Abstract
The endoplasmic reticulum (ER) is highly plastic, and increased expression of distinct single ER-resident membrane proteins, such as HMG-CoA reductase (HMGR), can induce a dramatic restructuring of ER membranes into highly organized arrays. Studies on the ER-remodeling behavior of the two yeast HMGR isozymes, Hmg1p and Hmg2p, suggest that they could be mechanistically distinct. We examined the features of Hmg2p required to generate its characteristic structures, and we found that the molecular requirements are similar to those of Hmg1p. However, the structures generated by Hmg1p and Hmg2p have distinct cell biological features determined by the transmembrane regions of the proteins. In parallel, we conducted a genetic screen to identify HER genes (required for Hmg2p-induced ER Remodeling), further confirming that the mechanisms of membrane reorganization by these two proteins are distinct because most of the HER genes were required for Hmg2p but not Hmg1p-induced ER remodeling. One of the HER genes identified was PSD1, which encodes the phospholipid biosynthetic enzyme phosphatidylserine decarboxylase. This direct connection to phospholipid biosynthesis prompted a more detailed examination of the effects of Hmg2p on phospholipid mutants and composition. Our analysis revealed that overexpression of Hmg2p caused significant and specific growth defects in nulls of the methylation pathway for phosphatidylcholine biosynthesis that includes the Psd1p enzyme. Furthermore, increased expression of Hmg2p altered the composition of cellular phospholipids in a manner that implied a role for PSD1. These phospholipid effects, unlike Hmg2p-induced ER remodeling, required the enzymatic activity of Hmg2p. Together, our results indicate that, although related, Hmg2p- and Hmg1p-induced ER remodeling are mechanistically distinct.
INTRODUCTION
The endoplasmic reticulum (ER) is the largest endomembrane compartment in most eukaryotic cell types, and it is a site of many and diverse cellular processes, including protein folding, drug detoxification, phospholipid, and sterol biosynthesis. The ER is extremely plastic, having the capacity to adapt to changing cellular need. For example, when lymphocytes differentiate into antibody-producing plasma cells, the volume of the ER increases by greater than fourfold (Wiest et al., 1990). In addition to this complex cellular differentiation cascade, elevation of certain single ER-resident proteins, such as HMG-CoA reductase (HMGR) or cytochrome P450 (P450), cause profound alterations in ER structure that are conserved from yeast to mammalian cells (Jones and Fawcett, 1966; Black, 1972; Wright et al., 1988; Menzel et al., 1996). Despite many studies of ER expansion and reorganization, the molecular mechanisms that underlie these structural changes have remained elusive (for review, see Federovitch et al., 2005). In particular, it remains an important question whether the same or distinct mechanisms mediate the ER remodeling caused by various proteins. Additionally, despite the supposed role of phospholipid synthesis in ER remodeling by HMGR, no direct connection has been made between these two phenomena. We focused our attention on the single-protein initiator of ER remodeling, HMGR, to explore these topics.
HMGR catalyzes the conversion of HMG-CoA to mevalonate, a key step in sterol biosynthesis. This protein is composed of three distinct domains: an N-terminal transmembrane region, a linker, and the highly conserved C-terminal cytoplasmic domain (CD) (Figure 1A). The large transmembrane domain spans the ER membrane eight times and is connected to the cytoplasmic domain via a flexible linker. The C-terminal CD is composed of two distinct subregions, depicted in Figure 1A: a proximal small helical N-domain (hN-domain) and the more C-terminal tightly folded multimerization domain, which contains the essential enzymatic residues (Istvan et al., 2000).
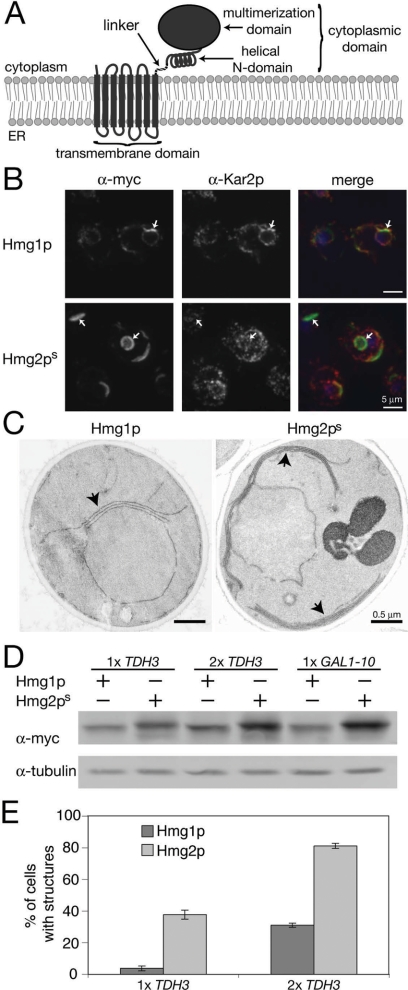
Figure 1.
Hmg1p and Hmg2ps make distinct structures. (A) Cartoon depicting the domains of HMGR. (B) Representative IF images of myc-tagged Hmg1p and Hmg2ps expressed at two copies per cell. HMGR is green, Kar2p is red, and cells are stained with DAPI. Arrows indicate characteristic structures. (C) Electron micrographs of membrane structures observed when two copies of Hmg1p or Hmg2ps are expressed from the TDH3pr. Arrows indicate membrane structures induced by Hmg1p and Hmg2ps. (D) Different expression levels of myc-tagged Hmg1p and Hmg2ps are shown by Western blot. Each protein is expressed from either the constitutive TDH3pr integrated at one (1× TDH3) or two (2× TDH3) genomic loci, or from the GAL1-10pr integrated at a single locus (1× GAL1-10). (E) Quantitation of cells with membrane structures expressing one or two TDH3pr-driven copies of Hmg1p or Hmg2ps. In three separate experiments, cells were harvested, prepared for IF, and a minimum of 200 cells was counted. Log phase cells were grown in YPD. Averages and standard deviations are shown.
Elevation of HMGR causes striking structural ER alterations, in both yeast and mammalian cells (Chin et al., 1982; Wright et al., 1988). Extensive arrays of smooth ER membranes are a prominent feature of cells in tissues that produce large quantities of sterols and hence have elevated HMGR expression (Sisson et al., 1967; Black, 1972). This cellular phenomenon has been recapitulated in a tissue culture line by increasing HMGR expression to >500-fold normal levels (Chin et al., 1982). Saccharomyces cerevisiae expresses two functional isozymes of HMGR, Hmg1p and Hmg2p (Basson et al., 1986). High levels of Hmg1p induce the formation of nuclear-associated stacks of membranes, called karmellae (Wright et al., 1988). Similarly, increased expression of Hmg2p causes the formation of cytoplasmic whorls and strips of membrane in addition to nuclear-associated stacks (Koning et al., 1996).
The Wright laboratory has extensively studied both the structural and genetic requirements for karmellae formation by Hmg1p. Their work revealed that both the transmembrane domain of Hmg1p and a large, properly folded cytoplasmic domain are required to generate karmellae (Parrish et al., 1995; Profant et al., 1999, 2000). Importantly, this cytoplasmic domain does not need to be the catalytically active, native cytoplasmic domain of Hmg1p; other folded domains that replace the CD of Hmg1p suffice. In addition, the Wright group has performed two genetic analyses of ER remodeling by Hmg1p. The first screen designed to identify genes required for karmellae formation uncovered a large group of Vacuolar Protein Sorting (VPS) genes that are required for vacuolar biogenesis (Koning et al., 2002). None of the genes identified affected Hmg2p-induced structures. The second screen was designed to identify viable null mutants that exhibited growth defects when Hmg1p was expressed at levels sufficient to induce karmellae. However, all nulls identified in this way were sensitive to the elevated HMGR enzymatic activity rather than the formation of karmellae (Wright et al., 2003; Loertscher et al., 2006).
Hmg1p and Hmg2p are similar both structurally and enzymatically. However, differences exist in how cells respond to each of these proteins. The cellular distribution of the structures made by Hmg1p and Hmg2p are distinct. Hmg2p causes characteristic nuclear-associated stacks of membrane, cytoplasmic strips, and whorls, whereas Hmg1p-induced karmellae are strictly nuclear-localized. Interestingly, Hmg1p-induced karmellae colocalize with the ER lumenal chaperone Kar2p, but the structures generated by Hmg2p exclude Kar2p (Koning et al., 1996). Finally, the genes required for Hmg1p-induced karmellae formation are not required for structures induced by Hmg2p. The above-mentioned studies from the Wright group indicated that the ER structures formed by the two HMGR isozymes have different structural, cell biological, and genetic features, which may manifest due to mechanistic differences.
In the current study, we have sought to further address these ideas by exploring the structural and genetic requirements for Hmg2p-induced ER remodeling. The specificity of the genes required for Hmg1p-induced karmellae led us to posit that a reciprocal analysis would uncover genes specific for Hmg2p-induced structures. We characterized the features of Hmg2p required to remodel the ER and in parallel ran a screen to discover the genes required for Hmg2p-induced structures, referred to as HER genes (Hmg2p-induced ER Remodeling). We have found that the structural features of Hmg2p required for ER remodeling are quite similar to those reported for Hmg1p. However, the genes that emerged from our screen were distinct from those involved in Hmg1p-induced karmellae formation. These HER genes were specifically required for Hmg2p-induced ER remodeling, but not for the remodeling that occurs in response to Hmg1p overexpression. Furthermore, among the genes discovered is the phospholipid biosynthetic enzyme phosphatidylserine decarboxylase 1 (PSD1). In S. cerevisiae, Psd1p generates ∼80% of the total phosphatidylethanolamine (PE) (Voelker, 1997), and as such it represents the first direct connection between ER remodeling and phospholipid biosynthesis. Psd1p is a key enzyme in one mode of phosphatidylcholine (PC) synthesis known as the methylation pathway. This connection led us to more fully investigate the role of phospholipid biosynthesis in Hmg2p-induced remodeling. We found that all of the enzymes of the methylation pathway have similar defects in Hmg2p-induced remodeling. Expression of Hmg2p at levels that caused ER remodeling also caused growth defects in nulls of the methylation pathway. Furthermore, overexpression of Hmg2p caused a significant alteration in phospholipid composition that suggested a role for PSD1. Surprisingly, these lipid effects were dependent both on the transmembrane region of Hmg2p implicated in ER remodeling and the enzymatic activity of Hmg2p. These studies define a specific, genetically distinct cellular response to Hmg2p that includes both changes to ER structure and total cellular lipid composition, connecting sterol and phospholipid pathways through expression of active Hmg2p.
MATERIALS AND METHODS
Strains, Recombinant DNA Methods, and Media
Yeast strains RHY471 (MATα ade2-101 met2 lys2-801 his3·200 ura3-52) and RHY2863 (MATa ade2-101 met2 lys2-801 his3·200 ura3-52 trp1·::hisG leu2·) were the parents for all strains in these experiments, except for RHY3013, which was used for the genetic screen. RHY3013 is derived from Y3656 (MATα lys2·0 ura3· leu2·0 his3·0 met15·0 can1·::MFA1pr-HIS3-MFα1pr-LEU2) (Tong et al., 2001). Plasmids were constructed using standard DNA techniques. Specific oligonucleotide sequences and plasmid construction methods will gladly be provided upon request. Hmg1p and Hmg2p expression plasmids were all integrating vectors. Plasmids were introduced into yeast according to standard techniques. Integrating plasmids were targeted to loci as follows: ADE2, and PflMI; URA3, StuI. Strains were verified for appropriate protein expression levels by flow cytometry for green fluorescent protein (GFP)-tagged constructs or by Western blot for epitope-tagged proteins. Gene knockouts were generated by polymerase chain reaction (PCR) amplification of KanMX or NatMX cassettes flanked by 50–300 bp of sequence to appropriately target for homologous recombination at the desired locus. Knockouts were verified by PCR amplification of the locus of interest. Strains were gown at 30°C in indicated media. Ethanolamine (E9508; Sigma-Aldrich, St. Louis, MO) or choline (C1879; Sigma-Aldrich) were prepared as 1 M stock solutions in water, sterile filtered, and added to media as indicated.
Electron Microscopy
Cells were prepared for electron microscopy (EM) as described by Wright (2000). Briefly, cells were grown to 0.65 ≤ OD600 ≥ 1.0 in YPD, fixed with 4% glutaraldehyde, postfixed with 2% potassium permanganate, en bloc stained with 1% uranyl acetate, dehydrated, and embedded in Spurr's resin. Eighty-nanometer-thick sections were cut and poststained with Sato lead before visualizing on a 1200 KeV electron microscope (JEOL, Tokyo, Japan).
Western Blot
Whole cell lysates were prepared via a general trichloroacetic acid (TCA) method. Two to 10 OD600 units of cells were collected and resuspended in 900 μl of water. Then, 100% TCA was added to 10% before a 30-min incubation on ice. Cells were pelleted, washed twice with acetone, and resuspended in 100 μl of TCA buffer (50 mM Tris, pH 7.5, 1 mM EDTA, 1% SDS, and 6 M urea). Glass beads (0.5 mm) were added before vortexing for 5 min. Lysates were heated to 50°C for 10 min before a 5-min spin at 13,000 rmp in a microfuge. Supernatant was transferred to a new tube, and protein was quantitated using the Bradford method. Ten micrograms of protein was loaded onto either 8 or 14% SDS-polyacrylamide gel electrophoresis gels, run electrophoretically, transferred onto nitrocellulose, and placed in Tris-buffered saline/Tween 20 (10 mM Tris, pH 8.0, 140 mM NaCl, and 0.05% Tween 20) with 2% Carnation nonfat dried milk. Blots were then incubated with either α-myc (9E10) 1:500 or rabbit α-tubulin (1:10,000), washed, and probed with goat anti-mouse horseradish peroxidase (HRP) or goat anti-rabbit HRP (1:12,500; Jackson ImmunoResearch Laboratories, West Grove, PA), rinsed with double-distilled H2O, and scanned on a Typhoon PhosphorImager after ECL Plus exposure (GE Healthcare, Chalfont St. Giles, Buckinghamshire, United Kingdom). Protein levels were then normalized to tubulin for expression level comparisons.
Immunofluorescence (IF)
The protocol for IF was adapted from Pringle et al. (1991). Briefly, 6–10 ml of cells was grown in YPD to 0.5 < OD600 >1.0. Formaldehyde was added to 4%, and cells were fixed for 1 h at room temperature. Cells were washed twice in 1 ml of IF buffer (1 mM KHPO4, pH 7.4, and 1.2 M sorbitol) and resuspended in 200 μl of IF buffer with 10 μg/ml Zymolyase 100T. Cells were incubated at 30°C for 20–25 min to spheroplast cells, followed by one wash with 1 ml of IF buffer and then a second wash with 1 ml of phosphate-buffered saline (PBS). Cells were resuspended in 1 ml of PBS and adhered to poly-l-lysine–coated preprinted slides (Carlson Scientific, Peotone, IL). Cells were blocked with 1% bovine serum albumin/0.5% Tween in PBS for 1 h, and then primary antibodies were added for 1 h at the following dilutions anti-myc ascites (9E10): 1:750, anti-hemagglutinin (HA) ascites 1:750, monoclonal anti-GFP (JL-8; Covance Research Products, Princeton, NJ), and anti-Kar2p: 1:1500. Wells were washed 5 × 5 min in block solution after each antibody incubation. Conjugated secondary antibodies were added at 1:500 for 40 min (goat α-rabbit Alexa Fluor 595, goat α-mouse Alexa Fluor 595, or goat α-mouse Alexa Fluor 488; Invitrogen, Carlsbad, CA). For the colocalization experiments with GFP-tagged HMGR, the GFP fluorescence was visualized, and IF of the epitope tag was used to visualize either 3myc-Cod1p/Spf1p, Hrd1p-HA, or Erg28p-HA. Due to the low abundance of Hrd1p-HA, cells were also incubated with rabbit α-mouse (1:100) and then mouse α-rabbit (1:100) unconjugated secondary antibodies (Jackson ImmunoResearch Laboratories), each for 40 min, before incubating with the appropriate conjugated secondary to amplify the signal. Wells were washed as described above, coated with Vectashield containing 4,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA), overlaid with a coverslip, and sealed with nail polish. Images were captured on a DM6000 spinning disk confocal unit (Leica, Wetzlar, Germany).
Genetic Screen
Strain RHY3013 expressed Hmg2ps-TM-hN-GFP from the inducible GAL1-10 promoter. This strain was crossed robotically to ∼4700 haploid deletion mutant collection, on glucose media, as described previously (Tong et al., 2001). The resultant collection of strains was patched onto fresh, selective glucose plates and grown for 24 h. We then replica plated onto raffinose, grew for 24 h more hours to eliminate glucose repression, and replica plated onto galactose plates and grew strains for another 18 h before scoring, to induce expression of GAL1-10pr-driven Hmg2ps-TM-hN-GFP. Cells from each strain were resuspended in a small volume of sterile double deionized (ddI) H2O and individually examined on an Optiphot II microscope (Nikon, Tokyo, Japan) equipped with GFP filters. Knockout strains that mislocalized Hmg2ps-TM-hN-GFP were noted as potential candidates.
Live Cell Imaging
Cells were grown to log phase 0.3 < OD600 > 0.6 in appropriate synthetic complete media, pelleted for 3 min at 6000 rpm in a tabletop microcentrifuge, and resuspended in 50 μl of PBS. Cells were then visualized on a DeltaVision microscope (Applied Precision, Issaquah, WA) equipped with appropriate filter sets. Images in Figure 7 were acquired on a DM6000 spinning disk confocal unit (Leica).
Figure 7.
Full-length active Hmg2ps causes a growth defect in strains deficient in the methylation pathway, but not the Kennedy pathway. (A) Diagram of the two major phospholipid biosynthetic pathways in S. cerevisiae. The Kennedy pathway is highlighted in blue, and the CDP-DAG/methylation pathway is highlighted in pink. (B) Live cell imaging of Hmg2ps-TM-hN-GFP in wild-type, cki1·, cpt1·, and ept1· null strains. Log phase cells were grown in SC-URA. Arrows indicate membrane structures. (C) Live cell imaging of Hmg2ps-TM-hN-GFP in wild-type, psd1·, cho2·, and opi3· null strains. Log phase cells were grown in SC-URA. Arrows indicate membrane structures. (D) Representative growth experiment of cells inoculated into synthetic complete liquid media lacking uracil and grown for ∼55 h at 30°C. Strains deficient in the methylation pathway do not grow to as high a density as wild-type strains in synthetic complete media (data not shown). When Hmg2ps is overexpressed (light bars) in psd1·, cho2·, or opi3· null strains, cells display a significant growth defect. This phenotype is not observed in cki1·, cpt1·, or ept1· null strains. Overexpression of Hmg2ps-TM-hN-GFP (dark bars) does not affect growth in any of the null strains tested.
Cell Growth Conditions for Phospholipid Analyses
Cells were grown in 100 ml of YPD for nine to 11 doublings before harvesting at 0.4 ≤ OD600 ≥ 0.6. Immediately before pelleting each culture, cell density was empirically determined using a hemacytometer. Cells were first pelleted in a KA-10 rotor at 5000 rpm at 4°C for 10 min. Cells were resuspended in 30 ml of ddI H2O (Mediatech, Herndon, VA), and 50 μl of cells was removed for determining cells per OD600 by hemacytometer. Cells were then pelleted three more times in a clinical centrifuge at 3/4 speed for 5 min, and resuspended in H2O volumes 25 and 5 ml, before decanting supernatant. Cell pellets were resuspended in the remaining water, and OD600 readings were taken. Briefly, 10 ODs of cells were collected in 2-ml Eppendorf tubes, and cells were flash frozen in liquid N2 and stored at −80°C until ready to harvest lipids.
Phospholipid Composition
Lipids were extracted as described previously (Parks et al., 1985), after cells were lysed in a mini-BeadBeater-8 (Biospec Instruments, Bartlesville, OK). Total lipids were dried under N2, resuspended in CHCl3, and injected onto a Zorbax RX-Sil 250 × 4.6 mm (5 μm) column (Agilent Technologies, Palo Alto, CA). Glycerophospholipids were separated basically as described previously (Stith et al., 2000). Solution A was CHCl3, MeOH, and ammonium hydroxide (80:19:1); solution B was CHCl3, MeOH, and ammonium hydroxide (60:39:1); and solution C was CHCl3, MeOH, H2O, and ammonium hydroxide (60:34:5:1). Solution A was set at 100% for 3 min, and then solution B was increased to 100% by 19 min and held at 100% for 5 min. Solution C was then increased to 100% over 9 min and held at 100% for 2 min. The column was allowed to re-equilibrate with solution A for 10 min before the each injection. Lipids were detected with a PL-ELS 2100 evaporative light scattering detector (Polymer Laboratories, Amherst, MA) with an evaporator temperature of 105°C, nebulizer temperature of 50°C, and a gas (N2) flow rate of 2 l/min. The peaks containing PC, PE, phosphatidylserine (PS), and phosphatidylinositol (PI) were identified by comparison to known standards (Avanti Polar Lipids, Alabaster, AL). Calibrations for mass amounts were based on external standards.
Liquid Culture Growth Assay
Strains were grown on synthetic complete agar plates lacking uracil for 2 d. A single colony was resuspended in 500 μl of sterile water, and OD600 was determined. Cells were diluted to OD600 = 0.35 before aliquoting into 3 ml of synthetic complete liquid media lacking uracil or YPD for a final OD600 = 0.005. As a control, fivefold serial dilutions were also plated onto synthetic complete agar plates lacking uracil. Liquid cultures were incubated with aeration at 30°C, and OD600 readings were measured every 12–18 h for 50–60 h; plates were grown for 48 h at 30°C. In this article, we have only reported the final time points. These experiments were repeated a minimum of three times.
RESULTS
Hmg2p, unlike Hmg1p, undergoes regulated degradation in response to increased sterol pathway signal. Mutating K6 to arginine completely prevents Hmg2p degradation (Gardner and Hampton, 1999), but it does not affect the ER structures formed when expressed at high levels (Figure 1, B and C; data not shown). To facilitate study of the effects of Hmg2p-induced ER remodeling and its comparison to the stable Hmg1p isozyme, we used the nondegraded, stable K6R mutant, referred to as Hmg2ps, throughout this work.
Although Hmg1p and Hmg2p are similar proteins, both structurally (Figure 1A) and functionally, they induce ER membrane arrays that have distinct features (Figure 1, B and C). Hmg1p-induced structures are only nuclear-localized, whereas the structures induced by Hmg2p are both perinuclear and cytoplasmic (Koning et al., 1996; Figure 1, B and C). The ER chaperone Kar2p colocalizes with Hmg1p-induced karmellae and is virtually excluded from membrane structures that result from Hmg2p overexpression (Koning et al., 1993; Figure 1B).
Hmg1p expressed from the strong, constitutive TDH3 promoter (TDH3pr) does not generate karmellae, whereas TDH3pr-expressed Hmg2p does generate membrane stacks and whorls (Hampton et al., 1996). We tested whether this was due to differences in protein levels, and we found that levels of Hmg1p and Hmg2ps were different when expressed from the same promoters (Figure 1D). Previous analyses of Hmg1p were primarily conducted using the strong, inducible GAL1-10 promoter (GAL1-10pr) (for examples, see Wright et al., 1988; Koning et al., 2002). To determine the relative expression levels of the TDH3pr and GAL1-10pr, we generated three sets of strains. Each harbored integrating plasmids of either myc-tagged Hmg1p or Hmg2ps expressed by the TDH3pr present at one or two copies per cell, or expressed from the inducible GAL1-10pr at one copy per cell (Figure 1D). Cells expressing the GAL constructs were first grown with raffinose as the carbon source to eliminate the glucose repression, and then galactose was added to 4% for 4 h to induce expression from the GAL1-10pr before analysis.
Expression from the TDH3pr was about half as strong as expression from the GAL1-10pr, based on relative signal from the myc epitope as determined by immunoblotting lysates (Figure 1D). Furthermore, expression of Hmg2ps was ∼45% higher than Hmg1p in all cases tested. These differences in protein expression levels were reflected when we scored cells for ER remodeling, demonstrating that comparable levels of Hmg1p or Hmg2ps induce ER remodeling to the same extent (Figure 1E). For example, >30% of cells had both nuclear and cytoplasmic stacks and whorls of membrane when expressing 1× TDH3pr-driven Hmg2ps, whereas two copies of TDH3pr-driven Hmg1p were required for comparable Hmg1p expression, thus inducing karmellae levels to 30% (Figure 1, D and E). These numbers from 2× TDH3pr-driven Hmg1p expression were consistent with Wright et al.'s observations when expressing Hmg1p from the GAL1-10pr (Parrish et al., 1995; Profant et al., 1999). The number of cells observed with stacks and whorls of membranes increased to >80% when expressing 2× TDH3pr-driven Hmg2ps. We do note that the number of layers of stacked membranes observed in Hmg1p-expressing cells was not as numerous as those reported previously with the GAL1-10pr, and they may be due to differences in acute versus chronic expression of Hmg1p.
Work by the Wright laboratory has demonstrated that both the transmembrane domain of Hmg1p and a folded CD are required for karmellae formation (Profant et al., 1999). The structural requirements for Hmg2p-induced ER remodeling have not been evaluated. Therefore, we generated a series of TDH3pr-driven Hmg2ps expression constructs to address those questions (Figure 2A).
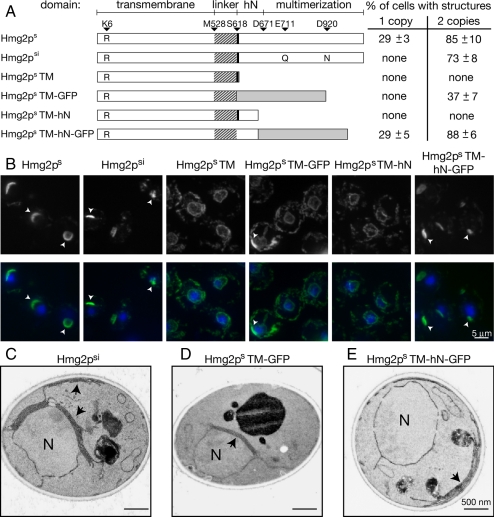
Figure 2.
The transmembrane domain of Hmg2p was not sufficient to remodel the ER. (A) Illustrations of Hmg2p variants examined. Amino acid positions at domain junctions are indicated, as well as the K6R stabilizing mutation in Hmg2p. Myc tag insertions in Hmg2p are indicated by the thick black bar immediately following the linker domain. TDH3pr-driven proteins were integrated at one or two loci. Cells were prepared for IF and examined for structures. At least 200 cells were counted in three independent experiments; averages and standard deviations are shown. (B) Representative IF images of constructs tested, present at two copies per cell. Hmg2p variants were visualized using either anti-myc or anti-GFP monoclonal antibodies. IF of the Hmg2 variants is shown in the top row, and overlaid with DAPI (blue) in the bottom row. Arrows indicate ER membrane structures. (C–E) Electron micrographs of the structures generated by the Hmg2p variants Hmg2psi (C), Hmg2ps-TM-GFP (D), and Hmg2ps-TM-hN-GFP (E). Nucleus is visualized with DAPI. Log phase cells were grown in YPD. Arrows indicate structures.
Enzymatic activity is not required for Hmg1p-induced karmellae (Parrish et al., 1995; Profant et al., 1999), so we began by determining whether the enzymatic activity was similarly dispensable for Hmg2p-induced ER remodeling. By mutating the crucial catalytic residues E711 and D920 to glutamine and asparagine, respectively, we generated an enzymatically inactive version of Hmg2p (Hmg2psi, Figure 2A; Istvan et al., 2000), and we verified the inactivity of this protein (data not shown). When we examined the expression level of the inactive version of Hmg2p, Hmg2psi, we found that it was not as strongly expressed as Hmg2ps (data not shown). Thus, at one copy per cell, TDH3pr expression of Hmg2psi did not generate any of the characteristic structures as indicated by IF. However, upon introducing a second expression plasmid, the number of cells with structures increased to ∼73%, demonstrating the catalytic activity was similarly not required for generating Hmg2p-induced structures (Figure 2, A and B). We confirmed by EM that these structures were similar to those generated by Hmg2ps (Figure 2C).
Next, we tested whether a tightly folded CD was required for Hmg2p-induced ER remodeling by completely removing the CD of Hmg2p, generating Hmg2ps-TM (Figure 2A). This protein has the entire transmembrane (TM) domain and linker region but lacks the C-terminal 425 amino acids that encode the CD and thus removed the enzymatic portion of the protein. Immunoblotting indicated that this protein was expressed at similar levels to Hmg2ps (data not shown). However, when we examined these cells by IF, even when two copies of TDH3pr-driven Hmg2ps-TM were present, we did not observe any structures (Figure 2, A and B). We further confirmed by EM that no ER membrane alterations were observable (data not shown), suggesting that the TM domain alone is not sufficient to generate membrane stacks and whorls.
Hmg1p requires a tightly folded CD to induce karmellae formation (Profant et al., 1999). To determine whether this was also the case for Hmg2p, we replaced the entire 425 amino acid CD with GFP (Hmg2ps-TM-GFP, Figure 2A). Multimerization of GFP itself does not seem to be a significant contributing factor of ER remodeling in this system (see below). When we expressed this protein in cells, we again did not observe any of the expected structures. However, introducing a second copy induced structures in ∼37% of cells (Figure 2A). By both IF (Figure 2B) and EM (Figure 2D), the ER structures looked similar to those generated by Hmg2ps. Hmg2ps-TM-GFP was expressed at comparable levels to Hmg2ps (data not shown), indicating that the decrease in membrane arrays observed was not due to decreased protein levels.
To better understand the role of the Hmg2p CD, we analyzed the subdomains of this region. The CD of HMGR is composed of two distinct regions: the small helical N-domain (hN-domain) followed by the large multimerization domain (Istvan et al., 2000; Figure 1A). Based on alignment, the hN-domain is encoded by the first 50 amino acids of the Hmg2p CD, and the remaining 375 amino acids encode the multimerization domain of Hmg2p. To determine whether the hN-domain contributes to ER remodeling, we generated two constructs with the Hmg2p hN-domain intact in the absence of the Hmg2p multimerization domain (Hmg2ps-TM-hN and Hmg2ps-TM-hN-GFP, Figure 3A). Previous work has shown that replacing the Hmg2p multimerization domain with GFP allows characteristic ER structures to form (Hampton et al., 1996; Koning et al., 1996), referred to as Hmg2p-GFP in those articles). When we tested the Hmg2ps-TM-hN-GFP construct, we found that the number of cells with structures was similar to that of full-length Hmg2ps, at both expression levels tested (Figure 2A). Examination of these structures by both IF (Figure 2B) and EM (Figure 2E) confirmed that they seem similar to structures that full-length Hmg2ps generates, and immunoblotting confirmed that protein levels are also comparable to full-length Hmg2ps (data not shown). The results from overexpression of Hmg2ps-TM-GFP and Hmg2ps-TM-hN-GFP suggest that although the Hmg2p multimerization domain is dispensable, the hN-domain of Hmg2p significantly contributes to Hmg2p-induced ER remodeling. Unlike the rest of the Hmg2p variants examined, Hmg2ps-TM-hN, (Hmg2ps truncated immediately after the hN-domain) was expressed at significantly lower levels, even upon introduction of the second expression construct (data not shown). Accordingly, we did not observe any structures when Hmg2ps-TM-hN was expressed at one copy or two copies per cell as assessed by IF (Figure 2, A and B) or by EM (data not shown). This suggests that a folded globular domain, such as the Hmg2p multimerization domain or GFP, may serve to stabilize Hmg2p when the hN-domain is present.
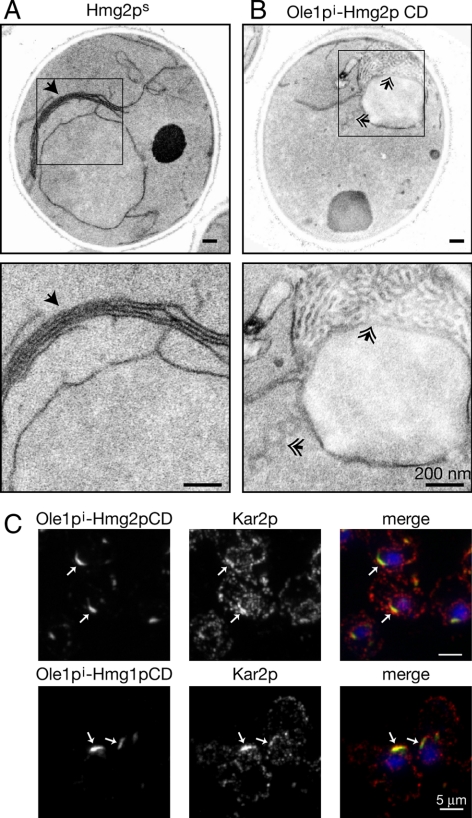
Figure 3.
The ER anchored CD of HMGR was sufficient to cause membrane reorganization. (A) Diagrams of Ole1pi variants examined. TDH3pr-driven triple-myc–tagged Ole1p with the H161A, H166A-inactivating mutations was fused to various soluble proteins as indicated. Nomenclature is as follows: Ole1pi, Ole1p with H161A and H166A mutations; Ole1pi-GFP, inactive Ole1p fused to GFP; Ole1pi-2hN-GFP, inactive Ole1p fused to the hN-domain of Hmg2p and GFP; Ole1pi-Hmg2p CD, inactive Ole1p fused to the CD of Hmg2p; and Ole1pi-Hmg1p CD, inactive Ole1p fused to the CD of Hmg1p. White- and gray-striped area corresponds to Hmg2p linker domain; hatched area corresponds to Hmg1p linker domain. Quantitation of cells harboring one or two copies of the expression plasmids are listed on the right. Expression plasmids were integrated at one or two loci. Cells were prepared for IF and examined for structures. A minimum of 200 cells was counted in three independent experiments; averages and standard deviations are shown. Nucleus is visualized with DAPI. (B) Representative IF of strains with two expression constructs is shown. Arrows indicate ER membrane structures. (C) Electron micrographs of cells with two copies of TDH3pr-driven Ole1pi-Hmg2p CD and Ole1pi-Hmg1p CD. Log phase cells were grown in YPD. Double arrowheads indicate structures.
In our analysis of ER remodeling by Hmg2p, we discovered that, like Hmg1p, Hmg2p required a folded CD. We next tested the intrinsic membrane-organizing features of these CDs. Previous reports indicated that attachment of a multimerizing GFP to a TM domain is sufficient to reorganize membranes in mammalian cells (Snapp et al., 2003), so we wondered whether multimerization through the cytoplasmic GFP was similarly sufficient to induce the structures that we were observing.
To test the effects of various CDs independent of the Hmg2p TM domain, we used the ER-localized stearoyl-CoA Δ9 desaturase Ole1p as an ER membrane anchor. Ole1p has four TM spans and has not been reported to cause ER remodeling (which we confirmed; see below), thus allowing us to test the ER-organizing effects of the CDs used above when fused to this “neutral” ER anchor. To eliminate any effects from enzymatic function, we used an inactive form of the protein that has histidines 161 and 166 mutated to alanine and a triple myc tag at the N terminus (Ole1pi, Figure 3A; Zhang et al., 1999). First we verified that overexpression of myc-tagged, inactive Ole1p did not confer any alteration of the ER membranes when expressed at one or two copies per cell from the TDH3pr (Figure 3B) and confirmed this observation by EM (data not shown). Then, we verified the expression level of each of the Ole1p fusion constructs in Figure 3A to the expression of Hmg2p by relative myc immunoreactivity (data not shown).
We generated fusion proteins between the Ole1pi and the various CDs tested in Figure 2 (Figure 3A). We first examined Ole1pi-GFP. When we evaluated the strain expressing this protein by IF and EM, we did not observe any ER membrane structures (Figure 3B and data not shown, respectively). These data indicated that, in this case, GFP could not alone induce membrane stacking. However, we cannot discount the possibility that Ole1p has features that prohibit GFP multimerization from occurring that would otherwise alter ER structure. Next we tested whether the Hmg2p cytoplasmic hN-domain fused to GFP, as in the ER altering Hmg2ps-TM-hN-GFP mentioned above, could stimulate membrane reorganization (Ole1pi-2hN-GFP). We again did not observe any structures, even when TDH3pr-driven Ole1pi-2hN-GFP was expressed at two copies per cell either by fluorescence (Figure 3B) or EM (data not shown). This demonstrated that the structures induced by expression of either Hmg2ps-TM-GFP or Hmg2ps-TM-hN-GFP were most likely not a result of an intrinsic activity of their respective CDs to remodel membranes.
The CD of mammalian HMGR forms homodimers and homotetramers, and biochemical evidence indicates that the same holds true for the highly conserved yeast CD (Qureshi et al., 1976). To determine whether the CDs of yeast HMGR have an intrinsic ability to reorganize membranes, potentially via multimerization, we generated a fusion between Ole1pi and the entire 425 amino acid CD of Hmg2p (Ole1pi-Hmg2p CD) and expressed it from the TDH3pr. In striking contrast to the other constructs tested, the complete CD of Hmg2p caused a reorganization of membranes when fused to Ole1pi; when present at only one copy, ∼72% of the population had visible membrane structures (Figure 3, A–C).
The CD of Hmg1p and Hmg2p are 93% identical, so we would expect the Hmg1p CD to similarly induce reorganization of ER membranes when anchored to the membrane. When we tested a construct that fused the CD of Hmg1p to Ole1pi (Ole1pi-Hmg1p CD), we found that the resultant ER structures were identical to those caused by the Ole1pi-Hmg2p CD fusion (Figure 3, B and C). The Ole1pi-Hmg1p CD fusion induced the formation of ER structures that were both nuclear-associated and cytoplasmic, like Ole1pi-Hmg2p CD (Figure 3C). Furthermore, when a single TDH3pr-driven copy was expressed, ∼69% of cells had visible ER arrays, and when the copy number was doubled, the percentage climbed to 77% (Figure 3A). These numbers were markedly higher than those achieved with the full-length Hmg1p. Our findings indicate that either HMGR CD, in its entirety, is sufficient to remodel ER membranes when anchored to the inert Ole1pi. This suggests that there may be two independent structural conditions that cause ER remodeling by Hmg2p. One is brought about by features of the Hmg2p transmembrane domain when fused to a folded cytoplasmic domain. The other is due to the strong multimerizing properties of the HMGR cytosolic domains, and is transferable to the normally nonproliferating Ole1p.
The membrane structures generated by both Hmg1p and Hmg2ps are smooth, closely associated stacks of unbranched membranes (Figure 1C). In contrast, we observed that the Ole1pi-HMGR CD fusions generated structures composed of branched networks of membranes (Figures 3C and 4B). In some cells expressing the Ole1pi-HMGR CD constructs, we saw that these bits of membrane were actually connected tubules that very closely resembled hexagonal arrays. One such panel is shown in Figure 4B of Ole1pi-Hmg2p CD. These highly branched networks are remarkably different from the tightly packed stacks of membrane generated by Hmg2ps (Figure 4A). We also verified that the catalytic activity of HMGR was not required to generate these structures by analyzing Ole1pi fused to the inactive CD of Hmg2p (data not shown). Finally, we made and examined an N-terminal Hmg2p CD fusion to Ole1pi (Hmg2p CD-Ole1pi) as opposed to the C-terminal fusion used above, and again we observed similarly branched membrane arrays by EM (data not shown).
Figure 4.
The Ole1pi-HMGR CD fusions make distinct membrane structures. (A and B) Electron micrographs of strains with two TDH3pr-driven copies of Ole1pi-Hmg2p CD, inactive Ole1p fused to the CD of Hmg2p (A); and Hmg2ps (B) at 20,000 (top) and 80,000 (bottom) magnification. Solid arrows indicate stacks of membrane characteristic of Hmg2p. Double arrows indicate tubular membrane arrays generated by Ole1pi-Hmg2p CD fusion constructs. (C) IF imaging of strains harboring two copies of Ole1pi-Hmg2p CD (top) and Ole1pi-Hmg1p CD (bottom). Ole1pi fusions are green and Kar2p is red; nucleus is visualized with DAPI. Log phase cells were grown in YPD. Arrows indicate structures.
Hmg1p and Hmg2p have nearly identical structural requirements for generating their characteristic ER arrays, and the resultant membrane structures seem similar (Figure 1, B and C). However, the arrays produced by each isozyme are distinct in cellular localization and the presence or absence of Kar2p (Koning et al., 1996; Figure 1, B and C). Our analysis revealed that either HMGR CD fused to Ole1pi generates both nuclear associated and cytoplasmic structures. This suggests that the TM domains of Hmg1p and Hmg2p determine the features of their induced structures. Using our Ole1pi-HMGR CD fusion proteins, we next examined Kar2p localization with respect to the generated structures. We would expect both Ole1pi-Hmg1p CD and Ole1pi-Hmg2p CD to behave similarly if the TM domain determines the characteristics of the structures. Both the Ole1pi-HMGR CD fusion proteins strongly colocalized with Kar2p (Figure 4C). This suggests that the TM domain determines both where the structures localize within the cell and whether Kar2p is present in these structures. Together, these findings suggest that although a cytoplasmic domain is necessary for either Hmg1p or Hmg2p-induced ER remodeling, the TM domain plays a critical role in dictating the characteristics of the structures generated.
The intriguing exclusion of Kar2p from Hmg2p-induced ER arrays implies that the Hmg2p structures have features that are distinct from the “typical” ER. The Wright group observed previously that Sec61p, an essential conserved ER membrane protein, colocalized with both Hmg1p and Hmg2p induced ER arrays (Koning et al., 1996). We decided to survey a group of ER proteins to further characterize the ER structures induced by the HMGR isozymes. To facilitate localization of the induced ER structures, we used GFP-tagged, ER-remodeling versions of each isozyme. Specifically, we used Hmg2ps-TM-hN-GFP (Hmg2ps with multimerization domain replaced by GFP) and Hmg1p·67-GFP, with the C-terminal 67 amino acids replaced with GFP (called Hmg1mem:Hmg1525-987:GFP; Profant et al., 1999) and then evaluated the presence of a given ER protein in the structures by comparing GFP fluorescence (HMGR) to the IF of an epitope-tagged ER protein.
We first tested the structures for the presence of ER-localized P-type ATPase, Cod1p/Spf1p, a multispanning ER transporter required for normal ER homeostasis (Cronin et al., 2000; Giaever et al., 2002). Our results were similar to those observed for Kar2p; Cod1p/Spf1p colocalized with Hmg1p-induced structures, but it was absent from those induced by Hmg2p (Figure 5A). This indicated that not all TM proteins colocalize with Hmg2p-induced structures. We next tested Hrd1p, the polytopic ER ubiquitin ligase required for regulated degradation of Hmg2p (Bays et al., 2001). Again, like Kar2p and Cod1p/Spf1p, Hrd1p colocalized with Hmg1p-induced structures, but not those generated by Hmg2p (Figure 5B). These results could provide insight into the regulated degradation of Hmg2p (see Discussion). Finally, we examined Erg28p, a transmembrane protein that plays a critical role in sterol biosynthesis by mediating protein-protein interactions between Erg26 and Erg27, and by tethering Erg6p to the ER membrane (Mo and Bard, 2005). Unlike the other proteins tested, Erg28p colocalized with structures induced by Hmg2p (Figure 5C). These data support the idea that the structures generated by Hmg2p may be a specialized subdomain of the ER.
Figure 5.
Structures induced by Hmg2p exclude Kar2p and Hrd1p, but they are enriched for Erg28p. (A and B) GFP-fluorescence of Hmg1p·67-GFP or Hmg2ps-GFP* and IF of (A) 3myc-Cod1p/Spf1p or (B) Hrd1p-HA. (C) GFP fluorescence of Hmg2ps-GFP* and IF of Erg28p-HA. Log phase cells were grown in YPD. Arrows indicate structures; nucleus is stained with DAPI.
Together, the above-mentioned studies strengthen the case that the structures produced by Hmg1p and Hmg2p are distinct. Furthermore, the Wright group's genetic analyses of Hmg1p identified genes required only for Hmg1p-induced ER structures, and not Hmg2p-induced ER remodeling, suggesting there might also be genes specifically required for Hmg2p-induced structures. Therefore, we conducted a screen to uncover HER genes required for Hmg2p-induced ER Remodeling. We used a strain expressing Hmg2ps-TM-hN-GFP, which generates characteristic Hmg2p structures with high fidelity (Figure 2). This protein is a stable, optical form of Hmg2p that allows for direct optical screening of live cells. The absence of the HMGR multimerization domain removes both the enzymatic activity of the protein and the independent ability of the CD to alter ER membranes, as determined with the Ole1p fusions described above.
Using the methods pioneered by the Boone group (Tong et al., 2001), Hmg2ps-TM-hN-GFP was introduced into the collection of ∼4700 viable deletion strains. A strain with our Hmg2ps-TM-hN-GFP coding region under the control of the inducible GAL1-10pr was robotically crossed to the viable haploid null collection (for strategy, see Tong et al., 2001). The entire crossing and selection process was conducted on plates with glucose so the expression of GAL1-10pr-driven Hmg2ps-TM-hN-GFP was repressed during the strain production phase. After induction on galactose, each null strain was individually examined for alterations or absence of the characteristic Hmg2ps-TM-hN-GFP structures. In this way, we identified 104 candidate nulls with altered structures from this primary screen. Of these nulls, ∼50 were manually remade in our strain background (genes are listed in Supplemental Table S2). We then introduced our TDH3pr driven Hmg2ps-TM-hN-GFP into this subset and scored the resulting strains for the HER phenotype.
All but one of the genes identified were separated into four distinct classes based on Hmg2ps-TM-hN-GFP localization as listed in Table 1. Class I nulls did not have any observable structures in >95% of the cells. Instead, Hmg2ps-GFP* localization was generally uniform throughout the ER, and it looked very similar to the distribution of numerous ER proteins in S. cerevisiae (Figure 6). The next class of mutants, class II, all similarly generated Hmg2ps-TM-hN-GFP–induced stacks of membranes that were completely nuclear associated, reminiscent of Hmg1p-induced karmellae (Figure 6). Membrane stacks were not observed away from the nucleus in any of these class II nulls when Hmg2ps-TM-hN-GFP was overexpressed. Class III nulls comprise the enzymes of the PC-synthesizing methylation pathway (Figure 7, A and B). The locus encoding Psd1p, the first enzyme of this pathway, was identified by our screen (see below), and the other genes were then directly tested. These class III nulls varied between having no proliferations to having only nuclear-associated stacks of membranes present in 20% of cells (Figure 7B). In addition, the cytoplasmic (nonnuclear) ER of class III nulls was aberrant, making it distinct from the class I and II null strains (Table 1). Class IV genes, NEM1 and SPO7, have been identified as generally altering nuclear and ER morphology (Siniossoglou et al., 1998; Santos-Rosa et al., 2005). Our initial screen identified NEM1, and based on the known role of this protein in ER and nuclear morphology, we tested its binding partner, SPO7, and we found that it similarly altered Hmg2ps-TM-hN-GFP localization. Class IV nulls were unique in that any ER protein that we expressed had a similar localization pattern of long smooth membrane stacks and misshapen nuclei (Figure 6). We confirmed by flow cytometry that the lack of structures observed in the null strains examined was not due to a decrease in Hmg2ps-TM-hN-GFP expression (data not shown). These genes are the first identified to alter the morphology of the structures generated by Hmg2p.
Table 1.
HER genes
| Gene | ORF | Protein description |
|---|---|---|
| Class I: no structures | ||
| BNI1 | YNL271C | Formin, involved in budding and actin dynamics |
| BNR1 | YIL159W | Formin, involved in budding and actin dynamics |
| HER2 | YMR293C | Verified ORF of unknown function |
| VMA2 | YBR127C | Subunit B of V-ATPase, proton pump found throughout endomembrane system |
| Class II: only nuclear structures | ||
| ERG28 | YER044C | ER membrane protein, facilitates protein–protein interactions |
| ERV25 | YML012W | Member of p24 family, involved in ER-to-Golgi transport |
| HER1 | YOR227W | Verified ORF of unknown function |
| HOF1 | YMR032W | Protein with SH3 domain, involved in cytokinesis and cytoskeletal dynamics |
| COD1/SPF1 | YEL031W | ER localized P-type ATPase |
| Class III: nuclear structures/altered cytoplasmic (non-nuclear) ER | ||
| PSD1 | YNL169C | Mitochondrial phosphatidyl serine decarboxylase, PC biosynthesis |
| CHO2 | YGR157W | PE methyltransferase (PEMT), PC biosynthesis |
| OPI3 | YJR073C | Phospholipid methyltransferase, PC biosynthesis |
| Class IV: general ER defect | ||
| NEM1 | YHR004C | Controls nuclear growth, negative regulator of phospholipid biosynthetic genes |
| SPO7 | YAL009W | Controls nuclear growth, negative regulator of phospholipid biosynthetic genes |
Figure 6.
HER genes fall into four distinct classes. Live cell imaging of Hmg2ps-TM-hN-GFP localization in four different strains is shown. Cells imaged have a single integrated copy of Hmg2ps-TM-hN-GFP. Wild type is shown for reference, and the representative classes are depicted as follows: her2· for class I, hof1· for class II, and nem1· for class IV. Log phase cells were grown in SC-URA. Arrows indicate distinct structures.
To further characterize the HER genes identified in our screen, we examined the localization of other ER proteins in the null strains and summarized our finding in Table 2. Our first objective was to determine whether the HER genes affected general ER morphology. To test this, we determined the localization of two classic ER markers, Sec63p, an ER membrane protein, and KGFP*, an ER lumenal protein, and we summarized our findings in Table 2. The localization patterns of these two proteins in the class I and II nulls were indistinguishable from wild type. Class III strains had a normal nuclear ER distribution; however, the cytoplasmic (nonnuclear) ER was aberrant. Lastly, the class IV nulls, looked comparable to the image depicted in Figure 6, of the nem1· strain expressing the Hmg2ps-TM-hN-GFP construct. These strains had long strips of membrane and misshapen nuclei as previously observed (Siniossoglou et al., 1998; Santos-Rosa et al., 2005).
Table 2.
Summary of protein localization by live cell imaging in HER mutant strains
| Protein expresseda |
|||||
|---|---|---|---|---|---|
| HER Hmg2ps-TM-hN-GFP | General ER morphology |
Other ER remodeling proteins |
|||
| Sec63p-GFP | KGFP* | Hmg1•67-GFP | P450Cm1-GFP | ||
| Class I | − | + | + | + | + |
| Class II | − | + | + | + | + |
| Class III | − | − | − | −b | −b |
| Class IV | − | − | − | − | − |
a +, wild-type protein localization; −, aberrant protein localization.
b Respective structures form, but the cytoplasmic (nonnuclear) ER is aberrant.
We next tested whether the HER genes were required for ER remodeling induced by other proteins, and tested the two known ER-altering proteins, Hmg1p and cytochrome P450. We used the karmellae-forming GFP-tagged Hmg1p described above, Hmg1p·67-GFP, and cytochrome P450 from Candida maltosa, P450CmA1, which also significantly alters ER membranes in S. cerevisiae (Menzel et al., 1996). The GFP-fusion that we used was confirmed to generate structures identical to the nontagged protein (data not shown). When we analyzed the localization of GFP-tagged Hmg1p and CYP450CmA1 in class I and II nulls, we found that none affected Hmg1p-induced karmellae formation or P450CmA1-induced ER remodeling (Table 2). Class III nulls, despite their unusual cytoplasmic (nonnuclear) ER patterns, still generated the respective structures for Hmg1p and CYP450CmA1. As expected, class IV nulls expressing Hmg1p·67-GFP or CYP450CmA1-GFP had the mutant ER localization typical of nem1· and spo7· null strains. These suggests that the genes that we identified are not generally required for all instances of ER membrane reorganization but that they are specifically required to generate the characteristic structures of Hmg2p.
We further examined the features of the HER genes to determine how they affect general ER function, in terms of protein folding. This is measured by the unfolded protein response (UPR), which is up-regulated when protein-folding capacity of the ER is compromised or otherwise overloaded (for review, see Hampton, 2000). We confirmed that the UPR is up-regulated greater than twofold when COD1/SPF1 is deleted (Cronin et al., 2002) and when HOF1 is deleted (Bicknell et al., 2007), and we determined that cells lacking ERV25 also have a significantly up-regulated UPR (data not shown). The other HER mutants showed no elevation in UPR.
In our screen, the uncharacterized open reading frame (ORF), YNL170W was identified as a HER gene. However, upon closer examination, we found that the deletion overlapped with YNL169C, the ORF of phosphatidylserine decarboxylase 1, PSD1. PSD1 catalyzes the conversion of phosphatidylserine to phosphatidylethanolamine in yeast, as part of the methylation pathway (see below; Voelker, 1997). To test which deleted ORF was responsible for the phenotype observed in our screen, we generated nonoverlapping deletions of each of these coding regions. In this way, we determined that the loss of PSD1 affected localization of, whereas loss of YNL170W did not (data not shown). We further verified that the phenotype was rescued by introducing a PSD1 expression plasmid (data not shown). These data demonstrated that PSD1 is required for Hmg2p-induced ER remodeling. Our identification of PSD1 as a HER gene led us to examine the other genes in the methylation biosynthesis pathway, OPI3 and CHO2, both of which affect Hmg2p ER remodeling (Figure 7A). Together, these three genes comprise the class III HER genes. As such, these are the first genes that link the membrane-altering effects of HMGR to phospholipid biosynthesis.
The discovery of the class III HER genes led us to investigate phospholipids in cells overexpressing Hmg2p. In S. cerevisiae, there are two pathways for generating PC: the methylation pathway and the Kennedy pathway (Figure 7A). The major pathway in yeast is the methylation pathway, in which PSD1 synthesizes PE by the decarboxylation of PS. Most of the PE generated is converted into the abundant phospholipid PC by the sequential methylation of PE by the Cho2p and Opi3p methyltransferases (for review, see Carman and Kersting, 2004). We tested whether the Kennedy pathway was also required for Hmg2p-induced ER remodeling, or whether this was specific to the methylation pathway. When we tested our Hmg2ps-TM-hN-GFP construct in cki1·, cpt1·, or ept1·, null strains, we did not observe a mutant phenotype (Figure 7B). These data suggested that in the medium used for the screen, Hmg2p-induced ER remodeling specifically required the methylation pathway.
We next examined the effects of overexpressing Hmg2p in the pathway nulls, by using both the Hmg2ps-TM-hN-GFP, and full-length, active Hmg2ps. We were surprised to find that strains lacking methylation pathway enzymes Psd1p, Cho2p, or Opi3p exhibited a significant growth defect in synthetic complete liquid minimal medium when active Hmg2ps was overexpressed (Figure 7C, dark bars). This growth defect was not observed in strains lacking the Kennedy pathway enzymes Ept1p, Cki1p or Cpt1p (Figure 7C, light bars). Because this phenotype was not observed in the Kennedy pathway nulls, these data further support a specific requirement for the methylation pathway when Hmg2p is elevated.
Surprisingly, the growth phenotype of the methylation pathway nulls was only observed when enzymatically active, full-length Hmg2p was overexpressed. When the same growth test was done with Hmg1p (data not shown), the optical version of Hmg2p used for the genetic screen, Hmg2ps-TM-hN-GFP (Figure 7B, light bars), or the inactive version of Hmg2p, Hmg2psi (Table 3), the growth effect was not observed. Given the fact that the enzymatic activity of Hmg2p is dispensable in Hmg2p-induced ER remodeling, these results were very surprising.
Table 3.
Effect of ethanolamine and choline on growth in SC-URA media
| OD600 after 50 h of growth |
|||
|---|---|---|---|
| No supplement | +ethanolamine | +choline | |
| psd1 • empty vector | 3.34 | 5.05 | 5.67 |
| psd1 • Hmg2ps | 0.24 | 5.68 | 5.73 |
| psd1 • Hmg2psi | 3.75 | 5.29 | 5.35 |
| PSD1 empty vector | 5.96 | 6.75 | 6.59 |
| PSD1 Hmg2ps | 6.13 | 7.27 | 6.54 |
Representative experiment shown.
The growth phenotype in the methylation nulls was not observed if cells from synthetic complete solid medium were inoculated into liquid YPD (data not shown). This is perhaps not surprising, because YPD provides many molecules that would otherwise be synthesized in leaner medium. For instance, ethanolamine auxotrophs grow in YPD but not synthetic complete medium, indicating that this rich medium provides sufficient ethanolamine (Storey et al., 2001; Prinz, personal communication). Consistent with this interpretation, addition of either ethanolamine or choline suppressed the synthetic complete medium growth phenotype in the psd1· strains expressing Hmg2ps (Table 3). These data are very interesting given the data reported by the Voelker laboratory. Storey et al. (2001) reported that psd1· null strains grow normally at 30°C without addition of ethanolamine; however, when grown at elevated temperatures (37°C) or on nonfermentable carbon sources, the growth of psd1· strains is severely compromised. Our data suggest that overexpression of active Hmg2ps causes a stress that severely compromises the psd1· null in growth conditions where this gene is responsible for PC synthesis.
In the synthetic medium used in the above-mentioned growth studies, the methylation pathway is the principle route for PC synthesis available to the cells. In YPD medium, the alternative head-group transfer pathway is also operative, thus allowing continued PC synthesis in the presence of the psd1· null. Thus, we wondered whether the PSD1 gene still played a role in Hmg2p-induced membrane alterations in this more permissive condition. Accordingly, we examined the effects of Hmg2p overexpression on phospholipid composition in YPD medium. We found that overexpression of a single copy of TDH3pr-driven Hmg2ps caused a significant increase in the relative amount of PC (Figure 8A). Furthermore, the relative amount of PS, the substrate for Psd1p, was significantly decreased when Hmg2ps was overexpressed (Figure 8A). Neither Hmg1p nor the soluble CD of Hmg2p caused a significant change in phospholipid composition (Figure 8A), despite each of these strains having very high HMGR enzymatic activity as determined by resistance to HMGR inhibitors (data not shown). This suggested that the increase in PC abundance that occurs when active Hmg2p is overexpressed is not simply due to increased sterol pathway products, but rather is caused by full-length, active Hmg2p.
Figure 8.
Increased Hmg2p expression alters phospholipid composition. (A) Phospholipid composition of cells with two integrated copies of empty vector, TDH3pr-driven Hmg2ps, Hmg1p, or the soluble CD of Hmg2p in a wild-type strain. (B) Phospholipid composition of psd1 null strains with or without two integrated copies of TDH3pr-driven Hmg2ps. Early log phase cells were grown in YPD. Averages and standard deviations are shown; n = 3.
The drop in PS that occurred concomitant with PC increase was consistent with a role for the methylation pathway in the effects of Hmg2p overexpression in YPD. Thus, we tested whether these effects on phospholipid composition required Psd1p. Previous studies have shown that psd1· null strains have PE levels that are significantly lower than wild-type strains (Trotter and Voelker, 1995). Consistent with this, the psd1· strains had the expected shift in phospholipid ratios (Figure 8B). However, overexpression of Hmg2p in this background had no effect on phospholipid composition of this strain (Figure 8B). These data suggest that elevating Hmg2p mobilizes the methylation pathway, even in strains where this in not the primary source of PC. However, a direct analysis of the effects on the separate routes of PC synthesis would be required to further test this possibility. Nevertheless, it is clear that phospholipid synthesis plays a key role in the response to elevated Hmg2p, and the methylation pathway might be a focal point for this response.
DISCUSSION
In this study, we have determined both the structural and genetic requirements for Hmg2p-induced ER remodeling. Specifically, we have delineated the features of Hmg2p that contribute to this response and have described a new set of genes, referred to as HER genes that are required for Hmg2p-induced ER remodeling. The majority of these genes are highly specific for this process, and they are not required for maintenance of ER structure or for the ER remodeling caused by Hmg1p or other membrane proteins.
Our studies revealed that Hmg2p has two separate features that are sufficient for Hmg2p-induced ER membrane remodeling. The first is through CD interactions, most likely via multimerization. We found that increased expression of Ole1pi alone did not alter ER membranes; however, fusing either HMGR CD to inactive Ole1p did cause striking membrane reorganization. Importantly, we found that although both the Hmg1p and Hmg2p CDs caused a profound and identical alteration in membrane structure when tethered to Ole1pi, these structures were not the same as the characteristic structures induced by Hmg1p or Hmg2p. Unlike the HMGR CDs, we found that identically fusing GFP to Ole1pi was not sufficient to reorganize membranes. This was surprising due to work in mammalian cells that demonstrated that dimerization between cytoplasmic GFP molecules fused to TM proteins was sufficient to cause membrane reorganization (Lippincott-Schwartz et al., 2000).
Increased expression of the TM domain of Hmg2p alone was not sufficient to remodel ER membranes, suggesting that, like Hmg1p, a folded domain attached to the TM domain was necessary. Elevated Hmg2p expression induced strips and whorls of membranes when the multimerization domain was replaced with GFP, or when the entire CD was replaced with GFP (Hmg2ps-TM-hN-GFP and Hmg2ps-TM-hN-GFP, respectively), indicating that the native CD of Hmg2p was not required to generate these structures. Our analysis with Ole1p demonstrated that fusing Ole1pi to GFP or the hN-domain of Hmg2p with GFP does not cause any observable alterations in ER membranes. Together, these data demonstrate that the characteristic ER remodeling caused by Hmg2p requires the Hmg2p TM domain in tandem with a folded CD. We do not know whether any folded CD would facilitate this interaction or whether there is a shared feature between GFP and the CD of Hmg2p. These findings illustrate another parallel between Hmg2p and Hmg1p, in that TM and CDs both contribute to structure formation.
The structures induced by increased expression of full-length Hmg1p or Hmg2p see, similar by simple observation; they each produce smooth, unbranched membranes that are closely associated into stacks. However, the structures have important differences that imply distinct molecular processes underlying their formation. This was first indicated by the Wright laboratory's surprising demonstration of Kar2p exclusion from Hmg2p-induced structures (Koning et al., 1996). Both of the Ole1pi fusions to the Hmg1p and Hmg2p CDs resulted in membrane arrays that colocalize with Kar2p. These findings demonstrated that exclusion of Kar2p is a unique feature of the Hmg2p-induced structures. Our analysis of the localization of other ER-resident proteins with respect to the structures generated by Hmg2p overexpression further supported the idea put forth from the Wright group that these structures may be a specialized ER subdomain in which only certain ER proteins reside. The P-type ATPase Cod1p/Spf1p and the ubiquitin ligase Hrd1p were virtually absent from Hmg2p-induced structures. However, we found that Erg28p, a scaffolding protein involved in mediating interactions between ergosterol biosynthetic enzymes, was abundant in the arrays generated by Hmg2p. This is particularly interesting because ERG28 was scored as a HER gene in the screen, Together, these findings suggest two interesting features of the structures generated by Hmg2p. The lack of Hrd1p, the ubiquitin ligase essential for Hmg2p degradation, indicates that Hmg2p degradation might be restricted to regions outside of these structures. The inclusion of Erg28p suggests that the structures may function in production of sterols by enriching the proteins involved in that process into subdomains of the ER. Whether these structures are the sites of emerging lipid droplets, the site of excess sterol and fatty acid storage, remains to be determined. It will be important to discover the mechanism by which Hmg2p is spatially maintained away from select ER proteins within the continuous membrane network of the ER.
The demonstration of genes specifically required for Hmg1p-induced structures (Koning et al., 2002; Wright et al., 2003), and the distinct features of Hmg2p-induced ER structures implied that a separate set of genes, which we call HER genes, would be required for Hmg2p-induced ER remodeling. Our screen confirmed this idea. Among the genes that were uncovered were two that have been previously identified as required for general ER morphology, NEM1 and SPO7, which we called class IV. These genes did not fit our stringent criteria as genes that were specifically required for Hmg2p-induced structures, as opposed to being involved in general ER morphology. However, all of the other genes identified were specifically required for Hmg2ps-TM-hN-GFP–induced ER alterations. They fell into three distinct classes, class I, II, and III, categorized using the localization of Hmg2ps-TM-hN-GFP by live cell imaging. In these conditions, class I nulls do not have observable membrane structures, whereas class II nulls seem to generate only nuclear-associated membrane arrays, similar to those caused by Hmg1p observed in both live cell imaging and IF. Class III nulls, the three enzymes of the methylation pathway, had an intermediate phenotype where a majority of the cells (80–100%) in a given experiment had no structures, whereas the remaining minority (20–0%) had only nuclear-associated stacks. Furthermore, even in the absence of overexpressed Hmg2ps-TM-hN-GFP, the cytoplasmic (nonnuclear) ER seemed altered in these mutants. This variability in the strength of the phenotype in the enzyme mutants is not currently understood, but it may be due to the fact that phospholipid synthesis can be impacted by subtle differences in growth medium, growth phase or other conditions in our experimental system.
One class of genes that did not occur in our screens was that of the reticulons that are required for tubular morphology of the ER in eukaryotes (Voeltz et al., 2006). Accordingly, we directly tested both overexpression of Rtn1p and the effect of the rtn1· rtn2· yop1· triple null on Hmg2p-induced ER remodeling. Neither of these manipulations had any observable effects (data not shown). It may be that the structures formed by Hmg2p are distinct from those that are shaped by reticulons, although this question requires more thorough study.
Importantly, we did not uncover any of the VPS genes that are necessary for karmellae formation by Hmg1p (Koning et al., 2002). In fact, only NEM1 and SPO7 have been previously implicated in ER remodeling. ER resident proteins were among the genes identified in our screen, including ERG28 and COD1/SPF1. Our observation that Erg28p is present in Hmg2p-induced structures and that the loss of ERG28 from the cells results in a loss of Hmg2p-induced structure formation, suggests that Erg28p may have a unique role in this process. Because Erg28p is included in the Hmg2p-induced structures, it could have a direct role on the formation or function of these membrane arrays. Erg28p, like HMGR, is highly conserved and loss of Erg28p results in sterol deficiencies across species (for examples, see Gachotte et al., 2001; Veitia and Hurst, 2001). Last, previous work demonstrated that human testis cells (Veitia et al., 1999) and adrenal cortical cells (Hu et al., 2000) both have increased expression of human Erg28p, as is observed for HMGR (for review, see Federovitch et al., 2005). This correlation is of particular interest due to the fact that both of these tissue types produce large amounts of sterols and have extended arrays of ER membrane, presumably from elevated HMGR needed for sterol production. Unraveling the roles of Erg28p and the other HER genes will provide information about the dynamics of ER membranes and the interplay between lipid and protein.
Our identification of PSD1 as a HER gene led us to directly examine the roles of the methylation and Kennedy phospholipid biosynthetic pathways in Hmg2p-induced ER remodeling. In synthetic complete medium, where the PSD1-dependent methylation pathway is the principle source of PC, overexpression of Hmg2p caused a strong growth phenotype in the absence of PSD1, implicating a need for PC synthesis upon formation of Hmg2p-induced structures. Furthermore, in YPD medium, where PC can be obtained by other routes, the effects of Hmg2p on phospholipid composition caused changes that were consistent with the activation of the pathway, and dependent on the PSD1 gene. These findings suggest that the overexpression of active Hmg2p requires or activates the phospholipid synthesis to effect changes in membrane structure, and it could possibly effect these changes by use of the methylation pathway. Further work will focus on testing this model. Intriguingly, we have so far not observed changes in the transcript levels of any phospholipid synthetic enzymes (Federovitch and Hampton, personal observations), implying that Hmg2p may be able to directly stimulate or alter PC synthesis. Our future studies will focus on the role of phospholipid synthesis in the mechanism of Hmg2p-induced ER remodeling and the effects of Hmg2p on cellular phospholipids.
PSD1 is one of two isozymes that are expressed in yeast, along with PSD2 (for reviews, see Voelker, 1997, 2000). Although the psd2· was included in the collection of nulls screened, it was not identified as a candidate in our screen. Because the two encoded proteins occupy distinct subcellular locations, it will be interesting and important to evaluate the degree to which Hmg2p-induced effects on ER remodeling and phospholipid composition are specific for PSD1.
An important new aspect of the ER effects of HMGR was unveiled in these studies. So far, our and other laboratories have agreed that the catalytic activity of HMGR is not required for ER remodeling (Koning et al., 1996; Profant et al., 1999). However, the requirement of the catalytic activity for the phospholipid effects we observed demands reevaluation of the role of the enzymatic activity in the full effects of HMGR, as would be expected to occur in the natural cases of ER remodeling caused by this enzyme, such as in adrenal cortical cells, or other professional sterol-synthesizing cells (Ross et al., 1958; Sisson and Fahrenbach, 1967; Black, 1972). Similarly, recent work from the Wright group has found a connection to HMGR enzyme activity in the cellular growth responses to elevated Hmg1p (Loertscher et al., 2006; Wright et al., 2003). These studies demand evaluation of active HMGR in the understanding of its effects on ER dynamics and structure. This requirement for HMGR catalytic activity for the effects we observed may underlie the known coupling of sterol synthesis to phospholipid synthesis that has been observed in mammalian cells (Ridgway et al., 1999). Our future studies will address the role of Hmg2p membrane and catalytic determinants on the various effects on the ER.
Supplementary Material
ACKNOWLEDGMENTS
We thank Scott Emr for use of the DeltaVision microscope, Gentry Patrick for use of the Leica DM6000 microscope, and National Center for Microscopy and Imaging Research at UCSD for use of the JEOL 1200 microscope. Plasmid pIU2589 was a kind gift from Martin Bard (Indiana University-Purdue University Indianapolis, Indianapolis, IN). Mark Rose (Princeton University, Princeton, NJ) provided anti-Kar2p antibody. We thank Dennis Voelker (University of Colorado Health Sciences Center, Denver, CO) for insightful discussions and the rescue plasmid YCp50-PSD1. CMF wishes to thank Matthew Kinseth, Alicia Bicknell, Danielle Huffman, and Renee Garza for many scientific discussions regarding this work. RYH wishes to thank the Hampton laboratory for patience, flexibility, and independent spirit during the past (sabbatical) year. These studies were supported by National Institutes of Health grant 5 R01 DK051996-15 (to R.Y.H.). C.M.F. was a trainee under the National Institutes of Health genetic training grant 5T32GM008666.
Abbreviations used:
- CD
cytoplasmic domain
- EM
electron microscopy
- ER
endoplasmic reticulum
- HER
Hmg2p-induced ER remodeling
- HMGR
Hmg-CoA reductase
- hN
helical N
- IF
immunofluorescence
- P450
cytochrome P450
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PS
phosphatidylserine
- TM
transmembrane.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-11-1188) on July 30, 2008.
REFERENCES
- Basson M. E., Thorsness M., Rine J. Saccharomyces cerevisiae contains two functional genes encoding 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proc. Natl. Acad. Sci. USA. 1986;83:5563–5567. doi: 10.1073/pnas.83.15.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bays N. W., Gardner R. G., Seelig L. P., Joazeiro C. A., Hampton R. Y. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat. Cell Biol. 2001;3:24–29. doi: 10.1038/35050524. [DOI] [PubMed] [Google Scholar]
- Bicknell A. A., Babour A., Federovitch C. M., Niwa M. A novel role in cytokinesis reveals a housekeeping function for the unfolded protein response. J. Cell Biol. 2007;177:1017–1027. doi: 10.1083/jcb.200702101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black V. H. The development of smooth-surfaced endoplasmic reticulum in adrenal cortical cells of fetal guinea pigs. Am. J. Anat. 1972;135:381–417. doi: 10.1002/aja.1001350307. [DOI] [PubMed] [Google Scholar]
- Carman G. M., Kersting M. C. Phospholipid synthesis in yeast: regulation by phosphorylation. Biochem. Cell Biol. 2004;82:62–70. doi: 10.1139/o03-064. [DOI] [PubMed] [Google Scholar]
- Chin D. J., Luskey K. L., Anderson R. G., Faust J. R, Goldstein J. L., Brown M. S. Appearance of crystalloid endoplasmic reticulum in compactin-resistant Chinese hamster cells with a 500-fold increase in 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proc. Natl. Acad. Sci. USA. 1982;79:1185–1189. doi: 10.1073/pnas.79.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin S. R., Khoury A., Ferry D. K., Hampton R. Y. Regulation of HMG-CoA reductase degradation requires the P-type ATPase Cod1p/Spf1p. J. Cell Biol. 2000;148:915–924. doi: 10.1083/jcb.148.5.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin S. R., Rao R., Hampton R. Y. Cod1p/Spf1p is a P-type ATPase involved in ER function and Ca2+ homeostasis. J. Cell Biol. 2002;157:1017–1028. doi: 10.1083/jcb.200203052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federovitch C. M., Ron D., Hampton R. Y. The dynamic ER: experimental approaches and current questions. Curr. Opin. Cell Biol. 2005;17:409–414. doi: 10.1016/j.ceb.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Gachotte D., Eckstein J., Barbuch R., Hughes T., Roberts C., Bard M. A novel gene conserved from yeast to humans is involved in sterol biosynthesis. J. Lipid Res. 2001;42:150–154. [PubMed] [Google Scholar]
- Gardner R. G., Hampton R. Y. A ‘distributed degron’ allows regulated entry into the ER degradation pathway. EMBO J. 1999;18:5994–6004. doi: 10.1093/emboj/18.21.5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Hampton R. Y. ER stress response: getting the UPR hand on misfolded proteins. Curr. Biol. 2000;10:R518–R521. doi: 10.1016/s0960-9822(00)00583-2. [DOI] [PubMed] [Google Scholar]
- Hampton R. Y., Koning A., Wright R. In vivo examination of membrane protein localization and degradation with green fluorescent protein. Proc. Natl. Acad. Sci. USA. 1996;93:828–833. doi: 10.1073/pnas.93.2.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R. M., et al. Gene expression profiling in the human hypothalamus-pituitary-adrenal axis and full-length cDNA cloning. Proc. Natl. Acad. Sci. USA. 2000;97:9543–9548. doi: 10.1073/pnas.160270997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Istvan E. S., Palnitkar M., Buchanan S. K., Deisenhofer J. Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis. EMBO J. 2000;19:819–830. doi: 10.1093/emboj/19.5.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A. L., Fawcett D. W. Hypertrophy of the agranular endoplasmic reticulum in hamster liver induced by phenobarbital (with a review on the functions of this organelle in liver) J. Histochem. Cytochem. 1966;14:215–232. doi: 10.1177/14.3.215. [DOI] [PubMed] [Google Scholar]
- Koning A. J., Larson L. L., Cadera E. J., Parrish M. L., Wright R. L. Mutations that affect vacuole biogenesis inhibit proliferation of the endoplasmic reticulum in Saccharomyces cerevisiae. Genetics. 2002;160:1335–1352. doi: 10.1093/genetics/160.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koning A. J., Lum P. Y., Williams J. M., Wright R. DiOC6 staining reveals organelle structure and dynamics in living yeast cells. Cell Motil. Cytoskeleton. 1993;25:111–128. doi: 10.1002/cm.970250202. [DOI] [PubMed] [Google Scholar]
- Koning A. J., Roberts C. J., Wright R. L. Different subcellular localization of Saccharomyces cerevisiae HMG-CoA reductase isozymes at elevated levels corresponds to distinct endoplasmic reticulum membrane proliferations. Mol. Biol. Cell. 1996;7:769–789. doi: 10.1091/mbc.7.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J., Roberts T. H., Hirschberg K. Secretory protein trafficking and organelle dynamics in living cells. Annu. Rev. Cell Dev. Biol. 2000;16:557–589. doi: 10.1146/annurev.cellbio.16.1.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loertscher J., Larson L. L., Matson C. K., Parrish M. L., Felthauser A., Sturm A., Tachibana C., Bard M., Wright R. Endoplasmic reticulum-associated degradation is required for cold adaptation and regulation of sterol biosynthesis in the yeast Saccharomyces cerevisiae. Eukaryot. Cell. 2006;5:712–722. doi: 10.1128/EC.5.4.712-722.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R., Kärgel E., Vogel F., Böttcher C., Schunck W. H. Topogenesis of a microsomal cytochrome P450 and induction of endoplasmic reticulum membrane proliferation in Saccharomyces cerevisiae. Arch. Biochem. Biophys. 1996;330:97–109. doi: 10.1006/abbi.1996.0230. [DOI] [PubMed] [Google Scholar]
- Mo C., Bard M. Erg28p is a key protein in the yeast sterol biosynthetic enzyme complex. J. Lipid Res. 2005;46:1991–1998. doi: 10.1194/jlr.M500153-JLR200. [DOI] [PubMed] [Google Scholar]
- Parks L. W., Bottema C. D., Rodriguez R. J., Lewis T. A. Yeast sterols: yeast mutants as tools for the study of sterol metabolism. Methods Enzymol. 1985;111:333–346. doi: 10.1016/s0076-6879(85)11020-7. [DOI] [PubMed] [Google Scholar]
- Parrish M. L., Sengstag C., Rine J. D., Wright R. L. Identification of the sequences in HMG-CoA reductase required for karmellae assembly. Mol. Biol. Cell. 1995;6:1535–1547. doi: 10.1091/mbc.6.11.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle J. R., Adams A. E., Drubin D. G., Haarer B. K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Profant D. A., Roberts C. J., Koning A. J., Wright R. L. The role of the 3-hydroxy 3-methylglutaryl coenzyme A reductase cytosolic domain in karmellae biogenesis. Mol. Biol. Cell. 1999;10:3409–3423. doi: 10.1091/mbc.10.10.3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Profant D. A., Roberts C. J., Wright R. L. Mutational analysis of the karmellae-inducing signal in Hmg1p, a yeast HMG-CoA reductase isozyme. Yeast. 2000;16:811–827. doi: 10.1002/1097-0061(20000630)16:9<811::AID-YEA579>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Qureshi N., Dugan R. E., Nimmannit S., Wu W. H., Porter J. W. Purification of beta-hydroxy-beta-methylglutaryl-coenzyme A reductase from yeast. Biochemistry. 1976;15:4185–4190. doi: 10.1021/bi00664a009. [DOI] [PubMed] [Google Scholar]
- Ridgway N. D., Byers D. M., Cook H. W., Storey M. K. Integration of phospholipid and sterol metabolism in mammalian cells. Prog. Lipid Res. 1999;38:337–360. doi: 10.1016/s0163-7827(99)00007-7. [DOI] [PubMed] [Google Scholar]
- Ross M. H., Pappas G. D., Lanman J. T., Lind J. Electron microscope observations on the endoplasmic reticulum in the human fetal adrenal. J. Biophys. Biochem. Cytol. 1958;4:659–661. doi: 10.1083/jcb.4.5.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Leung J., Grimsey N., Peak-Chew S., Siniossoglou S. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 2005;24:1931–1941. doi: 10.1038/sj.emboj.7600672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Santos-Rosa H., Rappsilber J., Mann M., Hurt E. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 1998;17:6449–6464. doi: 10.1093/emboj/17.22.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisson J. K., Fahrenbach W. H. Fine structure of steroidogenic cells of a primate cutaneous organ. Am. J. Anat. 1967;121:337–367. doi: 10.1002/aja.1001210211. [DOI] [PubMed] [Google Scholar]
- Snapp E. L., Hegde R. S., Francolini M., Lombardo F., Colombo S., Pedrazzini E., Borgese N., Lippincott-Schwartz J. Formation of stacked ER cisternae by low affinity protein interactions. J. Cell Biol. 2003;163:257–269. doi: 10.1083/jcb.200306020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stith B. J., Hall J., Ayres P., Waggoner L., Moore J. D., Shaw W. A. Quantification of major classes of Xenopus phospholipids by high performance liquid chromatography with evaporative light scattering detection. J. Lipid Res. 2000;41:1448–1454. [PubMed] [Google Scholar]
- Storey M. K., Clay K. L., Kutateladze T., Murphy R. C., Overduin M., Voelker D. R. Phosphatidylethanolamine has an essential role in Saccharomyces cerevisiae that is independent of its ability to form hexagonal phase structures. J. Biol. Chem. 2001;276:48539–48548. doi: 10.1074/jbc.M109043200. [DOI] [PubMed] [Google Scholar]
- Tong A. H., et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Trotter P. J., Voelker D. R. Identification of a non-mitochondrial phosphatidylserine decarboxylase activity (PSD2) in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1995;270:6062–6070. doi: 10.1074/jbc.270.11.6062. [DOI] [PubMed] [Google Scholar]
- Veitia R. A., Hurst L. D. Accelerated molecular evolution of insect orthologues of ERG28/C14orf 1, a link with ecdysteroid metabolism? J. Genet. 2001;80:17–21. doi: 10.1007/BF02811414. [DOI] [PubMed] [Google Scholar]
- Veitia R. A., Ottolenghi C., Bissery M. C., Fellous A. A novel human gene, encoding a potential membrane protein conserved from yeast to man, is strongly expressed in testis and cancer cell lines. Cytogenet. Cell Genet. 1999;85:217–220. doi: 10.1159/000015296. [DOI] [PubMed] [Google Scholar]
- Voelker D. R. Interorganelle transport of aminoglycerophospholipids. Biochim. Biophys. Acta. 2000;1486:97–107. doi: 10.1016/s1388-1981(00)00051-2. [DOI] [PubMed] [Google Scholar]
- Voelker D. R. Phosphatidylserine decarboxylase. Biochim. Biophys. Acta. 1997;1348:236–244. doi: 10.1016/s0005-2760(97)00101-x. [DOI] [PubMed] [Google Scholar]
- Voeltz G. K., Prinz W. A., Shibata Y., Rist J. M., Rapoport T. A. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- Wiest D. L., Burkhardt J. K., Hester S., Hortsch M., Meyer D. I., Argon Y. Membrane biogenesis during B cell differentiation: most endoplasmic reticulum proteins are expressed coordinately. J. Cell Biol. 1990;110:1501–1511. doi: 10.1083/jcb.110.5.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R. Transmission electron microscopy of yeast. Microsc. Res. Tech. 2000;51:496–510. doi: 10.1002/1097-0029(20001215)51:6<496::AID-JEMT2>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Wright R., Basson M., D'Ari L., Rine J. Increased amounts of HMG-CoA reductase induce “karmellae”: a proliferation of stacked membrane pairs surrounding the yeast nucleus. J. Cell Biol. 1988;107:101–114. doi: 10.1083/jcb.107.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright R., Parrish M. L., Cadera E., Larson L., Matson C. K., Garrett-Engele P., Armour C., Lum P. Y., Shoemaker D. D. Parallel analysis of tagged deletion mutants efficiently identifies genes involved in endoplasmic reticulum biogenesis. Yeast. 2003;20:881–892. doi: 10.1002/yea.994. [DOI] [PubMed] [Google Scholar]
- Zhang S., Skalsky Y., Garfinkel D. J. MGA2 or SPT23 is required for transcription of the delta9 fatty acid desaturase gene, OLE1, and nuclear membrane integrity in Saccharomyces cerevisiae. Genetics. 1999;151:473–483. doi: 10.1093/genetics/151.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.