Abstract
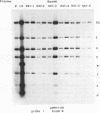
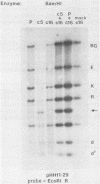
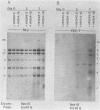
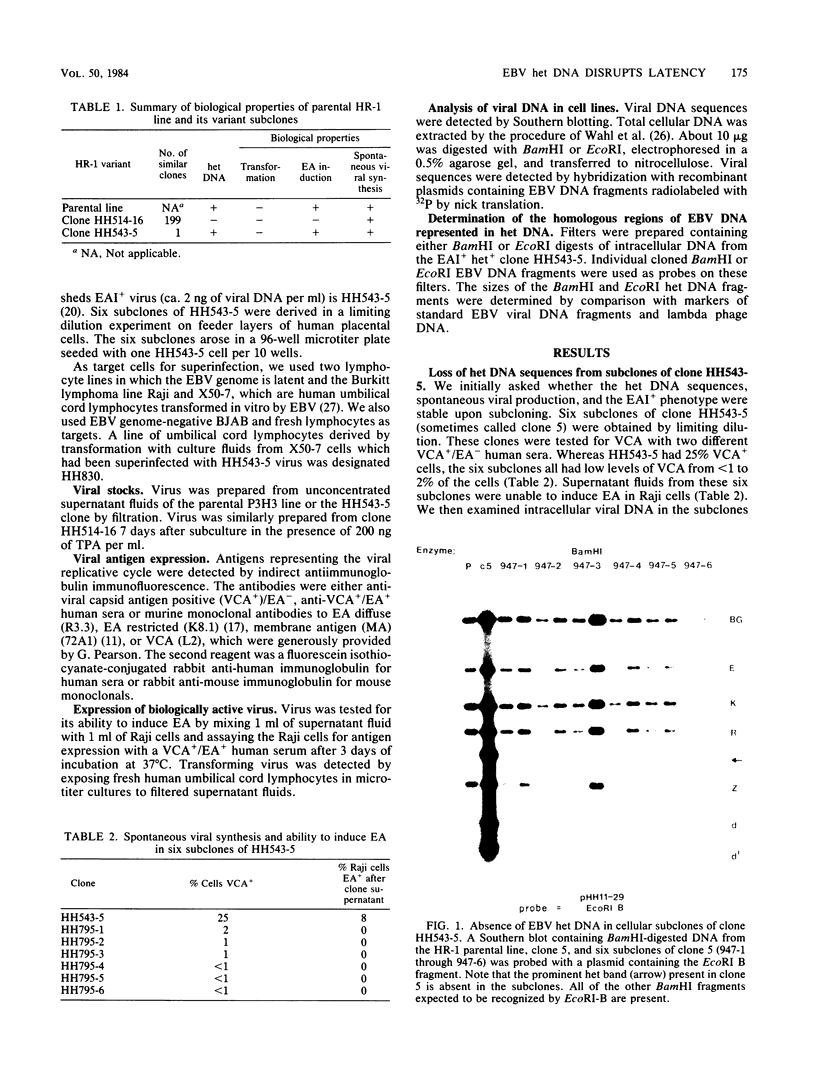
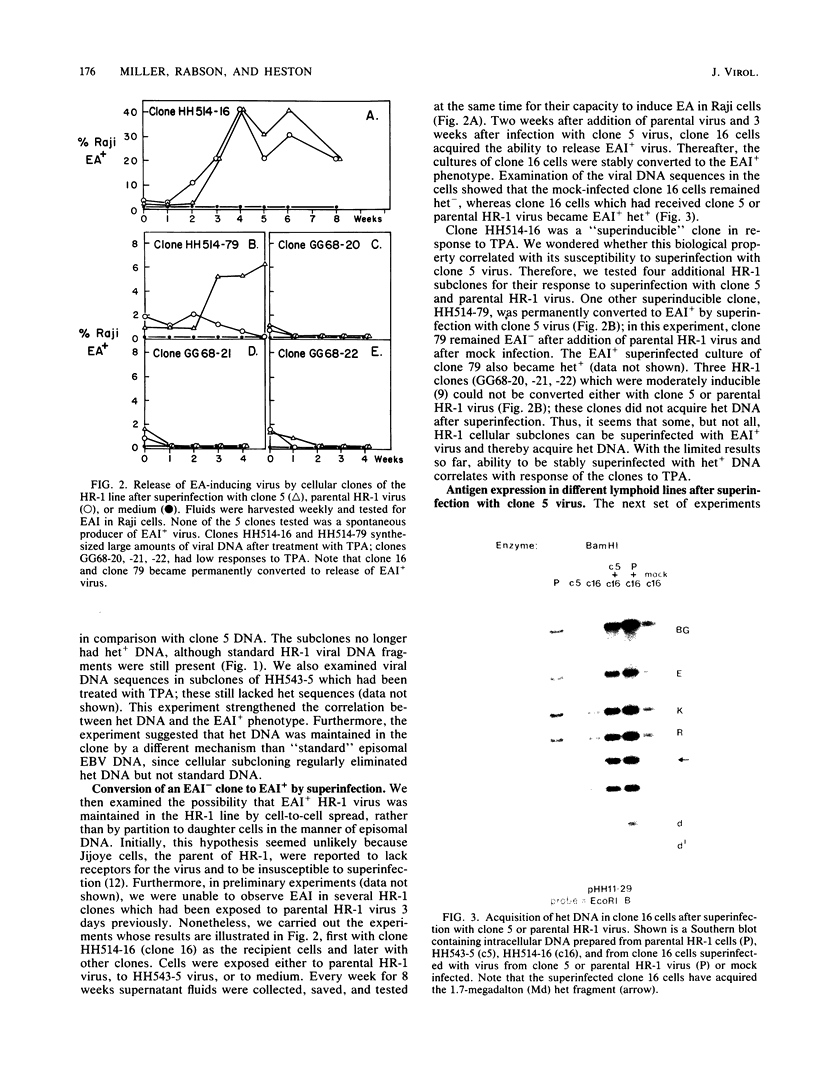
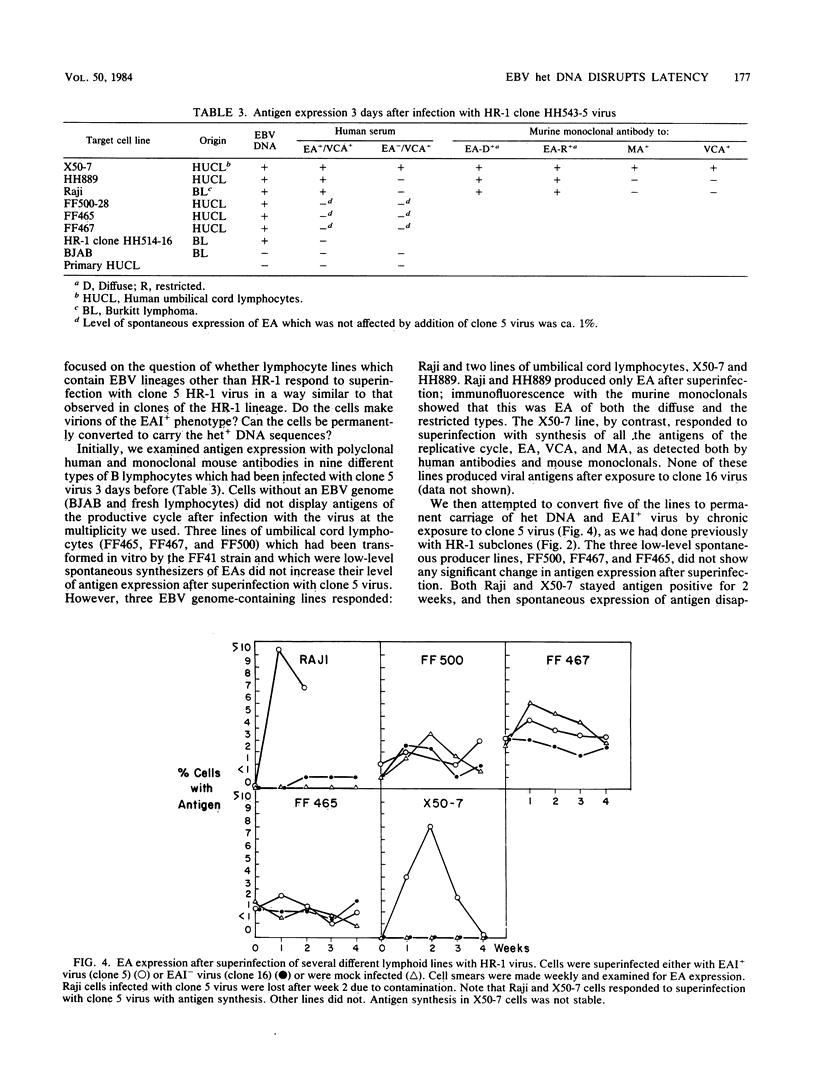
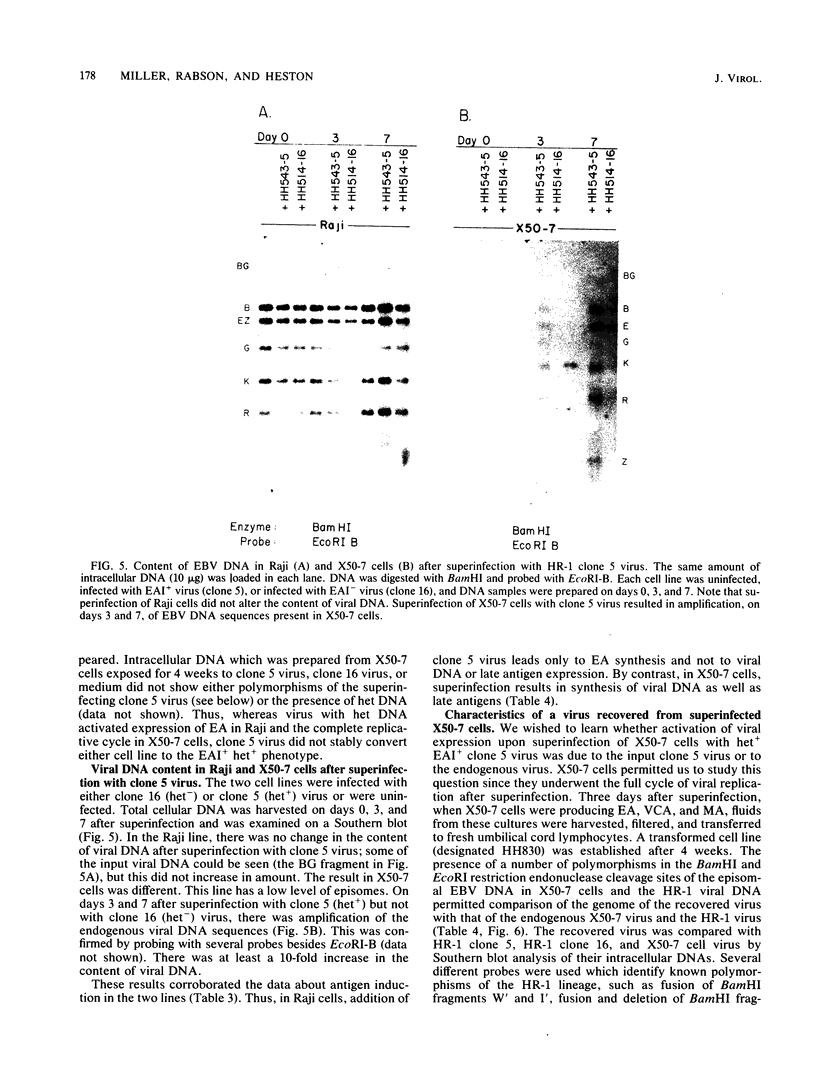
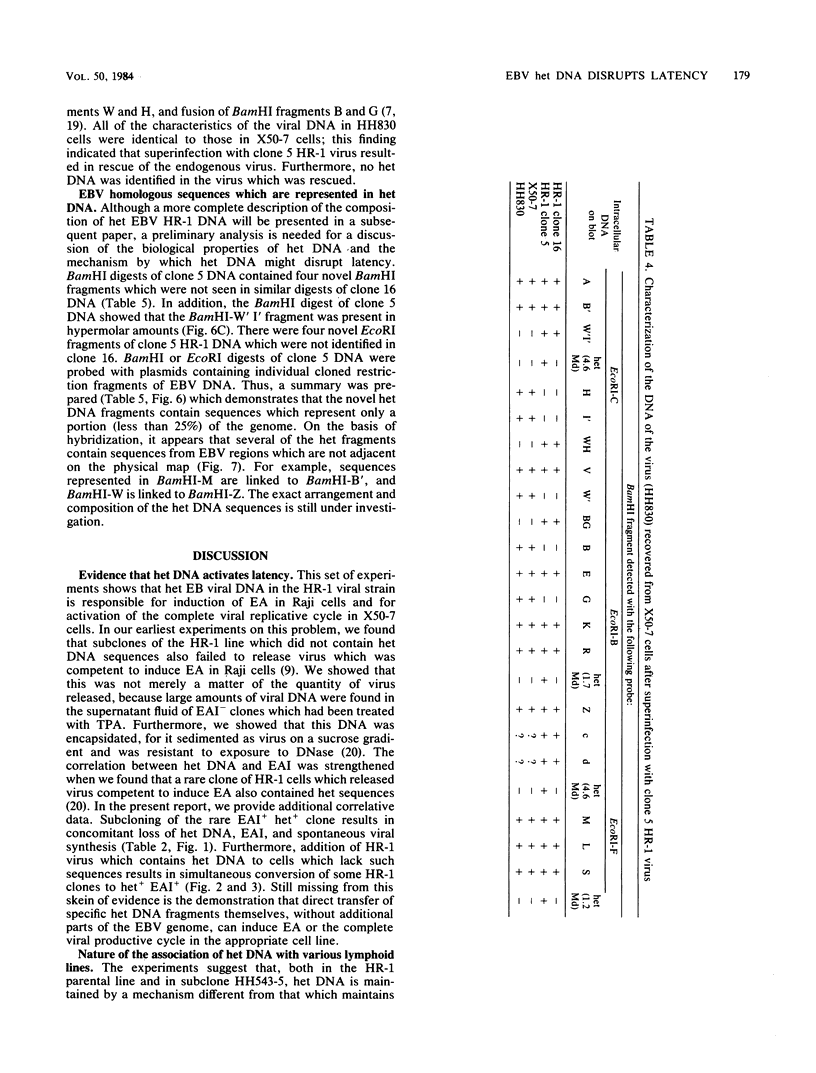
By cloning the HR-1 Burkitt lymphoma line, we previously uncovered two distinct biological variants of nontransforming Epstein-Barr virus (EBV). The most commonly cloned variant has a low rate of spontaneous viral synthesis and is unable to induce early antigen in Raji cells (EAI-). A rare variant spontaneously releases virus which is capable of inducing early antigen in Raji cells (EAI+). Since EAI- virus lacks heterogeneous DNA (het-) and EAI+ virus contains heterogeneous DNA (het+), we suggested that spontaneous viral synthesis and induction of early antigen are biological properties which correlate with the presence of het sequences. The present experiments provide three new lines of experimental evidence in favor of this hypothesis. (i) Revertant subclones of the EAI+ het+ variant which have lost the het DNA concomitantly lost EAI ability. Thus, het DNA is not stably associated with the cells as are the episomes. (ii) het DNA was acquired by two het- subclones of the HR-1 line after superinfection with EAI+ virus. After superinfection, these clones synthesized EAI+ het+ virus. Thus, het DNA may be maintained in the HR-1 line by cell-to-cell spread. (iii) Virus with het DNA activated full expression of endogenous latent EBV of the transforming phenotype in a line of immortalized neonatal lymphocytes designated X50-7. By use of restriction endonuclease polymorphisms unique to both the superinfecting and endogenous genomes, we show that the genome of the activated virus resembles that of the virus which was endogenous to X50-7 cells. This result suggests that het sequences result in transactivation of the latent EBV. het DNA had homology with EBV sequences which are not normally contiguous on the physical map of the genome. het DNA was always accompanied by the presence of DNA of nonheterogenous HR-1. Thus, het DNA is a form of "defective" EBV DNA. However, the biological effect of this defective DNA is to enhance rather than to interfere with EBV replication. This is a novel property of defective virus.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bornkamm G. W., Delius H., Zimber U., Hudewentz J., Epstein M. A. Comparison of Epstein-Barr virus strains of different origin by analysis of the viral DNAs. J Virol. 1980 Sep;35(3):603–618. doi: 10.1128/jvi.35.3.603-618.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornkamm G. W., Hudewentz J., Freese U. K., Zimber U. Deletion of the nontransforming Epstein-Barr virus strain P3HR-1 causes fusion of the large internal repeat to the DSL region. J Virol. 1982 Sep;43(3):952–968. doi: 10.1128/jvi.43.3.952-968.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delius H., Bornkamm G. W. Heterogeneity of Epstein-Barr virus. III. Comparison of a transforming and a nontransforming virus by partial denaturation mapping of their DNAs. J Virol. 1978 Jul;27(1):81–89. doi: 10.1128/jvi.27.1.81-89.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R. A., Schaffer P. A. Fine-structure mapping and functional analysis of temperature-sensitive mutants in the gene encoding the herpes simplex virus type 1 immediate early protein VP175. J Virol. 1980 Oct;36(1):189–203. doi: 10.1128/jvi.36.1.189-203.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller M., Dambaugh T., Kieff E. Epstein-Barr virus DNA. IX. Variation among viral DNAs from producer and nonproducer infected cells. J Virol. 1981 May;38(2):632–648. doi: 10.1128/jvi.38.2.632-648.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henle W., Henle G., Zajac B. A., Pearson G., Waubke R., Scriba M. Differential reactivity of human serums with early antigens induced by Epstein-Barr virus. Science. 1970 Jul 10;169(3941):188–190. doi: 10.1126/science.169.3941.188. [DOI] [PubMed] [Google Scholar]
- Heston L., Rabson M., Brown N., Miller G. New Epstein-Barr virus variants from cellular subclones of P3J-HR-1 Burkitt lymphoma. Nature. 1982 Jan 14;295(5845):160–163. doi: 10.1038/295160a0. [DOI] [PubMed] [Google Scholar]
- Hinuma Y., Konn M., Yamaguchi J., Wudarski D. J., Blakeslee J. R., Jr, Grace J. T., Jr Immunofluorescence and herpes-type virus particles in the P3HR-1 Burkitt lymphoma cell line. J Virol. 1967 Oct;1(5):1045–1051. doi: 10.1128/jvi.1.5.1045-1051.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman G. J., Lazarowitz S. G., Hayward S. D. Monoclonal antibody against a 250,000-dalton glycoprotein of Epstein-Barr virus identifies a membrane antigen and a neutralizing antigen. Proc Natl Acad Sci U S A. 1980 May;77(5):2979–2983. doi: 10.1073/pnas.77.5.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jondal M., Klein G., Oldstone M. B., Bokish V., Yefenof E. Surface markers on human B and T lymphocytes. VIII. Association between complement and Epstein-Barr virus receptors on human lymphoid cells. Scand J Immunol. 1976;5(4):401–410. doi: 10.1111/j.1365-3083.1976.tb00294.x. [DOI] [PubMed] [Google Scholar]
- King W., Dambaugh T., Heller M., Dowling J., Kieff E. Epstein-Barr virus DNA XII. A variable region of the Epstein-Barr virus genome is included in the P3HR-1 deletion. J Virol. 1982 Sep;43(3):979–986. doi: 10.1128/jvi.43.3.979-986.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locker H., Frenkel N. Structure and origin of defective genomes contained in serially passaged herpes simplex virus type 1 (Justin). J Virol. 1979 Mar;29(3):1065–1077. doi: 10.1128/jvi.29.3.1065-1077.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes J., Leibold W., Klein G. Biological differences between Epstein-Barr virus (EBV) strains with regard to lymphocyte transforming ability, superinfection and antigen induction. Exp Cell Res. 1975 May;92(2):478–484. doi: 10.1016/0014-4827(75)90404-8. [DOI] [PubMed] [Google Scholar]
- Miller G., Robinson J., Heston L., Lipman M. Differences between laboratory strains of Epstein-Barr virus based on immortalization, abortive infection, and interference. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4006–4010. doi: 10.1073/pnas.71.10.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson G. R., Vroman B., Chase B., Sculley T., Hummel M., Kieff E. Identification of polypeptide components of the Epstein-Barr virus early antigen complex with monoclonal antibodies. J Virol. 1983 Jul;47(1):193–201. doi: 10.1128/jvi.47.1.193-201.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston C. M. Control of herpes simplex virus type 1 mRNA synthesis in cells infected with wild-type virus or the temperature-sensitive mutant tsK. J Virol. 1979 Jan;29(1):275–284. doi: 10.1128/jvi.29.1.275-284.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabson M., Gradoville L., Heston L., Miller G. Non-immortalizing P3J-HR-1 Epstein-Barr virus: a deletion mutant of its transforming parent, Jijoye. J Virol. 1982 Dec;44(3):834–844. doi: 10.1128/jvi.44.3.834-844.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabson M., Heston L., Miller G. Identification of a rare Epstein-Barr virus variant that enhances early antigen expression in Raji cells. Proc Natl Acad Sci U S A. 1983 May;80(9):2762–2766. doi: 10.1073/pnas.80.9.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rixon F. J., Ben-Porat T. Structural evolution of the DNA of pseudorabies-defective viral particles. Virology. 1979 Aug;97(1):151–163. doi: 10.1016/0042-6822(79)90381-7. [DOI] [PubMed] [Google Scholar]
- Robinson R. A., Tucker P. W., Dauenhauer S. A., O'Callaghan D. J. Molecular cloning of equine herpesvirus type 1 DNA: analysis of standard and defective viral genomes and viral sequences in oncogenically transformed cells. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6684–6688. doi: 10.1073/pnas.78.11.6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rymo L., Forsblom S. Cleavage of Epstein-Barr virus DNA by restriction endonucleases EcoRI, HindIII and BamI. Nucleic Acids Res. 1978 Apr;5(4):1387–1402. doi: 10.1093/nar/5.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuhlmann H., Jähner D., Jaenisch R. Infectivity and methylation of retroviral genomes is correlated with expression in the animal. Cell. 1981 Oct;26(2 Pt 2):221–232. doi: 10.1016/0092-8674(81)90305-6. [DOI] [PubMed] [Google Scholar]
- Sugden B., Summers W. C., Klein G. Nucleic acid renaturation and restriction endonuclease cleavage analyses show that the DNAs of a transforming and a nontransforming strain of Epstein-Barr virus share approximately 90% of their nucleotide sequences. J Virol. 1976 May;18(2):765–775. doi: 10.1128/jvi.18.2.765-775.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl G. M., Padgett R. A., Stark G. R. Gene amplification causes overproduction of the first three enzymes of UMP synthesis in N-(phosphonacetyl)-L-aspartate-resistant hamster cells. J Biol Chem. 1979 Sep 10;254(17):8679–8689. [PubMed] [Google Scholar]
- Wilson G., Miller G. Recovery of Epstein-Barr virus from nonproducer neonatal human lymphoid cell transformants. Virology. 1979 Jun;95(2):351–358. doi: 10.1016/0042-6822(79)90490-2. [DOI] [PubMed] [Google Scholar]
- Yano S., Tanaka A., Takada K., Fujiwara S., Osato T., Nonoyama M. Analysis of epstein-Barr virus (EBV) of P3HR-1: isolation of EBV with EBNA induction ability in human cord lymphocytes and without EA induction ability in Raji cells. Virology. 1982 Jun;119(2):392–398. doi: 10.1016/0042-6822(82)90098-8. [DOI] [PubMed] [Google Scholar]
- zur Hausen H., O'Neill F. J., Freese U. K., Hecker E. Persisting oncogenic herpesvirus induced by the tumour promotor TPA. Nature. 1978 Mar 23;272(5651):373–375. doi: 10.1038/272373a0. [DOI] [PubMed] [Google Scholar]