic peptide representing residues 667–684 of Chlamydomonas MetE; an E residue at position 679 in this sequence functions as one of four zinc ligands important for MetE activity (Pejchal and Ludwig, 2005).
Figure 2A shows the distribution of MetE in fractions generated from isolated flagella as determined by immunoblot analysis using these antibodies. MetE from Chlamydomonas has a predicted molecular mass of 86.5 kDa, and affinity-purified antibodies to MetE identified a major band at this position in a sample of intact flagella (arrow, Figure 2). The antibodies also detected two other minor bands migrating at slightly larger relative masses. These minor isoforms remained bound to the axoneme upon extraction of isolated flagella with 0.05% NP-40. Because MetE from tobacco, which migrates on a 2D gel as seven isoforms, can be phosphorylated on several different residues (Moscatelli et al., 2005), it is possible these two minor bands detected by the MetE antibodies represent MetE that has been phosphorylated. However, the two minor isoforms can still be detected after treatment of axonemal and membrane/matrix fractions with lambda protein phosphatase, an enzyme with activity directed at phosphorylated serine, threonine, and tyrosine residues. Thus, either the two minor bands are refractory to phosphatase treatment, or they derive from another mechanism such as alternative splicing of the MetE primary transcript in Chlamydomonas.
Figure 2.
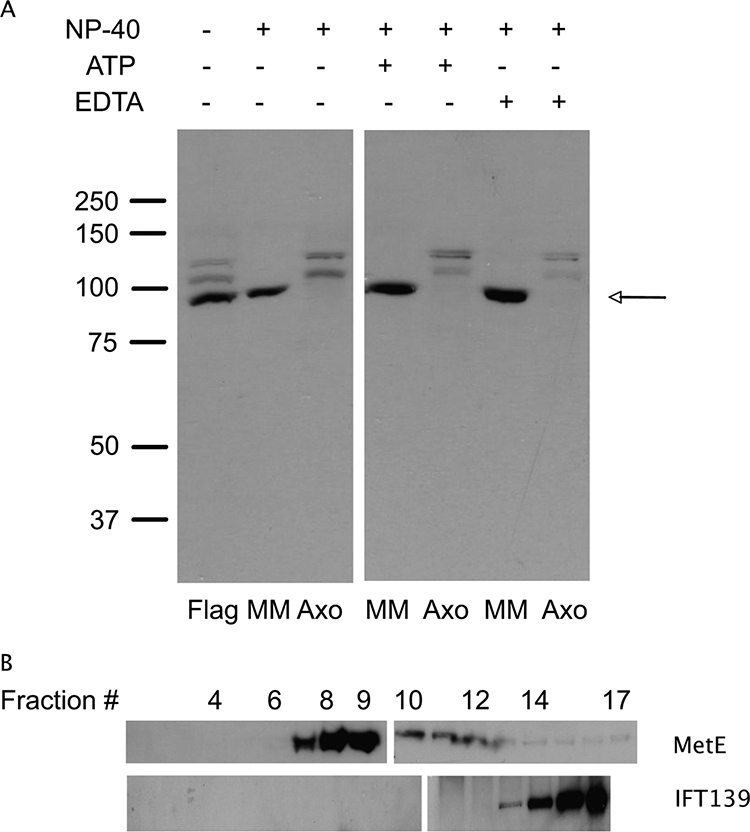
Immunoblot analysis of flagellar samples. (A) Flagellar (Flag), membrane/matrix (MM), and axonemal (Axo) fractions were probed with affinity purified antibodies to MetE. In whole flagella, MetE (∼86.5 kDa) runs as a major band (arrow), with two more slowly migrating isoforms that may be phosphorylated forms of the enzyme. The images shown here are from two halves of the same immunoblot. (B) Sucrose gradient analysis of MetE from the MM fraction indicates MetE runs at ∼9 S and is hence not associated with IFT particles, which appear in fractions 14–16, i.e., at 15–16.5 S. The two sections of each gradient image indicate the samples were run on two different gels (fractions 1–9 and 10–17 for MetE and fractions 1–10 and 11–16 for IFT139).
Because microtubule-based motors are dissociated from the microtubule subunit lattice in the presence of ATP (Gilbert et al., 1985; Lasek and Brady, 1985; Scholey et al., 1985; Vale et al., 1985) and because EDTA disrupts the structure of Zn-binding proteins (Matt et al., 2004), we next tested the effect of these reagents on the interaction of MetE with the axoneme. Under either of these conditions the major isoform of MetE was still found in the membrane/matrix fraction, and the modified isoforms remained associated with the axoneme. Although the bulk of the MetE is in the membrane/matrix fraction in the presence of ATP, as are IFT particles (Cole et al., 1998), the MetE in the membrane/matrix fraction is not tightly associated with IFT particles (Figure 2B). The immunoblots in Figure 2B show the fractionation on a 10–30% sucrose gradient of the membrane/matrix components extracted from purified flagella with 0.05% NP-40 and 10 mM Mg-ATP. The major MetE isoform sedimented at ∼9 S (fractions 7–9), whereas IFT complexes A and B sedimented at ∼15–16 S (fractions 14–16; see also Cole et al., 1998).
Despite these data, MetE antibodies stained intact Chlamydomonas flagella in a punctate pattern reminiscent of the staining of IFT particles. Figure 3A shows a DIC image of isolated flagella stained with antibodies to IFT139 (an IFT complex A component) and MetE. Both antibodies exhibit a punctate staining pattern. Higher magnification views (Figure 3, B and C) in color identify individual MetE-positive regions (red), individual IFT139-positive regions (green), and regions where the two signals are coincident (yellow). Thus, although the bulk of the MetE is in the membrane-matrix fraction, it is distributed along the length of the flagellum in a discontinuous manner similar to that of IFT particles, but neither entirely coincident with them (Figure 3) nor tightly bound to them (Figure 2B).
Figure 3.
Double label immunofluorescence microscopy. (A) Isolated flagella stained with antibodies to IFT139 (an IFT complex A component) and MetE. (B) Two color overlay showing IFT139 in green and MetE in red. (C) Collage of double labeled flagella to show more clearly both individual (green and red) and coincident (yellow) localization of IFT139 and MetE, respectively.
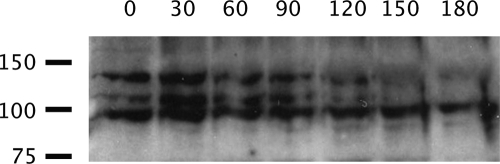
The higher molecular weight isoforms of MetE that are associated with the axoneme (Figure 2A) are lost from flagella in the absence of anterograde IFT, as is MetE, though to a lesser extent. When fla10 cells are incubated at the restrictive temperature (32°C), anterograde IFT ceases due to a temperature-sensitive mutation in the gene for one of the motor subunits of kinesin-2, the heterotrimeric anterograde IFT motor. Figure 4 shows the results obtained when cells were incubated at the restrictive temperature for 3 h. The two higher molecular weight isoforms of MetE were lost from flagella with increasing time at the restrictive temperature, i.e., in the absence of anterograde IFT. Therefore, both minor isoforms have the ability to dissociate from the axoneme, associate with IFT particles, and move out of the flagellum via retrograde IFT. It may be that the coincident staining of MetE and IFT particles noted in Figure 3 represents this interaction. The major isoform at ∼87 kDa also decreases in amount as a function of time at the restrictive temperature.
Figure 4.
MetE requires an active anterograde IFT system for its presence in flagella. fla10 cells were shifted to the restrictive temperature (32°C) to inactive anterograde IFT; the flagella were isolated every 30 min after the temperature shift and analyzed for MetE via immunoblotting. Time (min) after the temperature shift is shown across the top, and the positions of molecular weight standards (×10−3) are shown to the left.
The distribution of MetE in flagellar fractions varied also with the assembly state of the flagella. Control, full-length, wild-type flagella were isolated, frozen, and then thawed. This procedure damages the membrane, allowing soluble flagellar components to leak out, as has been shown for the microtubule plus end tracking protein EB1 in Chlamydomonas flagella (Pedersen et al., 2003). Proteins released by freeze-thaw were separated from insoluble material by centrifugation to produce a freeze-thaw supernatant. The sedimented flagella were then resuspended, extracted with 0.05% NP-40, and sedimented to produce membrane and axoneme fractions, respectively. These three samples from full-length flagella were then analyzed by immunoblotting with MetE antibodies and compared with the same fractions produced from flagella that had either been regenerated to a third of the wild-type length or resorbed to the same length before flagellar isolation.
Figure 5 shows the results of such an analysis. In full-length flagella, some MetE was released to the supernatant after freeze-thaw and/or detergent extraction. The bulk of the MetE, however, remained associated with the axoneme. Regenerating flagella, by contrast, contained more MetE than full-length flagella, and much of this MetE was not tightly associated with the axoneme, as it was found in the supernatant after freeze-thaw and/or detergent extraction. Resorbing flagella contained the highest levels of MetE, and in this case, a majority of the MetE was released to the supernatant after freeze-thaw. In addition, note that the axonemal fraction of resorbing flagella lacked the modified forms of MetE that were detectable in the axonemal fractions of full-length and regenerating flagella.
Figure 5.

Analysis of MetE in samples of flagella at varying stages. Flagella were isolated from full-length, regenerating, and resorbing flagella. Each sample was then fractionated into freeze-thaw (FT), membrane (M), and axoneme (Axo) fractions and analyzed by immunoblotting with affinity purified antibodies to MetE. For a given flagellar fraction, immunoblotting with tubulin antibody was used to determine that the same amount of total protein was loaded for each sample. The results shown here are lanes from a single immunoblot, cut, and pasted into a collage, indicated by the spaces between the respective sections.
What might be the function of MetE in flagella? MetE produces methionine from homocysteine, and methionine can then be converted to SAM by SAM synthase; SAM serves as the methyl donor for posttranslational protein modification reactions via protein arginine methyl transferases; SAM synthase and methyltransferases have been identified in the flagellar proteome (Pazour et al., 2005). To determine if methylated proteins are present in flagella, we used antibodies specific for asymmetric dimethyl arginine residues (Boisvert et al., 2003). Asym24 antibodies recognize asymmetric dimethyl arginine, in which both methyl groups are associated with only one of the two amines of the guanidino moiety of arginine. Samples of full-length, regenerating, and resorbing flagella were analyzed by immunoblotting to detect polypeptides containing asymmetric dimethyl arginine residues. Consistent with an increase in MetE in resorbing flagella (Figure 5), four proteins were preferentially methylated in resorbing flagella (Figure 6); these proteins migrated between ∼55 and 75 kDa and are labeled A–D in Figure 6. The methylated form of the 75-kDa protein (labeled A) was barely detected in the axoneme fraction of full-length flagella. Similarly, methylated protein A was present at a low level in the axonemes of regenerating flagella, as was a second methylated protein of ∼71-kDa, labeled B in Figure 6. Proteins A and B, plus two other proteins (C and D), were strongly methylated in the axonemal fraction of resorbing flagella, whereas some methylated protein B, and a lesser amount of methylated protein A, were found in the membrane fraction of resorbing flagella.
Figure 6.
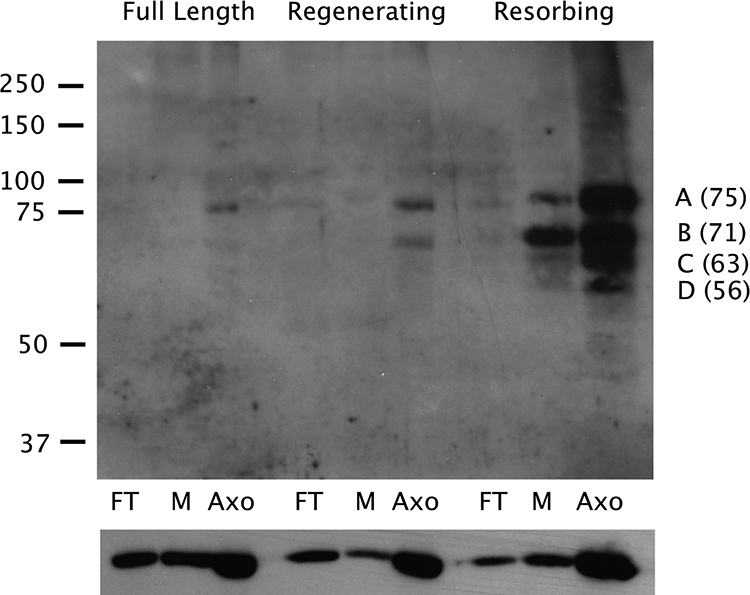
Protein methylation varies with the state of the flagella. Flagella were isolated from full-length, regenerating, and resorbing flagella, fractionated into freeze-thaw (FT), membrane (M), and axoneme (Axo) fractions, and analyzed by immunoblotting using antibodies to asymmetrically dimethylated arginine residues (top panel). The migration of standards is shown to the left (×10−3), and the masses (kDa) of proteins A–D, estimated from the migrations of the standards, are shown to the right. The blot was then stripped and probed with antibodies to α-tubulin to serve as a loading control (bottom panel).
Finally, asymmetric dimethyl arginine residues were detected associated with the FTC of resorbing flagella that appeared by DIC microscopy to have normal flagellar morphology (Figure 7). However, note that only some flagella (29.6%) in a given preparation reacted with asymmetric dimethyl arginine antibodies only in the region of the FTC. The majority of resorbing flagella (70.4%) clearly did not. Rather, the staining of the majority of resorbing flagella was punctate and similar to the staining of IFT particles and MetE in full-length flagella (see Figure 3).
Figure 7.
Localization of proteins containing asymmetric dimethyl arginine residues in resorbing flagella. Cells were stained with Asym24 antibodies demonstrating two classes of label: enrichment of proteins containing dimethyl arginine at the FTC (top row) or numerous puncta along the length of the flagellum, similar to IFT particle staining.
DISCUSSION
Using a proteomic screen for FTC proteins based on a DIGE analysis of long versus short flagella, we have identified the cobalamin (vitamin B12) independent form of methionine synthase (MetE; EC 2.1.1.14) as a flagellar protein in Chlamydomonas. MetE was previously identified as a Chlamydomonas gene whose transcription rate was up-regulated during the conversion of vegetative cells to gametes (Kurvari et al., 1995) and was also identified in the Chlamydomonas flagellar proteome (Pazour et al., 2005). Antibodies directed against a peptide epitope specific for the Chlamydomonas form of MetE recognize three isoforms; the major isoform is a component of the flagellar membrane-matrix fraction, whereas two minor isoforms are tightly bound to the flagellar axoneme (Figure 2). Although anti-MetE antibodies stain flagella in a punctate pattern, the localization of flagellar MetE is not entirely coincident with IFT particles, and the enzyme is not tightly bound to them because it does not fractionate with IFT complex A or B on sucrose gradients (Figures 2 and 3). In addition, observations with fla10 cells at the restrictive temperature suggest that retrograde IFT can translocate the two more slowly migrating isoforms out of flagella. This observation, combined with the localization of MetE shown in Figure 2, suggests that the subset of MetE that localizes to IFT particles may represent the two more slowly migrating isoforms of MetE. If this is correct, then a modification of MetE to produce the two more slowly migrating forms may induce MetE to associate with retrograde IFT particles. By inference, the MetE modifying enzyme(s) might themselves be components of the FTC.
MetE catalyzes the synthesis of methionine, the penultimate step in the production of SAM. Because SAM is the methyl donor in protein methylation reactions we also analyzed flagella at various stages of growth and resorption to identify proteins containing methylated arginine. To do this we used antibodies specific for asymmetric, dimethyl arginine residues, and these results (Figure 6) showed that resorbing flagella contain at least four proteins whose level of methylation increases dramatically during flagellar resorption.
Methylation of histones has been studied for many years, but only recently has methylation of cytoplasmic proteins been appreciated as an important posttranslational modification event involved in the regulation of cellular activities. Myelin was one of the first nonnuclear proteins to be identified as a target for methylation (Baldwin and Carnegie, 1971; Brostoff and Eylar, 1971). Protein methylation is now recognized as a widespread cellular activity, critical not only for gene transcription, but also responsible for regulating various steps in signal transduction and protein targeting mechanisms (reviewed in McBride and Silver, 2001). For example, a number of small GTPases and the catalytic subunit of protein phosphatase 2A are methylated during various cellular events (Mumby, 2001). The present study adds to this list the involvement of arginine methylation during flagellar resorption.
It is important to note two aspects of protein methylation that make this posttranslational modification similar, but not identical, to protein phosphorylation. First, O-linked methyl groups on Asp and Glu residues can be removed by demethylating enzymes such as histone demethylase. However, no known class of enzymes exists that can demethylate the N-linked methyl groups of arginine, i.e., the class of methylation modifications reported here, although an amine oxidase can function to demethylate N-linked methyl groups on Lys4 in histone H3 (Shi et al., 2004). Second, although methylation of arginine residues does not affect the positive charge on arginine, altered protein tertiary structure may result from the disruption of H-bonds caused by steric hindrance due to the addition of methyl groups to arginine residues.
Disrupted H-bonding, increased hydrophobic interactions, and steric hindrance are events potentially involved in flagellar resorption, as these could alter key protein–protein interactions or protein tertiary structures, events that might be required as a prelude to axoneme disassembly. Alternatively, arginine methylation might promote the association, via hydrophobic interactions, of disassembled axonemal components with retrograde IFT particles. Because dimethyl arginine modifications are most likely irreversible and because protein methylation is induced during resorption, the methylated proteins generated are most likely not reused when resorption conditions are reversed to allow flagella regeneration to occur. For example, it has been shown previously that when flagella are induced to resorb and then the conditions are reversed to allow flagella regeneration in the presence of protein synthesis inhibitors, the resorbed flagellar proteins are reutilized to regenerate full-length flagella (Rosenbaum et al., 1969; Lefebvre et al., 1978). Given these data and the results reported here, the cytoplasm must contain a pool of the nonmethylated forms of proteins A–D (Figure 7) to meet the requirement of these proteins for flagellar regeneration. One or more of these methylated axonemal proteins may represent the limiting component of the precursor pool first noted by Rosenbaum et al. (1969).
Previous work indicated that histone H1, which migrates at 34 kDa on SDS gels, is a component of the flagellar axoneme where it plays a role in microtubule stability (Multigner et al., 1992). A protein with axonemal stabilizing activity would be a likely target for an inactivating modification such as methylation during flagellar disassembly. We did not detect asymmetric dimethyl arginine residues in a protein of this size, but perhaps during flagellar resorption the axonemal form of H1 undergoes a methyl modification different from that reported here. This has yet to be determined.
Methylation of flagellar proteins, although a new observation with respect to flagellar dynamics, is not the first demonstration of posttranslational protein modification in flagella. For example, numerous phosphorylated proteins have been identified in Chlamydomonas flagella, including α-tubulin (Piperno and Luck, 1976), radial spoke stalk proteins (Piperno et al., 1981), outer arm dynein (King and Witman, 1994), and a number of membrane/matrix components (Bloodgood, 1992). Experiments using metabolic labeling of flagellar proteins with 32P indicate that protein phosphorylation levels change with alterations in flagellar activity (Bloodgood and Salomonsky, 1994); indeed, the flagellum contains >80 phosphoproteins (Piperno et al., 1981). Phosphorylation has recently been shown to be important in the control of flagellar length, as is IFT itself (Marshall and Rosenbaum, 2001; Marshall et al., 2005). Variations in flagellar length in Chlamydomonas have been correlated with the activity of a novel MAP kinase encoded by the LF4 gene (Berman et al., 2003), a NIMA-related kinase (Bradley and Quarmby, 2005), and glycogen synthase kinase 3 (Wilson and Lefebvre, 2004), although the target proteins for these kinases have not yet been identified.
Axonemal tubulin undergoes several other modifications in addition to phosphorylation, including glycylation, acetylation, and polyglutamylation. Glycylation in ciliary axonemes (Redeker et al., 1994) is an essential modification. Tetrahymena cilia lacking all α-tubulin glycylation sites are normal, whereas cilia lacking three of five β-tubulin glycylation sites are 9 + 0 and immobile (Xia et al., 2000). Surprisingly, it is not the position of the glycylation modifications on tubulin but the amount of total glycylation that is important. Acetylation occurs on the ε-amino group of Lys 40 in α-tubulin, and a deacetylase removes this group during flagellar resorption (L'Hernault and Rosenbaum, 1985). However, this modification is nonessential in Tetrahymena cilia (Gaertig et al., 1995) and probably in Chlamydomonas flagella as well (Kozminski et al., 1993a), as cells in which Lys 40 has been changed to arginine have no noticeable phenotype. However, arginine residues can also be acetylated, although it is not clear if the tubulin acetylase can also act on an arginine residue at position 40. In contrast, other deacetylation and phosphorylation reactions are important in the disassembly of primary cilia in tissue culture cells. For example, HDAC6, a tubulin deacetylase, is activated by phosphorylation via aurora kinase (Pan et al., 2004), and this in turn promotes ciliary disassembly (Pugacheva et al., 2007). Polyglutamylation of α-tubulin (Edde et al., 1990; Fouquet et al., 1994) is catalyzed by a specific polyglutamylase (Tt116), and the extent of glutamylation is modulated by the fleer gene in zebrafish (Pathak et al., 2007). Polyglutamylation occurs on the C-terminal tail of α-tubulin, a domain that is itself acidic and is involved in the binding and processivity of kinesin (Skiniotis et al., 2004). Moreover, polyglutamylation of this domain enhances dynein-based motility in sperm (Gagnon et al., 1996).
Flagella in Chlamydomonas (and primary cilia in mammalian cells) are dynamic structures; they assemble during G0/G1, and when they reach a mature length continue to turn over subunits at the tip (Marshall and Rosenbaum, 2001). Flagella and cilia are resorbed before the start of mitosis. The data reported here provide the first example of protein methylation at arginine residues that occurs coincident with flagellar resorption. These data also indicate a requirement for methionine and SAM during flagellar resorption. Protein methylation may be a necessary step in the disassembly of axonemal structures or may be required to promote the association of disassembled axonemal proteins with the retrograde IFT machinery. Importantly, because most flagellated and ciliated cells resorb these organelles before cell division (Bloodgood, 1974; Rieder et al., 1979), and because flagellar protein methylation occurs coincident with flagellar resorption, this work proposes a link between flagellar protein methylation and progression through the cell cycle.
ACKNOWLEDGMENTS
We thank Rita Werner-Peterson for technical assistance, Susan A. Schwarz (Dartmouth Research Computing) for writing the image file conversion program in MatLab, and Joel Rosenbaum and Dennis Diener for their generous gift of antibodies to IFT139. This work was supported by National Institutes of Health Grant DK071720 (R.D.S.) and National Science Foundation Grant MCB 0418877 (R.D.S.).
Abbreviations used:
- DIGE
difference gel electrophoresis
- IFT
intraflagellar transport
- FTC
flagellar tip complex
- MetE
methionine synthase (EC 2.1.1.14)
- SAM
S-adenosyl methionine.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-05-0470) on August 13, 2008.
REFERENCES
- Baldwin G. S., Carnegie P. R. Isolation and partial characterization of methylated arginines from the encephalitogenic basic protein of myelin. Biochem. J. 1971;123:69–74. doi: 10.1042/bj1230069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman S. A., Wilson N. F., Haas N. A., Lefebvre P. A. A novel MAP kinase regulates flagellar length in Chlamydomonas. Curr. Biol. 2003;13:1145–1149. doi: 10.1016/s0960-9822(03)00415-9. [DOI] [PubMed] [Google Scholar]
- Bloodgood R. A. Resorption of organelles containing microtubules. Cytobios. 1974;9:142–161. [PubMed] [Google Scholar]
- Bloodgood R. A. Calcium-regulated phosphorylation of proteins in the membrane-matrix compartment of the Chlamydomonas flagellum. Exp. Cell Res. 1992;198:228–236. doi: 10.1016/0014-4827(92)90375-i. [DOI] [PubMed] [Google Scholar]
- Bloodgood R. A., Salomonsky N. L. The transmembrane signaling pathway involved in directed movements of Chlamydomonas flagellar membrane glycoproteins involves the dephosphorylation of a 60-kD phosphoprotein that binds to the major flagellar membrane glycoprotein. J. Cell Biol. 1994;127:803–811. doi: 10.1083/jcb.127.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert F. M., Cote J., Boulanger M. C., Richard S. A proteomic analysis of arginine-methylated protein complexes. Mol. Cell Proteon. 2003;2:1319–1330. doi: 10.1074/mcp.M300088-MCP200. [DOI] [PubMed] [Google Scholar]
- Bradley B. A., Quarmby L. M. A NIMA-related kinase, Cnk2p, regulates both flagellar length and cell size in Chlamydomonas. J. Cell Sci. 2005;118:3317–3326. doi: 10.1242/jcs.02455. [DOI] [PubMed] [Google Scholar]
- Brostoff S., Eylar E. H. Localization of methylated arginine in the A1 protein from myelin. Proc. Natl. Acad. Sci. USA. 1971;68:765–769. doi: 10.1073/pnas.68.4.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole D. G., Diener D. R., Himelblau A. L., Beech P. L., Fuster J. C., Rosenbaum J. L. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J. Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft M. T., Lawrence A. D., Raux-Deery E., Warren M. J., Smith A. G. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature. 2005;438:90–93. doi: 10.1038/nature04056. [DOI] [PubMed] [Google Scholar]
- Dentler W. L. Structures linking the tips of ciliary and flagellar microtubules to the membrane. J. Cell Sci. 1980;42:207–220. doi: 10.1242/jcs.42.1.207. [DOI] [PubMed] [Google Scholar]
- Dentler W. L., Rosenbaum J. L. Flagellar elongation and shortening in Chlamydomonas. III. structures attached to the tips of flagellar microtubules and their relationship to the directionality of flagellar microtubule assembly. J. Cell Biol. 1977;74:747–759. doi: 10.1083/jcb.74.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher S. K., Huang B., Luck D. J. Genetic dissection of the central pair microtubules of the flagella of Chlamydomonas reinhardtii. J. Cell Biol. 1984;98:229–236. doi: 10.1083/jcb.98.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edde B., Rossier J., Le Caer J. P., Desbruyeres E., Gros F., Denoulet P. Posttranslational glutamylation of alpha-tubulin. Science. 1990;247:83–85. doi: 10.1126/science.1967194. [DOI] [PubMed] [Google Scholar]
- Fouquet J. P., Edde B., Kann M. L., Wolff A., Desbruyeres E., Denoulet P. Differential distribution of glutamylated tubulin during spermatogenesis in mammalian testis. Cell Motil. Cytoskelet. 1994;27:49–58. doi: 10.1002/cm.970270106. [DOI] [PubMed] [Google Scholar]
- Gaertig J., Cruz M. A., Bowen J., Gu L., Pennock D. G., Gorovsky M. A. Acetylation of lysine 40 in alpha-tubulin is not essential in Tetrahymena thermophila. J. Cell Biol. 1995;129:1301–1310. doi: 10.1083/jcb.129.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon C., White D., Cosson J., Huitorel P., Edde B., Desbruyeres E., Paturle-Lafanechere L., Multigner L., Job D., Cibert C. The polyglutamylated lateral chain of alpha-tubulin plays a key role in flagellar motility. J. Cell Sci. 1996;109(Pt 6):1545–1553. doi: 10.1242/jcs.109.6.1545. [DOI] [PubMed] [Google Scholar]
- Gibbons B. H., Asai D. J., Tang W. J., Hays T. S., Gibbons I. R. Phylogeny and expression of axonemal and cytoplasmic dynein genes in sea urchins. Mol. Biol. Cell. 1994;5:57–70. doi: 10.1091/mbc.5.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert S. P., Allen R. D., Sloboda R. D. Translocation of vesicles from squid axoplasm on flagellar microtubules. Nature. 1985;315:245–248. doi: 10.1038/315245a0. [DOI] [PubMed] [Google Scholar]
- Gorman D. S., Levine R. P. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iomini C., Babaev-Khaimov V., Sassaroli M., Piperno G. Protein particles in Chlamydomonas flagella undergo a transport cycle consisting of four phases. J. Cell Biol. 2001;153:13–24. doi: 10.1083/jcb.153.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. A., Rosenbaum J. L. Polarity of flagellar assembly in Chlamydomonas. J. Cell Biol. 1992;119:1605–1611. doi: 10.1083/jcb.119.6.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S. M., Witman G. B. Multiple sites of phosphorylation within the alpha heavy chain of Chlamydomonas outer arm dynein. J. Biol. Chem. 1994;269:5452–5457. [PubMed] [Google Scholar]
- Kozminski K. G., Beech P. L., Rosenbaum J. L. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J. Cell Biol. 1995;131:1517–1527. doi: 10.1083/jcb.131.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski K. G., Diener D. R., Rosenbaum J. L. High level expression of nonacetylatable alpha-tubulin in Chlamydomonas reinhardtii. Cell Motil. Cytoskelet. 1993a;25:158–170. doi: 10.1002/cm.970250205. [DOI] [PubMed] [Google Scholar]
- Kozminski K. G., Johnson K. A., Forscher P., Rosenbaum J. L. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc. Natl. Acad. Sci. USA. 1993b;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurvari V., Qian F., Snell W. J. Increased transcript levels of a methionine synthase during adhesion-induced activation of Chlamydomonas reinhardtii gametes. Plant Mol. Biol. 1995;29:1235–1252. doi: 10.1007/BF00020465. [DOI] [PubMed] [Google Scholar]
- L'Hernault S. W., Rosenbaum J. L. Reversal of the posttranslational modification on Chlamydomonas flagellar alpha-tubulin occurs during flagellar resorption. J. Cell Biol. 1985;100:457–462. doi: 10.1083/jcb.100.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lasek R. J., Brady S. T. Attachment of transported vesicles to microtubules in axoplasm is facilitated by AMP-PNP. Nature. 1985;316:645–647. doi: 10.1038/316645a0. [DOI] [PubMed] [Google Scholar]
- Lefebvre P. A., Nordstrom S. A., Moulder J. E., Rosenbaum J. L. Flagellar elongation and shortening in Chlamydomonas. IV. Effects of flagellar detachment, regeneration, and resorption on the induction of flagellar protein synthesis. J. Cell Biol. 1978;78:8–27. doi: 10.1083/jcb.78.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W. F., Qin H., Rodrigo Brenni M., Rosenbaum J. L. Flagellar length control system: testing a simple model based on intraflagellar transport and turnover. Mol. Biol. Cell. 2005;16:270–278. doi: 10.1091/mbc.E04-07-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W. F., Rosenbaum J. L. Intraflagellar transport balances continuous turnover of outer doublet microtubules: implications for flagellar length control. J. Cell Biol. 2001;155:405–414. doi: 10.1083/jcb.200106141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matt T., Martinez-Yamout M. A., Dyson H. J., Wright P. E. The CBP/p300 TAZ1 domain in its native state is not a binding partner of MDM2. Biochem. J. 2004;381:685–691. doi: 10.1042/BJ20040564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayfield S. P., Schirmer-Rahire G., Frank H., Zuber H., Rochaix J. D. Analysis of the genes of the oee1 and oee3 proteins of the photo system II photosystem 2 complex of Chlamydomonas reinhardtii. Plant Mol. Biol. 1989;12:683–693. doi: 10.1007/BF00044159. [DOI] [PubMed] [Google Scholar]
- McBride A. E., Silver P. A. State of the arg: protein methylation at arginine comes of age. Cell. 2001;106:5–8. doi: 10.1016/s0092-8674(01)00423-8. [DOI] [PubMed] [Google Scholar]
- Moscatelli A., Scali M., Prescianotto-Baschong C., Ferro M., Garin J., Vignani R., Ciampolini F., Cresti M. A methionine synthase homolog is associated with secretory vesicles in tobacco pollen tubes. Planta. 2005;221:776–789. doi: 10.1007/s00425-005-1487-7. [DOI] [PubMed] [Google Scholar]
- Multigner L., Gagnon J., Van Dorsselaer A., Job D. Stabilization of sea urchin flagellar microtubules by histone H1. Nature. 1992;360:33–39. doi: 10.1038/360033a0. [DOI] [PubMed] [Google Scholar]
- Mumby M. A new role for protein methylation: switching partners at the phosphatase ball. Sci STKE. 2001;2001:PE1. doi: 10.1126/stke.2001.79.pe1. [DOI] [PubMed] [Google Scholar]
- Pan J., Wang Q., Snell W. J. An aurora kinase is essential for flagellar disassembly in Chlamydomonas. Dev. Cell. 2004;6:445–451. doi: 10.1016/s1534-5807(04)00064-4. [DOI] [PubMed] [Google Scholar]
- Pathak N., Obara T., Mangos S., Liu Y., Drummond I. A. The zebrafish fleer gene encodes an essential regulator of cilia tubulin polyglutamylation. Mol. Biol. Cell. 2007;18:4353–4364. doi: 10.1091/mbc.E07-06-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Agrin N., Leszyk J., Witman G. B. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour G. J., Dickert B. L., Witman G. B. The DHC1b (DHC2) isoform of cytoplasmic dynein is required for flagellar assembly. J. Cell Biol. 1999;144:473–481. doi: 10.1083/jcb.144.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen L. B., Geimer S., Rosenbaum J. L. Dissecting the molecular mechanisms of intraflagellar transport in Chlamydomonas. Curr. Biol. 2006;16:450–459. doi: 10.1016/j.cub.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Pedersen L. B., Geimer S., Sloboda R. D., Rosenbaum J. L. The Microtubule plus end-tracking protein EB1 is localized to the flagellar tip and basal bodies in Chlamydomonas reinhardtii. Curr. Biol. 2003;13:1969–1974. doi: 10.1016/j.cub.2003.10.058. [DOI] [PubMed] [Google Scholar]
- Pejchal R., Ludwig M. L. Cobalamin-independent methionine synthase (MetE): a face-to-face double barrel that evolved by gene duplication. PLoS Biol. 2005;3:e31. doi: 10.1371/journal.pbio.0030031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G., Huang B., Ramanis Z., Luck D. J. Radial spokes of Chlamydomonas flagella: polypeptide composition and phosphorylation of stalk components. J. Cell Biol. 1981;88:73–79. doi: 10.1083/jcb.88.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G., Luck D. J. Phosphorylation of axonemal proteins in Chlamydomonas reinhardtii. J. Biol. Chem. 1976;251:2161–2167. [PubMed] [Google Scholar]
- Piperno G., Mead K., Henderson S. Inner dynein arms but not outer dynein arms require the activity of kinesin homologue protein KHP1FLA10 to reach the distal part of flagella in Chlamydomonas. J. Cell Biol. 1996;133:371–379. doi: 10.1083/jcb.133.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter M. E., Bower R., Knott J. A., Byrd P., Dentler W. Cytoplasmic dynein heavy chain 1b is required for flagellar assembly in Chlamydomonas. Mol. Biol. Cell. 1999;10:693–712. doi: 10.1091/mbc.10.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugacheva E. N., Jablonski S. A., Hartman T. R., Henske E. P., Golemis E. A. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129:1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redeker V., Levilliers N., Schmitter J. M., Le Caer J. P., Rossier J., Adoutte A., Bre M. H. Polyglycylation of tubulin: a posttranslational modification in axonemal microtubules. Science. 1994;266:1688–1691. doi: 10.1126/science.7992051. [DOI] [PubMed] [Google Scholar]
- Rieder C. L., Jensen C. G., Jensen L. C. The resorption of primary cilia during mitosis in a vertebrate (PtK1) cell line. J. Ultrastruct. Res. 1979;68:173–185. doi: 10.1016/s0022-5320(79)90152-7. [DOI] [PubMed] [Google Scholar]
- Rosenbaum J. L., Moulder J. E., Ringo D. L. Flagellar elongation and shortening in Chlamydomonas. The use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J. Cell Biol. 1969;41:600–619. doi: 10.1083/jcb.41.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sale W. S., Satir P. The termination of the central microtubules from the cilia of Tetrahymena pyriformis. Cell Biol. Intl. Rep. 1977;1:56–63. doi: 10.1016/0309-1651(77)90008-x. [DOI] [PubMed] [Google Scholar]
- Scholey J. M., Porter M. E., Grissom P. M., McIntosh J. R. Identification of kinesin in sea urchin eggs, and evidence for its localization in the mitotic spindle. Nature. 1985;318:483–486. doi: 10.1038/318483a0. [DOI] [PubMed] [Google Scholar]
- Shi Y., Lan F., Matson C., Mulligan P., Whetstine J. R., Cole P. A., Casero R. A., Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Signor D., Wedaman K. P., Orozco J. T., Dwyer N. D., Bargmann C. I., Rose L. S., Scholey J. M. Role of a class DHC1b dynein in retrograde transport of IFT motors and IFT raft particles along cilia, but not dendrites, in chemosensory neurons of living Caenorhabditis elegans. J. Cell Biol. 1999;147:519–530. doi: 10.1083/jcb.147.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skiniotis G., Cochran J. C., Muller J., Mandelkow E., Gilbert S. P., Hoenger A. Modulation of kinesin binding by the C-termini of tubulin. EMBO J. 2004;23:989–999. doi: 10.1038/sj.emboj.7600118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloboda R. D., Howard L. Localization of EB1, IFT polypeptides, and kinesin-2 in Chlamydomonas flagellar axonemes via immunogold scanning electron microscopy. Cell Motil. Cytoskelet. 2007;64:446–460. doi: 10.1002/cm.20195. [DOI] [PubMed] [Google Scholar]
- Snow J. J., Ou G., Gunnarson A. L., Walker M. R., Zhou H. M., Brust-Mascher I., Scholey J. M. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat. Cell Biol. 2004;6:1109–1113. doi: 10.1038/ncb1186. [DOI] [PubMed] [Google Scholar]
- Song L., Dentler W. L. Flagellar protein dynamics in Chlamydomonas. J. Biol. Chem. 2001;276:29754–29763. doi: 10.1074/jbc.M103184200. [DOI] [PubMed] [Google Scholar]
- Talian J. C., Olmsted J. B., Goldman R. D. A rapid procedure for preparing fluorescein-labeled specific antibodies from whole antiserum: its use in analyzing cytoskeletal architecture. J. Cell Biol. 1983;97:1277–1282. doi: 10.1083/jcb.97.4.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale R. D., Reese T. S., Sheetz M. P. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan S., Unlu M., Minden J. S. Two-dimensional difference gel electrophoresis. Nat. Protocol. 2006;1:1351–1358. doi: 10.1038/nprot.2006.234. [DOI] [PubMed] [Google Scholar]
- Wargo M. J., Dymek E. E., Smith E. F. Calmodulin and PF6 are components of a complex that localizes to the C1 microtubule of the flagellar central apparatus. J. Cell Sci. 2005;118:4655–4665. doi: 10.1242/jcs.02585. [DOI] [PubMed] [Google Scholar]
- Wilson N. F., Lefebvre P. A. Regulation of flagellar assembly by glycogen synthase kinase 3 in Chlamydomonas reinhardtii. Eukaryot. Cell. 2004;3:1307–1319. doi: 10.1128/EC.3.5.1307-1319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman G. B., Carlson K., Berliner J., Rosenbaum J. L. Chlamydomonas flagella. I. Isolation and electrophoretic analysis of microtubules, matrix, membranes, and mastigonemes. J. Cell Biol. 1972;54:507–539. doi: 10.1083/jcb.54.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman G. B., Plummer J., Sander G. Chlamydomonas flagellar mutants lacking radial spokes and central tubules. Structure, composition, and function of specific axonemal components. J. Cell Biol. 1978;76:729–747. doi: 10.1083/jcb.76.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia L., Hai B., Gao Y., Burnette D., Thazhath R., Duan J., Bre M. H., Levilliers N., Gorovsky M. A., Gaertig J. Polyglycylation of tubulin is essential and affects cell motility and division in Tetrahymena thermophila. J. Cell Biol. 2000;149:1097–1106. doi: 10.1083/jcb.149.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]