Abstract
The PhoQ sensor-kinase is essential for Salmonella typhimurium virulence for animals, and a homolog exists in the environmental organism and opportunistic pathogen Pseudomonas aeruginosa. S. typhimurium PhoQ (ST-PhoQ) is repressed by millimolar concentrations of divalent cations and activated by antimicrobial peptides and at acidic pH. ST-PhoQ has a periplasmic PAS domain, a fold commonly employed for ligand binding. However, substrate binding is instead accomplished by an acidic patch in the periplasmic domain that interacts with the inner membrane through divalent cation bridges. The DNA sequence encoding this acidic patch is absent from Pseudomonas phoQ (PA-PhoQ). Here, we demonstrate that PA-PhoQ binds and is repressed by divalent cations, and can functionally complement a S. typhimurium phoQ mutant. Mutational analysis and NMR spectroscopy of the periplasmic domains of ST-PhoQ and PA-PhoQ indicate distinct mechanisms of binding divalent cation. The data are consistent with PA-PhoQ binding metal in a specific ligand-binding pocket. PA-PhoQ was partially activated by acidic pH but not by antimicrobial peptides. S. typhimurium expressing PA-PhoQ protein were attenuated for virulence in a mouse model, suggesting that the ability of Salmonella to sense host environments via antimicrobial peptides and acidic pH is an important contribution to pathogenesis.
Introduction
The ability of organisms to sense and respond to their environments is central to survival. In bacteria, this is in part accomplished by two-component sensor-response regulator systems (West and Stock, 2001; Ulrich et al., 2005). These systems are generally composed of a sensor histidine kinase and a response regulator that controls gene expression. In response to appropriate environmental stimuli, the sensor kinase trans-autophosphorylates within a dimer and subsequently transfers phosphate to the response regulator, which then results in transcriptional activation or repression of specific genes.
Gram-negative bacteria have an outer and an inner membrane, and many sensor kinases have periplasmic sensing domains. Among the characterized sensor kinases is the citrate receptor, which is activated on binding of citrate (Bott, 1997). The structure of the citrate receptor periplasmic region is comprised of a Per-ARNT-Sim (PAS) domain, a widely conserved fold involved in binding small molecules via a pocket that binds a ligand with high specificity. (Reinelt et al., 2003). The citrate receptor has such a defined pocket where it binds citrate, and then in an unknown manner transfers that information across the inner membrane to control kinase activity in the cytoplasmic domain of the molecule. Thus, sensor kinases, and their sensing domains in particular, allow bacteria to sense and respond to specific components of their environments.
Another well-studied bacterial sensor kinase for which structural information is available is PhoQ, which functions to regulate the response regulator PhoP. This system is important for the virulence of a number of Gram-negative pathogens for animals and plants (Miller et al., 1989; Moss et al., 2000; Oyston et al., 2000; Llama-Palacios et al., 2003; Derzelle et al., 2004). Salmonella typhimurium PhoPQ is required for virulence for animals and intracellular survival within phagocytes. Salmonella PhoQ is activated within acidified macrophage phagosomes, suggesting that it is a sensor of the intracellular environment of its host (Alpuche-Aranda et al., 1992). Upon activation, PhoP controls transcription of a large group of genes (Miller et al., 1989; Belden and Miller, 1994; Soncini et al., 1996), including those involved in resistance to antimicrobial peptides and survival in an acidic environment (Fields et al., 1989; Miller et al., 1990; Foster and Hall, 1990). Thus, PhoPQ appears to be central to S. typhimurium survival within a host by sensing the intracellular phagosome environment and mediating an appropriate response.
PhoQ is an inner membrane sensor protein with a periplasmic domain that is responsible for sensing specific signals in the environment, and a cytoplasmic domain responsible for intermolecular autophosphorylation upon activation and subsequent phosphotransfer to PhoP. PhoQ is repressed by millimolar concentrations of the divalent cations Mg2+, Ca2+, and Mn2+, and the periplasmic domain directly binds these cations (Garcia Vescovi et al., 1997; Bader et al., 2005). Repression can be alleviated by growth in low micromolar concentrations of these cations; however, the Mg2+ concentration inside macrophage phagosomes has been measured at 1 mM (Martin-Orozco et al., 2006), suggesting that other signals may be required to activate PhoPQ in host tissues. Indeed, cationic antimicrobial peptides (CAMPs) and acidic pH, both of which are expected to be present in the phagosomal environment, have recently been shown to directly activate PhoPQ in the presence of millimolar divalent cation (Bader et al., 2005; Prost et al., 2007).
The crystal structure of the PhoQ periplasmic domain in the repressed state reveals a PAS domain, like the citrate receptor. However, in the case of PhoQ, no ligand-binding pocket is present. Instead, divalent cations are bound to aspartic and glutamic acid residues in an acidic patch of PhoQ that is adjacent to two novel α-helices and is hypothesized to face the membrane (Cho et al., 2006). The protein is thus held in the repressed state by divalent cation bridges between the acidic patch and the inner membrane. Several experiments have suggested that antimicrobial peptides also bind to the acidic patch, presumably displacing the divalent cations and causing a conformational change associated with activation (Bader et al., 2005). This has made it difficult to dissect the role of each signal during infection because CAMP and divalent cation sensing do not appear to be separable. By contrast, acidic pH is hypothesized to activate PhoQ by causing loss of structural rigidity in the core of the protein (Prost et al., 2007). Mutations resulting in a low-pH like conformation, as measured by NMR spectroscopy, also result in de-repression in high divalent cation, again making it difficult to determine the relative importance of each sensing capability to infection.
PhoPQ homologues have also been identified in Pseudomonas aeruginosa, a ubiquitous environmental organism commonly found in soil and water. Like in Salmonella, Pseudomonas PhoQ-dependent gene expression is repressed by divalent cations (Macfarlane et al., 1999). There is additional functional evidence that divalent cation is a ligand for Pseudomonas PhoQ, as growth in low divalent cation media causes specific changes in the Pseudomonas lipopolysaccharide that are PhoQ-dependent (Ernst et al., 1999). In addition, a phoQ-null mutation results in loss of transcriptional regulation of the PmrAB two-component system, which controls resistance to the antimicrobial peptide polymyxin B in response to growth in low divalent cation media (McPhee et al., 2003). The purified Pseudomonas PhoQ periplasmic domain exhibits a conformational change upon addition of Mg2+ as measured by fluorescence spectroscopy, suggesting that it directly binds divalent cation (Lesley and Waldburger, 2001). In previous experiments, a Pseudomonas-Salmonella PhoQ chimera containing the Pseudomonas periplasmic domain was expressed in Salmonella and PhoP-dependent gene expression in this system was activated by growth in low divalent cation, but not by antimicrobial peptides (Bader et al., 2005). This suggested that S. typhimurium PhoQ evolved a mechanism of CAMP sensing as a function of a transition to colonization of mammals. Furthermore, Pseudomonas PhoQ is lacking the amino acids that comprise the acidic patch and adjacent α-helices involved in the Salmonella PhoQ CAMP activation mechanism. These observations suggested that the primary function of Pseudomonas PhoQ was to act as a divalent cation sensor, while in contrast Salmonella PhoQ, as a pathogen sensor intimately associated with animals, may sense host signals such as CAMP and pH.
In the present study, the sensing capabilities of Pseudomonas PhoQ and the mechanism of ligand binding are examined and compared to those of Salmonella PhoQ. This comparison defines differences in the environmental sensing requirements of a pathogen versus an environmental organism, and elucidates how the PhoQ sensor protein itself may have evolved to fulfill different roles. In addition, these different capabilities shed light on the relative importance of each signal sensed by Salmonella PhoQ to virulence for multicellular hosts.
Results
P. aeruginosa PhoQ can Functionally Replace S. typhimurium PhoQ for Growth in Low Divalent Cation Environments but not Response to Antimicrobial Peptides
Previous work evaluating the signaling differences between the periplasmic domains of S. typhimurium PhoQ (ST-PhoQ) and P. aeruginosa PhoQ (PA-PhoQ) utilized multicopy plasmids. To better define the signals recognized by PA-PhoQ, a chromosomal reporter system was constructed using a chimeric and full length PA-PhoQ. A S. typhimurium strain expressing a reporter fusion to the PhoP-controlled gene encoding acid phosphatase (phoN) was used as a wild type ST-PhoQ reporter strain background. The chromosomal copy of PhoQ was then removed to make a PhoQ-null reporter strain, and it was replaced by two different PA-PhoQ constructs to test activation by full length PA-PhoQ (FL-PA-PhoQ) and a chimera (Chim-PhoQ) consisting of ST-PhoQ with its periplasmic domain replaced by that of PA-PhoQ (Fig 1A). The chimeric strain has the advantage of containing the ST-PhoQ cytoplasmic domain but, as an artificial chimera, it could exhibit unknown differences in transducing signals from the PA-PhoQ periplasmic domain to the ST-PhoQ transmembrane regions. Both strains are used throughout these experiments to most reliably determine the sensing responses of the PA-PhoQ periplasmic domain.
Figure 1. PA-PhoQ is repressed by divalent cation but does not recognize CAMP.
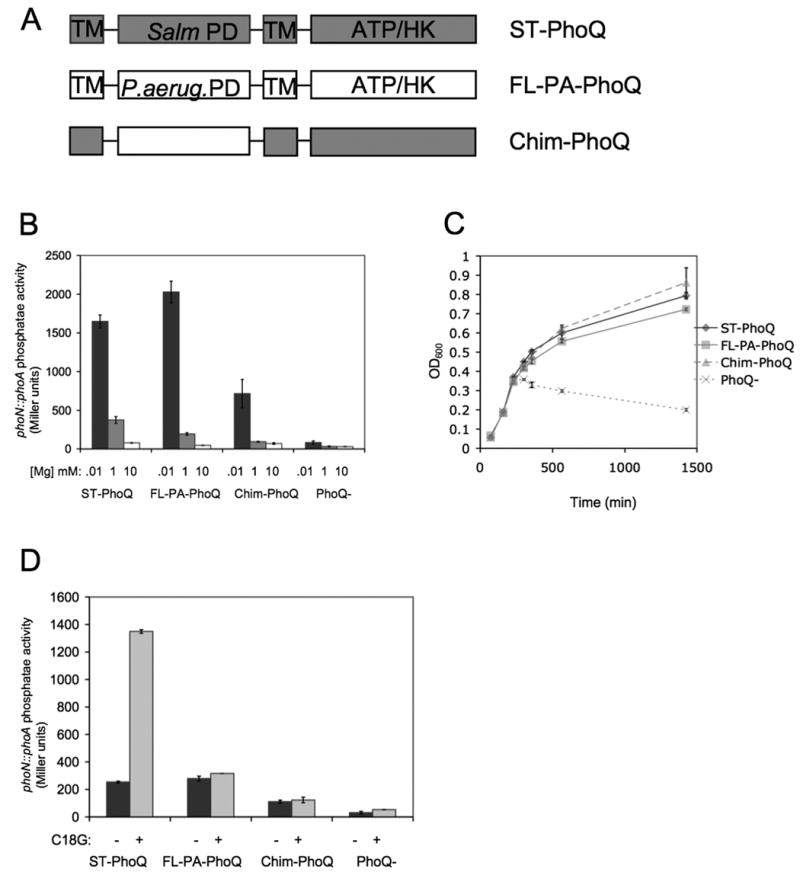
(A) A schematic of the domain organization of the PhoQ constructs used in this study. The PhoQ periplasmic domain (PD) is flanked by two transmembrane regions (TM) and contains a cytoplasmic ATP-binding domain with catalytic histidine kinase activity (ATP/HK). Chim-PhoQ consists of full-length ST-PhoQ except that the PD has been replaced with that of PA-PhoQ.
(B) PhoP-dependent gene activation decreases with increasing concentration of Mg2+ for all PhoQ constructs tested. A reporter fusion between PhoP-dependent acid phosphatase (PhoN) and PhoA was used to measure activation, and PhoQ constructs are expressed chromosomally. Cultures were grown in N minimal medium containing 10 μM (black bars), 1 mM (gray bars), or 10 mM (white bars) MgCl2.
(C) PA-PhoQ constructs can complement a PhoQ-null strain for growth in low divalent cation medium. Strains were grown in N minimal medium supplemented with 10 μM MgCl2 and growth was measured by OD600 over time.
(D) PA-PhoQ cannot activate PhoP-dependent gene activation in response to CAMP. Cultures were grown in N minimal medium supplemented with 1 mM MgCl2. Where indicated, C18G was added at 5 μg/ml. All graphed values are mean ± standard deviation of cultures grown in duplicate and are representative of at least three independent trials.
As shown in Fig 1B, ST-PhoQ strongly activated PhoP-dependent gene expression upon growth in divalent cation limited medium but was strongly repressed in divalent cation replete medium at 10 mM Mg2+, and no reporter activation was observed in the PhoQ-null strain. FL-PA-PhoQ exhibited a very similar response to ST-PhoQ, remarkably demonstrating that the cytoplasmic domain of PA-PhoQ is capable of interacting appropriately with S. typhimurium PhoP in order to control downstream gene activation. Chim-PhoQ also exhibited robust activation by divalent cation limitation though it appeared to be more easily repressed, as activation was lower than observed for ST-PhoQ or FL-PA-PhoQ in media containing 1 mM and 10 μM Mg2+. Similar results were obtained for a pagC∷phoA reporter (data not shown). A relevant functional assay for divalent cation repression is growth in divalent cation limited N minimal medium, which is dependent on PhoQ. As shown in Fig 1C and as previously described, the PhoQ null strain is severely inhibited for growth under these conditions (Garcia-Vescovi et al., 1996). However, both FL-PA-PhoQ and Chim-PhoQ restored growth equivalently to wild type levels. Thus, PA-PhoQ can respond to divalent cation limitation to the same extent as ST-PhoQ in terms of its ability to replicate.
An important physiological response of ST-PhoQ is its recognition of CAMP. The response to CAMP was further tested using the strains containing PA-PhoQ. Transcriptional activation assays of phoN and pagC were conducted in N minimal medium with 1 mM divalent cation with or without C18G, a synthetic cationic α-helical peptide with demonstrated antimicrobial activity (Darveau et al., 1992). As shown in Fig 1D, ST-PhoQ activated the phoN reporter in the presence of C18G. However, neither FL-PA-PhoQ nor Chim-PhoQ activated under this condition. Similar results were obtained for the pagC reporter (data not shown). Thus, the CAMP response is specific to the presence of the ST-PhoQ periplasmic domain and is not observed with PA-PhoQ. As Pseudomonas is an environmental organism not typically associated with animals, this PhoQ sensing difference could represent an important adaptation of Samonellae to animals, and an essential function of pathogenesis.
The Purified PA-PhoQ Periplasmic Domain Exhibits Unique Conformational Changes Upon Binding Ca2+ and Mg2+
To further demonstrate that the PA-PhoQ periplasmic domain mediates divalent cation binding and repression, this domain was purified and tested by fluorescence spectroscopy, which is based on intrinsic fluorescence of tryptophan residues. The PA-PhoQ periplasmic domain contains only a single tryptophan residue, simplifying the fluorescence signal and making this a convenient technique for following conformational changes. Changes in peak fluorescence (335 nm) were followed upon addition of increasing concentrations of Mg2+ and Ca2+. As shown in Fig 2A, binding saturation was reached at lower cation concentration for Ca2+ than Mg2+. These data were fit to a standard equilibrium model to estimate the binding affinities for each cation at an apparent KD of 37 μM Ca2+ and 207 μM Mg2+. Thus, PA-PhoQ exhibits a greater than five-fold difference in binding affinity for the two cations. Garcia Vescovi et al. (1997) measured the apparent KD of purified ST-PhoQ periplasmic domain for Mg2+ as 52.6 mM and stated that similar results were obtained for Ca2+. However, we estimate the binding affinity in the low millimolar range for Mg2+ and Ca2+ because ST-PhoQ appears fully saturated by NMR in the presence of 20 mM of either cation, and at least 5 mM Ca2+ or Mg2+ is required to out compete micromolar concentrations of CAMP for binding to ST-PhoQ, as measured by fluorescence spectroscopy and reporter assays (Bader et al., 2005). Overall, PA-PhoQ has a substantially higher binding affinity for divalent cation than ST-PhoQ.
Figure 2. Purified PA-PhoQ-PD exhibits distinct responses to Mg2+ vs. Ca2+.
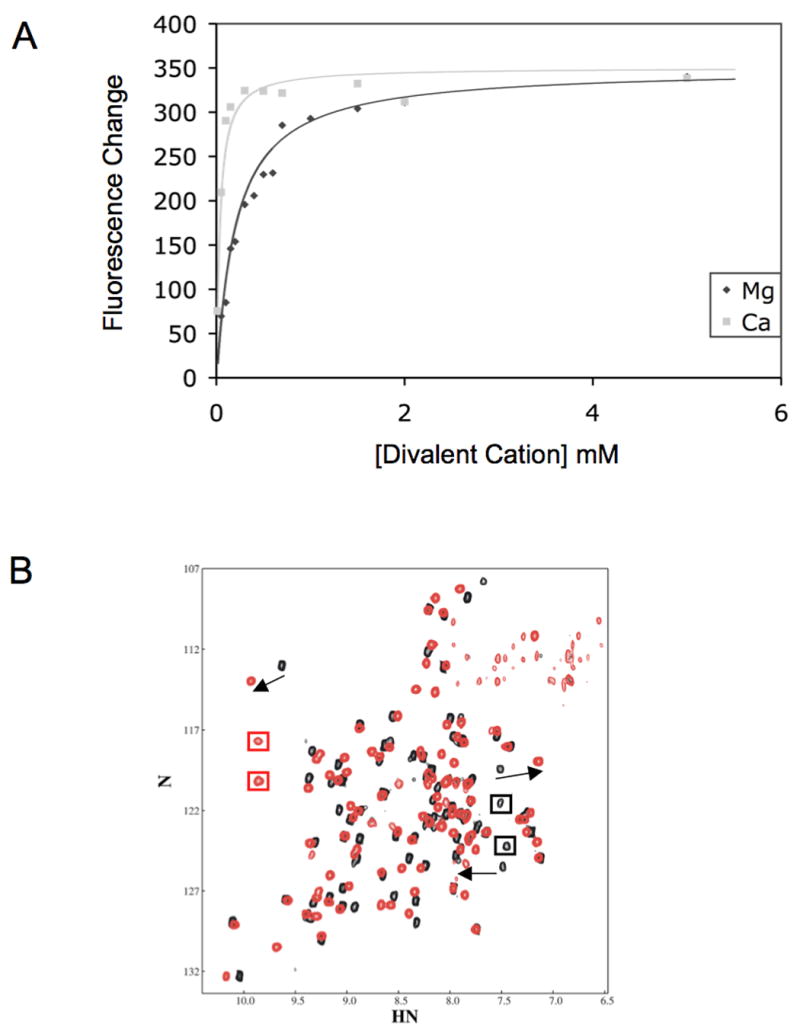
(A) PA-PhoQ binds to Ca2+ more strongly than Mg2+. MgCl2 or CaCl2 were added at indicated concentrations to 1 μM purified PA-PhoQ-PD. Peak fluorescence values were fit to a simple equilibrium model.
(B) PA-PhoQ assumes different conformations when bound to Mg2+ vs. Ca2+. Superimposed 15N-TROSY-HSQC spectra of the PA-PhoQ-PD in the presence of 20 mM MgCl2 (black) or 20 mM CaCl2 (red). Arrows identify several peaks that undergo significant shift between the Mg2+-bound state and the Ca2+-bound state. Black boxes illustrate Mg2+-specific resonances and red boxes illustrate Ca2+-specific resonances
To study conformational effects of each divalent cation on the purified PA-PhoQ periplasmic domain, NMR spectra were collected in the presence of saturating concentrations of Mg2+ or Ca2+. HSQC spectra of purified 15N-labeled PA-PhoQ periplasmic domain collected under different conditions are sensitive indicators of alterations in the immediate chemical environment of each amino acid residue, including binding or dissociation of a ligand. As shown in Fig 2B, there are significant differences in the NMR spectrum depending on the cation present. By titrating Ca2+ into a sample containing Mg2+-bound PA-PhoQ, we could follow some resonances from the Mg2+ state (black spectrum) to the Ca2+ state (red spectrum), indicated by arrows. In other cases, resonances that correspond to the Mg2+ state disappeared while new peaks that correspond to the Ca2+ state appeared, illustrated by black and red boxes, respectively. Both behaviors indicate that there are backbone amide protons in the protein that undergo chemical shift differences of 0.5 ppm or more. In general, changes in chemical shift may either be a direct effect of a ligand itself or an indirect effect due to a ligand-induced conformational change in a protein. The strong chemical and electronic similarities between Mg2+ and Ca2+ are inconsistent with the large observed differences they cause being attributable to direct effects. Therefore, the large and numerous chemical shift differences elicited by the two cations indicate that their binding results in a significantly different conformation of the sensor domain. The spectra could not be compared to a spectrum of the apo PA-PhoQ periplasmic domain because it is insoluble at NMR concentrations in the absence of divalent cations. These results are in contrast to ST-PhoQ, which is stable and soluble in its apo-state and which exhibits nearly identical spectra in the presence of either Mg2+ or Ca2+ (Cho et al., 2006). Taken together, the data demonstrate that PA-PhoQ, unlike ST-PhoQ, adopts different conformations dependent on the identity of cation bound, indicating higher ligand specificity.
Single Amino Acid Mutations in PA-PhoQ Result in Loss of Divalent Cation Repression
The observed divalent cation specificity and the lack of a defined acidic patch suggest that PA-PhoQ may bind divalent cation by a different mechanism than ST-PhoQ, perhaps in a pocket more analogous to the citrate receptor. To begin to elucidate the PA-PhoQ divalent cation binding mechanism, a genetic screen was undertaken to identify residues on the PA-PhoQ periplasmic domain that are important for repression by divalent cations (see Experimental Procedures). As shown in Fig 3A, ten mutations were found that result in a loss of repression by 10 mM divalent cation. Five of these residues are either Asp or Glu, the amino acids that expected to bind Ca2+ and Mg2+ ions in proteins (more information on cation binding proteins can be found at http://structbio.vanderbilt.edu/cabp_database/seq/index.html). Remarkably, several of these mutations result in reporter activation to about the same level as growth in divalent cation limited medium. This is in strong contrast to a similar screen performed with ST-PhoQ (Cho et al., 2006), where single mutations do not result in such strong levels of de-repression, presumably because of redundancy in divalent cation binding conferred by the acidic patch and divalent cation bridges to the membrane.
Figure 3. Mutations in specific PA-PhoQ residues lead to loss of Mg2+-mediated repression.

(A) Mutations in PA-PhoQ result in loss of repression of the PhoN reporter. Strain MB147, which carries a PhoQ-null allele and the plasmid pBAD24-FL-PA-PhoQ, was used as the wild type strain. The indicated mutations were made in pBAD24-FL-PA-PhoQ by Quickchange mutagenesis. Cultures were grown in N minimal medium supplemented with 10 mM MgCl2.
(B) Several mutants still show Mg2+-mediated repression of the PhoN reporter. A subset of the mutant strains from Fig 3A were grown in N minimal medium supplemented with 10 mM (black bars) or 10 μM (gray bars) MgCl2.
(C) Several mutations result in complete loss of Mg2+-mediated repression of the PhoN reporter. The remaining mutant strains from Fig 3A were grown in N minimal medium supplemented with 10 mM (black bars) or 10 μM (gray bars) MgCl2. All graphed values are mean ± standard deviation of cultures grown in duplicate and are representative of at least three independent trials.
(D) Mutations in PA-PhoQ alter Mg2+-binding saturation. MgCl2 was added at indicated concentrations to 1 μM purified wild type or mutant PA-PhoQ-PD. Peak fluorescence values were fit to a simple equilibrium model.
To further characterize the PA-PhoQ cation repression deficient mutants, they were also tested for activity upon growth in divalent cation limited medium. Based on these results, the mutations can be clustered into two classes: those for which a moderate level of de-repression was observed at growth in medium with 10 mM Mg2+ but growth in low divalent cation medium increased activation (Fig 3B), and those that demonstrated higher or maximal de-repression and for whom the presence of divalent cation in the growth medium did not alter gene expression (Fig 3C). PhoQ mutants with an alanine replacing glutamate at position 77 (E77A) and 115 (E115A), a glycine at position 139 (G139A), and a glycine replacing alanine at position 143 (A143G) still had some divalent cation mediated repression and therefore likely have some ability to bind cations, as growth in low divalent cation medium still results in an increase in PhoQ-mediated gene activation (Fig 3B). In contrast, PhoQ mutants with an alanine replacing aspartate at positions 110 (D110A) and 131 (D131A), a glutamate at position 133 (E133A), a leucine at positions 137 (L137A) and 158 (L158A), and a serine at 145 (S145A) exhibited strong de-repression in medium containing 10 mM Mg2+, and activity was approximately the same regardless of the cation content in the growth medium (Fig 3C), indicating that they are not able to respond to divalent cation. These mutated aspartate and glutamate residues represent good candidates for positions involved directly in specific divalent cation binding in PA-PhoQ.
Purified PA-PhoQ Periplasmic Domain Mutants Exhibit Altered Affinity for Mg2+
To further characterize the Mg2+-binding properties of the mutants described above, PA-PhoQ periplasmic domains were purified carrying a mutation from each class, as well as wild type. The purified proteins were tested for ability to bind Mg2+ via fluorescence spectroscopy. As shown in Fig 3D, wild type PA-PhoQ showed a change in fluorescence with increasing Mg2+ concentration that saturated by the low mM range, with an apparent KD of 207 μM. In contrast, purified mutant PhoQ E115A showed a much smaller fluorescence change that required higher concentrations of Mg2+ to saturate, with an apparent Kd of 1.3 mM. This result is consistent with the biological predictions from growth in low divalent cation medium that the mutation lowered affinity but did not abolish PhoQ cation binding. In contrast, purified mutant PhoQ D131A showed no fluorescence change well into the mM range of Mg2+, suggesting that this mutation renders the protein incapable of binding Mg2+. Circular dichroism spectra were collected on each purified mutant protein in the presence of Mg2+ to confirm that the PhoQ mutant proteins were folded similarly to wild type protein (data not shown). Therefore observed differences in the fluorescence spectra are highly likely to reflect biologically relevant differences in binding divalent cation and not non-specific alterations in protein structure.
A Limited Number of Residues in the PA-PhoQ Periplasmic Domain are Involved in Binding Divalent Cation
NMR spectroscopy was used to provide additional insight into the divalent cation binding mechanism of PA-PhoQ. In the presence of Mg2+, the NMR spectrum contains roughly 108 peaks, each corresponding to a specific amino acid residue. Lanthanide ions are known to bind competitively with Mg2+ and Ca2+ in many proteins (Lee and Sykes, 1983). We therefore performed competition experiments with Mg2+-bound PA-PhoQ periplasmic domain and the lanthanide Gd3+. The paramagnetic Gd3+ ion broadens NMR resonances in a distance-dependent manner. At very low occupancy, the observed effects of Gd3+ will be limited to the resonances of those nuclei that are in closest proximity to the bound ion. Addition of 50 μM Gd3+ to a 0.5 mM sample of PA-PhoQ in the presence of 20 mM Mg2+ results in the broadening of specific peaks in the NMR spectrum. Five peaks are broadened beyond detection, identifying these as derived from the amide groups that are closest to a Gd3+ ion. These peaks are highlighted by red boxes in the spectral region plotted in Figure 4, i.e., peaks in the black (no Gd3+ spectrum) that are absent in the 50 μM Gd3+ spectrum (red). An additional ten peaks disappear or are severely broadened in the presence of 100 μM Gd3+, identifying a second group of resonances that arise from groups that are near a Gd3+, although not as close as the first group. These peaks disappear or lose significant intensity in the blue spectrum relative to the red spectrum and are highlighted by blue boxes in Figure 4. These results are in stark contrast to those obtained when the same experiment was carried out on ST-PhoQ, which binds divalent cations via an acidic patch as opposed to a specific structural binding site. In this case, even the lowest concentration of Gd3+ produced general spectral broadening (data not shown), consistent with the existence of multiple cation binding sites of approximately equal affinity. Taken together, the data support a model in which cation binding occurs in a discrete, well-defined site in the PA-PhoQ periplasmic domain.
Figure 4. A discrete set of residues on PA-PhoQ are involved in binding Mg2+.
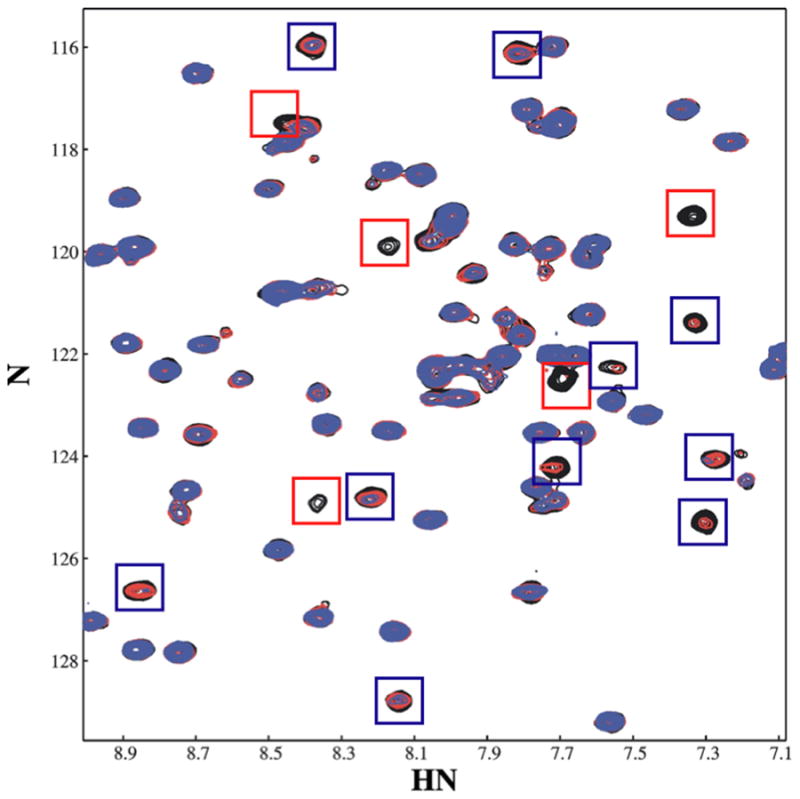
A region of 15N-TROSY-HSQC spectra of the PA-PhoQ-PD collected in the presence of increasing concentrations of Gd3+ is superimposed. The starting spectrum was collected in the presence of 20 mM MgCl2 (black); GdCl3 was added to a concentration of 50 μM (red) and 100 μM (blue). Resonances most affected by Gd3+ displacement of Mg2+ are shown in red boxes; those affected at higher Gd3+ concentrations, which are therefore further from the ion, are highlighted in blue boxes.
Secondary Structure Predictions for PA-PhoQ are Consistent with a PAS Domain Structural Binding Pocket
Secondary structural elements of PA-PhoQ were predicted using PsiPred (Jones, 1999; Bryson et al., 2005) and are shown in Figure 5. The experimentally observed structural elements of the sensor domains of ST-PhoQ and the Klebsiella pneumoniae CitA citrate receptor, both of which are PAS domains, are also shown (Cho et al., 2006; Reinelt et al., 2003). There is strong concordance between many of the secondary structural elements in ST-PhoQ and those predicted for PA-PhoQ – α1, β1, β2, β3, β4, β5, β6, β7 and α6. Among these conserved elements are those that comprise the ST-PhoQ PAS domain, though α2 and α3 appear to be replaced by an unstructured region in PA-PhoQ. As shown in Figure 6, the CitA and ST-PhoQ PAS domain cores (boxed) have very similar topology, and PA-PhoQ contains all the β-strands that comprise this domain. On the basis of this analysis, it is likely that the sensor domain of PA-PhoQ also takes on a PAS fold.
Figure 5. Sequence and structure alignments of PAS domain proteins.
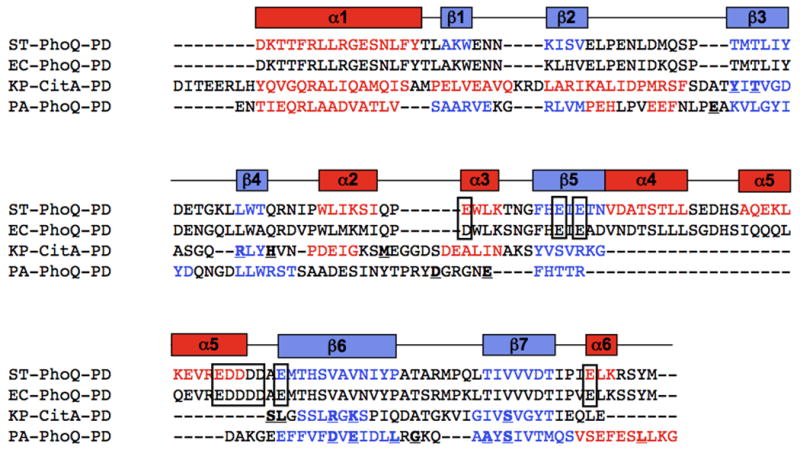
The primary sequences of ST-PhoQ and E. coli PhoQ are aligned according to sequence homology, and Klebsiella pneumoniae CitA and PA-PhoQ are aligned to ST-PhoQ according to secondary structural elements. Secondary structural elements of ST-PhoQ are depicted above the sequences, and the corresponding amino acid residues in each sequence are colored red (helices) or blue (strands). Residues that make up the ST-PhoQ acidic patch are shown in boxes. Residues involved in binding citrate are bolded and underlined on the CitA sequence. The PA-PhoQ sequence is colored according to predicted secondary structure, and mutations described in Figure 3 are bolded and underlined.
Figure 6. Topology maps of PAS domain proteins.
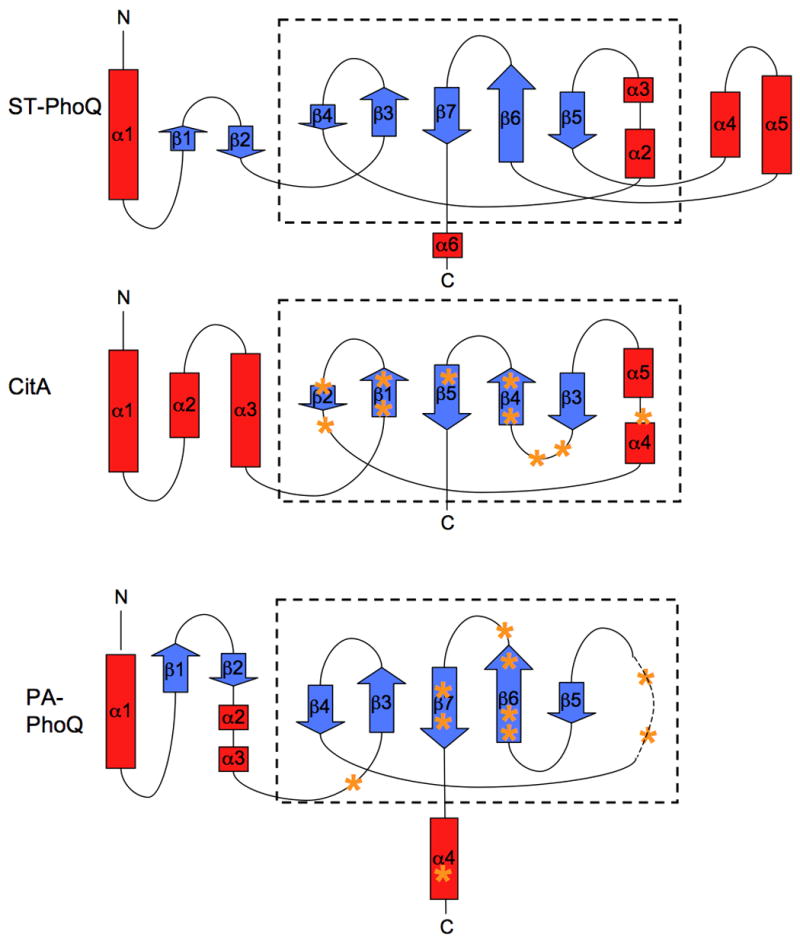
Helices are depicted as red boxes and strands as blue arrows. The size of each box/arrow is proportional to the number of amino acid residues that compose that element. The core PAS domain structure is boxed. The approximate locations of residues involved in binding citrate or Mg2+ are marked with orange asterisks on the CitA and PA-PhoQ maps, respectively.
As mentioned previously, PA-PhoQ lacks the helices α4 and α5 and the acidic domains that have been implicated in divalent cation and CAMP binding in ST-PhoQ. These motifs are present in PhoQ sequences from other animal-associated organisms and pathogens, such as E. coli (see Fig 5), but they are absent from CitA, which has a more conventional ligand-binding pocket associated with its PAS domain. Residues involved in binding citrate are shown in bold on the CitA sequence in Figure 5, and residues predicted by our mutagenesis studies to be involved in binding divalent cation are shown in bold on the PA-PhoQ sequence. The positions of these residues are also indicated on the topology maps shown in Figure 6. Notably, many of the citrate-binding residues fall in the citrate β4-β5 region, which is analogous to the PA-PhoQ β5-β6 region where many of the PA-PhoQ mutations are located. In CitA, the residues in the β4-β5 region are part of a defined binding pocket for citrate observed in the crystal structure, so it is highly likely that the structurally similar region in PA-PhoQ will also comprise a binding pocket. In addition, when the mutated PA-PhoQ residues are mapped onto the ST-PhoQ structure, they cluster to the same region on the central β-sheet, consistent with the hypothesis that the residues are in close proximity in the PA-PhoQ three-dimensional structure. Taken together, the structure predictions and alignments are consistent with the hypothesis that the PA-PhoQ sensor domain folds into a PAS domain with a traditional small molecule binding pocket comprised of a discrete set of cation binding residues.
PA-PhoQ, Like ST-PhoQ, is Activated by Acidic pH
In addition to its regulation by divalent cation and CAMP, ST-PhoQ has been shown to directly respond to acidic pH to activate PhoP (Prost et al., 2007). pH is hypothesized to activate PhoQ via a different mechanism than low cation or CAMP because activation by pH involves a partial loss of structural rigidity in the core of the protein. As this is independent of the acidic patch, it is possible that pH activation may also be a property of PA-PhoQ. To test this, the phoN reporter system was used and cultures were grown in N minimal medium buffered at either pH 7.5 or pH 5.5. As shown in Fig 7A, ST-PhoQ showed increased activation of the reporter when grown in medium at pH 5.5 as compared to pH 7.5. Both FL-PA-PhoQ and Chim-PhoQ showed similar responses: activity was robustly increased at pH 5.5 compared to 7.5. However, total activation at pH 5.5 was lower for the two PA-PhoQ constructs than for ST-PhoQ, by a factor of approximately two-fold. Similar results were also obtained with a pagC∷phoA reporter (data not shown). These data clearly demonstrate that PA-PhoQ is activated by acidic pH, though the protein may not be as fully activated as ST-PhoQ at equivalent pH.
Figure 7. PA-PhoQ recognizes changes in pH.
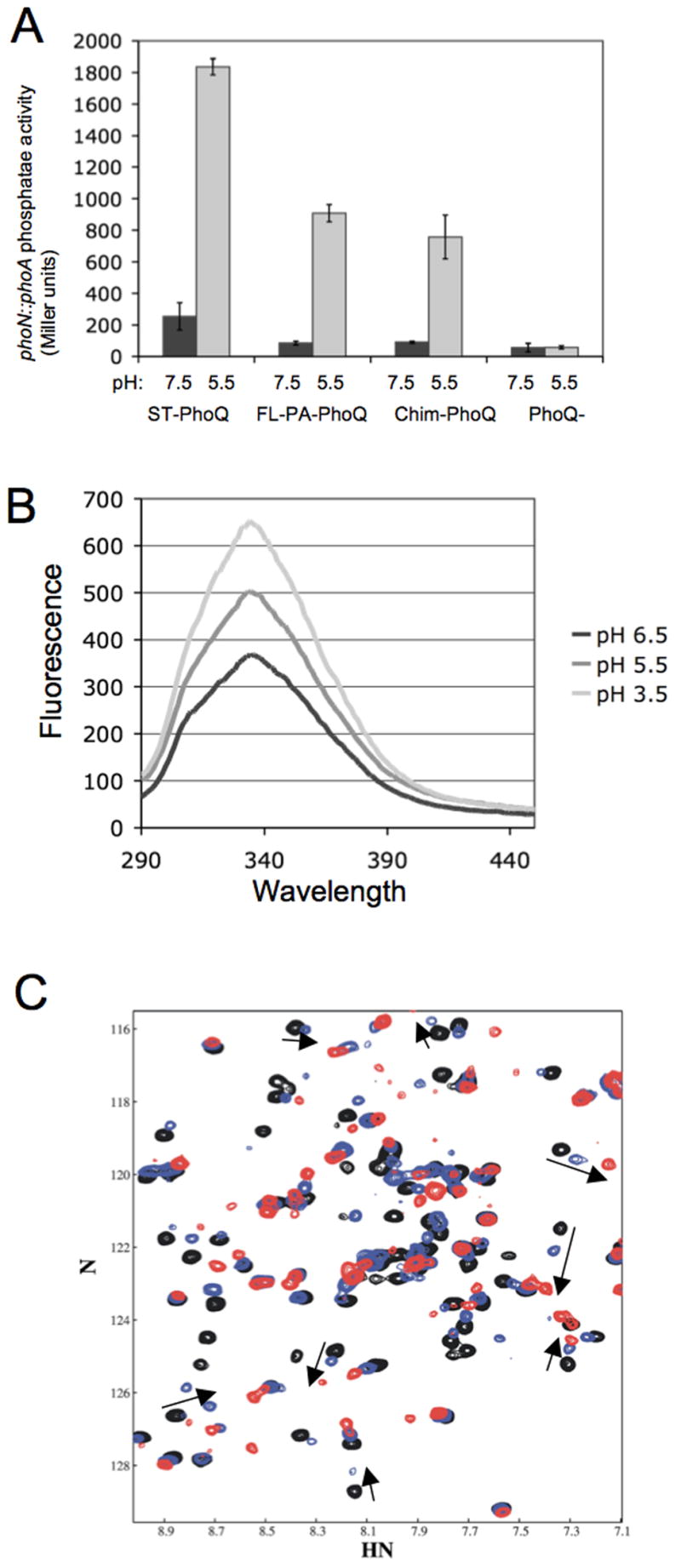
(A) PA-PhoQ activates PhoP-dependent gene activation in Salmonella when grown in acidic pH medium. The PhoN reporter system was used to measure activation, and PhoQ constructs are expressed chromosomally. Cultures were grown in N minimal medium buffered with MES to either pH 7.5 (black bars) or 5.5 (gray bars), and supplemented with 1 mM MgCl2.
(B) PA-PhoQ-PD undergoes a conformational change upon acidification. Fluorescence spectra were collected of 1 μM purified PA-PhoQ-PD in MES buffered at pH 6.5, 5.5, or 3.5, as indicated.
(C) Some conformational changes occur in the PA-PhoQ-PD upon acidification. Superimposed 15N-TROSY-HSQC spectra of the PA-PhoQ-PD in the presence of 20 mM MgCl2 at pH 7.7 (black), 6.6 (blue), and 5.5 (red). The arrows follow pH shifts of several peaks that were affected by Gd3+ (Fig 4).
To confirm that PA-PhoQ directly mediates pH activation, conformational changes in the purified PA-PhoQ periplasmic domain in response to acidification were studied by fluorescence spectroscopy. As shown in Figure 7B, fluorescence intensity increased with acidification, demonstrating a structural change. An increase in fluorescence intensity is consistent with tryptophan burial, so it does not appear that the protein is unfolding in the acidic environment at the low concentration used for fluorescence. To further study conformational changes associated with pH, an NMR titration was performed in which NMR spectra were collected on purified 15N-labeled PA-PhoQ periplasmic domain at pH values ranging from pH 7.7 to 5.5 (Fig 7C). Several peaks shift as a function of pH. Some resonances shifts in the pH range from 7.7 to 6.6, and additional shifts occurred at pH 6.6 and 5.5. As with the fluorescence, the effects observed with decreasing pH were not those associated with protein unfolding.
Many of the peaks that shift with pH are also those that are most affected by addition of Gd3+, suggesting that loss of cation binding may be involved in pH sensing (see arrows, Fig 7C). However, there are also shifts observed in peaks that are not hypothesized to bind cation. Mg2+ and Ca2+ ions are expected to bind to glutamate and aspartate side chains, and with average pKa values of 3.7 and 4.3, respectively, Asp and Glu residues are not expected to be protonated at pH values around 5.5. This allows for the possibility of structural changes specific to pH in addition to loss of cation binding. Overall, these data demonstrate structural changes in the PA-PhoQ periplasmic domain in an acidic environment that presumably lead to activation, though it is unclear the extent to which loss of cation binding contributes to these structural changes.
Salmonella Strains Expressing PA-PhoQ Have a Defect in Systemic Virulence for Mice
PA-PhoQ can functionally replace ST-PhoQ for promotion of growth in low divalent cation medium, but cannot respond to CAMP and is not activated as strongly by acidic pH. To provide evidence for whether these sensing differences affect virulence, we compared systemic animal virulence of a S. typhimurium strain expressing wild type ST-PhoQ to strains expressing FL-PA-PhoQ or Chim-PhoQ. The PA-PhoQ strains, as well as a PhoQ-null strain, were mixed in equal proportion to the ST-PhoQ strain and injected intraperitoneally into mice to determine competitive indices. As shown in Fig 8, all the mutant strains tested exhibited a virulence defect compared to wild type. The competitive index for ST-PhoQ was 1.01, for FL-PA-PhoQ was 0.26, for Chim-PhoQ was .54, and for PhoQ-null was .01. Neither FL-PA-PhoQ nor Chim-PhoQ was as avirulent as PhoQ-null. This result is consistent with intracellular activation of PA-PhoQ by pH and the observed basal activation of PhoP-dependent gene activation at the expected intra-phagosomal Mg2+ concentration of 1 mM. However, the defects are statistically significant and substantial enough to demonstrate that the altered sensing abilities of PA-PhoQ have a functional effect on systemic virulence. Since both PA-PhoQ strains allow for full repression of the PhoP-regulon on growth in high divalent cation medium (see Fig 1B) and promote growth equivalently in divalent cation limited medium (Fig 1C), the attenuation in virulence is not likely to be related to divalent cation sensing. These results are consistent with PhoQ activation by CAMP and pH as playing an important role in systemic pathogenesis.
Figure 8. PA-PhoQ expressed in Salmonella confers a systemic virulence defect.
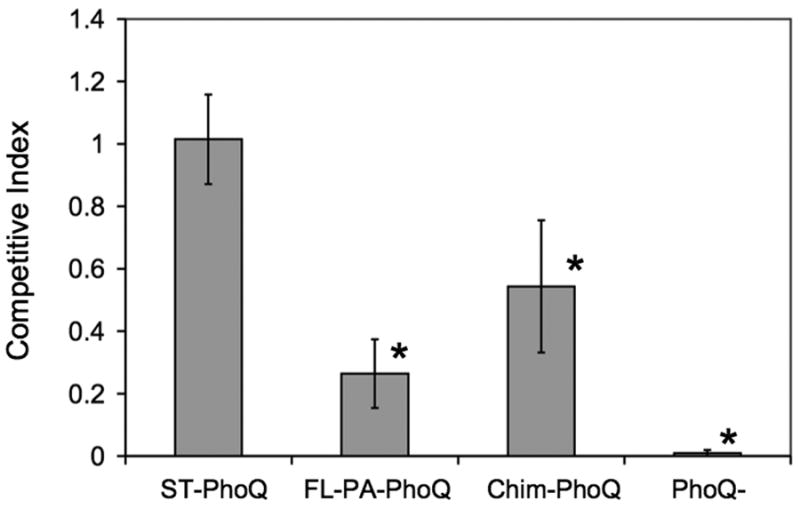
Mice were inoculated intraperitoneally with a 1:1 mixture of 5 × 104 Salmonella organisms each of two strains, wild type ST-PhoQ and the strain indicated below each bar. The CI was calculated by dividing the ratio of strain 1 to strain 2 bacteria in the output by the ratio of strain 1 to strain 2 bacteria in the input. The graph represents the mean ± standard deviation from at least 10 mice that were infected in three separate experiments. Asterisks denote CI values significantly different from wild type ST-PhoQ vs. wild type ST-PhoQ (p-value < 0.001).
Discussion
In the present study, we compare the ligands and sensing mechanisms of ST-PhoQ to those of PA-PhoQ, a highly similar protein from an environmental organism that can function as an opportunistic pathogen. Both PhoQ proteins have similar divalent cation-mediated repression, but they appear to bind cations via different structural mechanisms. The data suggest that, similar to the citrate receptor, PA-PhoQ binds divalent cations in a specific ligand-binding pocket. Unlike ST-PhoQ, PA-PhoQ is not activated by CAMPs, indicating that it does not have the metal bridge mechanism of activation employed by ST-PhoQ and that its main function is the recognition of cations. PA-PhoQ is also activated by acidic pH environments, though not as strongly as ST-PhoQ on expression in S. typhimurium. These sensing differences have an important functional outcome as S. typhimurium expressing PA-PhoQ had a significant virulence defect, despite the fact that it can function to promote growth of the bacteria in low divalent cation medium.
PA-PhoQ is a Cation Sensor that Likely Binds Divalent Cations via a PAS Domain with a Pocket
It is interesting that such closely related proteins as PA- and ST-PhoQ appear to bind divalent cations via distinct mechanisms. The sequence and topology comparisons with CitA shown in Figures 5 and 6 and several lines of experimental evidence suggest that PA-PhoQ binds divalent metals by a ligand binding pocket. NMR spectra demonstrate that PA-PhoQ undergoes distinct conformational changes when bound to Mg2+ versus Ca2+, in contrast to ST-PhoQ where Ca2+ and Mg2+ binding produce the same conformational effects. This is consistent with PA-PhoQ containing a well-defined binding site or pocket that adjusts its structure depending on the nature of the divalent cation bound. This is in contrast to the acidic patch cation-binding mechanism employed by ST-PhoQ, in which different equivalent cations can function as metal bridges that are equally able to bind to the acidic patch with very similar structural consequences for protein conformation.
Further consistent with the idea of a discrete metal binding pocket for PA-PhoQ was the observation of specific NMR resonances that respond to cation binding. In particular, NMR-based analysis of Gd3+ competition for Mg2+ binding to PA-PhoQ identified a set of residues affected due to proximity to a Gd3+ ion. The data are consistent with Gd3+ binding to a discrete site on the protein, thereby sequestering it from the majority of the protein. This is in contrast to divalent cation binding by ST-PhoQ, which involves a large surface of the protein, as evidenced by the general broadening effects that Gd3+ binding has on this domain. Though we have no direct structural proof that the residues identified in the mutational analysis comprise a pocket, it seems likely that some of these residues cooperate to form such a pocket and many of these residues map to sites on a predicted structure where the pocket for binding citrate in CitA is located. Therefore, all results in this work are consistent with a mechanism of binding and sensing ligands that is different for the two receptors PA-PhoQ (binding pocket) and ST-PhoQ (metal bridges to the membrane).
Activation of PA-PhoQ by proton concentration
This work also demonstrated that PA-PhoQ was activated by increased H+ ion concentration when expressed in S. typhimurium, though the mechanism by which this is sensed is not yet clearly defined. NMR spectra at pH 5.5 indicate some conformational changes as compared to pH 7.7 indicating that, similar to ST-PhoQ, PA-PhoQ can adopt a specific conformation at acidic pH. One possibility for this conformational change in PA-PhoQ could be a decreased ability to bind divalent cation in acidic environments. In this regard, NMR spectra of PA-PhoQ cannot be collected in the absence of cation because the protein precipitates at NMR concentrations without ligand present. When the PA-PhoQ NMR sample was titrated below pH 5.5, a considerable amount of protein precipitated, perhaps indicating some loss of ability to bind Mg2+. However, the pH range over which peak shifts are observed in Figure 7C suggests that the protein is able to adopt a pH-specific conformation independent of cation binding because at this pH most amino acids that coordinate metal should not be effectively titrated to lose metal binding. Thus the mechanism by which PA-PhoQ senses pH is unclear, but may involve both loss of divalent cation binding as well as some specific conformational change.
The physiological role of PhoQ in Pseudomonas is unknown and few experiments have been performed to address this issue. As an environmental organism, it may be important to identify environments that are low in divalent cation concentration, perhaps to turn on specific transporters to support growth. As acidic pH can also be encountered in soil environments, PA-PhoQ may also recognize this signal to regulate specific adaptive mechanisms, or it may be activated because of greater difficulty in binding divalent cations in an acidic environment. The PhoPQ regulon has not been well defined for P. aeruginosa, so it is difficult to speculate on the relevance of PhoQ activation by these ions to this organism. The ability to distinguish both acidic environments as well as divalent cation concentrations could be valuable to the organism in some environments, as conditions of low metal and low pH are often found together in environmental soils and waters.
An Evolutionary Model for PhoQ Ligand Recognition
PA-PhoQ can respond with high structural specificity to different concentrations of divalent cations and protons but, in contrast to ST-PhoQ, it does not respond to CAMPs. This is somewhat surprising in that P. aeruginosa has mechanisms to induce resistance to antimicrobial peptides in response to growth in low divalent cation environments, including specific modifications of lipid A (Selgrade, S. and Miller, S.I., unpublished observations). Therefore P. aeruginosa likely evolved a different mechanism or receptor specifically to sense antimicrobial peptides. In contrast, ST-PhoQ is a sensor of mammalian environments for an organism that persists in the intestinal tract lumen and within host cells, and thus may have evolved to specifically recognize CAMPs, which are found in much greater diversity in different species of mammals than in the soil, where they are largely produced as polymyxins by Gram-positive bacteria. As CAMP and acidic pH are encountered inside a host phagosome and in the animal intestinal tract, these sensing data suggest that ST-PhoQ evolved to identify host environments. This evolutionary hypothesis is supported by a sequence alignment of ST-PhoQ and PA-PhoQ, which demonstrates that PA-PhoQ lacks the acidic residues and α-helices involved in the ST-PhoQ acidic patch mechanism of CAMP sensing. Other organisms associated with animal hosts, including E. coli, Yersinia pestis and Photorhabdus luminescens, also contain the acidic residues and the α-helices upon aligning with ST-PhoQ, and are likely to have acidic patches, metal bridges, and to function as antimicrobial peptide sensors. In contrast, the largely environmental sensor PA-PhoQ most likely contains a PAS domain with a conventional binding pocket this is responsible for binding divalent cations, and may encode separate receptors for soil-specific peptides. This may also be the case for some other environmental organisms with large genomes such as Rhizoboium spp.. Thus, ST-PhoQ appears to have evolved specifically as a sensor for the mammalian environment (reviewed further in Prost and Miller, 2008).
The ST-PhoQ Sensing Capabilities Allow for Better Recognition of a Mammalian Host to Promote Bacterial Virulence
In support of the evolutionary model for PhoQ ligand sensing, S. typhimurium strains expressing FL-PA-PhoQ or Chim-PhoQ were deficient in systemic virulence for mice. This demonstrates that ST-PhoQ has evolved to optimally sense the host environment, and replacement with PA-PhoQ, from an environmental organism, confers a decrease in the ability to compete effectively within a host. In addition, this experiment provides a system to begin to dissect the relevance of each signal sensed by ST-PhoQ to animal virulence. First, by comparing ST-PhoQ to FL-PA-PhoQ, it appears that divalent cation repression and its functional aspects of growth in low metal are not enough for full virulence. Strains with these two different PhoQ proteins exhibit nearly identical levels of reporter activation when grown in medium containing 10 mM, 1 mM, and 10 μM Mg2+. The results for 10 mM and 10 μM Mg2+ were also confirmed by RT-PCR for phoN and four additional PhoP-controlled genes (data not shown), so this phenotype does not appear to be specific to the reporter system used. Since FL-PA-PhoQ, as well as the chimeric receptor with the PhoQ periplasmic domain, clearly have a competitive disadvantage upon infection of mice, some aspects of signaling required for virulence are not mediated by divalent cation repression.
It is worth noting that Chim-PhoQ does not confer as strong a defect as FL-PA-PhoQ. It is possible that there are biochemical differences in the cytoplasmic domains that would allow the chimera to partially make up for sensing deficiencies on the periplasmic side. It is also plausible that as yet unidentified signals could be differentially sensed by FL-PA-PhoQ and Chim-PhoQ and lead to the observed difference in virulence observed or that our assays are not sensitive to minor differences in regulation mediated by the cytoplasmic domain of PhoQ. However, given that both strains allow growth to the same extent in low divalent cation media, the observed virulence difference does not appear to be due to differences in metal acquisition or gene regulation.
Acidic pH and CAMP are expected to activate PhoPQ during infection, as both would be encountered inside host macrophage phagosomes. As such, they may represent redundant signals that work together to fully activate PhoPQ in vivo. Indeed, previous work demonstrated that reporter activation was greater in the presence of both signals than either signal alone (Prost et al., 2007). Our virulence data is consistent with this type of redundancy. The PA-PhoQ constructs lack the ability to activate in response to CAMP, and this may contribute to their virulence defects. However, even without the ability to sense CAMP, the PA-PhoQ strains are not as defective for virulence as a PhoQ-null strain, suggesting that the ability to sense pH provides at least some contribution to virulence. Since previous work indicates that vacuolar acidification is essential to PhoQ activation (Alpuche-Aranda et al., 1992), we favor a model in which sensing acidification is essential and CAMP-sensing provides additional activation. As the PA-PhoQ constructs exhibit a lower level of activation by acidic pH compared to ST-PhoQ, it is difficult to definitively conclude whether this lower pH activation or the lack of CAMP sensing is more important to virulence. It is plausible that the two signals and perhaps others within the phagosome are redundant and work together to fully activate ST-PhoQ to promote virulence.
In summary, we have presented evidence that the PhoQ sensor kinase of the environmental organism Pseudomonas aeruginosa functions as a sensor of divalent cation concentrations via a discrete set of cation-binding residues, and that this protein can also function as a sensor for acidic pH. The PhoQ homologue in Salmonella typhimurium exhibits an additional capacity for recognizing CAMPs via structural alterations – novel helices and acidic residues – that still allow for divalent cation binding and may allow for increased activation by acidification. Finally, these differences in ligand recognition and binding mechanism appear to have important functional implications to Salmonella virulence, suggesting that PhoQ sensing in Salmonella facilitates a pathogenic lifestyle.
Experimental Procedures
Bacterial strains and genetic methods
All bacterial strains and primers used in this study are listed in Tables 1 and 2. ATCC strain 14028s was used as the wild type S. typhimurium strain background. The chromosomal deletion of phoQ was constructed using the λ red recombinase system. The KanR gene and FLP recognition target (FRT) sites from pKD4 were PCR amplified with primers LP89 and LP90, which also contain flanking regions of homology to phoQ. The resulting purified PCR product was electroporated into CS093 containing pKD46, the culture was induced with arabinose, plated on LB-Kan, and candidate colonies were confirmed by PCR. The resulting strain was then grown at 42C to remove the KanR cassette, resulting in strain LP47. Chromosomal phoQ∷FL-PA-phoQ and phoQ∷Chim-phoQ constructs were generated using selection for tetracycline sensitivity. First, a phoQ∷tet strain was generated via λ red recombination using primers MB153 and MB154 in a CS093 background. The TetR cassette was then replaced by FL-PA-phoQ by λ red using PCR amplification from pMB147 with primers MB183 and MB184 to yield strain MB175. In parallel, the cassette was also replaced by Chim-phoQ by λ red using PCR amplication from pMB224 with primers LP87 and LP88 to yield strain LP46. Finally, the phoN∷TnphoA reporter was transduced from CS120 into strains MB175, LP46, and LP47 by generalized transduction using P22 HT/int bacteriophage to generate strains LP56, LP57, and LP58, respectively. To express multicopy FL-PA-PhoQ in Salmonella, full length phoQ was PCR amplified from P. aeruginosa using primers MB52 and MB88. The purified PCR product was digested with EcoRV and XbaI and ligated into pBAD24 digested NcoI, blunted, and digested with XbaI. The resulting plasmid, pMB147, was expressed in the MB101 reporter strain to make strain MB147. To express PA-PhoQ periplasmic domain for purification, residues 37-165 of PA-PhoQ were PCR amplified using primers MB89 and MB90. The PCR product was digested with enzymes HindIII and BamHI and ligated into pQE30 digested with the same enzymes. The resulting plasmid, pMB149, was expressed in E. coli BL21 as strain MB149. To make site-directed mutants, plasmids pMB147 and pMB149 served as templates for Quickchange mutagenesis (Stratagene) using the forward primers listed in Table 2 and corresponding reverse primers.
Table 1. Strains Used in This Study.
| Strain | Characteristics | Source |
|---|---|---|
| CS093 | 14028s wild type S. typhimurium | ATCC |
| CS120 | 14028s phoN105∷TnphoA, KanR | Miller et al., 1989 |
| MB101 | CS120 phoQ∷Tn10d(T-POP), TetR | Bader et al., 2005 |
| MB147 | MB101 pBAD24-FL-PA-phoQ, AmpR | This work |
| MB149 | BL21 pQE30-PA-phoQ-PD, AmpR | This work |
| MB175 | 14028s phoQ∷FL-PA-phoQ, AmpR | This work |
| MB224 | MB101 pBAD24-Chim-PhoQ, AmpR | Bader et al., 2005 |
| LP4 | MB101 pBAD24-FL-PA-phoQ D110A, AmpR | This work |
| LP5 | MB101 pBAD24-FL-PA-phoQ E133A, AmpR | This work |
| LP6 | MB101 pBAD24-FL-PA-phoQ D131A, AmpR | This work |
| LP7 | MB101 pBAD24-FL-PA-phoQ E115A, AmpR | This work |
| LP9 | BL21 pQE30-PA-phoQ-PD D131A, AmpR | This work |
| LP14 | BL21 pQE30-PA-phoQ-PD E115A, AmpR | This work |
| LP19 | MB101 pBAD24-FL-PA-phoQ E77A, AmpR | This work |
| LP20 | MB101 pBAD24-FL-PA-phoQ L137A, AmpR | This work |
| LP21 | MB101 pBAD24-FL-PA-phoQ G139A, AmpR | This work |
| LP22 | MB101 pBAD24-FL-PA-phoQ S145A, AmpR | This work |
| LP23 | MB101 pBAD24-FL-PA-phoQ L158A, AmpR | This work |
| LP25 | MB101 pBAD24-FL-PA-phoQ A143G, AmpR | This work |
| LP46 | CS093 phoQ∷Chim-phoQ | This work |
| LP47 | CS093 ΔphoQ | This work |
| LP48 | CS093 pWSK129, KanR | This work |
| LP49 | CS093 pWSK29, AmpR | This work |
| LP50 | MB175 pWSK29, AmpR | This work |
| LP51 | LP46 pWSK29, AmpR | This work |
| LP52 | LP47 pWSK29, AmpR | This work |
| LP56 | MB175 phoN105∷TnphoA, KanR | This work |
| LP57 | LP46 phoN105∷TnphoA, KanR | This work |
| LP58 | LP47 phoN105∷TnphoA, KanR | This work |
Table 2. Primers Used in This Study.
| Primer# | Name | Sequence |
|---|---|---|
| MB52 | PA PhoQ B | GCTCTAGATCAGACTGTAGCGAAACGTATGCGG |
| MB88 | PA PhoQ A | GGATATCCGTTCCCTGCGCATC |
| MB89 | PA PhoQ HisB | GGAAGCTTTTACTGCTCGCGGAACCCCTTGAG |
| MB90 | PA PhoQ HisA | CGGGATCCGAGAATCTTTATTTTCAGGGCGAGAACACCATCGAGCAGC |
| MB153 | PhoQ TetA | CACCGTACGCGGACAAGGATATCTTTTTGAATTGCGCTAATGTTAAGACCCACTTTCACA |
| MB154 | PhoQ TetB | TGCTTAACGAGATGCGTGGAAGAACGCACAGAAATGTTTACTAAGCACTTGTCTCCTG |
| MB183 | PA PhoQ ChrB | TGCTTAACGAGATGCGTGGAAGAACGCACAGAAATGTTCAGACTGTAGGCGAAACGTATGC |
| MB184 | PA PhoQ ChrA | TCTTTTTGAATTGCGGACAAGGATATCTTTTTGAATTGCGCTAGTGATCCGTTCCCTGCGCATC |
| MB258 | D110A | CGCCGCGCTACGCAGGCCGCGGCAA |
| MB260 | D131A | GGAGTTCTTCGTGTTCGCAGTCGAGATCGACCTGC |
| MB264 | E115A | CCGCGGCAACGCATTCCACACCACC |
| MB268 | D133A | TCGTGTTCGACGTCGCAATCGACCTGCTGCG |
| MB285 | PA PD MutA | GAGAACACCATCGAGCAGCG |
| MB286 | PA PD MutB | CTGCTCGCGGAACCCCTTGA |
| LP3 | E77A | CAACCTGCCGGCCGCCAAGGTCCTC |
| LP5 | S145A | CAGGCGGCCTACGCCATCGTCACCATG |
| LP17 | L137A | GAGATCGACCTGGCGCGCGGCAAGC |
| LP19 | G139A | CCTGCTGCGCGCAAAGCAGGCGGCC |
| LP23 | L158A | CGAGTTCGAGAGCGCGCTCAAGGGGTTC |
| LP29 | A143G | GCAAGCAGGCGGGCTACAGCATCGTC |
| LP87 | Chim-PhoQ ChrA | CACCGTACGCGGACAAGGATATCTTTTTGAATTGCGCTAATGAATAAATTTGCTGCGCATTTTC |
| LP88 | Chim-PhoQ ChrB | TGCTTAACGAGATGCGTGGAAGAACGCACAGAAATGTTTATTCCTCTTTCTGTGTGGGATG |
| LP89 | PhoQ-null ChrA | CACCGTACGCGGACAAGGATATCTTTTTGAATTGCGCTAGTGTAGGCTGGAGCTGCTTC |
| LP90 | PhoQ-null ChrB | TGCTTAACGAGATGCGTGGAAGAACGCACAGAAATGTCATATGAATATCCTCCTTAG |
Bacterial growth conditions
Strains were grown in LB or N minimal medium supplemented with the following antibiotics, where appropriate: ampicillin, 100 μg/ml; kanamycin, 45 μg/ml; and tetracycline, 10 μg/ml. Where experiments were conducted in buffered pH, a modified N minimal medium (Nelson et al., 1971) containing 100 mM MES instead of 100 mM Tris was used (Prost et al., 2007). Activation of the PhoP regulon was studied using the alkaline phosphatase reporter phoN∷TnphoA. Strains were grown overnight in N minimal medium containing the indicated amounts of MgCl2 and, where indicated, buffered to the appropriate pH value. Where peptide was used, strains were grown overnight in LB, washed twice in N minimal medium, and diluted 1:100 into fresh medium. Strains were then grown to an OD600 of 0.2-0.3, C18G was added at the indicated concentrations, and strains were grown for an additional 60-90 minutes. Alkaline phosphatase activity assays were performed according to a standard protocol on cultures grown in duplicate in at least three independent trials.
Purification of proteins
PA-PhoQ periplasmic domain (residues 37-165) was purified from strain MB149 via a C-terminal 6×His tag. MB149 was grown in LB to an OD600 of 0.6-0.8, then protein expression was induced by the addition of 0.5 mM IPTG and the culture was grown for an additional four hours. Cells were collected by centrifugation, lysed in a French pressure cell, inclusion bodies were collected by centrifugation and rinsed once in 50 mM sodium phosphate (pH 8.0), 300 mM NaCl. Inclusion bodies were resuspended in 10 mM sodium phosphate (pH 8.0), 200 mM NaCl, 7 M urea and incubated on ice for 30-60 minutes. Samples were then ultracentrifuged at 50,000 rpm for 60 minutes. To refold, the supernatant was rapidly diluted ten-fold into cold 100 mM sodium phosphate (pH 8.0), 400 mM L-arginine. Samples were filtered and purified using a 5 mL HiTrap nickel column (Amersham) according to a standard protocol. Samples were then dialyzed overnight against 20 mM Tris (pH 8.0), 100 mM NaCl. For NMR analysis, a similar protocol was followed except strain MB149 was grown in MOPS-minimal medium supplemented with 1 g/l 15N-ammonium chloride and induced for 5-6 hours. Following purification via nickel column, samples were further purified by size exclusion using a Superdex-75 gel filtration column (Amersham) equilibrated with 20 mM Tris (pH 8.0), 100 mM NaCl.
Fluorescence spectroscopy
Purified PA-PhoQ periplasmic domain was diluted to 1 μM in 50 mM Tris (pH 8.0). MgCl2 or CaCl2 was added at the indicated concentrations, and the samples equilibrated at room temperature for ∼30 minutes. Fluorescence measurements were performed in a Perkin Elmer LS55 luminescence spectrometer. Samples were excited at 280 nm and spectra were recorded from 290-450 nm. To estimate binding affinities, peak fluorescence (335 nm) for the metal free sample was subtracted from the peak fluorescence for each metal concentration tested to yield fluorescence change values. These data were then fit to a simple equilibrium model F = Fmax × Mg /(KD + Mg), where F is the observed fluorescence change, Fmax is the maximal fluorescence change, Mg is the concentration of Mg2+, and KD is the dissociation constant for the protein-Mg2+ complex.
NMR spectroscopy
The NMR samples of the PA-PhoQ periplasmic domain (37-165) used in all NMR experiments contained from 0.26 – 0.46 mM uniformly 15N-labeled PA-PhoQ in 20 mM Tris buffer at pH 8.0, with 100 mM NaCl, 0.1 mM EDTA, 20 mM MgCl2 or CaCl2 as described, and 10% (v/v) D2O. For the titration with Gd3+, microliter aliquots of 10 mM GdCl3 were added to the stated concentrations. For the pH titration, the protein was prepared in 20 mM MES buffer at pH 7.7, with 100 mM NaCl, 20 mM MgCl2, and 10% (v/v) D2O. The pH of the sample was lowered ∼0.5 units at a time with the addition of microliter aliquots of 500 mM DCl. All NMR experiments were performed at 25 °C on a Bruker DMX 500 MHz spectrometer equipped with a triple-resonance, triple-axis gradient probe. Data were processed and analyzed using the programs NMRPipe/NMRDraw (Delaglio et al., 1995) and NMRView (Johnson and Blevins, 1994).
Mutagenesis and screen for de-repression
A plate assay for PhoP-dependent gene activation using the phoN∷TnphoA reporter was employed to isolate mutants in the PA-PhoQ periplasmic domain. Activity of this reporter is readily visualized by growth on LB plates containing the alkaline phosphatase substrate XP. The background strain used, MB101, contains the phoN∷TnphoA reporter fusion and is phoQ null. When plasmid pMB147, expressing FL-PA-PhoQ, was expressed in this background and grown on LB+XP+Amp100 plates with 10 mM MgCl2, the colonies were white due to repression of the reporter. pMB147 was then randomly mutagenized using the GeneMorph II Mutagenesis Kit (Stratagene) via PCR mutagenesis with primers MB285 and MB286 to target only the periplasmic domain. The pool of mutagenized plasmid was transformed into MB101 and plated on LB+XP+Amp100+10 mM MgCl2, and approximately 3,000 colonies were screened. 52 blue colonies were chosen and sequenced, thus identifying 19 amino acids apparently involved in Mg2+-mediated repression. These 19 residues of interest were mutated to alanine by Quickchange mutagenesis (Stratagene). Only ten of these mutations resulted in a de-repressed phenotype, and these ten are shown in Figure 3.
Competitive index virulence assay
All animal experiments were performed with IACUC approval. Virulence assays were conducted by competitive index (CI) using 6- to 8-week old female BALB/c mice. The mice were inoculated intraperitoneally with a mixture of 5 × 104 organisms each of two S. typhimurium strains for a total of 105 bacteria in 0.1 mL saline. Each strain contained a stable plasmid-based antibiotic marker (pWSK129 or pWSK29) and was diluted from an overnight culture grown in LB. The ratio of the two strains in the inoculum was approximately 1:1 and was confirmed by plating serial dilutions on selective media. 48 hours after infection, mice were euthanized by CO2 asphyxiation, and the spleens were dissected and homogenized in saline using a tissue homogenizer. Samples were then serial diluted and plated on selective media. Ratios of the two strains were determined by counting colonies from these plates. The CI was calculated by dividing the ratio of strain 1 to strain 2 isolated from the spleen by the ratio of strain 1 to strain 2 inoculated into the mouse. CI results are the mean ± standard deviation of at least 10 mice that were infected in three independent experiments. Statistical significance (p-value) was determined using a Student's two-tailed t test.
Acknowledgments
We would like to thank Richard Pfuetzner for protein chemistry expertise and Loren Kinman for assistance with animal experiments. This work was supported by grant AI030479 from the NIAID to S.I.M. L.R.P. was supported in part by PHS NRSA T32 GM07270 from NIGMS. M.E.D. was supported by a postdoctoral fellowship from the CIHR. M.W.B. was supported by an Emmy-Noether fellowship from Deutsche Forschungsgemeinschaft.
References
- Alpuche-Aranda CM, Swanson JA, Loomis WP, Miller SI. Salmonella typhimurium activates virulence gene transcription within acidified macrophage phagosomes. PNAS. 1992;89:10079–10083. doi: 10.1073/pnas.89.21.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, LeMoual H, Miller SI. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- Belden WJ, Miller SI. Further characterization of the PhoP regulon: identification of new PhoP-activated virulence loci. Infect Immun. 1994;62:5095–5101. doi: 10.1128/iai.62.11.5095-5101.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bott M. Anaerobic citrate metabolism and its regulation in enterobacteria. Arch Microbiol. 1997;167:78–88. [PubMed] [Google Scholar]
- Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Protein structure prediction servers at University College London. Nucl Acids Res. 2005;33(Web Server):W36–38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho US, Bader MW, Amaya MF, Daley ME, Klevit RE, Miller SI. Metal bridges between the PhoQ sensor domain and the membrane regulate transmembrane signaling. J Mol Biol. 2006;356:1193–1206. doi: 10.1016/j.jmb.2005.12.032. [DOI] [PubMed] [Google Scholar]
- Darveau R, Blake J, Seachord CL, Cosand WL, Cunningham MD, Cassiano-Clough L, Maloney G. Peptides related to the carboxyl terminus of human platelet factor IV with antibacterial activity. J Clin Invest. 1992;90:447–455. doi: 10.1172/JCI115880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe – a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Derzelle S, Turlin E, Duchaud E, Pages S, Kunst F, Givaudun A, Danchin A. The PhoP-PhoQ two-component system of Photorhabdus luminescens is essential for virulence in insects. J Bacteriol. 2004;186:1270–1279. doi: 10.1128/JB.186.5.1270-1279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst RK, Yi EC, Guo L, Lim KB, Burns JL, Hackett M, Miller SI. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aearuginosa. Science. 1999;286:1561–1565. doi: 10.1126/science.286.5444.1561. [DOI] [PubMed] [Google Scholar]
- Fields PI, Groisman EA, Heffron F. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science. 1989;243:1059–1062. doi: 10.1126/science.2646710. [DOI] [PubMed] [Google Scholar]
- Foster JW, Hall HK. Adaptive acidification tolerance response of Salmonella typhimurium. J Bacteriol. 1990;172:771–778. doi: 10.1128/jb.172.2.771-778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Vescovi E, Soncini FC, Groisman EA. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- Garcia-Vescovi E, Ayala YM, Di Cera E, Groisman EA. Characterization of the bacterial sensor protein PhoQ: evidence for distinct binding sites for Mg2+ and Ca2+ JBC. 1997;272:1440–1443. doi: 10.1074/jbc.272.3.1440. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. NMRView – a computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Lee L, Sykes BD. Use of lanthanide-induced nuclear magnetic resonance shifts for determination of protein structure in solution: EF calcium binding site of carp parvalbumin. Biochemistry. 1983;22:4366–73. doi: 10.1021/bi00288a004. [DOI] [PubMed] [Google Scholar]
- Lesley JA, Waldburger CD. Comparison of the Pseudomonas aeruginosa and Escherichia coli PhoQ sensor domains. J Biol Chem. 2001;276:30827–30833. doi: 10.1074/jbc.M104262200. [DOI] [PubMed] [Google Scholar]
- Llama-Palacios A, Lopez-Solanilla E, Poza-Carrion C, Garcia-Olmeda F, Rodriguez-Palenzuela P. The Erwinia chrysanthemi phoP-phoQ operon plays an important role in growth at low pH, virulence and bacterial survival in plant tissue. Mol Microbiol. 2003;49:347–357. doi: 10.1046/j.1365-2958.2003.03583.x. [DOI] [PubMed] [Google Scholar]
- Macfarlane ELA, Kwasnicka A, Ochs MM, Hancock REW. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol Microbiol. 1999;34:305–316. doi: 10.1046/j.1365-2958.1999.01600.x. [DOI] [PubMed] [Google Scholar]
- Martin-Orozco N, Touret N, Zaharik ML, Park E, Kopelman R, Miller SI, Finlay BB, Gros P, Grinstein S. Visualization of vacuolar acidification-induced transcription of genes of pathogens inside macrophages. Mol Biol Cell. 2006;17:498–510. doi: 10.1091/mbc.E04-12-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhee JB, Lewenza S, Hancock REW. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol. 2003;50:205–217. doi: 10.1046/j.1365-2958.2003.03673.x. [DOI] [PubMed] [Google Scholar]
- Miller SI, Kukral AM, Mekalanos JJ. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. PNAS. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SI, Pulkkinen WS, Selsted ME, Mekalanos JJ. Characterization of defensin resistance phenotypes associated with mutations in the phoP virulence regulon of Salmonella typhimurium. Infect Immun. 1990;58:3706–3710. doi: 10.1128/iai.58.11.3706-3710.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss JE, Fisher PE, Vick B, Groisman EA, Zychlinsky A. The regulatory protein PhoP controls susceptibility to the host inflammatory response in Shigella flexneri. Cell Microbiol. 2000;2:443–452. doi: 10.1046/j.1462-5822.2000.00065.x. [DOI] [PubMed] [Google Scholar]
- Nelson DL, Kennedy EP. Magnesium transport in E. coli: inhibition by cobaltous ion. J Biol Chem. 1971;246:3042–3049. [PubMed] [Google Scholar]
- Oyston PC, Dorrell N, Williams K, Li SR, Green M, Titball RW, Wren BW. The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis. Infect Immun. 2000;68:3419–3425. doi: 10.1128/iai.68.6.3419-3425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prost LR, Daley ME, Le Sage V, Bader MW, Le Moual H, Klevit RE, Miller SI. Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol Cell. 2007;26:165–174. doi: 10.1016/j.molcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Prost LR, Miller SI. The Salmonellae PhoQ sensor: mechanisms of detection of phagosome signals. Cell Micro. 2008;10:576–582. doi: 10.1111/j.1462-5822.2007.01111.x. [DOI] [PubMed] [Google Scholar]
- Reinelt S, Hofmann E, Gerharz T, Bott M, Madden DR. The structure of the periplasmic ligand-binding domain of the sensor kinase CitA reveals the first extracellular PAS domain. J Biol Chem. 2003;278:39189–39196. doi: 10.1074/jbc.M305864200. [DOI] [PubMed] [Google Scholar]
- Soncini FC, Garcia-Vescovi E, Solomon F, Groisman EA. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J Bacteriol. 1996;178:5092–5099. doi: 10.1128/jb.178.17.5092-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich LE, Koonin EV, Zhulin IB. One-component systems dominate signal transduction in prokaryotes. TRENDS Microbiol. 2005;13:52–56. doi: 10.1016/j.tim.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AH, Stock AM. Histidine kinases and response regulator proteins in two-component systems. TRENDS Biochem Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]


