Abstract
Autophagy is a diverse family of processes that transport cytoplasm and organelles into the lysosome/vacuole lumen for degradation. During macroautophagy cargo is packaged in autophagosomes that fuse with the lysosome/vacuole. During microautophagy cargo is directly engulfed by the lysosome/vacuole membrane. Piecemeal microautophagy of the nucleus (PMN) occurs in Saccharomyces cerevisiae at nucleus-vacuole (NV) junctions and results in the pinching-off and release into the vacuole of nonessential portions of the nucleus. Previous studies concluded macroautophagy ATG genes are not absolutely required for PMN. Here we report using two biochemical assays that PMN is efficiently inhibited in atg mutant cells: PMN blebs are produced, but vesicles are rarely released into the vacuole lumen. Electron microscopy of arrested PMN structures in atg7, atg8, and atg9 mutant cells suggests that NV-junction–associated micronuclei may normally be released from the nucleus before their complete enclosure by the vacuole membrane. In this regard PMN is similar to the microautophagy of peroxisomes (micropexophagy), where the side of the peroxisome opposite the engulfing vacuole is capped by a structure called the “micropexophagy-specific membrane apparatus” (MIPA). The MIPA contains Atg proteins and facilitates terminal enclosure and fusion steps. PMN does not require the complete vacuole homotypic fusion genes. We conclude that a spectrum of ATG genes is required for the terminal vacuole enclosure and fusion stages of PMN.
INTRODUCTION
Eukaryotic cells are populated with numerous organelles and vesicles whose numbers increase or decrease, often dramatically, in response to specific physiological cues. Organelle biogenesis is balanced by turnover even in the steady state. In general, organelles are turned-over in the lysosome/vacuole by either macroautophagy or microautophagy. Macroautophagy begins with the enclosure of cytoplasm within a double membrane vesicle called the autophagosome. Bulk cytosol is often captured in autophagosomes alone or together with organelles. Fusion of the outer autophagosome membrane with the lysosome/vacuole membrane releases an autophagic body surrounded by the inner autophagosome membrane into the lumen. In contrast, organelles degraded by microautophagy are directly engulfed or encircled by the lysosome/vacuole membrane. Inward fusion of the vacuole membrane releases a microautophagic vesicle into the lumen that is surrounded by vacuole membrane.
Autophagic processes, including macroautophagy and chaperone mediated autophagy, are now understood to play important roles in major human diseases (Rubinsztein et al., 2007; Levine and Kroemer, 2008). This progress has been spurred by enormous recent progress on the identification and characterization of the factors that mediate these processes. In yeast, 31 autophagy (ATG) genes mediate macroautophagy (Klionsky et al., 2003), and most of them are conserved in mammals. Virtually all of the Atg proteins that are essential for macroautophagy localize to the preautophagosomal structure (PAS), which is a platform for the biogenesis of autophagosomes (Suzuki et al., 2007; Xie and Klionsky, 2007). In yeast a specialized form of macroautophagy called the Cvt pathway selectively targets the hydrolase proaminopeptidase I to the vacuole (Xie and Klionsky, 2007). The Cvt pathway is active under nutrient-rich conditions and shares a core set of Atg proteins with macroautophagy. Both the Cvt pathway and macroautophagy require additional process-specific Atg proteins.
Microautophagy is best characterized in the methylotrophic yeast Pichia pastoris (Farre and Subramani, 2004; Dunn et al., 2005; Sakai et al., 2006). A shift of these cells from methanol to glucose induces the selective microautophagic degradation of peroxisomes. Though macroautophagy and micropexophagy are morphologically very different processes, both depend on the same set of core Atg proteins. Micropexophagy requires an additional set of specific Atg proteins. A possible explanation for the use of a common machinery for macro- and microautophagy comes from fluorescence microscopic studies on micropexophagy, which point to the predominant localization of Atg proteins at two sites. First the perivacuolar structure, which might be analogous to the PAS and is thought to play a role in the formation of the sequestering vacuolar membrane (Chang et al., 2005), and second the micropexophagy-specific membrane apparatus (MIPA; Mukaiyama et al., 2004). The MIPA, a double membrane cap-like structure, is deposited at the far end of the encircled peroxisome, where it probably mediates the final vacuole membrane fusion event leading to the release of the microautophagic vesicle into the vacuole.
Micropexophagy has not been observed in Saccharomyces cerevisiae so far; however, a unique microautophagic process in this yeast mediates the pinching-off and degradation of nonessential portions of the nucleus. Piecemeal microautophagy of the nucleus (PMN) occurs at Velcro-like nucleus-vacuole (NV) junctions formed by interactions between phalanxes of Vac8 in the vacuole membrane and Nvj1 in the outer nuclear membrane (Pan et al., 2000; Roberts et al., 2003). During PMN a portion of the nucleus is extruded along the NV junctions into an invagination of the vacuolar membrane, forming a tethered bleb. Scission of the ER and fusion of the vacuolar membrane then releases a PMN-vesicle into the vacuole lumen, where it is degraded by resident hydrolases (Kvam and Goldfarb, 2007). The intravacuolar PMN vesicles are limited by three membrane layers. The outer membrane is derived from the vacuolar membrane, and the two inner layers are from the nuclear envelope. PMN is induced by nutrient depletion and degrades nonessential nuclear components. Nvj1 recruits at least two additional proteins to the NV junctions. Tsc13, an enoyl-CoA reductase needed for the synthesis of very-long-chain fatty acids (Kvam et al., 2005) and Osh1, which is homologous to mammalian oxysterol-binding protein (OSBP) and may function in nonvesicular lipid trafficking (Kvam and Goldfarb, 2004).
PMN has been categorized as a microautophagic process based on the morphological distinction that the nuclear bleb is directly engulfed by the vacuole rather than being initially packaged in autophagosome-like vesicles (Kvam and Goldfarb, 2007). The mechanisms governing the growth of PMN blebs and their release into the lumen as PMN vesicles are not known. We now show using two independent assays that the efficient production of PMN vesicles depends on the core Atg proteins and some subtype specific Atg proteins. PMN does not require the homotypic vacuole fusion machinery. In S. cerevisiae a distinct microautophagy-like process involves the formation of “autophagic tubes” (Muller et al., 2000). We report that PMN requires a set of proteins that is distinct from those needed for microautophagy via autophagic tube formation and a second, possibly distinct, microautophagy-like process that occurs during the recovery of cells from rapamycin treatment (Dubouloz et al., 2005). Significantly, an electron microscopic analysis of arrested PMN structures in atg mutant cells suggests that release of a micronuclear vesicle occurs before fusion of the vacuole membrane. Therefore, the latter stages of PMN vesicle formation in S. cerevisiae and micropexophagic vesicle formation in P. pastoris may occur by mechanistically similar pathways.
MATERIALS AND METHODS
Strains, Media, and Growth Conditions
Standard media were used according to Ausubel et al. (1987). Cells were starved in SD(-N) medium (0.17% yeast nitrogen base without amino acids and ammonium sulfate, 2% glucose).
Antibodies.
Horseradish peroxidase (HRP)-conjugated goat anti-rabbit (Medac, Hamburg, Germany); HRP-conjugated goat anti-mouse (Dianova, Hamburg, Germany); anti-proaminopeptidase I (Barth et al., 2002); green fluorescent protein (GFP) antibodies (Roche, Mannheim, Germany).
Chemicals.
PMSF (Sigma, Deisenhofen, Germany); oligonucleotides (Operon Technologies, Cologne, Germany), other analytical chemicals were from Sigma or Merck (Darmstadt, Germany). The ECL Plus detection kit (Amersham, Braunschweig, Germany) and a Fuji LAS-3000 imaging system (Tokyo, Japan) was used for immunoblots.
Strains.
Strains used in this study were derived from the wild-type (WT) strain WCG4a Mata ura3 his3-11,15 leu2-3112 (Thumm et al., 1994). Deletions from the WCG4a were created by using PCR-based knockout strategies (Güldener et al., 1996) and confirmed by Southern blotting or PCR: YYW07 WCG4a atg7Δ::HIS3; YYW08 WCG4a nvj1Δ::HIS3; YYW09 WCG4a vac8Δ::HIS3; YYW10 WCG4a atg11Δ::HIS3; and YRK897 atg29Δ::NATNT2; YRK899 atg31Δ::NATNT2. As indicated commercially available yeast deletion strains in the background BY4741 Mata his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 (Euroscarf Collection, Frankfurt, Germany) were used. RSY269 Matα sec17-1 ura3-52 his4-619 and RSY 272 Mata sec18-1 ura3-52 his4-619 are described in Kaiser and Schekman (1990).
Plasmids
The plasmid for the constitutive expression of GFP-Osh1 (pRS416-GFP-Osh1) is described in Kvam and Goldfarb (2004), those for inducible expression of NVJ1-EGFP under CUP1 promoter control (pCUP1-NVJ1-EGFP) in Pan et al. (2000). The centromeric plasmid pGFP-Atg8 (Suzuki et al., 2001) carries a GFP-Atg8 fusion under control of the native Atg8 promoter. For expression of mcherry-NLS the plasmid pNab2-NLS (TPI1::Nab2NLS-2mcherry) obtained from B. Timney and M. Rout (Rockefeller University, New York) was used. The plasmid used for the overexpression of Nvj1p was pRS316 with MET25 promoter and EGFP tag down stream of NVJ1.
Degradation Analysis of GFP-Osh1 and Nvj1-GFP
Cells expressing GFP-Osh1 were grown in SC-medium without uracil to the stationary growth phase, harvested, washed with SD(-N) medium, and then shaken at 30°C in SD(-N) medium. Two OD units of cell culture were harvested after 0, 2, 4, 6, and 8 h of starvation, respectively. Protein extracts were prepared by TCA precipitation, and samples were separated by SDS-PAGE and analyzed by immunoblotting with antibodies against GFP after wet-blotting overnight at 4°C onto PVDF membranes.
Cells expressing Nvj1-GFP were grown in SC-medium without uracil to exponential phase, and then 5 μm Cu2+ was added to the medium and the cells were shaken at 30°C for additional 2 h. The cells were then processed as described above.
Fluorescence Microscopy
Cells grown to stationary phase were washed with SD(-N) medium, then shifted to SD(-N) medium with or without 1 mM PMSF for the indicated times, and then visualized with a Zeiss Axioscope2 microscope and an Axiocam digital camera (Thornwood, NY). For nuclear staining 2 ml of the cell culture was incubated for 15 min at 30°C with 0.5 μl of 81 mM Hoechst 33342 before shift to SD(-N) medium. The pictures were pseudocolorized.
Electron Microscopy
For electron microscopy the cells were fixed with permanganate and embedded in Epon as described previously (Epple et al., 2003). A Jeol JEM1200EX-II transmission electron microscope (Tokyo, Japan) was used to take photographs.
RESULTS
PMN Throughput Can Be Monitored after the Vacuolar Breakdown of GFP-Osh1 or Nvj1-GFP
During PMN a section of the NV junction is targeted to the vacuole lumen and degraded. Because the components of the NV junction are concentrated within PMN vesicles, their degradation provides a good measure of PMN throughput. Epitope-tagged Nvj1 reporters have been used to monitor PMN rates; however, tagging of the NV junction apparatus could disturb the normal process. We have developed the use of GFP-Osh1 as a PMN cargo. Osh1 is not required for normal PMN (Kvam and Goldfarb, 2004), and its degradation in the vacuole results in the release of free GFP, which accumulates because it is resistant to vacuolar hydrolases. This strategy also circumvents the potential contribution of nonvacuolar proteases to the turnover of NV junction–associated proteins, which may be increased in mutant backgrounds. The accumulation of free GFP in the vacuole lumen has been used to monitor the degradation of GFP-Atg8 by macroautophagy (Meiling-Wesse et al., 2002). By analogy, the rate of PMN can be assessed by monitoring the accumulation of free GFP derived from the degradation of GFP-Osh1. The accumulation of small amounts of free GFP is easier to detect than the loss by degradation of a small percentage of the total GFP-Osh1 pool.
The selectivity of GFP-Osh1 degradation by PMN was assessed in various mutants after the induction of PMN by nitrogen starvation. To induce PMN we shifted S. cerevisiae cells expressing GFP-Osh1 to nitrogen-free SD-(N) medium and after the indicated times analyzed cell lysates by Western blotting with antibodies to GFP. As shown in Figure 1A, free GFP accumulates in WT cells within 2 h of starvation. As expected, GFP accumulation is largely dependent on the NV junction apparatus proteins Nvj1 and Vac8. Only minor amounts of GFP (quantification of the blots showed 10–20% of WT levels) were detected in nvj1Δ and vac8Δ cells (Figure 1A). GFP-Osh1 degradation was also dependent on the vacuolar proteinase A (Pep4) and Atg15. These results indicate that the degradation of GFP-Osh1 in starved cells occurs primarily in the vacuole lumen. We conclude that Atg15, a putative lipase required for the lysis of autophagic bodies (Epple et al., 2001), which are surrounded by the inner autophagosome membrane, is similarly used in the degradation of PMN-derived microautophagic vesicles, which are surrounded by vacuole membrane.
Figure 1.
Determination of the PMN rate by Western blots. As detailed in the text GFP is rather resistant against vacuolar proteolysis. The level of GFP generated by breakdown of the NV junction proteins GFP-Osh1 (A) or Nvj1-GFP (B and C) therefore corresponds to the PMN rate. Cells of the stationary growth phase were shifted to nitrogen-free SD(-N) medium, at the indicated times aliquots were taken, and crude extracts were prepared. The samples were then separated by SDS-PAGE and after electroblotting onto PVDF membranes were probed with antibodies against GFP and a secondary antibody coupled to peroxidase. Chemiluminescence was detected with a Fuji LAS3000. The absence of free GFP in cells lacking vacuolar proteinase A (pep4Δ) confirms the vacuolar origin of GFP. As a loading control the blots were reprobed with antibodies to cytosolic 3-phosphoglycerate kinase (PGK). (B) Nvj1-GFP is expressed under control of the copper-inducible CUP1 promotor. The expression level was checked in the presence of increasing copper concentrations. (C) Expression of CUP1::Nvj1-GFP was induced for 2 h with 5 μM Cu2+, and then the cells were shifted to starvation medium and treated as above.
An important question is whether the core ATG genes are required for PMN. It has been previously reported that some free PMN blebs and vesicles containing Nvj1-GFP, GFP-Osh1, and Tsc13-GFP were observed in atg7Δ cells (Kvam and Goldfarb, 2007). Atg7 is absolutely required for macroautophagy in S. cerevisiae and micropexophagy in P. pastoris (Xie and Klionsky, 2007). In addition, degradation of epitope-tagged Nvj1 was observed at nearly normal levels in atg7Δ cells by Western blot (Kvam and Goldfarb, 2004). These observations led to the conclusion that the core ATG genes are not absolutely required for PMN. However, the number of PMN vesicles in atg7Δ cells was greatly reduced, so it remained a possibility that ATG genes are, in fact, required in vivo for normal PMN. Therefore, we next analyzed GFP-Osh1 degradation in atg7Δ cells. Within our detection limit no free GFP was formed in these cells during starvation (Figure 1A), indicating that the core ATG apparatus is required for efficient PMN. The virtually complete absence of free GFP in starved atg7Δ cells suggests that the small amount of degradation we observed in nvj1Δ and vac8Δ cells is likely due to nonselective macroautophagy.
To corroborate these results using a different reporter, we used Nvj1-GFP expressed with the copper-inducible CUP1-promotor as an independent reporter. We checked Nvj1-GFP levels after 1 and 2 h in the presence of increasing Cu2+ concentrations and found that a 2-h incubation with 5 μM Cu2+ provided adequate levels of Nvj1-GFP (Figure 1B). The use of Nvj1-GFP confirmed our findings with Osh1-GFP that Vac8 and Atg7 are required for efficient PMN (Figure 1C).
Efficient PMN Requires a Full Battery of Core Atg Proteins
We extended the genetic analysis of PMN to include other ATG genes. Two ubiquitin-like conjugation systems are essential for the biogenesis of autophagosomes and Cvt vesicles. One system consisting of the E1-like Atg7 and the E2-like Atg10 covalently links Atg12 to Atg5. The Atg12–Atg5 conjugate then oligomerizes together with Atg16 (Suzuki and Ohsumi, 2007; Xie and Klionsky, 2007; Figure 2A). Atg8 is first processed at its carboxy-terminus by the cysteine-proteinase Atg4 and is then covalently coupled to phosphatidylethanolamine via Atg7 and the E2-like Atg3 (Suzuki and Ohsumi, 2007; Xie and Klionsky, 2007; Figure 2A). As shown in Figure 2A we followed by Western blot the accumulation of free GFP from GFP-Osh1 in cells chromosomally deleted for these ATG genes. We found that the core autophagy genes ATG3, 4, 5, 7, 10, 12, and 16 are all required for efficient degradation of GFP-Osh1 (Figure 2A; Table 1). Also extended starvation for 24 h did not lead to the generation of free GFP in atg7Δ or atg15Δ cells (Figure 2A). To further confirm the requirement of ATG genes, we used WT and mutant strains of two different genetic backgrounds. PMN-mediated degradation levels were consistently higher in WT cells of our standard lab background WCG compared with the commercially available BY-background (Figure 2A). Because of the large number of mutant strains analyzed, the experiments were done in batches, each time including the appropriate WT and vac8Δ strain.
Figure 2.
Efficient PMN requires the core autophagic machinery, consisting of (A) the two ubiquitin-like conjugation systems, coupling Atg12 to Atg5 and Atg8 to phosphatidylethanolamine; (B) the Atg9 cycling system except for Atg23 and Atg27; and (C) the phosphatidylinositol 3-kinase complex I. PMN further requires Atg1, Atg13, and most of the accessory Atg proteins (D). Atg17, Atg29, and Atg31 are required only for unselective macroautophagy, but not the selective Cvt pathway. Atg11 and the sorting nexins Atg20/Snx42 and Atg24/Snx4 are specifically required for the Cvt pathway. Please note that not all interactions in the drawing must take place simultaneously. (E) Atg21 and the proaminopeptidase I-receptor Atg19 are Cvt pathway–specific components. The function of the sterol glucosyltransferase Atg26 is restricted to pexophagy in P. pastoris. Further details are given in the text. Cells chromosomally deleted for the indicated genes and expressing GFP-Osh1 were starved in SD(-N) medium, and the increase in free GFP levels was monitored in Western blots as described in Figure 1. Cytosolic 3-phosphoglycerate kinase (PGK) was detected as a loading control. As noted cells from two different strain backgrounds (BY4741 or WCG) were analyzed.
Table 1.
Requirements for efficient PMN
| Required for efficient PMN | |
|---|---|
| Autophagy proteins | |
| Ubi-like conjugation systems | |
| Atg3 | Yes |
| Atg4 | Yes |
| Atg5 | Yes |
| Atg7 | Yes |
| Atg8 | Yes |
| Atg10 | Yes |
| Atg12 | Yes |
| Atg16 | Yes |
| Atg9-cycling system | |
| Atg1 | Yes |
| Atg2 | Yes |
| Atg9 | Yes |
| Atg13 | Yes |
| Atg18 | Yes |
| Atg23 | No |
| Atg27 | No |
| PtdIns 3-kinase complexes | |
| Atg14 | Yes |
| Vps30/Atg6 | Yes |
| Vps38 | No |
| PtdIns(3)P5-kinase | |
| Fab1 | No |
| Macroautophagy-specific proteins | |
| Atg17 | Yes |
| Atg29 | Yes |
| Atg31 | Yes |
| Lysis of autophagic bodies | |
| Atg15 | Yes |
| Cvt-specific proteins | |
| Atg11 | Yes |
| Atg19 | No |
| Atg20 | No |
| Atg21 | Yesa |
| Atg24 | Yesa |
| Atg26 | No |
| Trs85 | Yes |
| Vps45 | No |
| Vps51 | No |
| Vps52 | No |
| Vps53 | No |
| Tlg2 | No |
| Homotypic vacuole fusion | |
| Sec17 | Yes |
| Sec18 | Yes |
| Vam7 | Yes |
| Vam3 | Yes |
| Nyv1 | No |
| Ypt7 | Yes |
| Vps33 | Yes |
| Vps39 | Yes |
| Vps41 | Yes |
| Vtc1 | No |
| Vtc2 | No |
| Vtc3 | No |
| Vtc4 | No |
| Vph1 | No |
| Pbi2 | No |
| Vma2 | No |
| Recovery from rapamycin treatment | |
| Ego1 | No |
| Ego3 | No |
| ER-phagy induction | |
| Ire1 | No |
| Hac1 | No |
a PMN is significantly reduced.
A second set of ATG genes mediates cycling of the membrane protein Atg9 between the PAS and a peripheral pool at or near mitochondria (Reggiori et al., 2005). Atg9 remains restricted to the PAS in atg1, 2, 13, and 18 mutant cells (Reggiori et al., 2004). All of these factors are essential for both macroautophagy and the Cvt pathway. We found that all these factors are also required for the degradation of GFP-Osh1p in starved cells (Figure 2B). The retrieval of Atg9 from the peripheral pool back to the PAS depends on Atg23 and Atg27. Both proteins are essential for the Cvt pathway, but macroautophagy proceeds in their absence, albeit at a somewhat lower rate (Xie and Klionsky, 2007). GFP-Osh1 degradation is not blocked in either atg23Δ or atg27Δ cells, which indicates that PMN is more similar in this regard to macroautophagy than to the Cvt pathway (Figure 2B).
Macroautophagy depends on phosphatidylinositol-3-phosphate, which in S. cerevisiae is generated by two protein complexes that share Vps15, Vps30, and Vps34 (Figure 2C). The Atg14-containing complex I is essential for macroautophagy and the Cvt pathway, whereas the Vps38-containing complex II functions in vacuole biogenesis (Obara et al., 2006). VPS38 was not required for GFP-Osh1 breakdown. However, no degradation was detected in atg14Δ and vps30Δ cells (Figure 2C). These results further support our finding that efficient PMN depends on factors that are required for both the Cvt pathway and macroautophagy. We also checked whether PtdIns(3,5)P2 is required for PMN. In S. cerevisiae PtdIns(3,5)P2 is generated by Fab1. As shown in Figure 2C GFP-Osh1 degradation was normal in fab1Δ cells, indicating that PtdIns(3,5)P2, per se, is not required for PMN.
A Subset of Cvt- and Macroautophagy-specific Atg Proteins Is Important for PMN
Atg17 and its interacting proteins Atg29 and Atg31 are needed for normal macroautophagy, but are dispensable for the Cvt pathway (Kawamata et al., 2005; Kabeya et al., 2007). Atg17 further interacts with Atg13, a regulator of the serine/threonine kinase Atg1 and acts as a basal scaffold for the organization of the PAS during starvation-induced macroautophagy (Suzuki and Ohsumi, 2007; Xie and Klionsky, 2007; Figure 2D). Consistent with its important role in PAS organization, Atg17 was found to be essential for normal PMN levels (Figure 2D). It has been reported that the autophagic defects of atg29Δ cells are strongly dependent on the genetic background (Kawamata et al., 2005). We independently deleted ATG29 and ATG31, in our lab background WCG (see Materials and Methods). As a control, we monitored in atg29Δ and atg31Δ cells the starvation-induced breakdown of GFP-Atg8, an established procedure to quantify the autophagic rate. GFP-Atg8 breakdown was strongly affected in both atg29Δ and atg31Δ cells (Supplemental Figure S1). As shown in Figure 2D, PMN is also severely affected in atg29Δ and atg31Δ cells.
Some ATG genes are required for the Cvt pathway, but have only mild effects on macroautophagy when deleted. One of these genes, ATG11, plays a central role in the Cvt pathway in S. cerevisiae and in selective macro- and micropexophagy in P. pastoris (Farre and Subramani, 2004; Dunn et al., 2005; Sakai et al., 2006; Xie and Klionsky, 2007). During the Cvt pathway the cargo proaminopeptidase I assembles together with its receptor Atg19 into the Cvt complex, which is then linked via Atg11 to the vesicle-forming machinery at the PAS. Atg11 further serves as an organizing scaffold at the PAS during the Cvt pathway, (Figure 2D) and is thought to play similar functions during macro- and micropexophagy in P. pastoris (Shintani and Klionsky, 2004; Yorimitsu and Klionsky, 2005). As shown in Figure 2D Atg11 is also required for normal PMN levels.
The sorting nexin Atg24/Snx4 (Figure 2D) functions in retrograde transport from the early endosome to the late Golgi (Hettema et al., 2003) and is also required for the Cvt pathway and macro- and micropexophagy in P. pastoris. Atg24/Snx4 probably mediates the fusion between the MIPA and the vacuolar membrane during micropexophagy and fusion of pexophagosomes with the vacuole during macropexophagy (Ano et al., 2005; Sakai et al., 2006). Our experiments showed that Atg24 is required for normal operation of PMN (Figure 2D). S. cerevisiae Atg24 has been shown to interact with Atg20/Snx42, another sorting nexin specifically involved in the Cvt pathway (Nice et al., 2002; Yorimitsu and Klionsky, 2005). atg20Δ cells showed no reduced PMN rate (Figure 2D), suggesting that Atg20 may be exclusively required for the Cvt pathway. As expected, the lack of Snx41, another member of the sorting nexin family, which is not required for the Cvt pathway or other specific autophagic processes, did not affect vacuolar degradation of GFP-Osh1 (Figure 2D).
Atg21 is a phosphatidylinositol-3-phosphate binding protein that is required for the Cvt pathway but not macroautophagy (Dove et al., 2004; Meiling-Wesse et al., 2004; Stromhaug et al., 2004; Krick et al., 2006). As shown in Figure 2E, GFP-Osh1 degradation is significantly reduced but not blocked in atg21Δ cells. Atg26/Ugt51, a sterol glucosyltransferase, is required for micro- and macropexophagy in P. pastoris, but in S. cerevisiae it is required neither for the Cvt pathway nor macroautophagy (Oku et al., 2003; Cao and Klionsky, 2007; Nazarko et al., 2007). Consistently, we found that Atg26 is dispensable for PMN (Figure 2E). Also Atg19 the receptor of proaminopeptidase I in the Cvt pathway (Xie and Klionsky, 2007) is not needed for PMN (Figure 2E).
Intravacuolar PMN Vesicles Can Be Visualized in Fluorescence Microscopy with a NLS-mcherry Fusion Protein
The results described above indicate that the core macroautophagic apparatus and a number of Cvt pathway factors are required for the proper execution of PMN. To test this, we sought to directly visualize the targeting of PMN cargo in various mutant strains. We used a NLS-mcherry reporter consisting of the nuclear localization sequence of Nab2 fused to a tandem repeat of the red fluorescent mcherry protein (Materials and Methods). This brightly fluorescent reporter localizes strongly to the nucleus and is readily monitored by fluorescence microscopy (Figure 3, A–D). Wild-type and atg15Δ cells expressing NLS-mcherry were starved for 4 h in nitrogen-free medium to induce PMN and then checked by fluorescence microscopy. atg15Δ cells accumulated red fluorescent, free-floating vesicles in their vacuoles (see Supplemental Video 1 and Figure 3A), which were mostly absent in WT cells (not shown). This result is consistent with the proposed role of Atg15 in the lysis of intravacuolar autophagic bodies (Epple et al., 2003). Treatment of WT cells with the proteinase B inhibitor PMSF led to the accumulation of similar red fluorescent vesicles in the vacuole lumen (Figure 3D).
Figure 3.
Detection of intravacuolar free-floating PMN vesicles by fluorescence microscopy. To trace the nuclear content of PMN vesicles a NLS-mcherry fusion protein consisting of the nuclear localization sequence of Nab2 and a tandem repeat of the fluorescent mcherry protein was used. (A) The lipase-like Atg15 protein is required for intravacuolar lysis of autophagic bodies. atg15Δ cells expressing NLS-mcherry were starved for 4 h in SD(-N) and examined with a Zeiss Axioscope2 microscope (see also Supplemental Video 1). The red fluorescent vesicles were absent in atg1Δ atg15Δ cells (A and Supplemental Video 2); bar, 5 μm. atg15Δ cells expressing NLS-mcherry and the autophagosomal marker GFP-Atg8 (B) or the cytosolic marker 3-phosphoglycerate kinase (PGK; C) showed after 4-h starvation in SD-(N) medium significantly more green than red fluorescent intravacuolar vesicles. This suggests that the red fluorescent PMN vesicles, which contain nuclear material are distinct from autophagic bodies. The images shown in B and C were taken simultaneously with a Leica TCS SP2 AOBS confocal laser scan microscope. (D) The indicated mutant cells coexpressing mcherry-NLS and the autophagic marker GFP-Atg8 were starved for 4 h in SD(-N) medium in the presence of 1 mM of the proteinase B inhibitor PMSF. The accumulation of red fluorescent PMN vesicles is in accordance with the Western blot analysis of PMN. The absence of intravacuolar PMN vesicles in vac8Δ cells, with the synchronous appearance of GFP-Atg8 in their vacuoles further confirms that PMN vesicles are distinct from autophagic bodies. Bar, 10 μm.
The absence of intravacuolar NLS-mcherry containing vesicles in atg1Δ atg15Δ double mutant cells confirmed that their biogenesis depends on the autophagic machinery (Supplemental Video 2 and Figure 3A). The autophagosomal marker GFP-Atg8 was simultaneously expressed in these cells to confirm that the red fluorescent vesicles were, in fact, not autophagic bodies. As shown in Figure 3B the more numerous GFP-Atg8-containing vesicles did not colocalize with NLS-mcherry-containing vesicles. As an additional marker for autophagic bodies, we expressed a GFP-tagged version of cytosolic 3-phosphoglycerate kinase (PGK-GFP) under the control of its endogenous promotor on a centromeric plasmid. atg15Δ cells coexpressing PGK-GFP, and NLS-mcherry were starved 4 h in nitrogen-free medium and then analyzed by confocal fluorescence microcopy. As expected, more green fluorescent autophagic bodies were observed within the vacuoles of these cells than red-fluorescent PMN vesicles (Figure 3C).
Vac8 is essential for PMN, but not for macroautophagy. Indeed, in the vacuoles of starved PMSF-treated vac8Δ cells GFP-Atg8-tagged autophagic bodies accumulated, but no NLS-mcherry–containing vesicles (Figure 3D). Taken together, these results indicate that NLS-mcherry is a useful reporter to monitor PMN by fluorescence microscopy. As shown in Figure 3D the analysis of a set of autophagy-deficient mutants using NLS-mcherry fully confirmed the findings of the Western blot analysis using GFP-Osh1 and Nvj1-GFP reported above. All these results are consistent with the hypothesis that the biogenesis of PMN vesicles depends on the core autophagic machinery as well as additional specific Atg-components such as Atg11 and Atg17.
Requirement of the TRAPP and GARP/VFT-tethering Complexes for PMN
The TRAPPI-tethering complex functions in ER-to-Golgi traffic, whereas TRAPPII is implicated in intra-Golgi traffic and in sorting from the endosome to the late Golgi (Cai et al., 2007). Trs33, Trs65, and Trs85 are subunits of both TRAPP-tethering complexes, and unlike other TRAPP subunits they are not essential for viability of yeast cells. Trs85, but not Trs33 or Trs65, is essential for the biogenesis of Cvt vesicles and for efficient pexophagy in S. cerevisiae (Meiling-Wesse et al., 2005). Similarly, we found that only Trs85, but not Trs33 or Trs65, is required for efficient PMN (Figure 4A).
Figure 4.
Requirement of Cvt pathway specific factors in PMN. Cells expressing GFP-Osh1 were starved in SD(-N) medium, and aliquots taken at the indicated times were analyzed in Western blotting as detailed in Figure 1. The blots were reprobed with antibodies to 3-phosphoglycerate kinase (PGK) as a loading control. (A) Trs33, Trs65 and Trs85 are part of the TRAPP tethering complexes. Only Trs85 has a Cvt-specific function and as shown is also needed for PMN. As an additional control the blots were reprobed with antibodies to aminopeptidase I. proApeI, proaminopeptidase I; mApeI, mature aminopeptidase I. (B) The GARP/VFT-tethering complex consisting of Vps51, Vps52, Vps53, and Vps54, the SNARE Tlg2 and the SM (Sec1/Munc18 related) protein Vps45 are specifically needed for biogenesis of Cvt-vesicles. PMN does not depend on these proteins.
The GARP/VFT-complex consisting of Vps51, Vps52, Vps53, and Vps54, is another tethering complex that acts together with the Qa-SNARE (soluble NSF [N-ethylmaleimide-sensitive factor]) attachment protein Tlg2 and the SM (Sec1/Munc18 related) protein Vps45 in the retrieval of proteins from endosomes to the late Golgi (Cai et al., 2007). All six proteins are essential for the formation of Cvt-vesicles but not autophagosomes (Abeliovich et al., 1999; Reggiori et al., 2003; Reggiori and Klionsky, 2006). For cells lacking one of the GARP/VFT subunits, a defect in Atg9 cycling and a defect in PAS targeting of proaminopeptidase I, Atg19, and Atg11 was reported, suggesting a function in cargo recruitment during the Cvt pathway (Reggiori et al., 2003; Reggiori and Klionsky, 2006). Consistent with this specialized role, none of the components of the GARP/VFT complex nor Tlg2 or Vps45 were needed for normal PMN levels (Figure 4B).
The Standard Homotypic Vacuole Fusion Apparatus Is Not Required for the Biogenesis of PMN Vesicles
PMN requires as a final step the fusion of the engulfing vacuolar membrane to release a PMN vesicle into the vacuole lumen (Figure 10). Homotypic vacuolar fusion in S. cerevisiae has been studied in-depth genetically and by in vitro reconstitution (Wickner, 2002; Ostrowicz et al., 2008) and is divided into distinct stages. During priming cis-SNARE complexes are disassembled by the AAA-ATPase Sec18, an NSF homologue, and Sec17/α-SNAP. Tethering is mediated by the HOPS complex, consisting of Vps11/Pep5, Vps16, Vps18/Pep3, Vps33, Vps41, and Vam6/Vps39, together with the small Rab GTPase Ypt7. In the final docking and fusion step trans-SNARE complexes are formed most likely between the Q-SNAREs Vam3, Vam7, and Vti1 on one membrane and the R-SNARE Nyv1 on the other membrane. A number of additional factors are required for homotypic vacuole fusion. The LMA1 complex composed of the proteinase B inhibitor Pbi1 and thioredoxin most likely binds via Sec18 to Vam3 and probably stabilizes the activated state of the cis-SNARE complexes (Ostrowicz et al., 2008). Members of the vacuolar transport chaperone (VTC) family are also required for homotypic vacuolar fusion. Vtc proteins associate with the SNARE Nyv1 and the V0-part of the vacuolar H+-ATPase. Vtc1 and Vtc4 are involved in priming of SNAREs and HOPS and LMA1 recruitment (Muller et al., 2002). Vtc3 is necessary for fusion and LMA1 release (Muller et al., 2002). The vacuolar H+-ATPase consists of integral membrane V0 and peripheral V1 subunits. Surprisingly, the V0 part has a specific function in fusion, and it was shown that the Vo subunit Vph1 must be present on both fusing vacuoles (Bayer et al., 2003). To learn more about the molecular mechanism of PMN, we checked the relevance of the components needed for homotypic vacuole fusion.
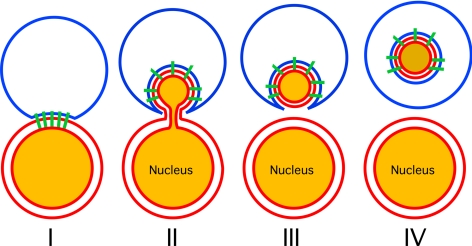
Figure 10.
Schematic illustration of PMN. In stage I nucleus-vacuole (NV) junctions form. Then the nuclear ER bulges into invaginations of the vacuolar membrane (stage II), followed by fission of an ER-deduced vesicle (stage III). After fusion at the tips of the vacuolar membrane extensions (stage IV), a PMN vesicle is released into the vacuolar lumen, where it is finally degraded.
Using the same approach described above, we investigated the possible role of these genes in PMN. Because Sec17 and Sec18 are essential for growth of S. cerevisiae cells, we used temperature-sensitive strains. As shown in Figure 5A (top panel) at the permissive temperature (23°C) breakdown of GFP-Osh1 proceeded normally in sec17ts and sec18ts mutant cells. However, GFP-Osh1 breakdown was significantly affected after shift to the nonpermissive temperature (34°C), indicating that these factors are required for normal PMN levels. As a loading control the Western blots were reprobed with antibodies to PGK. It has previously been shown that Sec17 and Sec18 are essential for autophagy and that Sec18 affects the fusion of autophagosomes with the vacuole, but not their biogenesis. To confirm the efficiency of the temperature-induced block of Sec17 and Sec18 functions in macroautophagy, we monitored in a simultaneous experiment the autophagic rate by following the breakdown of the macroautophagy marker GFP-Atg8 (Figure 5A, bottom panel).
Figure 5.
Involvement of the homotypic vacuolar fusion apparatus in PMN. (A) Top panel, temperature-sensitive sec17ts and sec18ts mutant cells expressing GFP-Osh1 were grown in YPD at 23°C, shifted to SD(-N) medium, and incubated at either 23 or 34°C. At the indicated times aliquots were withdrawn and processed for Western blots with antibodies to GFP as detailed in Figure 1. At the nonpermissive temperature (34°C) PMN is affected in sec17 and sec18 mutant cells. Bottom panel, as a control for the effective block of Sec17 and Sec18 function at the nonpermissive temperature in parallel cells expressing GFP-Atg8 were processed, showing the published block in macroautophagy (Ishihara et al., 2001). As a loading control the blots were reprobed with antibodies to 3-phosphoglycerate kinase (PGK). (B) The SNAREs Vam3 and Vam7, the HOPS tethering complex components Vps33, Vps39, and Vps41, and the small GTPase Ypt7, but not the SNARE Nyv1, are needed for PMN. Cells expressing GFP-Osh1 and chromosomally deleted for the listed genes were starved in SD(-N) medium, and aliquots taken at the indicated times were analyzed in Western blots as detailed in Figure 1. (C) The vacuolar transport chaperones Vtc1, Vtc2, Vtc3, and Vtc4, the LMA1 subunit Pbi2, and the vacuolar H+-ATPase subunits Vph1 and Vma2 are dispensable for PMN. Cells expressing GFP-Osh1 and chromosomally deleted for these genes were starved in SD(-N) medium, and aliquots were processed for Western blots as detailed in Figure 1.
As shown in Figure 5B, GFP-Osh1 degradation was affected in ypt7Δ cells, in cells lacking components of the HOPS-tethering complex (vps33Δ, vps41Δ, and vps39Δ) and in cells lacking the Q-SNAREs Vam3 or Vam7. The R-SNARE Nyv1 was not required, possibly because redundant SNAREs may substitute for its function in vivo (Ostrowicz et al., 2008). Finally, Vtc1, Vtc2, Vtc3, Vtc4, the LMA1 subunit Pbi2, the V0-subunit Vph1, or the V1-subunit Vma2 were not required for efficient PMN (Figure 5C). Because GFP-Osh1 breakdown takes place in the absence of these factors, we conclude that the vacuolar fusion step during PMN is distinct from classical homotypic fusion and instead requires an alternate machinery. We expect that the requirement of Sec17, Sec18, the HOPS complex, and the Rab-GTPase Ypt7 in PMN is due to the role of these components in vacuolar biogenesis.
Is PMN Related to Other Microautophagy-like Processes?
Other microautophagic processes have been described in yeast that may or may not share components with PMN. Ego1, Ego3, and the GTPase Gtr2 were identified as subunits of a protein complex required for exit from rapamycin-induced growth arrest (Dubouloz et al., 2005) and for trafficking of the general amino acid permease Gap1 out of the endosome (Gao and Kaiser, 2006). During recovery from rapamycin treatment this complex is also required for induction of a microautophagy-like process thought to compensate for the membrane influx to the vacuole (Dubouloz et al., 2005). We found that neither Ego1 nor Ego3 are needed for efficient PMN (Figure 6).
Figure 6.
PMN does not require Ego1 and Ego3, which are needed for a microautophagy-like process during recovery from rapamycin treatment. ego1Δ and ego3Δ cells expressing GFP-Osh1 were starved in SD(-N) medium, and samples taken at the indicated times subjected to Western blot analysis with antibodies to GFP. Reprobing of the blots with antibodies to 3-phosphoglycerate kinase (PGK) serves as a loading control.
Another microautophagy-like process in yeast involves the formation of large tubular invaginations (autophagic tubes) of the vacuolar membrane (Muller et al., 2000). This process may be required to assure the membrane homeostasis after induction of macroautophagy by recycling extra membrane through the vacuole lumen. In vitro reconstitution of microautophagic tube formation showed that it is independent of Sec17, Sec18, Vam3, Vam7, Ypt7, and Nyv1 (Sattler and Mayer, 2000), but is dependent on the Vtc protein complex (Uttenweiler et al., 2007). Autophagic tube formation can also proceed in the absence of Atg proteins, albeit at a somewhat reduced rate, which might be explained by the dependence of this process on the delivery of membrane components via macroautophagy. As shown in Figure 5, efficient PMN depends on Sec17, Sec18, Vam3, Vam7, and Ypt7, but not on Nyv1 and the Vtc protein complex. We therefore conclude that PMN is at least partially mechanistically distinct from these microautophagy-like processes in S. cerevisiae.
ER-Phagy But Not PMN Requires Ire1 and Hac1
Recently, it was reported that drug-induced ER-stress leads to the incorporation of parts of the ER into autophagosomes by a process called ER phagy. ER phagy depends on Ire1 and Hac1 (Bernales et al., 2006; Yorimitsu et al., 2006). The ER membrane protein Ire1 is activated by the accumulation of unfolded proteins within the ER and then splices Hac1 mRNA, which after translation induces the unfolded protein response. During PMN also parts of the nuclear ER are incorporated into the PMN vesicles. We therefore checked for a possible involvement of Ire1 and Hac1 in PMN. As shown in Figure 7 in cells lacking either Ire1 or Hac1 PMN still proceeds. We conclude that PMN and ER phagy mediate the breakdown of ER membranes by different mechanisms.
Figure 7.
Ire1 and Hac1 needed for induction of ER-Phagy are not required for PMN. ire1Δ and hac1Δ cells expressing GFP-Osh1 were starved for nitrogen and the generation of free GFP followed in Western blots. 3-Phosphoglycerate kinase (PGK) is a loading control.
NV Junctions Form Independent from Autophagy Proteins
To gain further insight into the role of the autophagic machinery in PMN, we determined at which step PMN is blocked in autophagy-deficient mutant cells. The formation of NV junctions can be visualized by fluorescence microscopy using the GFP-Osh1 reporter (Kvam and Goldfarb, 2004). We tallied the percentage of cells that exhibited an accumulation of GFP-Osh1 at the NV junctions. As shown in Figure 8, NV junctions (white arrowheads) were detectable in 49% of 538 WT cells and 5% of 409 nvj1Δ cells. As a representative of the core autophagic machinery, we analyzed the requirement for Atg18 in the formation of NV junctions. Because 45% of 371 atg18Δ cells showed NV junctions, we conclude that the autophagic machinery is not needed for their formation (Figure 8).
Figure 8.
Formation of nucleus-vacuole (NV) junctions does not depend on Atg18, a core autophagy protein. In fluorescence microscopy NV junctions can be detected by the accumulation of GFP-Osh1 in regions between the nucleus and the vacuolar membrane (white arrowheads). The frequency of cells with NV junctions was counted and expressed as percentage of the total number of cells. In atg18Δ cells the frequency of NV junctions was WT like. Bar, 10 μm. From bottom to top, Nomarski optics, merge, nuclear staining with Hoechst 33342, and GFP fluorescence.
Autophagy-deficient Mutant Cells Are Blocked in Formation of PMN Vesicles after Formation of Nuclear Envelope–derived Vesicles
We studied PMN in autophagy-deficient mutant cells using transmission electron microscopy (TEM) to investigate the morphological basis for the PMN defects. Autophagy-deficient mutant cells were starved for 4 h in nitrogen-free medium and prepared for TEM. Tethered blebs of the nuclear ER into invaginations of the vacuolar membrane were still formed in atg7Δ, atg8Δ, and atg9 mutant cells (Figure 9, C–E). A closer examination of these images suggests that nuclear envelope-derived vesicles had budded-off from NV junctions (black arrowheads). Remarkably, in these mutant cells the vacuolar extensions had not fused to release a PMN vesicle into the vacuole lumen (Figure 9, C–E, white arrowheads). With a diameter of ∼100–150 nm these structures are not well resolved by fluorescence microscopy. Previous studies showed that the size of PMN blebs depends on the Nvj1 expression levels (Roberts et al., 2003). We therefore analyzed the localization of the NLS-mcherry reporter in nvj1Δ and atg7Δ cells expressing Nvj1-GFP from the strong MET25 promotor in fluorescence microscopy. As shown in Figure 9F (left) after 4-h starvation in nitrogen-free medium, free-floating intravacuolar PMN vesicles of enlarged size were visible in nvj1Δ cells expressing Nvj1-GFP. In atg7Δ cells part of the nucleus appear sequestered into PMN bleb-like structures, which were surrounded by Nvj1-GFP. These PMN-like structures generally remained in close proximity to the NV junctions (Figure 9F, right, white arrowhead).
Figure 9.
Autophagy-deficient mutant cells are blocked in PMN after formation of ER-deduced vesicles. Wild type (WT), pep4Δ, atg7Δ, atg8Δ, and atg9 cells were starved for 4 h in SD(-N) medium, fixed with permanganate, and processed for transmission electron microscopy (A–E). Bar, 500 nm. Black arrowheads, ER-deduced vesicles; white arrowheads, nonfused vacuolar extensions. (F) nvj1Δ (left) and atg7Δ cells (right) of the logarithmic growth phase expressing Nvj1-GFP from the strong MET25 promotor and mcherry-NLS were grown 4 h in selective medium lacking methionine and then shifted for 3 h on SD-(N). Images were taken with a Leica TCS NT laser confocal microscope. PMN structures were captured by scanning the Z-axis of the cell for the section that yielded the largest surface area of colocalization between Nvj1p-GFP and mcherry-NLS.
Both TEM and fluorescence microscopic images are consistent with the hypothesis that these Atg proteins are required for final engulfment and vacuole fusion steps. In these atg mutants it appears that the formation of PMN blebs occurs normally, and proceeds through to a novel intermediate—the release of tiny micronuclei from the nucleus—before the process is arrested before vacuole fusion and the production of PMN vesicles. This supports the following hypothesis. PMN micronuclei are normally released first in order to allow the vacuole membrane to completely encircle the structure and fuse to produce a PMN vesicle into the lumen. Thus the Atg apparatus may function at a late step that is directly analogous to its role in the MIPA during micropexophagy in P. pastoris. Such a function is consistent with our finding that the homotypic vacuole fusion apparatus is not required for efficient PMN (see above).
DISCUSSION
PMN takes place through a series of morphologically distinct intermediates (Figure 10). It demands the formation of protrusions of the nuclear ER into the vacuole (stage II), and then a vesicle filled with nuclear material and covered by both membranes of the ER bud off (stage III). Finally, a fusion event at the vacuolar membrane releases a PMN vesicle into the vacuole (stage IV), where it is degraded dependent on the presence of vacuolar proteinase A.
To date, only components required for the formation of NV junctions have been well characterized (Kvam and Goldfarb, 2007), but the molecular machinery required for further stages of PMN remained elusive and its classification as a microautophagic process was based solely on morphological criteria. We here established two novel assays that allow us to measure PMN in various genetic backgrounds. The first assay uses a fluorescent NLS-mcherry fusion protein, which allows us to observe the targeting of nucleoplasm-containing blebs and vesicles into the vacuole. These vesicles were shown to accumulate in the vacuole lumen when vacuole hydrolases were inhibited by the proteinase B inhibitor PMSF (Figure 3). In a second assay we took advantage of the resistance of GFP against vacuolar hydrolases. Using Nvj1-GFP and GFP-Osh1 reporters, the rate of PMN-mediated proteolysis was followed by detecting by Western blot over time the accumulation of free GFP (Figure 1, 2). Both assays consistently showed the requirement of autophagic components for efficient PMN (Figures 1–3).
In previous studies the degradation of Nvj1 reporters was monitored by fluorescence microscopy and Western blot, and Atg7 was reported not to be required for PMN (Roberts et al., 2003). Therefore we confirmed our findings in two different genetic backgrounds (Figures 1 and 2). Detecting the decreases in the levels of a reporter is less sensitive than detecting the appearance of free GFP. The discrepancy between these previous studies could be due to the nonvacuole proteolysis of the Nvj1 reporters.
Among the autophagic components needed for effective PMN we identified all components shared between the selective Cvt pathway and unselective macroautophagy (summarized in Table 1). These components seem to constitute the core autophagic machinery, which is common even between macroautophagy in S. cerevisiae and micropexophagy in P. pastoris. Our work therefore identifies PMN as a true microautophagic process, which can also be termed micronucleophagy and further helps to define the core set of autophagy proteins.
Atg18 is a member of the core autophagy machinery. In contrast to most other Atg proteins Atg18 has two separable functions. Its first function during the biogenesis of autophagosomes depends on PtdIns3P, but not on PtdIns(3,5)P2. Its second function in retrograde transport from the vacuole back to late endosomes is strictly dependent on its binding to PtdIns(3,5)P2 at the vacuolar membrane (Dove et al., 2004; Krick et al., 2006). In S. cerevisiae PtdIns(3,5)P2 is exclusively synthesized by the PtdIns-3P 5-kinase Fab1. Because PMN proceeds in fab1Δ cells (Figure 2C), we conclude that PMN requires the autophagic function of Atg18, but not its function in retrograde vacuolar transport. In line with these findings PMN depends only on the phosphatidylinostitol 3-kinase complex I, which is required for macro- and microautophagy, but not on complex II, which affects vacuolar biogenesis.
Additional sets of autophagy proteins adopt the core machinery to the specific necessity of the individual autophagic subtype. Interestingly, Atg11 and Atg17 are both required for PMN. Atg17 is supposed to act as a basic scaffold for the organization of the PAS under starvation conditions, when macroautophagy is active. For the selective Cvt pathway, which is active under nutrient-rich conditions, it is not required (Suzuki et al., 2007). PMN occurs in dividing cells in rich medium, but, like macroautophagy, is induced to higher levels during nutrient depletion (Roberts et al., 2003). The requirement of Atg17 for PMN is therefore consistent with the idea that a PAS-like structure is involved in PMN. Atg11 is thought to play a similar role in PAS organization during selective subtypes of autophagy such as the Cvt pathway and micropexophagy in P. pastoris (Xie and Klionsky, 2007; Farre and Subramani, 2004; Dunn et al., 2005; Sakai et al., 2006). The requirement of Atg11 groups PMN with the Cvt pathway and micropexophagy in P. pastoris. This is in agreement with morphological studies, showing the specific engulfment of the nuclear ER protrusions by the vacuole, which excludes cytosolic material (Figure 9; Roberts et al., 2003). Among the other Cvt-specific components tested, such as the GARP/VFT-tethering complex (Vps51, Vps52, Vps53, and Vps54), the SNARE Tlg2 and the SM-protein Vps45 only Trs85, a subunit of the TRAPP-tethering complexes, the sorting nexin Atg24/Snx4 and Atg21 were found to affect the PMN rates (Figures 2, D and E, and 4). This suggests that these proteins play a more central role in specific subtypes of autophagy.
Interestingly, the Cvt-specific proteins Atg20, Atg23, and Atg27 are not needed for normal PMN. This assigns to these proteins a more peripheral role in PAS organization and groups them into the family of Atg proteins needed to adopt the autophagic machinery for a specific subtype. Taken together the requirement of PMN for macroautophagy-specific Atg17 and a subset of Cvt-specific Atg proteins places PMN between these two processes and defines it as a selective, starvation-induced autophagic subtype, whose genetic requirements resemble micropexophagy in P. pastoris. In S. cerevisiae only rare cases of microautophagy-like processes were reported such as during recovery from rapamycin treatment (Dubouloz et al., 2005), the formation of autophagic tubes (Muller et al., 2000), or during mitophagy (Kissova et al., 2007). We have shown that the molecular machinery, which is required for formation of autophagic tubes and for the microautophagy-like process during recovery from rapamycin is distinct from those of PMN.
The last stage of PMN is the breakdown of PMN vesicles within the vacuole. This stage requires the presence of vacuolar proteinase A and Atg15 and is affected by the proteinase B inhibitor PMSF (Figures 1 and 3). Atg15 is a putative lipase that is required for intravacuolar lysis of autophagic bodies and MVB vesicles (Epple et al., 2003).
Fusion of the vacuolar membrane extensions (Figure 10, stage IV) is the crucial step in release of a PMN vesicle into the vacuole. Because several components essential for homotypic vacuole fusion such as the Vtc proteins, LMA1 and Vph1, are not needed for PMN (Figure 5C), we conclude that the fusion of the vacuolar extensions during PMN is not a classical homotypic fusion event. We expect that the requirement of PMN for other components of the vacuole fusion machinery such as Sec17, Sec18, the HOPS-complex, Ypt7, and the SNAREs Vam3 and Vam7 is due to their function in vacuolar biogenesis. TEM analysis of several autophagy-deficient mutants shows that the nuclear envelope ER blebs can still bud off in these cells to produce small micronuclei in the absence of complete vacuolar engulfment (Figure 9). These micronuclear vesicles appeared partially engulfed by indentations of the vacuolar membrane. Remarkably, the tips of the vacuolar indentations were not fused, suggesting a function of the autophagic machinery in completing the sequestration and/or fusion of the vacuolar membranes (Figure 9). Our finding that the fusion of the vacuolar membranes does not depend on the classical homotypic vacuolar fusion machinery further supports a role for the Atg apparatus in the terminal engulfment and fusion stages of PMN.
In this regard, PMN both morphologically and genetically resembles micropexophagy in P. pastoris. Peroxisomes are bulky cargoes, which experimentally allowed the spatial separation of events during micropexophagy, and the actual placement of autophagy proteins at two locations the perivacuolar structure (PVS) and the MIPA (Farre and Subramani, 2004; Mukaiyama et al., 2004; Chang et al., 2005; Dunn et al., 2005; Sakai et al., 2006). Although the PVS has been implicated in formation of the vacuolar sequestration membranes, the MIPA is thought to heterotypically fuse with both vacuolar arms, thus mediating their fusion. We speculate that during PMN a membranous structure containing Atg proteins analogous to the MIPA might fuse with the vacuolar tips. Further detailed work will be necessary to test this hypothesis.
Why might microautophagy employ such a sophisticated and complex apparatus to achieve membrane fusion? A closer look at homotypic fusion of vacuoles shows that it does not take place at a small fusion pore, which then expands radially, but at vertex regions (Wang et al., 2002). Probably, several SNARE-complexes must form simultaneously to generate enough force to achieve fusion, therefore requiring a larger surface area. For the same reason fusion of the vacuolar invagination should not occur at a single point. This problem can be overcome by using a membranous helper structure, which heterotypically fuses with the surrounding vacuolar membrane. Indeed, Atg8 has been shown to function in tethering and hemifusion of membranes (Nakatogawa et al., 2007; Subramani and Farre, 2007).
In conclusion, our results establish PMN as a bona fide microautophagic process that is dependent on the core autophagic machinery and opens the possibility of addressing conserved aspects of microautophagy using this model process. So far PMN has only been detected in S. cerevisiae, but most likely an analogous process also exist in mammalian cells. Further work will be needed to identify such a process and to get deeper insight into the physiological relevance of micronucleophagy both in lower and higher eukaryotes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Benjamin Timney and Michael Rout (Rockefeller University, New York) for providing the NLS-mcherry construct, R. Schekman (University of California, Berkeley, California) for strains, M. Bredschneider and U. D. Epple for help and discussions. We further thank the Electron Microscopy Unit of the Institute of Biotechnology, University of Helsinki, for providing laboratory facilities. This work was supported by National Science Foundation Grant MCB-072064 to D.G.
Abbreviations used:
- Atg
autophagy
- PAS
preautophagosomal structure
- PMN
piecemeal microautophagy of the nucleus
- PMSF
phenylmethylsulfonyl fluoride.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-04-0363) on August 13, 2008.
REFERENCES
- Abeliovich H., Darsow T., Emr S. D. Cytoplasm to vacuole trafficking of aminopeptidase I requires a t-SNARE- Sec1p complex composed of Tlg2p and Vps45p. EMBO J. 1999;18:6005–6016. doi: 10.1093/emboj/18.21.6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ano Y., Hattori T., Oku M., Mukaiyama H., Baba M., Ohsumi Y., Kato N., Sakai Y. A sorting nexin PpAtg24 regulates vacuolar membrane dynamics during pexophagy via binding to phosphatidylinositol-3-phosphate. Mol. Biol. Cell. 2005;16:446–457. doi: 10.1091/mbc.E04-09-0842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D. Current Protocols in Molecular Biology. New York: Greene Publishing Associates; 1987. [Google Scholar]
- Barth H., Meiling-Wesse K., Epple U. D., Thumm M. Mai1p is essential for maturation of proaminopeptidase I but not for autophagy. FEBS Lett. 2002;512:173–179. doi: 10.1016/s0014-5793(02)02252-4. [DOI] [PubMed] [Google Scholar]
- Bayer M. J., Reese C., Buhler S., Peters C., Mayer A. Vacuole membrane fusion: V0 functions after trans-SNARE pairing and is coupled to the Ca2+-releasing channel. J. Cell Biol. 2003;162:211–222. doi: 10.1083/jcb.200212004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S., McDonald K. L., Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H., Reinisch K., Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev. Cell. 2007;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Cao Y., Klionsky D. J. Atg26 is not involved in autophagy-related pathways in Saccharomyces cerevisiae. Autophagy. 2007;3:17–20. doi: 10.4161/auto.3371. [DOI] [PubMed] [Google Scholar]
- Chang T., Schroder L. A., Thomson J. M., Klocman A. S., Tomasini A. J., Stromhaug P. E., Dunn W. A., Jr PpATG9 encodes a novel membrane protein that traffics to vacuolar membranes, which sequester peroxisomes during pexophagy in Pichia pastoris. Mol. Biol. Cell. 2005;16:4941–4953. doi: 10.1091/mbc.E05-02-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove S. K., et al. Svp1p defines a family of phosphatidylinositol 3,5-bisphosphate effectors. EMBO J. 2004;23:1922–1933. doi: 10.1038/sj.emboj.7600203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouloz F., Deloche O., Wanke V., Cameroni E., De Virgilio C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol. Cell. 2005;19:15–26. doi: 10.1016/j.molcel.2005.05.020. [DOI] [PubMed] [Google Scholar]
- Dunn W. A., Jr, Cregg J. M., Kiel J. A., van der Klei I. J., Oku M., Sakai Y., Sibirny A. A., Stasyk O. V., Veenhuis M. Pexophagy: the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- Epple U. D., Eskelinen E. L., Thumm M. Intravacuolar membrane lysis in Saccharomyces cerevisiae. Does vacuolar targeting of Cvt17/Aut5p affect its function? J. Biol. Chem. 2003;278:7810–7821. doi: 10.1074/jbc.M209309200. [DOI] [PubMed] [Google Scholar]
- Epple U. D., Suriapranata I., Eskelinen E. L., Thumm M. Aut5/Cvt17p, a putative lipase essential for disintegration of autophagic bodies inside the vacuole. J. Bacteriol. 2001;183:5942–5955. doi: 10.1128/JB.183.20.5942-5955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre J. C., Subramani S. Peroxisome turnover by micropexophagy: an autophagy-related process. Trends Cell Biol. 2004;14:515–523. doi: 10.1016/j.tcb.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Gao M., Kaiser C. A. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat. Cell Biol. 2006;8:657–667. doi: 10.1038/ncb1419. [DOI] [PubMed] [Google Scholar]
- Güldener U., Heck S., Fielder T., Beinhauer J., Hegemann J. H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema E. H., Lewis M. J., Black M. W., Pelham H. R. Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 2003;22:548–557. doi: 10.1093/emboj/cdg062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N., Hamasaki M., Yokota S., Suzuki K., Kamada Y., Kihara A., Yoshimori T., Noda T., Ohsumi Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol. Biol. Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y., Kawamata T., Suzuki K., Ohsumi Y. Cis1/Atg31 is required for autophagosome formation in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 2007;356:405–410. doi: 10.1016/j.bbrc.2007.02.150. [DOI] [PubMed] [Google Scholar]
- Kaiser C. A., Schekman R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
- Kawamata T., Kamada Y., Suzuki K., Kuboshima N., Akimatsu H., Ota S., Ohsumi M., Ohsumi Y. Characterization of a novel autophagy-specific gene, ATG29. Biochem. Biophys. Res. Commun. 2005;338:1884–1889. doi: 10.1016/j.bbrc.2005.10.163. [DOI] [PubMed] [Google Scholar]
- Kissova I., Salin B., Schaeffer J., Bhatia S., Manon S., Camougrand N. Selective and non-selective autophagic degradation of mitochondria in yeast. Autophagy. 2007;3:329–336. doi: 10.4161/auto.4034. [DOI] [PubMed] [Google Scholar]
- Klionsky D. J., et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- Krick R., Tolstrup J., Appelles A., Henke S., Thumm M. The relevance of the phosphatidylinositolphosphat-binding motif FRRGT of Atg18 and Atg21 for the Cvt pathway and autophagy. FEBS Lett. 2006;580:4632–4638. doi: 10.1016/j.febslet.2006.07.041. [DOI] [PubMed] [Google Scholar]
- Kvam E., Goldfarb D. S. Nvj1p is the outer-nuclear-membrane receptor for oxysterol-binding protein homolog Osh1p in Saccharomyces cerevisiae. J. Cell Sci. 2004;117:4959–4968. doi: 10.1242/jcs.01372. [DOI] [PubMed] [Google Scholar]
- Kvam E., Gable K., Dunn T. M., Goldfarb D. S. Targeting of Tsc13p to nucleus-vacuole junctions: a role for very-long-chain fatty acids in the biogenesis of microautophagic vesicles. Mol. Biol. Cell. 2005;16:3987–3998. doi: 10.1091/mbc.E05-04-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvam E., Goldfarb D. S. Nucleus-vacuole junctions and piecemeal microautophagy of the nucleus in S. cerevisiae. Autophagy. 2007;3:85–92. doi: 10.4161/auto.3586. [DOI] [PubMed] [Google Scholar]
- Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiling-Wesse K., Barth H., Thumm M. Ccz1p/Aut11p/Cvt16p is essential for autophagy and the cvt pathway. FEBS Lett. 2002;526:71–76. doi: 10.1016/s0014-5793(02)03119-8. [DOI] [PubMed] [Google Scholar]
- Meiling-Wesse K., Barth H., Voss C., Eskelinen E. L., Epple U. D., Thumm M. Atg21 is required for effective recruitment of Atg8 to the preautophagosomal structure during the Cvt pathway. J. Biol. Chem. 2004;279:37741–37750. doi: 10.1074/jbc.M401066200. [DOI] [PubMed] [Google Scholar]
- Meiling-Wesse K., Epple U. D., Krick R., Barth H., Appelles A., Voss C., Eskelinen E. L., Thumm M. Trs85 (Gsg1), a component of the TRAPP complexes, is required for the organization of the preautophagosomal structure during selective autophagy via the Cvt pathway. J. Biol. Chem. 2005;280:33669–33678. doi: 10.1074/jbc.M501701200. [DOI] [PubMed] [Google Scholar]
- Mukaiyama H., Baba M., Osumi M., Aoyagi S., Kato N., Ohsumi Y., Sakai Y. Modification of a ubiquitin-like protein Paz2 conducted micropexophagy through formation of a novel membrane structure. Mol. Biol. Cell. 2004;15:58–70. doi: 10.1091/mbc.E03-05-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller O., Bayer M. J., Peters C., Andersen J. S., Mann M., Mayer A. The Vtc proteins in vacuole fusion: coupling NSF activity to V(0) trans-complex formation. EMBO J. 2002;21:259–269. doi: 10.1093/emboj/21.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller O., Sattler T., Flotenmeyer M., Schwarz H., Plattner H., Mayer A. Autophagic tubes: vacuolar invaginations involved in lateral membrane sorting and inverse vesicle budding. J. Cell Biol. 2000;151:519–528. doi: 10.1083/jcb.151.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H., Ichimura Y., Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- Nazarko T. Y., Polupanov A. S., Manjithaya R. R., Subramani S., Sibirny A. A. The requirement of sterol glucoside for pexophagy in yeast is dependent on the species and nature of peroxisome inducers. Mol. Biol. Cell. 2007;18:106–118. doi: 10.1091/mbc.E06-06-0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nice D. C., Sato T. K., Stromhaug P. E., Emr S. D., Klionsky D. J. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J. Biol. Chem. 2002;277:30198–30207. doi: 10.1074/jbc.M204736200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K., Sekito T., Ohsumi Y. Assortment of phosphatidylinositol 3-kinase complexes—Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol. Biol. Cell. 2006;17:1527–1539. doi: 10.1091/mbc.E05-09-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku M., Warnecke D., Noda T., Muller F., Heinz E., Mukaiyama H., Kato N., Sakai Y. Peroxisome degradation requires catalytically active sterol glucosyltransferase with a GRAM domain. EMBO J. 2003;22:3231–3241. doi: 10.1093/emboj/cdg331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowicz C. W., Meiringer C. T., Ungermann C. Yeast vacuole fusion: a model system for eukaryotic endomembrane dynamics. Autophagy. 2008;4:5–19. doi: 10.4161/auto.5054. [DOI] [PubMed] [Google Scholar]
- Pan X., Roberts P., Chen Y., Kvam E., Shulga N., Huang K., Lemmon S., Goldfarb D. S. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol. Biol. Cell. 2000;11:2445–2457. doi: 10.1091/mbc.11.7.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Klionsky D. J. Atg9 sorting from mitochondria is impaired in early secretion and VFT-complex mutants in Saccharomyces cerevisiae. J. Cell Sci. 2006;119:2903–2911. doi: 10.1242/jcs.03047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Shintani T., Nair U., Klionsky D. J. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 2005;1:101–109. doi: 10.4161/auto.1.2.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F., Tucker K. A., Stromhaug P. E., Klionsky D. J. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- Reggiori F., Wang C. W., Stromhaug P. E., Shintani T., Klionsky D. J. Vps51 is part of the yeast Vps fifty-three tethering complex essential for retrograde traffic from the early endosome and Cvt vesicle completion. J. Biol. Chem. 2003;278:5009–5020. doi: 10.1074/jbc.M210436200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts P., Moshitch-Moshkovitz S., Kvam E., O'Toole E., Winey M., Goldfarb D. S. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:129–141. doi: 10.1091/mbc.E02-08-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein D. C., Gestwicki J. E., Murphy L. O., Klionsky D. J. Potential therapeutic applications of autophagy. Nat. Rev. Drug Discov. 2007;6:304–312. doi: 10.1038/nrd2272. [DOI] [PubMed] [Google Scholar]
- Sakai Y., Oku M., van der Klei I. J., Kiel J. A. Pexophagy: autophagic degradation of peroxisomes. Biochim. Biophys. Acta. 2006;1763:1767–1775. doi: 10.1016/j.bbamcr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- Sattler T., Mayer A. Cell-free reconstitution of microautophagic vacuole invagination and vesicle formation. J. Cell Biol. 2000;151:529–538. doi: 10.1083/jcb.151.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Klionsky D. J. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J. Biol. Chem. 2004;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromhaug P. E., Reggiori F., Guan J., Wang C. W., Klionsky D. J. Atg21 is a phosphoinositide binding protein required for efficient lipidation and localization of Atg8 during uptake of aminopeptidase I by selective autophagy. Mol. Biol. Cell. 2004;15:3553–3566. doi: 10.1091/mbc.E04-02-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S., Farre J. C. A ubiquitin-like protein involved in membrane fusion. Cell. 2007;130:18–20. doi: 10.1016/j.cell.2007.06.041. [DOI] [PubMed] [Google Scholar]
- Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Kubota Y., Sekito T., Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- Suzuki K., Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS Lett. 2007;581:2156–2161. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- Thumm M., Egner R., Koch B., Schlumpberger M., Straub M., Veenhuis M., Wolf D. H. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- Uttenweiler A., Schwarz H., Neumann H., Mayer A. The vacuolar transporter chaperone (VTC) complex is required for microautophagy. Mol. Biol. Cell. 2007;18:166–175. doi: 10.1091/mbc.E06-08-0664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Seeley E. S., Wickner W., Merz A. J. Vacuole fusion at a ring of vertex docking sites leaves membrane fragments within the organelle. Cell. 2002;108:357–369. doi: 10.1016/s0092-8674(02)00632-3. [DOI] [PubMed] [Google Scholar]
- Wickner W. Yeast vacuoles and membrane fusion pathways. EMBO J. 2002;21:1241–1247. doi: 10.1093/emboj/21.6.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Klionsky D. J. Autophagosome formation: core machinery and adaptations. Nat. Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T., Klionsky D. J. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol. Biol. Cell. 2005;16:1593–1605. doi: 10.1091/mbc.E04-11-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T., Nair U., Yang Z., Klionsky D. J. Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem. 2006;281:30299–30304. doi: 10.1074/jbc.M607007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.