Abstract
tER sites are specialized cup-shaped ER subdomains characterized by the focused budding of COPII vesicles. Sec16 has been proposed to be involved in the biogenesis of tER sites by binding to COPII coat components and clustering nascent-coated vesicles. Here, we show that Drosophila Sec16 (dSec16) acts instead as a tER scaffold upstream of the COPII machinery, including Sar1. We show that dSec16 is required for Sar1-GTP concentration to the tER sites where it recruits in turn the components of the COPII machinery to initiate coat assembly. Last, we show that the dSec16 domain required for its localization maps to an arginine-rich motif located in a nonconserved region. We propose a model in which dSec16 binds ER cups via its arginine-rich domain, interacts with Sar1-GTP that is generated on ER membrane by Sec12 and concentrates it in the ER cups where it initiates the formation of COPII vesicles, thus acting as a tER scaffold.
INTRODUCTION
A crucial step in the anterograde membrane transport is the packaging and exit of newly synthesized proteins from the endoplasmic reticulum (ER). This takes place at specialized ribosome-free ER subdomains, the ER exit sites, (also called tER sites), that are characterized by a cup-shaped ER membrane from which COPII vesicles bud. The COPII machinery comprises the small GTPase Sar1 that is recruited to the ER membrane by its guanine nucleotide exchange factor (GEF) Sec12, and the proteins forming the coat itself, the two complexes Sec23/24 and Sec13/31 (Bonifacino and Glick, 2004).
Throughout the eukaryotic kingdom, tER sites are characterized by the presence of COPII components. Except for Saccharomyces cerevisiae (Rossanese et al., 1999), the tER sites in animal species studied so far are focused and form a limited number of sites: 1 in trypanosome (He et al., 2005), 2–5 in Pichia pastoris (Bevis et al., 2002), more than 200 in HeLa cells (Hammond and Glick, 2000), and 17 ± 8 in nonsynchronized Drosophila S2 cells (Kondylis et al., 2007). What controls the focused organization and the number of tER sites remains largely unknown.
A genetic screen searching for factors influencing the organization of tER sites in P. pastoris has identified Sec16. A temperature-sensitive sec16 mutant causes fragmentation of tER sites at nonpermissive temperature (Connerly et al., 2005). Similarly, depletion of Sec16 by RNA interference (RNAi) in mammalian cells leads to disruption of tER sites and inhibition of anterograde transport, accompanied by the accumulation of cargo proteins in the ER (Watson et al., 2006; Bhattacharyya and Glick, 2007).
Sec16 is a large ∼240–280-kDa peripheral membrane protein that was originally characterized in S. cerevisiae as a factor involved in the formation of COPII vesicles (Kaiser and Schekman, 1990). In yeast, it is copurified with ER-derived COPII transport vesicles formed in vitro (Espenshade et al., 1995), but also plays an active role in their formation (Supek et al., 2002). Consistently, Sec16 displays biochemical interactions with the COPII subunits Sec23 (Espenshade et al., 1995; Bhattacharyya and Glick, 2007), Sec24 (Gimeno et al., 1996), Sec31 (Shaywitz et al., 1997), and possibly Sar1 (Supek et al., 2002). A genetic (Kaiser and Schekman, 1990) but not biochemical (Gimeno et al., 1995) interaction with Sec12 has also been shown.
As largely debated for the Golgi complex, tER sites have been proposed to be generated on a tER matrix (Shaywitz et al., 1997; Mogelsvang et al., 2003; Soderholm et al., 2004) in which Sec16 functions as a molecular platform upstream of COPII assembly, dictating where the focused budding of the COPII vesicles takes place. Alternatively, Sec16 could physically interact with several COPII coat components of the nascent COPII vesicles after the initial coat assembly on ER membrane (Espenshade et al., 1995; Gimeno et al., 1996; Shaywitz et al., 1997). Sec16 oligomerization properties (Bhattacharyya and Glick, 2007) would result in the clustering of COPII vesicles, thus forming distinct and focused tER sites. Supporting this Sec16 role downstream of COPII assembly, Sar1-GTP has been recently shown to recruit Sec16 to tER sites in human cells (Watson et al., 2006; Iinuma et al., 2007).
Here, using Drosophila S2 cells as a model system (Kondylis and Rabouille, 2003; Kondylis et al., 2005), we show that dSec16 acts as a tER scaffold dictating where the tER sites are formed. We first characterized the Drosophila Sec16 (dSec16), whose properties strictly conform to its orthologues in other species. Second, using a combination of light and immunoelectron microscopy, we show that dSec16 can associate to discrete ER domains in the absence of assembled tER sites, COPII vesicles and even Sar1, suggesting that it acts upstream of the COPII machinery. We find that Sar1 concentration to tER sites is Sec16-dependent. In dSec16-depleted cells, Sar1 is no longer concentrated on tER sites and is dispersed to the ER. Conversely, a form of dSec16 artificially localized to endosomes is able to recruit Sar1 but also other COPII subunits, indicating that Sec16 is part of the molecular machinery regulating the positioning and building of tER sites. Third, we identified the dSec16 domain involved in its localization to tER sites to a stretch of 65-amino acid arginine-rich domain in the N-terminal nonconserved region of the protein.
We propose a model for tER site biogenesis in which dSec16 acts as a tER matrix by binding ER cups through its arginine-rich domain and adjacent sequences followed by its oligomerization. Concomitantly, Sar1-GTP is recruited to ER membrane by Sec12 GEF activity and gets concentrated to ER cups by interacting with dSec16, initiating the assembly of the COPII coat.
MATERIALS AND METHODS
Antibodies
The polyclonal anti-dSec16 antibody 764 was generated by immunizing rabbits with a GST-fusion protein comprising amino acids 655–817 of dSec16. The other rabbit polyclonal antibodies used were: anti-Sec23 (clone PA-069, Affinity Bioreagents, Golden, CO; Kondylis and Rabouille, 2003), anti-green fluorescent protein (GFP; Abcam, Cambridge, United Kingdom), anti-Sec31 (gift from F. Gorelick, Yale University), anti-Sar1 (Fromme et al., 2007) from B. Kleizin (VU Amsterdam, The Netherlands), and the anti-dGMAP (Friggi-Grelin et al., 2006). The following monoclonal antibodies were from the indicated sources: anti-Delta (from the Developmental Studies Hybridoma Bank maintained by the University of Iowa, Iowa City, IA), anti-d120kd (Calbiochem, La Jolla, CA; Kondylis and Rabouille, 2003), anti-GFP (Roche, Woerden, The Netherlands), anti α-tubulin (Sigma, St. Louis, MO), anti-V5 (Invitrogen, Carlsbad, CA), and anti-vesicular stomatitis virus glycoprotein (VSVG; clone P5D4, Roche).
Double-stranded RNA
dSec16 cDNA RE70141 was used to PCR a 820-base pair fragment with flanking T7 RNA polymerase-binding sites (TTAATACGACTCACTATAGGGAGA) using the primers 5′ T7-CGTCTGGGACAAGGAGC and 3′ T7-CCTGCTGGAAAGTTGAC corresponding to the C-terminus of dSec16. Sec23 cDNA was used as a template to amplify by PCR a 793-base pair fragment with the primers 5′ T7-GTGCAGGATATGCTCGGAAT and 3′ T7-GTGGAGCTGGGATTCAATGT. Full-length Sar1 (nucleotides 1–465) was amplified using 5′ T7-GTTTTCCTCGCTTCCAGATG and 3′T7-CTTGCCGGTTGTTAGCTGAT as primers.
These fragments were subjected to in vitro transcription for generating double-stranded RNA (dsRNA) using MEGASCRIPT T7 transcription kit (Ambion, Austin, TX), according to manufacturer's protocol and as described (Kondylis and Rabouille, 2003).
Plasmids
The cDNA clone LP14866 was obtained from Flybase (http://flybase.org) and was used to generate reporter constructs. This clone encodes the short isoform of dSec16, except for the last 68 amino acids. All constructs were amplified by PCR and cloned in pMT/V5-HisA or HisB (Invitrogen). The restriction sites and the corresponding size of the resulting constructs are described in Supplementary Table S1. The pEGFP-2XFYVE containing the tandem FYVE domain of mouse Hrs was a gift from Dr. Harald Stenmark (Oslo, Norway). This construct was used to PCR the 2xFYVE domain, followed by subsequent ligation into the XbaI site of pMT/V5-dSec16 to generate dSec16-2xFYVE-V5. To obtain GST-dSec16, the dSec16 region comprising amino acids 655–817 was cloned into XhoI-NotI sites of pGEX-4T1. Sec23 was PCR amplified from the Drosophila cDNA clone RE35250 (containing the full-length open reading frame of the gene) and ligated in pRmeGFP vector to generate an in frame GFP-dSec23 construct. Sar1 was amplified by PCR from a reverse transcriptase reaction of a total RNA extract from Drosophila S2 cells and cloned into the pRmeGFP vector as a C-terminal GFP fusion. QuickChange Mutagenesis kit (Stratagene, La Jolla, CA) was used to create the NC23-CCD 807/861AAA, as well as the Sar1[H74G] and Sar1[T34N] mutants. Sec16-VSVG was made by replacement of the V5 epitope with VSVG (YTDIEMNRLGK) in the dSec16-containing pMT/V5-His plasmid. All constructs were checked by sequencing.
Cell Culture, RNAi, Transfection, and Drug Treatment
Wild-type S2 cells or stably transfected with Delta-WTNdeMYC, Fringe-GFP, dSar1[H74G]-GFP or GFP-Sec23 (from Drosophila) were grown and depleted by RNAi as previously described (Kondylis and Rabouille, 2003; Kondylis et al., 2007). Cells were analyzed after 5 d of RNAi treatment in case of Sec16 depletion or 4 d in case of Sar1.
For transient transfections, ∼1 × 106 S2 cells were plated on glass coverslips in 3.5-cm dishes and transfected the next day with 0.5–1 μg DNA using the Effectene transfection reagent (Qiagen, Hilden, Germany) according to the manufacturer's instructions. After 48–72 h, the protein synthesis was induced by adding 1 mM CuSO4 in the cell medium for 2–3 h at 27°C. The CuSO4 was washed out, and the cells were further incubated for 1–2 h in fresh medium before being fixed for immunofluorescence (IF) or immunoelectron microscopy (IEM). On average, 50% transfection efficiency was achieved. For biochemical experiments, cells were processed 2–3 h after induction.
For the brefeldin A (BFA) experiment, stably transfected Sar1[H74G]-GFP S2 cells were induced for expression by adding 1 mM CuSO4 for 1.5 h. The medium was changed, and the cells were mock-treated or treated with BFA (20 μM) for 2 h. Control S2 cells transiently transfected with Fringe-GFP were treated with BFA for 30 min before Fringe-GFP expression induced by addition of 1 mM CuSO4 for 1.5 h in the presence of BFA.
Immunofluorescence and Immunoelectron Microscopy
S2 cells grown on coverslips were fixed in 4% PFA in PBS and processed for IF as described (Kondylis and Rabouille, 2003). Cells were viewed under a Leica TCS-NT (Jena, Germany) or a Zeiss LSM-510 confocal microscope (Wetzlar, Germany). When a whole cell projection is presented, it is indicated as “proj.” When nothing is indicated, a confocal section is presented. Immunoelectron microscopy was performed as published (Kondylis and Rabouille, 2003).
Delta and Fringe Transport Assays
Delta and Fringe-GFP transport assays were done as described (Kondylis and Rabouille, 2003; Kondylis et al., 2007). A Fringe-GFP stable cell line was generated according to the manufacturer's protocol.
dSec16-FYVE and Sar1 Recruitment to Endosomes
To examine whether dSec16–2xFYVE-V5 localized to endosomes a BSA-gold uptake was included after the 2-h pulse with CuSO4 in the 1- or 2-h chase period of transfected S2 cells for the IEM or IF analyses, respectively. The effect of delocalizing dSec16 on the localization of dSar1 was determined by double transfection of dSec16-2xFYVE-V5 and Sar1[T34N]-GFP or Sar1[H74G]-GFP. The quantification of the colocalization between dSec16-FYVE and Sar1-GTP or Sar1-GDP is based on analyzing 113 and 104 endosomes, respectively, from 40 cells in two independent experiments.
Subcellular Fractionation
S2 cells (∼80 million) were harvested by centrifugation, resuspended in 1.2 ml homogenization buffer (20 mM HEPES, pH 7.5, 250 mM sucrose and protease inhibitors) and broken by three freeze-thaw cycles. The nuclei and nonbroken cells were pelleted by centrifugation 15 min at 800× g, and the resulting postnuclear supernatant (PNS) was applied to ultracentrifugation at 100,000× g for 1 h using a TLA 100.2 rotor to generate the cytosol (C) and the membrane fraction. To analyze the partition of dSec16 between the membrane soluble (S) and insoluble fraction (P), the total membranes were further resuspended and incubated for 1 h at 4°C in a PNS-equivalent volume of homogenization buffer to which NaCl and/or Triton X-100 was added to a final concentration of 0.5 M and 1%, respectively. The suspension was ultracentrifuged as above to separate the soluble from the insoluble material. The insoluble fraction was finally dissolved in Laemmli sample buffer. Equal volumes of PNS, C, S, and P were loaded on an SDS-PAA gel and analyzed by Western blot with anti-dSec16 antibodies.
In Vivo Interaction between dSec16 and Sar1
S2 cells cotransfected with Sar1[H74G]-GFP or Sar1[T34N]-GFP and dSec16-V5 were resuspended in lysis buffer (20 mM HEPES, pH7.5, 150 mM NaCl, 5 mM MgCl2, 0.2% NP-40, and protease inhibitors) for 30 min on ice. The lysates were centrifuged for 15 min, and the resulting supernatants were subjected to immunoprecipitation with a polyclonal anti-GFP antibody. The precipitated proteins were resolved by SDS-PAGE and analyzed by immunoblotting with monoclonal anti-GFP and anti-V5 antibodies.
dSec16 Oligomerization
S2 cells transfected with the indicated plasmids were lysed in 50 mM Tris, pH 7.5, 150 mM NaCl, and 1% Triton X-100 (immunoprecipitation buffer) supplemented with protease inhibitor cocktail (Roche) for 30 min on ice. The lysate was centrifuged for 15 min, and the resulting supernatant was subjected to immunoprecipitation using anti-V5 antibodies bound to protein A-agarose beads (Repligen, Minneapolis, MN). To improve the coupling of the V5 antibody to protein A-agarose beads, a bridging affinity-purified rabbit anti-mouse antibody (Jackson ImmunoResearch, West Grove, PA) was used. The incubation was carried out for 2 h at 40°C. The beads were then washed for three times with immunoprecipitation buffer. Immunoprecipitated proteins were eluted from the beads by heating in Laemmli reducing sample buffer (RSB) for 7 min at 95°C and analyzed by SDS-PAGE and Western blot using anti-VSVG- and anti-V5 antibodies.
RESULTS
Drosophila Sec16 Is the Orthologue of Yeast and Human Sec16
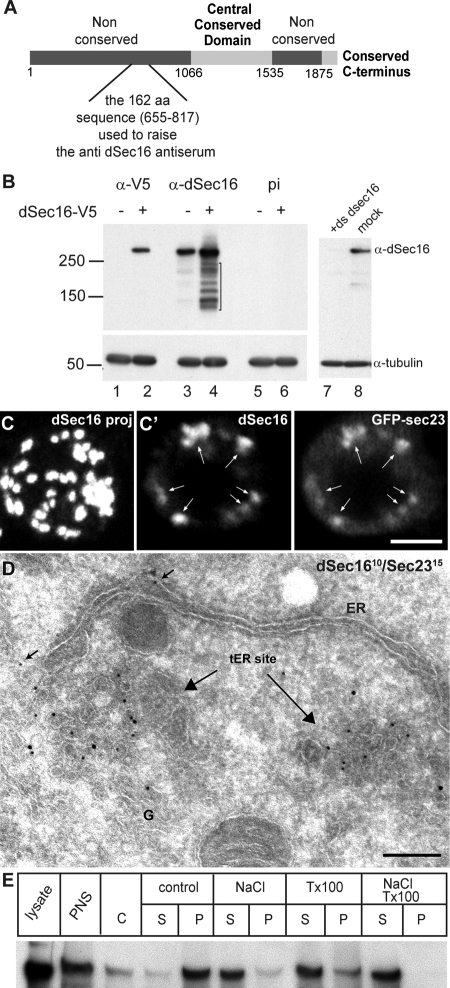
The Drosophila melanogaster homologue of human and yeast Sec16 is encoded by the gene CG32654 (flybase) that exhibits at least two predicted splice variants. CG32654-RD encodes a 2221-amino acid form that lacks the conserved C-terminus reported in other species, whereas CG32654-RE encodes a 2021-amino acid form that contains the C-terminus but has a shorter second nonconserved region (Figure 1A). Using an antibody we raised against the 162-amino acid sequence upstream of the central conserved domain (Figure 1A; see Materials and Methods) to blot a S2 cell lysate fractionated on SDS-PAGE, a single band was detected at the molecular weight of ∼280–300-kDa (Figure 1B, lane 3), similar to that of the human and yeast proteins (Espenshade et al., 1995; Watson et al., 2006; Bhattacharyya and Glick, 2007). This band had a migration identical to that of the transfected shorter form of V5-tagged Sec16 (Figure 1B, lane 4) and was very efficiently knocked down (up to 90–95%) using a double-stranded RNA corresponding to the C-terminus (+ds dsec16; Figure 1B, lanes 7 and 8). As previously reported for the yeast homologue (Espenchade 1995, Supek et al., 2002), overexpressed dSec16 was partially degraded, giving rise to several breakdown products (Figure 1B, lane 4, bracket). Blotting with the preimmune serum (pi) did not yield any band (Figure 1B, lanes 5 and 6), confirming the specificity of our antibody. This shows that S2 cells only express the short form of Sec16, and we will refer to it as dSec16.
Figure 1.
dSec16 is the Drosophila orthologue of human and yeast Sec16. (A) Schematic representation of dSec16. The conserved domains are in light gray, whereas the nonconserved ones in dark gray. The region used to raise antibodies against dSec16 is indicated. (B) Western blot of a lysate from S2 cells transfected (+) or not (−) with dSec16-V5 or depleted for dSec16 was probed with the anti-dSec16 antibody. One band of ∼280–300-kDa was specifically detected in both nontransfected and transfected cells, which was not detected by the preimmune serum (pi). The V5 antibody was used as a positive control for the transfected protein. dSec16 was depleted to 90–95% using dsRNA against the C-terminus. α-Tubulin served as a loading control. Note the degradation products in lane 4 corresponding to proteolysis from the C-terminus, therefore not detected by the V5 epitope but by the anti-dSec16 antibody. (C and C′) Localization of endogenous dSec16 was assessed by IF in one wild-type S2 cell (C, projection), and in one GFP-Sec23 expressing S2 cells (C′, confocal section). Note that both proteins colocalize significantly. (D) Double-labeling of endogenous dSec16 (10 nm) and Sec23 (15 nm) on S2 cells frozen sections shows that they both decorate tER sites next to a Golgi stack (G) and an ER cisterna. Arrows indicate a small pool of dSec16 on ER membrane. (E) S2 cell postnuclear supernatant (PNS) was fractionated in cytosol (C) and membranes (M). The membrane fraction was further separated into pellet (P) and supernatant (S) after treatment with 1% Triton X-100, 0.5 mM NaCl or combination of the two. dSec16 is tightly associated to the membrane fraction and is highly soluble in NaCl. Detergent lysate was loaded as control. Scale bars, (C) 5 μm and (D) 200 nm.
dSec16 is overall 23% identical and 35% similar to the long form of the human Sec16 protein, hSec16L (Bhattacharyya and Glick, 2007), with the highest conservation in the central conserved domain (CCD, Figure 1A; 33% identity and 50% similarity) and the C-terminus (28% identity and 39% similarity; using the EMBOSS align program, http://emboss.sourceforge.net/).
Human and Pichia Sec16 localize to tER sites (Connerly et al., 2005; Watson et al., 2006; Bhattacharyya and Glick, 2007). IF localization using the antiserum we generated showed that, as expected, endogenous dSec16 localizes to the 17 ± 8 tER sites in S2 cells (Figure 1C; Kondylis et al., 2007) and colocalizes with GFP-Sec23 (Figure 1C′) and Sar1-GFP (Supplementary Figure S2D).
As previously described (Kondylis and Rabouille, 2003), the Drosophila tER sites are in close proximity to the Golgi complex. Accordingly, dSec16 localization is juxtaposed to that of the integral Golgi membrane proteins Fringe (Kondylis et al., 2007; Figure 2D) and d120kd, the Drosophila homologue of the vertebrate 160-kDa medial Golgi sialoglycoprotein MG160 (Yano et al., 2005; Figure 3A). By IEM, dSec16 was found to decorate the typical pleiomorphic tubular/vesicular membrane marked by Sec23 and located near a Golgi stack, the characteristic morphology of tER sites (Figure 1D), as well as the adjacent ER cup albeit at much lower level (Figure 1D, arrow).
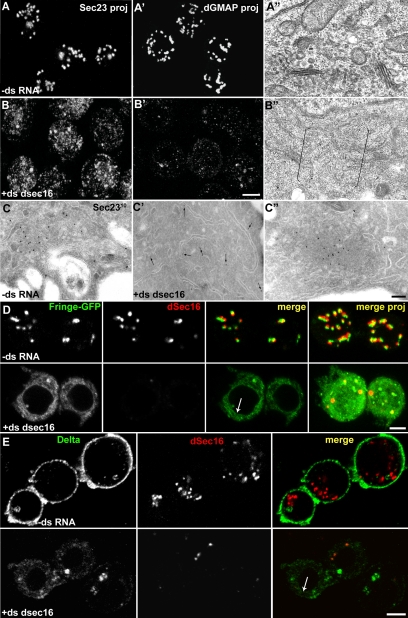
Figure 2.
dSec16 depletion affects the functional organization of the early exocytic pathway. (A–C″) S2 cells were mock-treated (−ds RNA, A–A″ and C) and dSec16 depleted for 5d (+ds dsec16, B–B″, C′, and C″), and processed for IF (A, A′, B, and B′), conventional EM (A″ and B″) or cryo-IEM (C–C″). Both the tER marker Sec23 (B) and the Golgi marker dGMAP (B′) are dispersed. The typical tER-Golgi organization (A″) is absent in Sec16-depleted cells (B″; brackets indicate a cytoplasmic area where a tER-Golgi unit would typically be present). IEM of Sec23 (10 nm gold) shows that a large majority of Sec23 is displaced from tER sites (C) and is found in the cytoplasm (C′, arrows), whereas a pool aggregates in abortive tER sites (C″). (D and E) Anterograde transport of Fringe-GFP (green, D) and Delta (green, E) in mock (−ds RNA) and dSec16-depleted cells (+ds dsec16). Endogenous dSec16 is labeled in red (D and E). The panels presented are confocal sections. Note that upon depletion, both Fringe-GFP and Delta are retained in the ER, exemplified by the nuclear envelope labeling (arrow in D and E). The level of Delta expression is reduced when dSec16 is depleted, suggesting that it is perhaps partially degraded. Scale bars, 5 μm (A, A′, B, B′, D, and E) and 200 nm (A″, B″, C–C″).
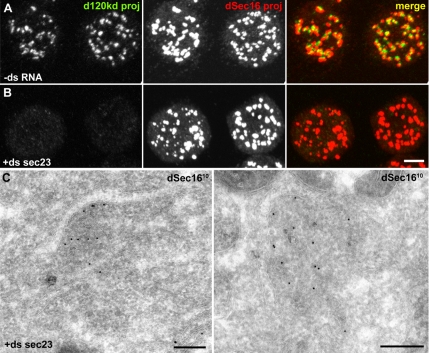
Figure 3.
dSec16 localizes to tER sites in Sec23-depleted cells. (A and B) IF localization of endogenous dSec16 (red) and the Golgi marker d120kd (green) in mock-treatedd (−ds RNA, A) and Sec23-depleted (+ds sec23, B) S2 cells. (C) IEM localization of dSec16 (10 nm gold) in Sec23-depleted cells. The panels presented are projections. Note that dSec16 localizes to tER sites in the absence of Sec23. Scale bars, (A and B) 5 μm and (C) 200 nm.
Finally, both human and S. cerevisiae Sec16 peripherally associate to the membrane fraction from which they can be extracted by high salt concentrations and pH, but not by Triton X-100 or urea (Espenshade et al., 1995; Watson et al., 2006). The subcellular fractionation of dSec16 shows that it is mostly associated to the membrane fraction (Figure 1E, control) in a salt-sensitive manner, consistent with the properties of yeast and human proteins. However, its solubility in detergents was significantly higher than in the above species, perhaps reflecting differences in the lipid membrane composition (Figure 1E).
Taken together, these data suggest that, as do human and yeast Sec16, dSec16 localizes to tER sites and exhibits similar membrane association properties.
Depletion of dSec16 Leads to Disruption of tER Sites
Human cells depleted of Sec16 and P. pastoris harboring a nonfunctional Sec16 show a severe disruption in the organization of tER sites (Connerly et al., 2005; Watson et al., 2006; Bhattacharyya and Glick, 2007). In agreement, depletion of dSec16 from S2 cells by RNAi (as above, Figure 1B) caused a significant fragmentation of tER sites. The typical Sec23 IF pattern (Figure 2A) was converted into a haze of numerous small fluorescence punctae (Figure 2B). By EM, the tER sites and the COPII vesicles were no longer visible (Figure 2B″; Supplementary Figure S1C′), and Sec23 was mostly cytoplasmic (Figure 2C′).
Few larger Sec23 fluorescent spots sometimes remained (Figure 2B) that correspond to Sec23 aggregates associated to clustered membrane remnants close to an ER cisterna, likely representing nonfunctional tER sites (Figure 2C″; compare to the tER sites in a nondepleted cell, Figure 2C).
In dSec16-depleted S2 cells, the Golgi stacks were absent, as observed by the redistribution of the peripheral cis-Golgi marker dGMAP throughout the cytoplasm (Figure 2B′). By EM, Golgi cisternae were not visible (compare Figure 2B″ to Figure 2A″). Furthermore, dSec16 depletion led to the proliferation of ER membrane, as exemplified by long aligned ER sheets that were only rarely seen in mock-depleted cells (not shown), a very reproducible and significant feature that resembles the S. cerevisiae sec16 mutant phenotype (Kaiser and Schekman, 1990).
In the absence of functional Sec16, the anterograde transport both in yeast and human cells is largely inhibited (Espenshade et al., 1995; Watson et al., 2006; Bhattacharyya and Glick, 2007). To test this in dSec16-depleted cells, we performed an ER-to-plasma membrane transport assay making use of the inducible expression of the plasma membrane marker Delta (Klueg et al., 1998; Kondylis and Rabouille, 2003) as well as the Golgi marker Fringe-GFP (Munro and Freeman, 2000; Kondylis et al., 2007). We found that both Delta (Figure 2E) and Fringe-GFP (Figure 2D) were retained in the ER in more than 80% of dSec16-depleted cells, indicating that ER export is largely inhibited. This was confirmed by the severe cell lethality in Drosophila tissues depleted of dSec16 by RNAi (not shown).
Taken all together, dSec16 has similar sequence homology, size, localization, biochemical properties, and role in tER site organization and anterograde transport as the other Sec16 proteins already described, demonstrating that it functions as an orthologue.
dSec16 Localization to tER Sites Is Independent of COPII-coated Vesicle Assembly
A wealth of data has proven the essential role of Sec16 in the organization of ER exit sites (Hughes and Stephens, 2008). However, the mechanism behind it is largely unknown. As outlined in the Introduction, one model proposes that Sec16 is recruited by the COPII coat components to the nascent vesicles and clusters them into a typical tER site due to its oligomerization properties (Connerly et al., 2005). If this is true, dSec16 localization should be clearly affected if COPII coat formation is prevented. To test this, we first depleted Sec23 from S2 cells, as observed by Western blot (Supplementary Figure S1A) and by the large down-regulation of GFP-Sec23 (Supplementary Figure S1C). As for dSec16 depletion, the IF pattern of the Golgi marker dGMAP (not shown) and d120kd (Figure 3B) was dispersed in the cytoplasm, and the ER was also significantly proliferated and dilated (Supplementary Figure S1, D and D′), in agreement with the ER patterns in fibroblasts of patients carrying a mutation in the sec23a gene (Boyadjiev et al., 2006; Fromme et al., 2007). By IEM, typical tER sites and Golgi stacks were no longer visible (Figure 3C). Remarkably, in Sec23-depleted cells, the IF pattern of endogenous dSec16 was indistinguishable from mock-treated cells (Figure 3, A and B). When investigated by IEM, dSec16 was found on distinct ER cups and dense material underneath that contains almost no vesicular profiles (Figure 3C). This suggests that COPII coat assembly is not necessary for dSec16 recruitment, and therefore its localization occurs upstream of the Sec23/Sec24 complex. In human cells, Sec16 has been reported to be recruited and stabilized to tER sites by Sar1-GTP (Watson et al., 2006; Iinuma et al., 2007). To investigate whether this is also the case in Drosophila, we first tested their interaction by immunoprecipitation using the Drosophila inactive GDP-locked Sar1[T34N] and the active GTP-locked Sar1[H74G] mutants. dSec16 was found to interact only with the active form of Sar1 (Figure 4A).
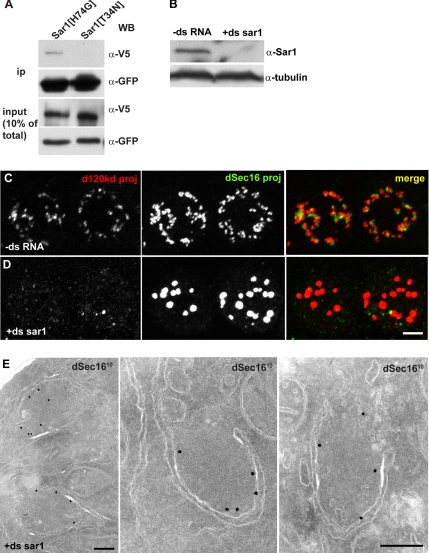
Figure 4.
Sar1 does not mediate dSec16 localization to tER sites. (A) Western blot of immunoprecipitates using an anti-GFP antibody on lysates of S2 cells cotransfected with dSec16-V5 and Sar1[H74G]-GFP and dSec16-V5 and Sar1[T34N]-GFP. Transfected dSec16 coimmunoprecipitates with the active form of Sar1. (B) Western blot of Sar1 in mock-treated (−ds RNA) or Sar1-depleted (+ds sar1) S2 cells. (C and D) IF localization of endogenous dSec16 (red) and the Golgi marker d120kd (green) in mock-treated (−ds RNA, C) and Sar1-depleted (+ds sar1, D) S2 cells. (E) IEM localization of dSec16 (10 nm gold) in Sar1-depleted cells. Note that dSec16 localizes to ER cup in the absence of Sar1. Scale bars, (C and D) 5 μm and (E) 200 nm.
To investigate the functional relevance of this interaction, we tested whether Sar1 recruits dSec16. We first displaced endogenous Sar1 from the tER sites by overexpressing the GDP-locked Sar1[T34N] mutant. This has been shown to potently inhibit the formation of COPII vesicles (Kuge et al., 1994; Aridor et al., 1995, 2001; Rowe et al., 1996) by binding and inhibiting the Sec12-mediated exchange activity on wild-type Sar1 (Weissman et al., 2001), thus blocking anterograde transport (Prescott et al., 2001). Accordingly, the Golgi complex was fragmented and dispersed as assessed by dGMAP and d120kd distribution (Supplementary Figure S2, A and B), the latter also partially found in the ER (not shown). In cells overexpressing Sar1[T34N], endogenous dSec16 was still localized in spots reminiscent of tER sites (Supplementary Figure S2C), though they were less numerous and larger than in cells transfected with wild-type Sar1 and Sar1[H74G] (Supplementary Figure 2, D and E; see Discussion). Surprisingly, although dSec16 and wild-type Sar1 colocalize perfectly at tER sites, dSec16 and Sar1[H74G]-GFP did not. Instead, their labeling seem to juxtapose, perhaps because, as COPII vesicles that are formed in the presence of the active Sar1 mutant do not uncoat, dSec16 is not accessible to its antibody by immunofluorescence. By IEM, however, dSec16 and Sar1[H74G]-GFP were shown localize to the same tER site (Supplementary Figure S2F).
To confirm that dSec16 is not released from the tER sites in the absence of functional Sar1, we depleted Sar1 from S2 cells for 4 d (Figure 4B) and assessed the localization of endogenous dSec16. Again, dSec16 was localized in spots (Figure 4D). As in cells overexpressing Sar1[T34N], their number was reduced and their size larger when compared with nondepleted cells (Figure 4C, see Discussion). Using IEM, the IF spots were shown to correspond to discrete ER cups decorated by endogenous dSec16. The rest of the ER was not labeled. As expected, most of the vesicle budding was inhibited (Figure 4E). Despite the tER site morphometric changes (see Discussion), these results suggest that dSec16 localization is independent of Sar1.
Taken together, these results indicate that dSec16 is able to localize to tER sites in the absence of COPII coated membrane (achieved either by Sec23 or Sar1 depletion). This shows that its localization is not mediated by binding to Sec23, the prebudding complex (Sar1-Sec23/24; Kuehn et al.,1998) or Sar1-GTP. Although dSec16 could associate to the tER sites by interacting with the Sec13/31 complex, this is very unlikely as recent data have shown that the tER localization of Sec13/31 critically depends on the Sar1-Sec23/24 complex with the Sec23 Phe380 residue central for this association (Bi et al., 2007; Fromme et al., 2007). Therefore, our results fit with the hypothesis that Sec16 functions upstream of the COPII coat formation.
dSec16 Mediates the Recruitment of Sar1 to tER Sites
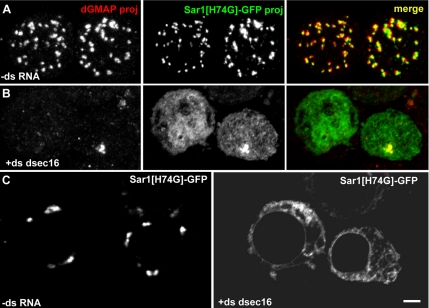
We next investigated whether Sar1 localization depends on dSec16. To do so, we localized Sar1[H74G] in dSec16-depleted S2 cells. As expected, in mock-treated cells, Sar1[H74G] localized to tER sites (Figure 5A), but depletion of dSec16 led to the complete relocalization of the GTPase in a dispersed pattern (Figure 5B), probably corresponding to the ER (Figure 5C), in agreement with the membrane association of GTP locked form of the small GTPAses (Novick and Zerial, 1997). This results shows that dSec16 recruits Sar1-GTP and concentrates it to tER sites.
Figure 5.
Sar1 localization to tER sites depends on dSec16. (A–C) IF localization of Sar1[H74G]-GFP (green) and the Golgi marker dGMAP (red) in mock-treated (−dsRNA, A) and dSec16-depleted S2 cells (+ds dsec16, B). Note that depletion of dSec16 completely delocalizes Sar1[H74G]-GFP from the tER sites to the ER (C). In A and B, the panels are projections. In C, the panels are confocal sections. Scale bars, 5 μm.
However, Sar1[H74G] dispersion could be an indirect effect of the transport inhibition caused by Sec16 depletion. To test this, we analyzed the distribution of Sar1[H74G] when ER-to-Golgi transport was inhibited by BFA, as shown by the localization of the Golgi protein Fringe-GFP in the ER (Supplementary Figure S3B). Under BFA treatment, the tER localization of Sar1[H74G]-GFP was largely unchanged when compared with mock-treated cells (Supplementary Figure S3A), though a faint nuclear envelope staining was visible.
This shows that its dispersion upon dSec16 depletion is not due to anterograde transport inhibition, but to the loss of dSec16 protein. This suggests that dSec16 functions upstream of Sar1-GTP, concentrating it from the ER to ER cups, where it can drive the local budding of COPII-coated vesicles, thus inducing tER biogenesis.
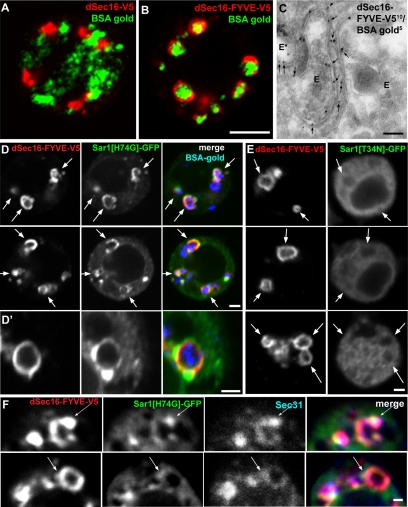
If this hypothesis is correct, dSec16 should be able recruit COPII components, including Sar1-GTP, to a membrane to which it is ectopically localized. To test this, dSec16-V5 was fused to a tandem FYVE domain, known to mediate localization to endosomes (Gillooly et al., 2000). Accordingly, dSec16-FYVE-V5 was found to significantly localize on large cytoplasmic ring-like structures (2–5 per cell confocal section, Figure 6B), shown by BSA-gold uptake and IEM to correspond to endosomes (Figure 6, B and C). As predicted, dSec16-V5 did not colocalize with BSA-gold (Figure 6A).
Figure 6.
Ectopically localized Sec16 recruits Sar1-GTP on endosomes. (A and B) IF localization of dSec16-V5 (A) or dSec16-FYVE-V5 (B) in S2 cells that have internalized BSA-gold (10 nm) using an anti-V5 antibody (red). BSA-gold is visualized by reflective light. Note that dSec16-FYVE-V5 localizes in ring-like compartments positive for BSA-gold. (C) IEM localization of dSec16-FYVE-V5 in S2 cells that have internalized 5 nm BSA-gold using an anti V5 antibody (10 nm). dSec16-FYVE-V5 localizes to endosomes (E), one of them marked by BSA-gold (E*). (D and E) Gallery of IF localization of Sar1[H74G]-GFP (green, D and D′) and Sar1[T34N]-GFP (E) in cells expressing dSec16-FYVE-V5 (red) fed with BSA-gold (blue, D and D′). D′ shows a single endosome at high magnification. The Sar1[H74G]/dSec16/BSA gold positive endosomes are marked by arrows. Note that there are no Sar1[T34N]-positive Sec16 rings. (F) Gallery of IF colocalization of dSec16-FYVE-V5 (red), Sar1[H74G]-GFP (green) and endogenous Sec31(blue) on endosomes (arrows). All the panels presented in this figures are confocal sections. Scale bars, (A and B) 5 μm, (C) 200 nm, and (D–F) 2 μm.
Sar1[H74G]-GFP was then cotransfected with dSec16-FYVE-V5. We chose this form of dSar1 to bypass the requirement for the GEF activity of Sec12. Sar1[H74G] was found to significantly colocalize to dSec16-FYVE/BSA-gold positive endosomes (Figure 6D, arrows). 44% of the endosomes labeled by ectopic dSec16 were also positive for Sar1[H74G]-GFP versus 2.8% for dSar1[T34N]-GFP (Figure 6E). Furthermore, Sar1[H74G] transfected to wild-type cells (Supplementary Figure 2E) or cotransfected with dSec16-V5 (not shown) was not localized to endosomes. Importantly, we also found that other COPII components, such as Sec23 (not shown) and Sec31 (Figure 5F) colocalized with Sar1[H74G] and ectopic dSec16 on endosomes. These results show that dSec16 is able to recruit COPII components, including activated Sar1 at the site of its localization.
Together with the loss of Sar1 localization upon dSec16 depletion and the fact that, conversely, dSec16 localization does not change upon Sar1 or Sec23 depletion, this experiment suggests that dSec16 recruits the COPII machinery, starting with the concentration of the GTP loaded Sar1 to ER cups (alone or as a prebudding complex), where it drives the biogenesis of tER sites.
A tER Specificity Motif Is Mapped to a 65-Amino Acid Arginine-Rich Domain
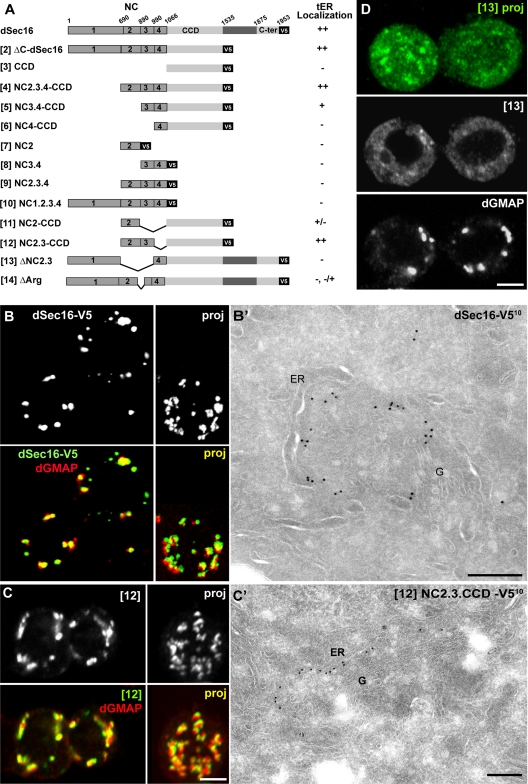
As dSec16 is not recruited to the tER through binding to COPII components, we set out to identify the domain mediating its localization. To do so, we generated a series of V5 tagged C-terminal truncations of dSec16 (Figure 7A) and analyzed their intracellular localization by IF and IEM.
Figure 7.
Mapping of the dSec16 domain that mediates its localization to tER sites. (A) Schematic representation of the truncation and deletion mutants of dSec16. The numbers these constructs are referred to in the text are indicated in between brackets. The degree of colocalization of these mutants with dGMAP was evaluated. ++, Strong colocalization; +, moderate colocalization; −, hardly or no colocalization. (B–D) IF localization of the indicated constructs tagged with V5 (green) and the Golgi marker dGMAP (red) (B–D) and by IEM using the anti-V5 antibody (10 nm) (B′ and C′). In B and C, the merge of both confocal sections and projections are presented, whereas in D only single channels are shown. NC2.3-CCD-V5 [12] and dGMAP colocalize extensively. Note the fragmented and dispersed pattern of ΔNC2.3-dSec16-V5 [13] and its very poor colocalization with dGMAP (D). Scale bars, (B–D) 5 μm, (B′ and C′) 200 nm.
dSec16-V5 localized very efficiently to tER sites in a pattern indistinguishable from the endogenous dSec16. By IF, it overlapped with the Golgi marker dGMAP (Figure 7B), and by IEM, it labeled tER sites (Figure 7B′). It appeared, though, that dSec16-V5 is more restricted to the ER cup outlining the tER sites when compared with the endogenous dSec16 in control cells that was found more prominently on the vesicular-tubular pleiomorphic membrane (compare Figures 7B′ and 1D, see Discussion). Removing the 400 C-terminal amino acids (ΔC-dSec16-V5), did not change the localization characteristics (Supplementary Figure S4, A and A′ construct [2]), showing that the C-terminus is dispensable. Accordingly, the C-terminal conserved domain alone localized to membrane in a nonspecific manner (ER, Plasma membrane, Golgi; not shown).
We then generated further truncations of ΔC-dSec16-V5 (Figure 7A). Alone, the CCD behaved as the C-terminus (Supplementary Figure S4B, [3]), indicating perhaps the presence of a general membrane-binding domain lacking specificity to confer tER localization. However, it is necessary for the localization to tER sites, as truncations lacking it were mislocalized (Figure 7A, [7, 8, 9, and 10]). We found that the CCD domain extended at its N-terminus by 376 amino acids (NC2.3.4-CCD) is sufficient to mediate localization to tER sites (Supplementary Figure S4C, [4]). An extension of 176 amino acids only (NC3.4-CCD) was less efficient (Supplementary Figure S4D, [5]), suggesting that NC2 is necessary.
To test this further, we designed a chimera encompassing the CCD extended at its N-terminus by NC2 (NC2-CCD) that was poorly localized to tER sites (Supplementary Figure S4E, [11]). When the CCD was extended by NC2.3 (NC2.3-CCD), we recovered full tER site localization and decoration of the ER cup (Figure 7, C and C′, [12]). To confirm the role of NC2.3 in the localization of dSec16, we deleted this region (aa 690–989) from dSec16-V5. This chimera (ΔNC2.3) did not properly localize to tER sites and became largely cytoplasmic (Figure 7D, [13], Figure 8C). Taken together, these results show that the domain mediating the localization of dSec16 to tER sites (NC2.3) is located in the nonconserved region. On its own, however, it does not confer the specificity for tER membrane binding that requires the presence of the CCD.
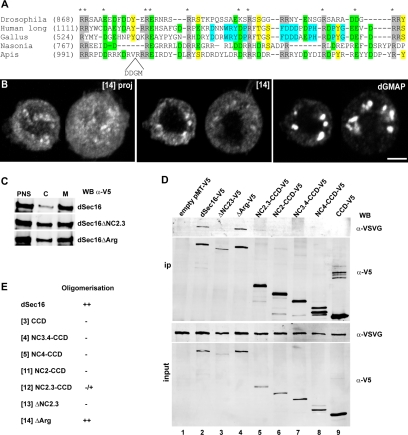
Figure 8.
The arginine-rich domain is involved in the dSec16 localization to tER sites. (A) Alignment of the arginine-rich domain of Sec16 in Drosophila (CG 32654E), in human (hSec16L, KIAA0310), in fowl (Gallus gallus, XP_415419), in wasp (Nasonia vitripennis, XP_001604818), and in bee (Apis, XP_393378). The number in brackets indicates the start of the motif in each sequence. The conserved arginine residues (R) are outlined in gray, the conserved acidic amino acids (D and E), in green, and the conserved S, T, and Y in yellow. In blue, amino acids conserved between human and Gallus (and Xenopus, not shown) are indicated. The conserved amino acids in all five species are marked by a star. (B) IF localization of ΔArg-dSec16-V5 [14] and the Golgi marker dGMAP. Their colocalization is very reduced and the ΔArg-dSec16-V5 labeling is mostly dispersed. The left-hand panel is a projection, whereas the two other panels are confocal sections. (C) Subcellular fractionation of dSec16-V5–, ΔNC2.3-dSec16-V5–, and ΔArg-dSec16-V5–expressing S2 cells as described in Materials and Methods. Both ΔNC2.3-dSec16-V5 and ΔArg-dSec16-V5 show an increase in the cytosolic pool compared with dSec16-V5. (D) Western blotting of the immunoprecipitated dSec16 oligomeric complex. dSec16-VSVG was cotransfected in S2 cells with the indicated V5-tagged dSec16 constructs. Cells were lysed in 1% Triton X-100–containing buffer and subjected to immunoprecipitation (ip) with an anti-V5 antibody. The ip was immunoblotted with the anti-VSVG and the anti-V5 antibodies. Scale bars, 5 μm (B).
hSec16 has been shown to oligomerize (Bhattacharyya and Glick, 2007; see below). To rule out that the observed localization of our truncated proteins is due to oligomerization with the endogenous dSec16, we monitored the localization of ΔC-dSec16-V5 (Supplementary Figure S4F, [2]) and NC2.3-CCD (Supplementary Figure S4G, [12]) in dSec16-depleted cells. Both localized in puncta that by IEM were shown to correspond to cupped ER membrane (Supplementary Figure S4F′), indicating that their tER site localization is not due to oligomerization with the endogenous protein.
The tER localization domain in hSec16L is also found in the nonconserved 300-amino acid sequence upstream of the CCD (924–1227; Bhattacharyya and Glick, 2007). When NC2.3 was aligned to this region, several residues appeared conserved, of which two consensus sequences for PKC sites at positions 807 and 861 and a 65-amino acid arginine-rich domain (aa 860–925) present in many species (Figure 8A). A role of a putative PKC-mediated phosphorylation in the dSec16 localization was proven negative. Although dSec16-V5 is phosphorylated (unpublished results), the mutation of the two putative PKC sites to alanine did not abrogate its localization to tER sites (unpublished results), even in dSec16-depleted cells to avoid oligomerization with the endogenous protein (see below).
The arginine-rich domain, however, has an important role. Expression of a dSec16-V5 mutant lacking the 65 amino acids covering this domain (860–925, ΔArg-V5, Figure 7A [14]) resulted in a strong cytoplasmic distribution of the protein (compare Figure 8B to 7B), though the tER sites localization was not completely abrogated. The residual localization of ΔArg-V5 to the tER sites can be explained by its binding to the localized coat components and their oligomerization to the endogenous protein (see below). The fractionation experiment confirmed a 30–40% increase the cytosolic pool of ΔArg-V5 and ΔNC2.3-V5 relative to the full-length protein (Figure 8C).
Taken together, these results show that the arginine-rich domain is involved in conferring the specificity for dSec16 localization to tER sites.
The Localization of dSec16 to tER Site Does Not Require its Oligomerization
The function of Sec16 in the organization of ER exit sites has been proposed to depend on its self-oligomeric properties (Connerly et al., 2005; Bhattacharyya and Glick, 2007). We first tested whether dSec16 also exhibits oligomeric properties in vivo. Transfected dSec16-V5 and dSec16-VSVG were efficiently coimmunoprecipitated from detergent extract of S2 cells, showing that dSec16 oligomerizes (Figure 8D, lane 2). However, dSec16-missing NC2.3 (ΔNC2.3-V5, [13]) did not (Figure 8D, lane 3), suggesting that NC2.3 is required for oligomerization.
We next asked whether dSec16 oligomerization and localization were linked. Because dSec16 missing the arginine-rich domain (ΔArg-V5, [14]) does not localize to tER sites very efficiently (Figure 8B), we tested whether ΔArg-V5 oligomerizes with dSec16-VSVG. They did, as strongly as dSec16 with itself (compare Figure 8D, lane 4 with lane 2). This suggests that oligomerization is not sufficient to mediate localization. Conversely, NC2.3-CCD-V5, which localizes very efficiently to tER sites (Figure 7, A, and C), oligomerized very poorly with dSec16-VSVG (Figure 8D, lane 5). A similar result was obtained with NC3.4-CCD, and importantly, the CCD alone did not oligomerize.
Taken together, these results show that NC2.3 is required but not sufficient for oligomerization and suggest that the tER site localization and oligomerization are two independent properties.
DISCUSSION
Here, using a combination of biochemical methods, RNAi depletion and retransfection followed by immunofluorescence and immunoelectron microscopy analyses, we present evidence that dSec16, the Drosophila orthologue of human and yeast Sec16, is part of a tER scaffold regulating where tER sites assemble on the ER. dSec16 binds to ER cups through an arginine-rich domain and concentrates the active form of Sar1, either alone or as a prebudding complex together with Sec23/24 (Kuehn et al., 1998), thus initiating COPII coat formation.
dSec16 shares at least four critical features with human and P. pastoris Sec16. They all are large hydrophilic proteins (above 250-kDa) that exhibit a CCD and a conserved C-terminus of ∼200 aa. They localize to tER sites and their inactivation by depletion or mutation leads to disruption of the functional organization of tER sites, causing in turn a severe inhibition in anterograde transport. Last, they display an increased solubility in ionic over detergent based buffers (this study and Supek et al., 2002; Watson et al., 2006).
dSec 16 Localization to tER Sites Is Specified by an Arginine-Rich Domain
As for the long form of human Sec16, hSec16L, the domain mediating dSec16 localization has been mapped to 300 amino acids present in the nonconserved region upstream of the CCD (this study and Bhattacharyya and Glick, 2007). For hSec16, this domain is both necessary and sufficient to mediate its tER site localization, but not for dSec16. In Drosophila, this domain has to be expanded by the conserved central domain that on its own localizes to all intracellular membrane. This indicates that the 300-amino acid domain confers tER specificity and that the two domains cooperate to bring about the binding of dSec16 to tER site membrane.
The dSec16 tER specificity motif was further narrowed to a 65-amino acid arginine-rich domain that is also present in hSec16L and other species. Arginine-rich domains have been shown to interact with ionic lipids (Hitz et al., 2006), to remodel membrane (Shaw et al., 2007), and even to cross membrane (Futaki, 2005), as it is the case for the twin arginine transport (TAT) pathway (Sargent, 2007). It remains to be tested whether the arginine-rich domain of dSec16 has such properties.
As hSec16, dSec16 also oligomerizes. However, oligomerization is not necessarily followed by efficient tER site localization, and conversely, localization can occur without oligomerization. This suggests that these two properties are independent from one another. However in vivo, they both are likely to be used in parallel or in a sequential manner. We propose that the recruitment of dSec16 to the ER cups could be followed by its oligomerization, which in turn strengthens its localization (see below). Alternatively, Sec16 could oligomerize in the cytoplasm and be recruited as a performed oligomer to the ER cups. What drives the initial recruitment needs to be further investigated (see below).
dSec16 Acts as a tER Matrix Upstream of COPII
We provide here several lines of evidence that dSec16 acts upstream of the COPII machinery in defining the ER subdomain at which tER sites are built. First, we show that dSec16 can still associate to distinct ER cups in the absence of COPII coat formation, either upon Sec23 depletion or in the absence of (functional) Sar1 (depletion and expression of the inactive Sar1[T34N] mutant).
Second, as its yeast homologue (Supek et al., 2002), dSec16 interacts with Sar1-GTP, and dSec16 is required to concentrate it to tER sites. Depletion of dSec16 led to a significant loss of Sar1-GTP from tER sites, and the ectopic localization of a chimeric form of dSec16 to endosomes results in the quantitative recruitment of Sar1 to the same compartment. These findings are not in strict agreement with the conclusions drawn from studies in human cells, suggesting that Sar1 recruits/maintains Sec16 to tER sites. The first study shows that overexpression of GTP-locked Sar1 leads to an apparent dispersal of endogenous hSec16, whereas transfected Venus-hSec16 appears enriched in crescent-like Sar1-positive structures reminiscent of tER sites (Watson et al., 2006). In a second study, the membrane association of hSec16 (measured in vitro) is shown to increase upon addition of GTP-loaded Sar1 (Iinuma et al., 2007). However, hSec16 was not dissociated from membrane in the presence of excess of GDP-loaded form of Sar1, suggesting that the initial binding of Sec16 to membrane does not depend on Sar1.
Taken together and in agreement with data showing that Sec16 stabilizes Sar1-GTP (Supek et al., 2002), our results demonstrate that Sec16 not only acts upstream of the structural COPII coat subunits, but also upstream of Sar1, suggesting that Sec16 is part of an initial molecular platform (or tER matrix) that regulates the biogenesis of the tER sites by nucleating COPII coat assembly and vesicle budding.
Our results, however, do not exclude a role for Sar1 in the tER site organization in influencing directly or indirectly dSec16 functions. Indeed, the number of tER sites labeled by dSec16 under conditions where Sar1 is inactivated or depleted is reduced and their size is larger when compared with control cells. Several explanations are possible: First, Sar1 has been shown to activate a PI4 kinase (Blumental-Perry et al., 2006), and as such, it indirectly contributes to the generation of phospholipids, such as PI4P on the ER membrane, that are required for tER site maintenance. Second, Sar1 depletion could impede on the dynamics of tER site fusion/fission (Bevis et al., 2002; Stephens, 2003), thus leading to larger and less numerous sites. Last, the binding of Sar1-GTP to dSec16 could change the properties of dSec16, for instance modulating its oligomeric state. On its own, Sec16 would form very large oligomers in a reduced number of spots, but the binding of Sar1-GTP would drive the dissociation of monomeric Sec16 from the oligomer, thus allowing its incorporation in the forming COPII vesicles (see below). This will have to be further investigated.
Localization Versus Function
Given the large size and multidomain structure of the protein, the role of Sec16 as a tER matrix does not exclude an additional role in COPII vesicle formation and budding (Kaiser and Schekman, 1990). In yeast and human, Sec16 C-terminus has been shown to bind Sec23 (Espenshade et al., 1995; Bhattacharyya and Glick, 2007). As this domain is also found and conserved in dSec16, it is likely that its interaction with Sec23 will also be conserved in Drosophila.
We propose that these two independent functions of Sec16 are reflected in its localization. dSec16 is located to the ER cup where COPII components are initially recruited to form nascent COPII vesicles that bud, uncoat, and form pleiomorphic membrane clusters, which also contain dSec16. In control cells, endogenous dSec16 is mostly found on this membrane, but under conditions where COPII budding is inhibited (in Sar1 depletion), the localization shifts toward the ER cup. dSec16 is never seen diffused in the entire ER. Whether the ER cups pre-exist as such or are formed upon dSec16-binding remains to be established.
Strikingly, transfected dSec16 constructs localize also preferentially to ER cups, suggesting that they recapitulate the initial association of dSec16 to ER membrane, but not its incorporation in COPII vesicles, resulting in the inhibition of anterograde transport. This could be explained by the fact that the function of Sec16 in transport depends drastically on its stoichiometry. Sec16 overexpression causes a lethal secretion defect in yeast (Espenshade et al., 1995) and inhibition of ER export in mammalian cells (Watson et al., 2006). The mapping of the domains required for transport by using the usual transfection techniques will therefore be difficult. Genome tagging under endogenous promoters, so far only feasible in yeast, will prevent this overexpression.
dSec16 Acts as a tER Scaffold Involved in the Biogenesis of Functional tER Sites
We propose a model for tER site biogenesis in which dSec16 dictates the site of COPII coat component recruitment and vesicle budding, before being incorporated in the vesicles themselves. First, dSec16 binds to ER cups upstream of the COPII coat and Sar1, through the dual activity of its central conserved domain and its arginine-rich domain. Second, localized dSec16 assembles into oligomeric complexes, possibly increasing its stability in the ER cup. Third, concomitant to dSec16 localizing in ER cups, Sar1-GTP is generated on ER membrane due to the GEF activity of Sec12, itself exhibiting an ER localization in S2 cells (not shown) and other organisms (Weissman et al., 2001). Fourth, Sar1-GTP diffuses in the plane of the ER and reaches ER cups enriched in dSec16 oligomers to which it binds, leading to its concentration and the initiation of the COPII coat assembly.
Alternatively, before binding to Sec16, Sar1 is perhaps already part of the prebudding complex with Sec23/24 (Kuehn et al., 1998). As dSec16 does not interact with the GDP loaded form of Sar1, we propose that Sec16 acts in parallel with Sec12, though Sec16 itself might also be capable of recruiting first Sec12 from the ER to the ER cup. In Pichia, Sec12 associates to the tER sites by binding to a partner protein (Soderholm et al., 2004), and it would be tempting to speculate that this protein is Sec16. Once the coat is formed, Sec16 associates to it, leading to the stabilization of the COPII coat and its incorporation into functional COPII vesicles. The Sec16 molecule that associates to the coat can either be recruited from the cytosol or dissociates from the oligomers.
Whether Sec16 is the most upstream factor driving the building of tER sites remains to be established. Although its putative membrane deformation properties and oligomerization properties (see above) could lead to its self-assembly to tER sites, other factors are likely to be involved. Phosphatidylinositol 4-phosphate has been recently shown to be locally regulated at tER sites (Blumental-Perry et al., 2006), and depletion of the phospholipase A1-like protein p125, specifically disrupts the organization of tER sites, but not their function (Shimoi et al., 2005). In addition, we cannot rule out the existence of a receptor. The first half of the dSec16 localization domain encompasses a short coiled-coil known to mediate protein–protein interactions. Furthermore, dSec16 overexpression does not cause tER site enlargement (this study and Connerly et al., 2005), suggesting a saturable binding. Identification of this potential receptor and other binding partners will shed light onto the mechanism of dSec16-membrane interaction.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Department of Cell Biology for helpful discussions, Dr. Fulvio Reggiori for critically reading the manuscript, and our colleagues for reagents mentioned in the Materials and Methods. V.I. is supported by an Aard en Levenswetenschappen grant from the Nederlandse Organisatie voor Wetenschappelijke Onderzoek (NWO, 816.02.004), and V.K. and G.d.V. by a ZonMw Grant NWO 912-04-009 to C.R.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-03-0246) on July 9, 2008.
REFERENCES
- Aridor M., Bannykh S. I., Rowe T., Balch W. E. Sequential coupling between COPII and COPI vesicle coats in endoplasmic reticulum to Golgi transport. J. Cell Biol. 1995;131:875–894. doi: 10.1083/jcb.131.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aridor M., Fish K. N., Bannykh S., Weissman J., Roberts T. H., Lippincott-Schwartz J., Balch W. E. The Sar1 GTPase coordinates biosynthetic cargo selection with endoplasmic reticulum export site assembly. J. Cell Biol. 2001;152:213–229. doi: 10.1083/jcb.152.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevis B. J., Hammond A. T., Reinke C. A., Glick B. S. De novo formation of transitional ER sites and Golgi structures in Pichia pastoris. Nat. Cell Biol. 2002;4:750–756. doi: 10.1038/ncb852. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya D., Glick B. S. Two mammalian Sec16 homologues have nonredundant functions in endoplasmic reticulum (ER) export and transitional ER organization. Mol. Biol. Cell. 2007;18:839–849. doi: 10.1091/mbc.E06-08-0707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumental-Perry A., Haney C. J., Weixel K. M., Watkins S. C., Weisz O. A., Aridor M. Phosphatidylinositol 4-phosphate formation at ER exit sites regulates ER export. Dev. Cell. 2006;11:671–682. doi: 10.1016/j.devcel.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Bonifacino J. S., Glick B. S. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- Boyadjiev S. A., et al. Cranio-lenticulo-sutural dysplasia is caused by a SEC23A mutation leading to abnormal endoplasmic-reticulum-to-Golgi trafficking. Nat. Genet. 2006;38:1192–1197. doi: 10.1038/ng1876. [DOI] [PubMed] [Google Scholar]
- Bi X., Mancias J. D., Goldberg J. Insights into COPII coat nucleation from the structure of Sec23.Sar1 complexed with the active fragment of Sec31. Dev. Cell. 2007;13:635–645. doi: 10.1016/j.devcel.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connerly P. L., Esaki M., Montegna E. A., Strongin D. E., Levi S., Soderholm J., Glick B. S. Sec16 is a determinant of transitional ER organization. Curr. Biol. 2005;15:1439–1447. doi: 10.1016/j.cub.2005.06.065. [DOI] [PubMed] [Google Scholar]
- Espenchade P., Gimeno P. E., Holzmacher E., Teung P., Kaiser C. A. Yeast SEC16 gene encodes a multidomain vesicle coat protein that interacts with sec23p. J. Cell Biol. 1995;131:311–324. doi: 10.1083/jcb.131.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friggi-Grelin F., Rabouille C., Therond P. The cis-Golgi Drosophila GMAP has a role in anterograde transport and Golgi organization in vivo, similar to its mammalian ortholog in tissue culture cells. Eur. J. Cell Biol. 2006;85:1155–1166. doi: 10.1016/j.ejcb.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Fromme J. C., Ravazzola M., Hamamoto S., Al-Balwi M., Eyaid W., Boyadjiev S. A., Cosson P., Schekman R., Orci L. The genetic basis of a craniofacial disease provides insight into COPII coat assembly. Dev. Cell. 2007;13:623–634. doi: 10.1016/j.devcel.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futaki S. Membrane-permeable arginine-rich peptides and the translocation mechanisms. Adv. Drug Deliv. Rev. 2005;57:547–558. doi: 10.1016/j.addr.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Gillooly D. J., Morrow I. C., Lindsay M., Gould R., Bryant N. J., Gaullier J. M., Parton R. G., Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno R. E., Espenshade P., Kaiser C. A. SED4 encodes a yeast endoplasmic reticulum protein that binds sec16p and participates in vesicle formation. J. Cell Biol. 1995;131:325–338. doi: 10.1083/jcb.131.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno R. E., Espenshade P., Kaiser C. A. COPII coat subunit interactions: Sec24p and Sec23p bind to adjacent regions of Sec16p. Mol. Biol. Cell. 1996;11:1815–1823. doi: 10.1091/mbc.7.11.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond A. T., Glick B. S. Dynamics of transitional endoplasmic reticulum sites in vertebrate cells. Mol. Biol. Cell. 2000;11:3013–3030. doi: 10.1091/mbc.11.9.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C. Y., Pypaert M., Warren G. Golgi duplication in Trypanosoma brucei requires Centrin2. Science. 2005;310:1196–1198. doi: 10.1126/science.1119969. [DOI] [PubMed] [Google Scholar]
- Hitz T., Iten R., Gardiner J., Namoto K., Walde P., Seebach D. Interaction of alpha-and beta-oligoarginine-acids and amides with anionic lipid vesicles: a mechanistic and thermodynamic study. Biochemistry. 2006;45:5817–5829. doi: 10.1021/bi060285d. [DOI] [PubMed] [Google Scholar]
- Hughes H., Stephens D. Assembly, organization, and function of the COPII coat. Histochem. Cell Biol. 2008;129:129–151. doi: 10.1007/s00418-007-0363-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iinuma T., Shiga A., Nakamoto K., O'Brien M. B., Aridor M., Arimitsu N., Tagaya M., Tani K. Mammalian Sec16/p250 plays a role in membrane traffic from the endoplasmic reticulum. J. Biol. Chem. 2007;282:17632–17639. doi: 10.1074/jbc.M611237200. [DOI] [PubMed] [Google Scholar]
- Kaiser C. A., Schekman R. Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell. 1990;61:723–733. doi: 10.1016/0092-8674(90)90483-u. [DOI] [PubMed] [Google Scholar]
- Klueg K. M., Parody T. R., Muskavitch M. A. T. Complex proteolytic processing acts on Delta, a transmembrane ligand for Notch, during Drosophila development. Mol. Biol. Cell. 1998;9:1709–1723. doi: 10.1091/mbc.9.7.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis V., Rabouille C. A novel role for dp115 in the organization of tER sites in Drosophila. J. Cell Biol. 2003;162:185–198. doi: 10.1083/jcb.200301136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis V., Spoorendonk K. M., Rabouille C. dGRASP localization and function in the early exocytic pathway in Drosophila S2 cells. Mol. Biol. Cell. 2005;16:4061–4072. doi: 10.1091/mbc.E04-10-0938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondylis V., van Nispen tot Pannerden H. E., Herpers B., Friggi-Grelin F., Rabouille C. The Golgi comprises a paired stack that is separated at G2 by modulation of the actin cytoskeleton through Abi and Scar/WAVE. Dev. Cell. 2007;12:901–915. doi: 10.1016/j.devcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Kuehn M. J., Herrmann J. M., Schekman R. COPII-cargo interactions direct protein sorting into ER-derived transport vesicles. Nature. 1998;391:187–190. doi: 10.1038/34438. [DOI] [PubMed] [Google Scholar]
- Kuge O., Dascher C., Orci L., Rowe T., Amherdt M., Plutner H., Ravazzola M., Tanigawa G., Rothman J. E., Balch W. E. SAR1p promotes vesicle budding from the endoplasmic reticulum but not the Golgi compartments. J. Cell Biol. 1994;125:51–66. doi: 10.1083/jcb.125.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogelsvang S., Gomez-Ospina N., Soderholm J., Glick B. S., Staehelin L. A. Tomographic evidence for continuous turnover of Golgi cisternae in Pichia pastoris. Mol. Biol. Cell. 2003;14:2277–2291. doi: 10.1091/mbc.E02-10-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S., Freeman M. The Notch signaling receptor Fringe acts in the Golgi apparatus and requires the glycosyltransferase signature motif DXD. Curr. Biol. 2000;10:813–820. doi: 10.1016/s0960-9822(00)00578-9. [DOI] [PubMed] [Google Scholar]
- Novick P., Zerial M. The diversity of Rab proteins in vesicle transport. Curr. Opin. Cell Biol. 1997;9:496–504. doi: 10.1016/s0955-0674(97)80025-7. [DOI] [PubMed] [Google Scholar]
- Prescott A. R., Farmaki T., Thomson C., James J., Paccaud J.-P., Tang B.-L., Hong W., Quinn M., Ponnambalam S., Lucocq J. Evidence for prebudding arrest of ER export in animal cell mitosis and its role in generating Golgi partitioning intermediates. Traffic. 2001;2:321–335. doi: 10.1034/j.1600-0854.2001.002005321.x. [DOI] [PubMed] [Google Scholar]
- Rossanese O. W., Soderholm J., Bevis B. J., Sears I. B., O'Connor J., Williamson E. K., Glick B. S. Golgi structure correlates with transitional endoplasmic reticulum organization in Pichia pastoris and Saccharomyces cerevisiae. J. Cell Biol. 1999;145:69–81. doi: 10.1083/jcb.145.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe T., Aridor M., McCaffery J. M., Plutner H., Nuoffer C., Balch W. E. COPII vesicles derived from mammalian endoplasmic reticulum microsomes recruit COPI. J. Cell Biol. 1996;135:895–912. doi: 10.1083/jcb.135.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent F. The twin-arginine transport system: moving folded proteins across membranes. Biochem. Soc. Trans. 2007;35:835–847. doi: 10.1042/BST0350835. [DOI] [PubMed] [Google Scholar]
- Shaw J. E., Epand R. F., Hsu J. C. Y., Mo G. C. H., Epand R. M., Yip C. M. Cationic peptide-induced remodelling of model membranes: direct visualization by in situ atomic force microscopy. J. Struct. Biol. 2008;162:121–138. doi: 10.1016/j.jsb.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Shaywitz D. A., Espenshade P. J., Gimeno R. E., Kaiser C. A. COPII subunit interactions in the assembly of the vesicle coat. J. Biol. Chem. 1997;272:25413–25416. doi: 10.1074/jbc.272.41.25413. [DOI] [PubMed] [Google Scholar]
- Shimoi W., Ezawa I., Nakamoto K., Uesaki S., Gabreski G., Aridor M., Yamamoto A., Nagahama M., Tagaya M., Tani K. p125 is localized in endoplasmic reticulum exit sites and involved in their organization. J. Biol. Chem. 2005;280:10141–10148. doi: 10.1074/jbc.M409673200. [DOI] [PubMed] [Google Scholar]
- Soderholm J., Bhattacharyya D., Strongin D., Markovitz V., Connerly P. L., Reinke C. A., Glick B. S. The transitional ER localization mechanism of Pichia pastoris Sec12. Dev. Cell. 2004;6:649–659. doi: 10.1016/s1534-5807(04)00129-7. [DOI] [PubMed] [Google Scholar]
- Stephens D. J. De novo formation, fusion and fission of mammalian COPII-coated endoplasmic reticulum exit sites. EMBO Rep. 2003;4:210–217. doi: 10.1038/sj.embor.embor736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek F., Madden D. T., Hamamoto S., Orci L., Schekman R. Sec16p potentiates the action of COPII proteins to bud transport vesicles. J. Cell Biol. 2002;158:1029–1038. doi: 10.1083/jcb.200207053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson P., Townley A. K., Koka P., Palmer K. J., Stephens D. J. Sec16 defines endoplasmic reticulum exit sites and is required for secretory cargo export in mammalian cells. Traffic. 2006;7:1678–1687. doi: 10.1111/j.1600-0854.2006.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman J. T., Plutner H., Balch W. E. The mammalian guanine nucleotide exchange factor mSec12 is essential for activation of the Sar1 GTPase directing endoplasmic reticulum export. Traffic. 2001;2:465–475. doi: 10.1034/j.1600-0854.2001.20704.x. [DOI] [PubMed] [Google Scholar]
- Yano H., Yamamoto-Hino M., Abe M., Kuwahara R., Haraguchi S., Kusaka I., Awano W., Kinoshita-Toyoda A., Toyoda H., Goto S. Distinct functional units of the Golgi complex in Drosophila cells. Proc. Natl. Acad. Sci. USA. 2005;102:13467–13472. doi: 10.1073/pnas.0506681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.